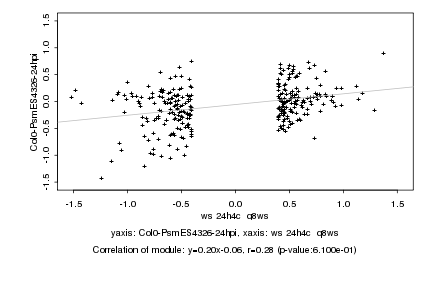
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
ws 24h4c q8ws |
Log2 signal ratio
Col0-PsmES4326-24hpi |
| 1 |
264400_at |
AT1G61800
|
ATGPT2, ARABIDOPSIS GLUCOSE-6-PHOSPHATE/PHOSPHATE TRANSLOCATOR 2, GPT2, glucose-6-phosphate/phosphate translocator 2 |
1.370 |
0.907 |
| 2 |
264511_at |
AT1G09350
|
AtGolS3, galactinol synthase 3, GolS3, galactinol synthase 3 |
1.288 |
-0.156 |
| 3 |
249848_at |
AT5G23220
|
NIC3, nicotinamidase 3 |
1.176 |
0.158 |
| 4 |
253595_at |
AT4G30830
|
unknown |
1.139 |
0.047 |
| 5 |
253293_at |
AT4G33905
|
[Peroxisomal membrane 22 kDa (Mpv17/PMP22) family protein] |
1.119 |
0.284 |
| 6 |
245749_at |
AT1G51090
|
[Heavy metal transport/detoxification superfamily protein ] |
0.977 |
0.254 |
| 7 |
251793_at |
AT3G55580
|
[Regulator of chromosome condensation (RCC1) family protein] |
0.974 |
-0.058 |
| 8 |
264940_at |
AT1G60470
|
AtGolS4, galactinol synthase 4, GolS4, galactinol synthase 4 |
0.933 |
0.257 |
| 9 |
245306_at |
AT4G14690
|
ELIP2, EARLY LIGHT-INDUCIBLE PROTEIN 2 |
0.920 |
-0.082 |
| 10 |
250135_at |
AT5G15360
|
unknown |
0.904 |
-0.011 |
| 11 |
265290_at |
AT2G22590
|
[UDP-Glycosyltransferase superfamily protein] |
0.899 |
0.105 |
| 12 |
254566_at |
AT4G19240
|
unknown |
0.896 |
0.099 |
| 13 |
246922_at |
AT5G25110
|
CIPK25, CBL-interacting protein kinase 25, SnRK3.25, SNF1-RELATED PROTEIN KINASE 3.25 |
0.889 |
0.027 |
| 14 |
258893_at |
AT3G05660
|
AtRLP33, receptor like protein 33, RLP33, receptor like protein 33 |
0.835 |
0.100 |
| 15 |
266532_at |
AT2G16890
|
[UDP-Glycosyltransferase superfamily protein] |
0.834 |
0.571 |
| 16 |
248729_at |
AT5G48010
|
AtTHAS1, Arabidopsis thaliana thalianol synthase 1, THAS, THALIANOL SYNTHASE, THAS1, thalianol synthase 1 |
0.831 |
0.139 |
| 17 |
265147_at |
AT1G51380
|
[DEA(D/H)-box RNA helicase family protein] |
0.815 |
-0.050 |
| 18 |
246419_at |
AT5G17030
|
UGT78D3, UDP-glucosyl transferase 78D3 |
0.788 |
0.099 |
| 19 |
257876_at |
AT3G17130
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.785 |
-0.184 |
| 20 |
253104_at |
AT4G36010
|
[Pathogenesis-related thaumatin superfamily protein] |
0.785 |
0.309 |
| 21 |
252316_at |
AT3G48700
|
ATCXE13, carboxyesterase 13, CXE13, carboxyesterase 13 |
0.774 |
0.139 |
| 22 |
254075_at |
AT4G25470
|
ATCBF2, CBF2, C-repeat/DRE binding factor 2, DREB1C, DRE/CRT-BINDING PROTEIN 1C, FTQ4, FREEZING TOLERANCE QTL 4 |
0.758 |
0.056 |
| 23 |
258321_at |
AT3G22840
|
ELIP, ELIP1, EARLY LIGHT-INDUCABLE PROTEIN |
0.756 |
0.148 |
| 24 |
256779_at |
AT3G13784
|
AtcwINV5, cell wall invertase 5, CWINV5, cell wall invertase 5 |
0.751 |
0.097 |
| 25 |
250826_at |
AT5G05220
|
unknown |
0.751 |
0.444 |
| 26 |
255958_at |
AT1G22150
|
SULTR1;3, sulfate transporter 1;3 |
0.739 |
0.165 |
| 27 |
260140_at |
AT1G66390
|
ATMYB90, MYB90, myb domain protein 90, PAP2, PRODUCTION OF ANTHOCYANIN PIGMENT 2 |
