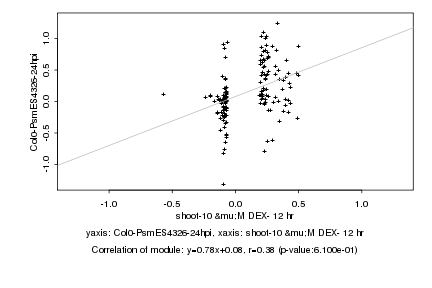
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
shoot-10 μM DEX- 12 hr |
Log2 signal ratio
Col0-PsmES4326-24hpi |
| 1 |
266142_at |
AT2G39030
|
NATA1, N-acetyltransferase activity 1 |
0.495 |
0.417 |
| 2 |
251770_at |
AT3G55970
|
ATJRG21, JRG21, jasmonate-regulated gene 21 |
0.493 |
0.883 |
| 3 |
245341_at |
AT4G16447
|
unknown |
0.487 |
-0.265 |
| 4 |
267357_at |
AT2G40000
|
ATHSPRO2, ARABIDOPSIS ORTHOLOG OF SUGAR BEET HS1 PRO-1 2, HSPRO2, ortholog of sugar beet HS1 PRO-1 2 |
0.483 |
0.456 |
| 5 |
253061_at |
AT4G37610
|
BT5, BTB and TAZ domain protein 5 |
0.434 |
0.227 |
| 6 |
256527_at |
AT1G66100
|
[Plant thionin] |
0.431 |
-0.027 |
| 7 |
260287_at |
AT1G80440
|
KFB20, Kelch repeat F-box 20, KMD1, KISS ME DEADLY 1 |
0.426 |
0.300 |
| 8 |
263539_at |
AT2G24850
|
TAT, TYROSINE AMINOTRANSFERASE, TAT3, tyrosine aminotransferase 3 |
0.418 |
0.452 |
| 9 |
265387_at |
AT2G20670
|
unknown |
0.418 |
-0.159 |
| 10 |
258402_at |
AT3G15450
|
[Aluminium induced protein with YGL and LRDR motifs] |
0.413 |
0.018 |
| 11 |
248164_at |
AT5G54490
|
PBP1, pinoid-binding protein 1 |
0.400 |
0.664 |
| 12 |
247543_at |
AT5G61600
|
ERF104, ethylene response factor 104 |
0.396 |
0.392 |
| 13 |
262986_at |
AT1G23390
|
[Kelch repeat-containing F-box family protein] |
0.394 |
-0.049 |
| 14 |
245250_at |
AT4G17490
|
ATERF6, ethylene responsive element binding factor 6, ERF-6-6, ERF6, ethylene responsive element binding factor 6 |
0.393 |
0.035 |
| 15 |
258487_at |
AT3G02550
|
LBD41, LOB domain-containing protein 41 |
0.375 |
0.335 |
| 16 |
248964_at |
AT5G45340
|
CYP707A3, cytochrome P450, family 707, subfamily A, polypeptide 3 |
0.374 |
-0.150 |
| 17 |
247925_at |
AT5G57560
|
TCH4, Touch 4, XTH22, xyloglucan endotransglucosylase/hydrolase 22 |
0.367 |
0.201 |
| 18 |
266385_at |
AT2G14610
|
ATPR1, PATHOGENESIS-RELATED GENE 1, PR 1, PATHOGENESIS-RELATED GENE 1, PR1, pathogenesis-related gene 1 |
0.346 |
0.356 |
| 19 |
256577_at |
AT3G28220
|
[TRAF-like family protein] |
0.345 |
-0.308 |
| 20 |
254954_at |
AT4G10910
|
unknown |
0.339 |
0.005 |
| 21 |
267238_at |
AT2G44130
|
KMD3, KISS ME DEADLY 3 |
0.333 |
0.499 |
| 22 |
264415_at |
AT1G43160
|
RAP2.6, related to AP2 6 |
0.330 |
1.253 |
| 23 |
261648_at |
AT1G27730
|
STZ, salt tolerance zinc finger, ZAT10 |
0.321 |
0.813 |
| 24 |
256589_at |
AT3G28740
|
CYP81D11, cytochrome P450, family 81, subfamily D, polypeptide 11 |
0.316 |
0.071 |
| 25 |
250793_at |
AT5G05600
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.315 |
0.560 |
| 26 |
256300_at |
AT1G69490
|
ANAC029, Arabidopsis NAC domain containing protein 29, ATNAP, NAC-LIKE, ACTIVATED BY AP3/PI, NAP, NAC-like, activated by AP3/PI |
