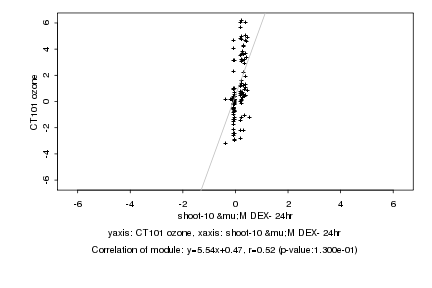
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
shoot-10 μM DEX- 24hr |
Log2 signal ratio
CT101 ozone |
| 1 |
245341_at |
AT4G16447
|
unknown |
0.527 |
-1.161 |
| 2 |
245250_at |
AT4G17490
|
ATERF6, ethylene responsive element binding factor 6, ERF-6-6, ERF6, ethylene responsive element binding factor 6 |
0.431 |
4.893 |
| 3 |
247543_at |
AT5G61600
|
ERF104, ethylene response factor 104 |
0.426 |
0.879 |
| 4 |
267357_at |
AT2G40000
|
ATHSPRO2, ARABIDOPSIS ORTHOLOG OF SUGAR BEET HS1 PRO-1 2, HSPRO2, ortholog of sugar beet HS1 PRO-1 2 |
0.419 |
3.391 |
| 5 |
249052_at |
AT5G44420
|
LCR77, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 77, PDF1.2, plant defensin 1.2, PDF1.2A, PLANT DEFENSIN 1.2A |
0.405 |
4.642 |
| 6 |
266142_at |
AT2G39030
|
NATA1, N-acetyltransferase activity 1 |
0.382 |
3.671 |
| 7 |
253061_at |
AT4G37610
|
BT5, BTB and TAZ domain protein 5 |
0.374 |
1.141 |
| 8 |
258487_at |
AT3G02550
|
LBD41, LOB domain-containing protein 41 |
0.366 |
1.973 |
| 9 |
261648_at |
AT1G27730
|
STZ, salt tolerance zinc finger, ZAT10 |
0.366 |
4.724 |
| 10 |
263539_at |
AT2G24850
|
TAT, TYROSINE AMINOTRANSFERASE, TAT3, tyrosine aminotransferase 3 |
0.365 |
6.075 |
| 11 |
248448_at |
AT5G51190
|
[Integrase-type DNA-binding superfamily protein] |
0.355 |
0.532 |
| 12 |
245692_at |
AT5G04150
|
BHLH101 |
0.355 |
0.481 |
| 13 |
248164_at |
AT5G54490
|
PBP1, pinoid-binding protein 1 |
0.350 |
5.111 |
| 14 |
258402_at |
AT3G15450
|
[Aluminium induced protein with YGL and LRDR motifs] |
0.347 |
1.346 |
| 15 |
251677_at |
AT3G56980
|
BHLH039, bHLH39, basic helix-loop-helix 39, ORG3, OBP3-RESPONSIVE GENE 3 |
0.331 |
0.427 |
| 16 |
247925_at |
AT5G57560
|
TCH4, Touch 4, XTH22, xyloglucan endotransglucosylase/hydrolase 22 |
0.326 |
3.220 |
| 17 |
260287_at |
AT1G80440
|
KFB20, Kelch repeat F-box 20, KMD1, KISS ME DEADLY 1 |
0.323 |
0.937 |
| 18 |
262986_at |
AT1G23390
|
[Kelch repeat-containing F-box family protein] |
0.322 |
-1.030 |
| 19 |
266292_at |
AT2G29350
|
SAG13, senescence-associated gene 13 |
0.318 |
2.925 |
| 20 |
253915_at |
AT4G27280
|
[Calcium-binding EF-hand family protein] |
0.304 |
4.225 |
| 21 |
265387_at |
AT2G20670
|
unknown |
0.295 |
-2.177 |
| 22 |
255807_at |
AT4G10270
|
[Wound-responsive family protein] |
0.294 |
1.265 |
| 23 |
248799_at |
AT5G47230
|
ATERF-5, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR- 5, ATERF5, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 5, AtMACD1, ERF102, ERF5, ethylene responsive element binding factor 5 |
0.280 |
4.318 |
| 24 |
262383_at |
AT1G72940
|
[Toll-Interleukin-Resistance (TIR) domain-containing protein] |
0.271 |
0.551 |
| 25 |
266821_at |
AT2G44840
|
ATERF13, ETHYLENE-RESPONSIVE ELEMENT BINDING FACTOR 13, EREBP, ERF13, ethylene-responsive element binding factor 13 |
0.271 |
2.273 |
| 26 |
256442_at |
AT3G10930
|
unknown |
0.270 |
3.659 |
| 27 |
262381_at |
AT1G72900
|
[Toll-Interleukin-Resistance (TIR) domain-containing protein] |
