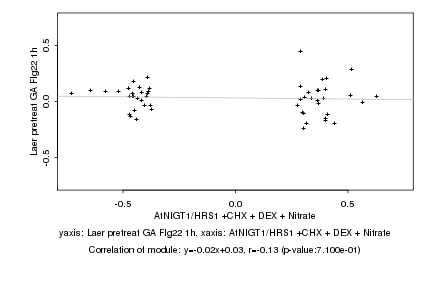
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
AtNIGT1/HRS1 +CHX + DEX + Nitrate |
Log2 signal ratio
Laer pretreat GA Flg22 1h |
| 1 |
249738_at |
AT5G24510
|
[60S acidic ribosomal protein family] |
0.623 |
0.051 |
| 2 |
266393_at |
AT2G41260
|
ATM17, M17 |
0.562 |
0.000 |
| 3 |
256243_at |
AT3G12500
|
ATHCHIB, basic chitinase, B-CHI, CHI-B, HCHIB, basic chitinase, PR-3, PATHOGENESIS-RELATED 3, PR3, PATHOGENESIS-RELATED 3 |
0.512 |
0.289 |
| 4 |
265316_at |
AT2G20400
|
[myb-like HTH transcriptional regulator family protein] |
0.511 |
0.057 |
| 5 |
258995_at |
AT3G01790
|
[Ribosomal protein L13 family protein] |
0.438 |
-0.192 |
| 6 |
250854_at |
AT5G04710
|
[Zn-dependent exopeptidases superfamily protein] |
0.408 |
-0.115 |
| 7 |
248704_at |
AT5G48450
|
sks3, SKU5 similar 3 |
0.401 |
0.207 |
| 8 |
247405_at |
AT5G62880
|
ARAC10, RAC-like 10, ATRAC10, ARABIDOPSIS THALIANA RAC-LIKE 10, ATROP11, RHO-RELATED PROTEIN FROM PLANTS 11, RAC10, RAC-like 10, ROP11, RHO-related protein from plants 11 |
0.399 |
-0.146 |
| 9 |
245279_at |
AT4G17270
|
[Mo25 family protein] |
0.399 |
0.110 |
| 10 |
266905_at |
AT2G34560
|
AtCCP1, CCP1, conserved in ciliated species and in the land plants 1 |
0.397 |
-0.168 |
| 11 |
266577_at |
AT2G23945
|
[Eukaryotic aspartyl protease family protein] |
0.388 |
0.033 |
| 12 |
261640_at |
AT1G49960
|
[Xanthine/uracil permease family protein] |
0.384 |
0.204 |
| 13 |
249775_at |
AT5G24160
|
SQE6, squalene monoxygenase 6 |
0.368 |
-0.014 |
| 14 |
256209_at |
AT1G50940
|
ETFALPHA, electron transfer flavoprotein alpha |
0.367 |
0.106 |
| 15 |
251708_at |
AT3G56580
|
AtRZF1, RZF1, RING Zinc Finger 1 |
0.364 |
0.013 |
| 16 |
266861_at |
AT2G26830
|
emb1187, embryo defective 1187 |
0.363 |
0.102 |
| 17 |
258689_at |
AT3G07940
|
[Calcium-dependent ARF-type GTPase activating protein family] |
0.335 |
0.033 |
| 18 |
263215_at |
AT1G30710
|
[FAD-binding Berberine family protein] |
0.323 |
0.082 |
| 19 |
260384_at |
AT1G74040
|
IMS1, 2-isopropylmalate synthase 1, IPMS2, SOPROPYLMALATE SYNTHASE 2, MAML-3 |
0.312 |
-0.193 |
| 20 |
250331_at |
AT5G11820
|
[Plant self-incompatibility protein S1 family] |
0.306 |
0.037 |
| 21 |
261112_at |
AT1G75420
|
[UDP-Glycosyltransferase superfamily protein] |
0.302 |
-0.234 |
| 22 |
266904_at |
AT2G34590
|
[Transketolase family protein] |
0.299 |
-0.105 |
| 23 |
246948_at |
AT5G25130
|
CYP71B12, cytochrome P450, family 71, subfamily B, polypeptide 12 |
0.294 |
-0.092 |
| 24 |
266489_at |
AT2G35190
|
ATNPSN11, NPSN11, novel plant snare 11, NSPN11 |
0.289 |
0.452 |
| 25 |
256322_at |
AT1G54990
|
AXR4, AUXIN RESISTANT 4, RGR, REDUCED ROOT GRAVITROPISM, RGR1, REDUCED ROOT GRAVITROPISM 1 |
0.288 |
0.021 |
| 26 |
247255_at |