0.313 |
0.436 |
| 27 |
248448_at |
AT5G51190
|
[Integrase-type DNA-binding superfamily protein] |
0.301 |
-0.002 |
| 28 |
256442_at |
AT3G10930
|
unknown |
0.288 |
0.879 |
| 29 |
266656_at |
AT2G25900
|
ATCTH, ATTZF1, A. THALIANA TANDEM ZINC FINGER PROTEIN 1 |
0.288 |
-0.609 |
| 30 |
255807_at |
AT4G10270
|
[Wound-responsive family protein] |
0.274 |
-0.142 |
| 31 |
266292_at |
AT2G29350
|
SAG13, senescence-associated gene 13 |
0.259 |
0.716 |
| 32 |
253161_at |
AT4G35770
|
ATSEN1, ARABIDOPSIS THALIANA SENESCENCE 1, DIN1, DARK INDUCIBLE 1, SEN1, SENESCENCE 1, SEN1, SENESCENCE ASSOCIATED GENE 1 |
0.257 |
0.487 |
| 33 |
262381_at |
AT1G72900
|
[Toll-Interleukin-Resistance (TIR) domain-containing protein] |
0.256 |
0.715 |
| 34 |
264379_at |
AT2G25200
|
unknown |
0.255 |
-0.140 |
| 35 |
247524_at |
AT5G61440
|
ACHT5, atypical CYS HIS rich thioredoxin 5 |
0.254 |
0.090 |
| 36 |
253915_at |
AT4G27280
|
[Calcium-binding EF-hand family protein] |
0.249 |
0.436 |
| 37 |
249052_at |
AT5G44420
|
LCR77, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 77, PDF1.2, plant defensin 1.2, PDF1.2A, PLANT DEFENSIN 1.2A |
0.248 |
-0.633 |
| 38 |
265276_at |
AT2G28400
|
unknown |
0.246 |
0.693 |
| 39 |
263161_at |
AT1G54020
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.246 |
0.786 |
| 40 |
245252_at |
AT4G17500
|
ATERF-1, ethylene responsive element binding factor 1, ERF-1, ethylene responsive element binding factor 1 |
0.245 |
0.900 |
| 41 |
245662_at |
AT1G28190
|
unknown |
0.245 |
1.045 |
| 42 |
248799_at |
AT5G47230
|
ATERF-5, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR- 5, ATERF5, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 5, AtMACD1, ERF102, ERF5, ethylene responsive element binding factor 5 |
0.244 |
0.198 |
| 43 |
262383_at |
AT1G72940
|
[Toll-Interleukin-Resistance (TIR) domain-containing protein] |
0.241 |
0.428 |
| 44 |
245275_at |
AT4G15210
|
AT-BETA-AMY, ATBETA-AMY, ARABIDOPSIS THALIANA BETA-AMYLASE, BAM5, beta-amylase 5, BMY1, RAM1, REDUCED BETA AMYLASE 1 |
0.240 |
0.101 |
| 45 |
259879_at |
AT1G76650
|
CML38, calmodulin-like 38 |
0.237 |
1.015 |
| 46 |
261567_at |
AT1G33055
|
unknown |
0.235 |
0.045 |
| 47 |
252367_at |
AT3G48360
|
ATBT2, BT2, BTB and TAZ domain protein 2 |
0.234 |
0.811 |
| 48 |
264788_at |
AT2G17880
|
DJC24, DNA J protein C24 |
0.231 |
-0.013 |
| 49 |
251745_at |
AT3G55980
|
ATSZF1, SZF1, salt-inducible zinc finger 1 |
0.231 |
0.362 |
| 50 |
246099_at |
AT5G20230
|
ATBCB, blue-copper-binding protein, BCB, BLUE COPPER BINDING PROTEIN, BCB, blue-copper-binding protein, SAG14, SENESCENCE ASSOCIATED GENE 14 |