0.260 |
3.852 |
| 28 |
252193_at |
AT3G50060
|
MYB77, myb domain protein 77 |
0.252 |
0.334 |
| 29 |
254926_at |
AT4G11280
|
ACS6, 1-aminocyclopropane-1-carboxylic acid (acc) synthase 6, ATACS6 |
0.238 |
3.155 |
| 30 |
245711_at |
AT5G04340
|
C2H2, CZF2, COLD INDUCED ZINC FINGER PROTEIN 2, ZAT6, zinc finger of Arabidopsis thaliana 6 |
0.234 |
3.735 |
| 31 |
247524_at |
AT5G61440
|
ACHT5, atypical CYS HIS rich thioredoxin 5 |
0.231 |
0.747 |
| 32 |
246825_at |
AT5G26260
|
[TRAF-like family protein] |
0.229 |
0.225 |
| 33 |
254954_at |
AT4G10910
|
unknown |
0.228 |
-0.147 |
| 34 |
259879_at |
AT1G76650
|
CML38, calmodulin-like 38 |
0.227 |
3.122 |
| 35 |
254120_at |
AT4G24570
|
DIC2, dicarboxylate carrier 2 |
0.226 |
5.012 |
| 36 |
263402_at |
AT2G04050
|
[MATE efflux family protein] |
0.225 |
1.409 |
| 37 |
245041_at |
AT2G26530
|
AR781 |
0.220 |
3.596 |
| 38 |
245252_at |
AT4G17500
|
ATERF-1, ethylene responsive element binding factor 1, ERF-1, ethylene responsive element binding factor 1 |
0.219 |
4.754 |
| 39 |
245777_at |
AT1G73540
|
atnudt21, nudix hydrolase homolog 21, NUDT21, nudix hydrolase homolog 21 |
0.217 |
1.281 |
| 40 |
251745_at |
AT3G55980
|
ATSZF1, SZF1, salt-inducible zinc finger 1 |
0.212 |
3.227 |
| 41 |
258653_at |
AT3G09870
|
SAUR48, SMALL AUXIN UPREGULATED RNA 48 |
0.209 |
0.083 |
| 42 |
266658_at |
AT2G25735
|
unknown |
0.203 |
6.262 |
| 43 |
247585_at |
AT5G60680
|
unknown |
0.201 |
-1.213 |
| 44 |
266259_at |
AT2G27830
|
unknown |
0.199 |
0.593 |
| 45 |
257774_at |
AT3G29250
|
AtSDR4, SDR4, short-chain dehydrogenase reductase 4 |
0.198 |
0.750 |
| 46 |
263799_at |
AT2G24550
|
unknown |
0.194 |
1.652 |
| 47 |
249337_at |
AT5G41080
|
AtGDPD2, GDPD2, glycerophosphodiester phosphodiesterase 2 |
0.189 |
1.258 |
| 48 |
259364_at |
AT1G13260
|
EDF4, ETHYLENE RESPONSE DNA BINDING FACTOR 4, RAV1, related to ABI3/VP1 1 |
0.188 |
0.479 |
| 49 |
252131_at |
AT3G50930
|
BCS1, cytochrome BC1 synthesis |
0.186 |
6.040 |
| 50 |
251443_at |
AT3G59940
|
KFB50, Kelch repeat F-box 50, KMD4, KISS ME DEADLY 4 |
0.181 |
-2.168 |
| 51 |
245275_at |
AT4G15210
|
AT-BETA-AMY, ATBETA-AMY, ARABIDOPSIS THALIANA BETA-AMYLASE, BAM5, beta-amylase 5, BMY1, RAM1, REDUCED BETA AMYLASE 1 |
0.178 |
-1.423 |
| 52 |
257623_at |
AT3G26210
|
CYP71B23, cytochrome P450, family 71, subfamily B, polypeptide 23 |
0.173 |
3.550 |
| 53 |
265724_at |
AT2G32100
|
ATOFP16, RABIDOPSIS THALIANA OVATE FAMILY PROTEIN 16, OFP16, ovate family protein 16 |
0.172 |
-2.804 |
| 54 |
247540_at |
AT5G61590
|
[Integrase-type DNA-binding superfamily protein] |
0.167 |
0.805 |
| 55 |
260037_at |
AT1G68840
|
AtRAV2, EDF2, ETHYLENE RESPONSE DNA BINDING FACTOR 2, RAP2.8, RELATED TO AP2 8, RAV2, related to ABI3/VP1 2, TEM2, TEMPRANILLO 2 |
0.166 |
1.200 |
| 56 |
255753_at |
AT1G18570
|
AtMYB51, myb domain protein 51, BW51A, BW51B, HIG1, HIGH INDOLIC GLUCOSINOLATE 1, MYB51, myb domain protein 51 |
0.165 |
4.885 |
| 57 |
254314_at |
AT4G22470
|