AT5G64780
|
[Uncharacterised conserved protein UCP009193] |
0.285 |
0.137 |
| 27 |
266050_at |
AT2G40770
|
[zinc ion binding] |
0.272 |
-0.034 |
| 28 |
263584_at |
AT2G17040
|
anac036, NAC domain containing protein 36, NAC036, NAC domain containing protein 36 |
-0.732 |
0.074 |
| 29 |
262510_at |
AT1G11270
|
[F-box and associated interaction domains-containing protein] |
-0.645 |
0.101 |
| 30 |
250388_at |
AT5G11310
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.578 |
0.090 |
| 31 |
246568_at |
AT5G14960
|
DEL2, DP-E2F-like 2, E2FD, E2L1 |
-0.521 |
0.097 |
| 32 |
252513_at |
AT3G46220
|
unknown |
-0.476 |
0.124 |
| 33 |
258354_at |
AT3G14320
|
[Zinc finger, C3HC4 type (RING finger) family protein] |
-0.475 |
0.046 |
| 34 |
267336_at |
AT2G19310
|
[HSP20-like chaperones superfamily protein] |
-0.473 |
-0.114 |
| 35 |
267556_at |
AT2G32810
|
BGAL9, beta galactosidase 9, BGAL9, beta-galactosidase 9 |
-0.467 |
-0.130 |
| 36 |
253948_at |
AT4G26940
|
[Galactosyltransferase family protein] |
-0.459 |
0.078 |
| 37 |
248657_at |
AT5G48570
|
ATFKBP65, FKBP65, ROF2 |
-0.458 |
0.045 |
| 38 |
250925_at |
AT5G03370
|
[acylphosphatase family] |
-0.456 |
0.045 |
| 39 |
265908_at |
AT4G00270
|
[DNA-binding storekeeper protein-related transcriptional regulator] |
-0.455 |
0.184 |
| 40 |
257051_at |
AT3G15270
|
SPL5, squamosa promoter binding protein-like 5 |
-0.450 |
-0.080 |
| 41 |
248541_at |
AT5G50180
|
[Protein kinase superfamily protein] |
-0.444 |
-0.156 |
| 42 |
258336_at |
AT3G16050
|
A37, ATPDX1.2, ARABIDOPSIS THALIANA PYRIDOXINE BIOSYNTHESIS 1.2, PDX1.2, pyridoxine biosynthesis 1.2 |
-0.437 |
0.028 |
| 43 |
258984_at |
AT3G08970
|
ATERDJ3A, TMS1, THERMOSENSITIVE MALE STERILE 1 |
-0.430 |
0.130 |
| 44 |
255547_at |
AT4G01920
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.422 |
0.084 |
| 45 |
246475_at |
AT5G16720
|
MyoB3, myosin binding protein 3 |
-0.420 |
0.011 |
| 46 |
253095_at |
AT4G37510
|
[Ribonuclease III family protein] |
-0.407 |
-0.034 |
| 47 |
259037_at |
AT3G09350
|
Fes1A, Fes1A |
-0.396 |
0.046 |
| 48 |
261069_at |
AT1G07410
|
ATRAB-A2B, ARABIDOPSIS RAB GTPASE HOMOLOG A2B, ATRABA2B, RAB GTPase homolog A2B, RAB-A2B, RAB GTPASE HOMOLOG A2B, RABA2b, RAB GTPase homolog A2B |
-0.393 |
0.218 |
| 49 |
248280_at |
AT5G52950
|
unknown |
-0.392 |
0.075 |
| 50 |
261201_at |
AT1G12850
|
[Phosphoglycerate mutase family protein] |
-0.389 |
0.098 |
| 51 |
266590_at |
AT2G46240
|
ATBAG6, ARABIDOPSIS THALIANA BCL-2-ASSOCIATED ATHANOGENE 6, BAG6, BCL-2-associated athanogene 6 |
-0.384 |
0.123 |
| 52 |
261016_at |
AT1G26560
|
BGLU40, beta glucosidase 40 |
-0.378 |
-0.028 |
| 53 |
245725_at |
AT1G73370
|
ATSUS6, ARABIDOPSIS THALIANA SUCROSE SYNTHASE 6, SUS6, sucrose synthase 6 |
-0.377 |
-0.070 |