0.230 |
0.380 |
| 51 |
247585_at |
AT5G60680
|
unknown |
0.229 |
-0.039 |
| 52 |
266658_at |
AT2G25735
|
unknown |
0.229 |
0.652 |
| 53 |
265724_at |
AT2G32100
|
ATOFP16, RABIDOPSIS THALIANA OVATE FAMILY PROTEIN 16, OFP16, ovate family protein 16 |
0.228 |
-0.789 |
| 54 |
254926_at |
AT4G11280
|
ACS6, 1-aminocyclopropane-1-carboxylic acid (acc) synthase 6, ATACS6 |
0.226 |
0.432 |
| 55 |
252193_at |
AT3G50060
|
MYB77, myb domain protein 77 |
0.226 |
-0.026 |
| 56 |
253829_at |
AT4G28040
|
UMAMIT33, Usually multiple acids move in and out Transporters 33 |
0.224 |
0.563 |
| 57 |
245711_at |
AT5G04340
|
C2H2, CZF2, COLD INDUCED ZINC FINGER PROTEIN 2, ZAT6, zinc finger of Arabidopsis thaliana 6 |
0.221 |
0.804 |
| 58 |
245264_at |
AT4G17245
|
[RING/U-box superfamily protein] |
0.220 |
0.178 |
| 59 |
266800_at |
AT2G22880
|
[VQ motif-containing protein] |
0.218 |
0.549 |
| 60 |
252136_at |
AT3G50770
|
CML41, calmodulin-like 41 |
0.218 |
0.681 |
| 61 |
252250_at |
AT3G49790
|
[Carbohydrate-binding protein] |
0.216 |
0.216 |
| 62 |
258277_at |
AT3G26830
|
CYP71B15, PAD3, PHYTOALEXIN DEFICIENT 3 |
0.216 |
1.098 |
| 63 |
264339_at |
AT1G70290
|
ATTPS8, ATTPSC, TPS8, trehalose-6-phosphatase synthase S8 |
0.214 |
0.049 |
| 64 |
249417_at |
AT5G39670
|
[Calcium-binding EF-hand family protein] |
0.213 |
0.634 |
| 65 |
249923_at |
AT5G19120
|
[Eukaryotic aspartyl protease family protein] |
0.210 |
0.087 |
| 66 |
245041_at |
AT2G26530
|
AR781 |
0.209 |
0.467 |
| 67 |
251443_at |
AT3G59940
|
KFB50, Kelch repeat F-box 50, KMD4, KISS ME DEADLY 4 |
0.209 |
0.119 |
| 68 |
266290_at |
AT2G29490
|
ATGSTU1, glutathione S-transferase TAU 1, GST19, GLUTATHIONE S-TRANSFERASE 19, GSTU1, glutathione S-transferase TAU 1 |
0.206 |
0.861 |
| 69 |
249337_at |
AT5G41080
|
AtGDPD2, GDPD2, glycerophosphodiester phosphodiesterase 2 |
0.204 |
0.746 |
| 70 |
259364_at |
AT1G13260
|
EDF4, ETHYLENE RESPONSE DNA BINDING FACTOR 4, RAV1, related to ABI3/VP1 1 |
0.204 |
0.103 |
| 71 |
250942_at |
AT5G03350
|
[Legume lectin family protein] |
0.202 |
0.037 |
| 72 |
255926_at |
AT1G22190
|
RAP2.4, related to AP2 4 |
0.202 |
0.113 |
| 73 |
257774_at |
AT3G29250
|
AtSDR4, SDR4, short-chain dehydrogenase reductase 4 |
0.202 |
1.038 |
| 74 |
266259_at |
AT2G27830
|
unknown |
0.200 |
0.423 |
| 75 |
254120_at |
AT4G24570
|
DIC2, dicarboxylate carrier 2 |
0.199 |
0.175 |
| 76 |
259682_at |
AT1G63040
|
[pseudogene] |
0.195 |
0.101 |
| 77 |
255753_at |
AT1G18570
|
AtMYB51, myb domain protein 51, BW51A, BW51B, HIG1, HIGH INDOLIC GLUCOSINOLATE 1, MYB51, myb domain protein 51 |
0.195 |
0.658 |
| 78 |
252483_at |
AT3G46600
|
[GRAS family transcription factor] |