[protease inhibitor/seed storage/lipid transfer protein (LTP) family protein] |
0.162 |
5.713 |
| 58 |
252250_at |
AT3G49790
|
[Carbohydrate-binding protein] |
0.162 |
0.672 |
| 59 |
252320_at |
AT3G48580
|
XTH11, xyloglucan endotransglucosylase/hydrolase 11 |
0.159 |
0.036 |
| 60 |
251428_at |
AT3G60140
|
BGLU30, BETA GLUCOSIDASE 30, DIN2, DARK INDUCIBLE 2, SRG2, SENESCENCE-RELATED GENE 2 |
-0.393 |
-3.188 |
| 61 |
249082_at |
AT5G44120
|
ATCRA1, CRUCIFERINA, CRA1, CRUCIFERINA, CRU1 |
-0.380 |
0.225 |
| 62 |
256814_at |
AT3G21370
|
BGLU19, beta glucosidase 19 |
-0.190 |
0.225 |
| 63 |
266098_at |
AT2G37870
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.137 |
0.138 |
| 64 |
255369_at |
AT4G04080
|
ATISU3, ISCU-LIKE 3, ISU3, ISCU-like 3 |
-0.135 |
0.225 |
| 65 |
261781_at |
AT1G76320
|
CPD25, CHLOROPLAST DIVISION MUTANT 25, FRS4, FAR1-related sequence 4 |
-0.129 |
0.306 |
| 66 |
266693_at |
AT2G19800
|
MIOX2, myo-inositol oxygenase 2 |
-0.105 |
3.172 |
| 67 |
256875_at |
AT3G26330
|
CYP71B37, cytochrome P450, family 71, subfamily B, polypeptide 37 |
-0.098 |
-0.390 |
| 68 |
248890_at |
AT5G46270
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.096 |
-0.977 |
| 69 |
265831_at |
AT2G14460
|
unknown |
-0.096 |
-0.479 |
| 70 |
263796_at |
AT2G24540
|
AFR, ATTENUATED FAR-RED RESPONSE |
-0.094 |
-2.113 |
| 71 |
254546_at |
AT4G19850
|
ATPP2-A2, ARABIDOPSIS THALIANA PHLOEM PROTEIN 2-A2, PP2-A2, phloem protein 2-A2, PP2A2 |
-0.093 |
-0.504 |
| 72 |
266778_at |
AT2G29090
|
CYP707A2, cytochrome P450, family 707, subfamily A, polypeptide 2 |
-0.092 |
-1.728 |
| 73 |
245024_at |
ATCG00120
|
ATPA, ATP synthase subunit alpha |
-0.091 |
-0.131 |
| 74 |
250261_at |
AT5G13400
|
[Major facilitator superfamily protein] |
-0.090 |
-2.411 |
| 75 |
263219_at |
AT1G30600
|
[Subtilase family protein] |
-0.088 |
-0.431 |
| 76 |
266465_at |
AT2G47750
|
GH3.9, putative indole-3-acetic acid-amido synthetase GH3.9 |
-0.085 |
-1.504 |
| 77 |
251513_at |
AT3G59220
|
ATPIRIN1, PRN, pirin, PRN1 |
-0.082 |
2.347 |
| 78 |
248413_at |
AT5G51600
|
ATMAP65-3, ARABIDOPSIS THALIANA MICROTUBULE-ASSOCIATED PROTEIN 65-3, MAP65-3, MICROTUBULE-ASSOCIATED PROTEIN 65-3, PLE, PLEIADE |
-0.082 |
-0.169 |
| 79 |
259596_at |
AT1G28130
|
GH3.17 |
-0.082 |
-0.606 |
| 80 |
261070_at |
AT1G07390
|
AtRLP1, receptor like protein 1, RLP1, receptor like protein 1 |
-0.081 |
1.052 |
| 81 |
247183_at |
AT5G65440
|
unknown |
-0.081 |
-0.762 |
| 82 |
262613_at |
AT1G13960
|
WRKY4, WRKY DNA-binding protein 4 |
-0.081 |
0.981 |
| 83 |
258423_at |
AT3G16730
|
HEB2, hypersensitive to excess boron 2 |
-0.081 |
0.437 |
| 84 |
252262_at |
AT3G49510
|
[F-box family protein] |
-0.081 |
-1.314 |
| 85 |
260124_at |
AT1G36340
|
UBC31, ubiquitin-conjugating enzyme 31 |
-0.079 |
0.225 |
| 86 |
265111_at |
AT1G62510
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.078 |
-2.593 |
| 87 |
255562_at |
AT4G02070
|
ATMSH6, ARABIDOPSIS THALIANA MUTS HOMOLOG 6, MSH6, MUTS homolog 6, MSH6-1, MUTS HOMOLOG 6-1 |
-0.077 |
-0.465 |
| 88 |
251851_at |
AT3G54670
|
ATSMC1, SMC1, STRUCTURAL MAINTENANCE OF CHROMOSOMES 1, TTN8, TITAN8 |
-0.076 |
0.145 |
| 89 |
254572_at |
AT4G19380
|
[Long-chain fatty alcohol dehydrogenase family protein] |
-0.076 |
0.368 |
| 90 |
266160_at |
AT2G28180
|
ATCHX8, CHX08, CHX8, CATION/H+ EXCHANGER 8 |
-0.075 |
-0.442 |
| 91 |
260556_at |
AT2G43620
|
[Chitinase family protein] |
-0.075 |
4.716 |
| 92 |
263658_at |
AT1G04490
|
unknown |
-0.074 |
4.101 |
| 93 |
262523_at |
AT1G17110
|
UBP15, ubiquitin-specific protease 15 |
-0.074 |
0.158 |
| 94 |
257105_at |
AT3G15300
|
[VQ motif-containing protein] |
-0.073 |
-0.720 |
| 95 |
266006_at |
AT2G37360
|
ABCG2, ATP-binding cassette G2 |
-0.072 |
0.602 |
| 96 |
266541_at |
AT2G35110
|
GRL, GNARLED, NAP1, NCK-ASSOCIATED PROTEIN 1, NAPP |
-0.071 |
-0.730 |
| 97 |
255345_at |
AT4G04460
|
[Saposin-like aspartyl protease family protein] |
-0.071 |
-0.042 |
| 98 |
262783_at |
AT1G10850
|
[Leucine-rich repeat protein kinase family protein] |
-0.071 |
-1.209 |
| 99 |
258660_at |
AT3G09850
|
[D111/G-patch domain-containing protein] |
-0.070 |
0.132 |
| 100 |
246572_at |
AT5G15010
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.070 |
-1.346 |
| 101 |
256364_at |
AT1G66550
|
ATWRKY67, WRKY67, WRKY DNA-binding protein 67 |
-0.070 |
3.154 |
| 102 |
257635_at |
AT3G26280
|
CYP71B4, cytochrome P450, family 71, subfamily B, polypeptide 4 |
-0.070 |
-2.964 |
| 103 |
256201_at |
AT1G58230
|
[binding] |
-0.069 |
0.397 |
| 104 |
248112_at |
AT5G55350
|
[MBOAT (membrane bound O-acyl transferase) family protein] |
-0.069 |
-0.147 |
| 105 |
261158_at |
AT1G34500
|
[MBOAT (membrane bound O-acyl transferase) family protein] |
-0.069 |
0.225 |
| 106 |
254938_at |
AT4G10770
|
ATOPT7, ARABIDOPSIS THALIANA OLIGOPEPTIDE TRANSPORTER 7, OPT7, oligopeptide transporter 7 |
-0.069 |
-2.829 |
| 107 |
253471_at |
AT4G32220
|
[gypsy-like retrotransposon family, has a 4.8e-57 P-value blast match to GB:AAD19359 polyprotein (gypsy_Ty3-element) (Sorghum bicolor)] |
-0.068 |
0.225 |
| 108 |
244999_at |
ATCG00190
|
RPOB, RNA polymerase subunit beta |
-0.068 |
0.038 |
| 109 |
256900_at |
AT3G24670
|
[Pectin lyase-like superfamily protein] |
-0.068 |
0.228 |
| 110 |
255326_at |
AT4G04300
|
[pseudogene] |
-0.066 |
0.225 |
| 111 |
256945_at |
AT3G19020
|
[Leucine-rich repeat (LRR) family protein] |
-0.066 |
1.034 |
| 112 |
263207_at |
AT1G10550
|
XET, XYLOGLUCAN:XYLOGLUCOSYL TRANSFERASE 33, XTH33, xyloglucan:xyloglucosyl transferase 33 |
-0.066 |
-0.665 |
| 113 |
260048_at |
AT1G73750
|
[Uncharacterised conserved protein UCP031088, alpha/beta hydrolase] |
-0.065 |
-2.376 |
| 114 |
249996_at |
AT5G18600
|
[Thioredoxin superfamily protein] |
-0.065 |
0.718 |
| 115 |
249676_at |
AT5G35960
|
[Protein kinase family protein] |
-0.065 |
0.400 |
| 116 |
266504_at |
AT2G47820
|
unknown |
-0.065 |
0.398 |
| 117 |
251700_at |
AT3G56640
|
SEC15A, exocyst complex component sec15A |
-0.065 |
-0.209 |