0.195 |
0.314 |
| 79 |
257615_at |
AT3G26510
|
[Octicosapeptide/Phox/Bem1p family protein] |
0.193 |
-0.029 |
| 80 |
249494_at |
AT5G39050
|
PMAT1, phenolic glucoside malonyltransferase 1 |
0.193 |
0.592 |
| 81 |
263799_at |
AT2G24550
|
unknown |
0.190 |
0.103 |
| 82 |
249082_at |
AT5G44120
|
ATCRA1, CRUCIFERINA, CRA1, CRUCIFERINA, CRU1 |
-0.576 |
0.120 |
| 83 |
256814_at |
AT3G21370
|
BGLU19, beta glucosidase 19 |
-0.243 |
0.072 |
| 84 |
253155_at |
AT4G35720
|
unknown |
-0.202 |
0.103 |
| 85 |
254482_at |
AT4G20370
|
TSF, TWIN SISTER OF FT |
-0.199 |
0.081 |
| 86 |
257947_at |
AT3G21720
|
ICL, isocitrate lyase |
-0.171 |
0.015 |
| 87 |
260727_at |
AT1G48100
|
[Pectin lyase-like superfamily protein] |
-0.144 |
-0.163 |
| 88 |
256766_at |
AT3G22231
|
PCC1, PATHOGEN AND CIRCADIAN CONTROLLED 1 |
-0.143 |
0.087 |
| 89 |
257876_at |
AT3G17130
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.142 |
-0.184 |
| 90 |
262643_at |
AT1G62770
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.138 |
0.048 |
| 91 |
251886_at |
AT3G54260
|
TBL36, TRICHOME BIREFRINGENCE-LIKE 36 |
-0.134 |
0.025 |
| 92 |
260483_at |
AT1G11000
|
ATMLO4, MILDEW RESISTANCE LOCUS O 4, MLO4, MILDEW RESISTANCE LOCUS O 4 |
-0.130 |
0.031 |
| 93 |
263382_at |
AT2G40230
|
[HXXXD-type acyl-transferase family protein] |
-0.125 |
-0.264 |
| 94 |
257262_at |
AT3G21890
|
BBX31, B-box domain protein 31 |
-0.123 |
0.038 |
| 95 |
267595_at |
AT2G32990
|
AtGH9B8, glycosyl hydrolase 9B8, GH9B8, glycosyl hydrolase 9B8 |
-0.120 |
-0.449 |
| 96 |
249191_at |
AT5G42760
|
[Leucine carboxyl methyltransferase] |
-0.115 |
-0.162 |
| 97 |
250103_at |
AT5G16600
|
AtMYB43, myb domain protein 43, MYB43, myb domain protein 43 |
-0.112 |
-0.030 |
| 98 |
266770_at |
AT2G03090
|
ATEXP15, ATEXPA15, expansin A15, ATHEXP ALPHA 1.3, EXP15, EXPANSIN 15, EXPA15, expansin A15 |
-0.111 |
-0.173 |
| 99 |
247878_at |
AT5G57760
|
unknown |
-0.107 |
-0.217 |
| 100 |
250746_at |
AT5G05880
|
[UDP-Glycosyltransferase superfamily protein] |
-0.106 |
0.017 |
| 101 |
260560_at |
AT2G43590
|
[Chitinase family protein] |
-0.106 |
0.406 |
| 102 |
264016_at |
AT2G21220
|
SAUR12, SMALL AUXIN UPREGULATED RNA 12 |
-0.104 |
0.010 |
| 103 |
253496_at |
AT4G31870
|
ATGPX7, GLUTATHIONE PEROXIDASE 7, GPX7, glutathione peroxidase 7 |
-0.101 |
-0.140 |
| 104 |
259736_at |
AT1G64390
|
AtGH9C2, glycosyl hydrolase 9C2, GH9C2, glycosyl hydrolase 9C2 |
-0.101 |
-0.825 |
| 105 |
267382_at |
AT2G44300
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.101 |
-0.067 |
| 106 |
265278_at |
AT2G28330
|
unknown |
-0.100 |
-0.088 |
| 107 |
258748_at |
AT3G05930
|
GLP8, germin-like protein 8 |
-0.097 |
0.104 |
| 108 |
264400_at |
AT1G61800
|
ATGPT2, ARABIDOPSIS GLUCOSE-6-PHOSPHATE/PHOSPHATE TRANSLOCATOR 2, GPT2, glucose-6-phosphate/phosphate translocator 2 |
-0.096 |
0.907 |
| 109 |
246983_at |
AT5G67200
|
[Leucine-rich repeat protein kinase family protein] |
-0.095 |
-0.298 |
| 110 |
252607_at |
AT3G44990
|
AtXTH31, ATXTR8, XTH31, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 31, XTR8, xyloglucan endo-transglycosylase-related 8 |
-0.095 |
-1.303 |
| 111 |
259101_at |
AT3G11640
|
unknown |
-0.095 |
0.046 |
| 112 |
251065_at |
AT5G01870
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.094 |
0.044 |
| 113 |
257185_at |
AT3G13100
|
ABCC7, ATP-binding cassette C7, ATMRP7, multidrug resistance-associated protein 7, MRP7, MRP7, multidrug resistance-associated protein 7 |
-0.094 |
0.851 |
| 114 |
250049_at |
AT5G17780
|
[alpha/beta-Hydrolases superfamily protein] |
-0.091 |
0.214 |
| 115 |
264039_at |
AT2G03740
|
[late embryogenesis abundant domain-containing protein / LEA domain-containing protein] |
-0.091 |
0.069 |
| 116 |
261059_at |
AT1G01250
|
[Integrase-type DNA-binding superfamily protein] |
-0.090 |
-0.232 |
| 117 |
266778_at |
AT2G29090
|
CYP707A2, cytochrome P450, family 707, subfamily A, polypeptide 2 |
-0.089 |
-0.253 |
| 118 |
265831_at |
AT2G14460
|
unknown |
-0.089 |
-0.176 |
| 119 |
261221_at |
AT1G19960
|
unknown |
-0.089 |
-0.409 |
| 120 |
267590_at |
AT2G39700
|
ATEXP4, ATEXPA4, expansin A4, ATHEXP ALPHA 1.6, EXPA4, expansin A4 |
-0.089 |
-0.762 |
| 121 |
262620_at |
AT1G06540
|
unknown |
-0.089 |
0.014 |
| 122 |
251928_at |
AT3G53980
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.088 |
0.063 |
| 123 |
262811_at |
AT1G11700
|
unknown |
-0.087 |
-0.226 |
| 124 |
266970_at |
AT2G39560
|
[Putative membrane lipoprotein] |
-0.087 |
-0.121 |
| 125 |
251968_at |
AT3G53100
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.086 |
-0.349 |
| 126 |
253815_at |
AT4G28250
|
ATEXPB3, expansin B3, ATHEXP BETA 1.6, EXPB3, expansin B3 |
-0.085 |
-0.641 |
| 127 |
259934_at |
AT1G71340
|
AtGDPD4, GDPD4, glycerophosphodiester phosphodiesterase 4 |
-0.085 |
0.001 |
| 128 |
265634_at |
AT2G25530
|
[AFG1-like ATPase family protein] |
-0.085 |
0.368 |
| 129 |
262234_at |
AT1G48270
|
GCR1, G-protein-coupled receptor 1 |
-0.084 |
0.091 |
| 130 |
261758_at |
AT1G08250
|
ADT6, arogenate dehydratase 6, AtADT6, Arabidopsis thaliana arogenate dehydratase 6 |
-0.083 |
0.159 |
| 131 |
248104_at |
AT5G55250
|
AtIAMT1, IAMT1, IAA carboxylmethyltransferase 1 |
-0.083 |
-0.101 |
| 132 |
248097_at |
AT5G55260
|
PPX-2, PROTEIN PHOSPHATASE X -2, PPX2, protein phosphatase X 2 |
-0.082 |
-0.084 |
| 133 |
259629_at |
AT1G56510
|
ADR2, ACTIVATED DISEASE RESISTANCE 2, WRR4, WHITE RUST RESISTANCE 4 |
-0.082 |
0.081 |
| 134 |
261781_at |
AT1G76320
|
CPD25, CHLOROPLAST DIVISION MUTANT 25, FRS4, FAR1-related sequence 4 |
-0.082 |
0.054 |
| 135 |
254096_at |
AT4G25150
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
-0.081 |
-0.013 |
| 136 |
250699_at |
AT5G06820
|
SRF2, STRUBBELIG-receptor family 2 |
-0.081 |
-0.018 |
| 137 |
250648_at |
AT5G06760
|
AtLEA4-5, Late Embryogenesis Abundant 4-5, LEA4-5, Late Embryogenesis Abundant 4-5 |
-0.081 |
0.710 |
| 138 |
261945_at |
AT1G64530
|
NLP6, NIN-like protein 6 |
-0.080 |
-0.219 |
| 139 |
253382_at |
AT4G33040
|
[Thioredoxin superfamily protein] |
-0.080 |
0.360 |
| 140 |
247498_at |
AT5G61810
|
APC1, ATP/phosphate carrier 1 |
-0.080 |
0.158 |
| 141 |
250577_at |
AT5G07910
|
[Leucine-rich repeat (LRR) family protein] |
-0.079 |
0.214 |
| 142 |
248694_at |
AT5G48340
|
unknown |
-0.077 |
0.130 |
| 143 |
258437_at |
AT3G16560
|
[Protein phosphatase 2C family protein] |
-0.077 |
-0.099 |
| 144 |
249693_at |
AT5G35750
|
AHK2, histidine kinase 2, HK2, histidine kinase 2 |
-0.076 |
-0.514 |
| 145 |
263982_at |
AT2G42860
|
unknown |
-0.076 |
0.021 |
| 146 |
251103_at |
AT5G01700
|
[Protein phosphatase 2C family protein] |
-0.075 |
0.025 |
| 147 |
260946_at |
AT1G06010
|
unknown |
-0.075 |
0.001 |
| 148 |
250344_at |
AT5G11930
|
[Thioredoxin superfamily protein] |
-0.074 |
-0.209 |
| 149 |
265837_at |
AT2G14560
|
LURP1, LATE UPREGULATED IN RESPONSE TO HYALOPERONOSPORA PARASITICA |
-0.074 |
0.151 |
| 150 |
264323_at |
AT1G04180
|
YUC9, YUCCA 9 |
-0.074 |
0.125 |
| 151 |
249622_at |
AT5G37550
|
unknown |
-0.073 |
0.070 |
| 152 |
245697_at |
AT5G04200
|
AtMC9, metacaspase 9, AtMCP2f, metacaspase 2f, MC9, metacaspase 9, MCP2f, metacaspase 2f |
-0.073 |
0.237 |
| 153 |
255876_at |
AT2G40480
|
unknown |
-0.072 |
-0.562 |
| 154 |
260354_at |
AT1G69330
|
[RING/U-box superfamily protein] |
-0.072 |
-0.085 |
| 155 |
250378_at |
AT5G11570
|
[Major facilitator superfamily protein] |
-0.071 |
0.084 |
| 156 |
248404_at |
AT5G51460
|
ATTPPA, TPPA, trehalose-6-phosphate phosphatase A |
-0.071 |
-0.328 |
| 157 |
262654_at |
AT1G14180
|
[RING/U-box superfamily protein] |
-0.071 |
-0.096 |
| 158 |
253500_at |
AT4G31920
|
ARR10, response regulator 10, RR10, response regulator 10 |
-0.071 |
0.174 |
| 159 |
249718_at |
AT5G35740
|
[Carbohydrate-binding X8 domain superfamily protein] |
-0.071 |
-0.140 |
| 160 |
248246_at |
AT5G53200
|
TRY, TRIPTYCHON |
-0.071 |
-0.526 |
| 161 |
246401_at |
AT1G57560
|
AtMYB50, myb domain protein 50, MYB50, myb domain protein 50 |
-0.070 |
0.948 |