0.728 |
0.682 |
| 28 |
262050_at |
AT1G80130
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.724 |
-0.674 |
| 29 |
256602_at |
AT3G28310
|
unknown |
0.719 |
0.083 |
| 30 |
246703_at |
AT5G28080
|
WNK9 |
0.699 |
-0.039 |
| 31 |
261187_at |
AT1G32860
|
[Glycosyl hydrolase superfamily protein] |
0.694 |
0.009 |
| 32 |
248160_at |
AT5G54470
|
BBX29, B-box domain protein 29 |
0.687 |
0.090 |
| 33 |
245904_at |
AT5G11110
|
ATSPS2F, sucrose phosphate synthase 2F, KNS2, KAONASHI 2, SPS1, SUCROSE PHOSPHATE SYNTHASE 1, SPS2F, sucrose phosphate synthase 2F, SPSA2, sucrose-phosphate synthase A2 |
0.680 |
0.621 |
| 34 |
247655_at |
AT5G59820
|
AtZAT12, RHL41, RESPONSIVE TO HIGH LIGHT 41, ZAT12 |
0.668 |
0.744 |
| 35 |
256631_at |
AT3G28320
|
unknown |
0.666 |
0.067 |
| 36 |
251753_at |
AT3G55760
|
unknown |
0.666 |
-0.239 |
| 37 |
246282_at |
AT4G36580
|
[AAA-type ATPase family protein] |
0.661 |
-0.134 |
| 38 |
260264_at |
AT1G68500
|
unknown |
0.660 |
0.245 |
| 39 |
256852_at |
AT3G18610
|
ATNUC-L2, nucleolin like 2, NUC-L2, nucleolin like 2, PARLL1, PARALLEL1-LIKE 1 |
0.636 |
-0.015 |
| 40 |
250504_at |
AT5G09840
|
[Putative endonuclease or glycosyl hydrolase] |
0.631 |
-0.233 |
| 41 |
247070_at |
AT5G66815
|
unknown |
0.624 |
0.021 |
| 42 |
245427_at |
AT4G17550
|
AtG3Pp4, glycerol-3-phosphate permease 4, G3Pp4, glycerol-3-phosphate permease 4 |
0.620 |
-0.007 |
| 43 |
264907_at |
AT2G17280
|
[Phosphoglycerate mutase family protein] |
0.616 |
-0.093 |
| 44 |
262421_at |
AT1G50290
|
unknown |
0.608 |
-0.071 |
| 45 |
253975_at |
AT4G26600
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.595 |
-0.335 |
| 46 |
260357_at |
AT1G69260
|
AFP1, ABI five binding protein |
0.593 |
0.524 |
| 47 |
252355_at |
AT3G48250
|
BIR6, Buthionine sulfoximine-insensitive roots 6 |
0.587 |
-0.321 |
| 48 |
248870_at |
AT5G46710
|
[PLATZ transcription factor family protein] |
0.584 |
0.188 |
| 49 |
262760_at |
AT1G10770
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.578 |
0.089 |
| 50 |
245770_at |
AT1G30240
|
[FUNCTIONS IN: binding] |
0.575 |
-0.337 |
| 51 |
265483_at |
AT2G15790
|
CYP40, CYCLOPHILIN 40, SQN, SQUINT |
0.571 |
0.032 |
| 52 |
247287_at |
AT5G64230
|
unknown |
0.570 |
0.466 |
| 53 |
250279_at |
AT5G13200
|
[GRAM domain family protein] |
0.564 |
0.482 |
| 54 |
246461_at |
AT5G16930
|
[AAA-type ATPase family protein] |
0.563 |
-0.209 |
| 55 |
264261_at |
AT1G09240
|
ATNAS3, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 3, NAS3, nicotianamine synthase 3 |
0.562 |
0.142 |
| 56 |
245734_at |
AT1G73480
|
[alpha/beta-Hydrolases superfamily protein] |
0.562 |
0.254 |
| 57 |
258270_at |
AT3G15650
|
[alpha/beta-Hydrolases superfamily protein] |
0.561 |
-0.042 |
| 58 |
258871_at |
AT3G03060
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.556 |
-0.022 |
| 59 |
254667_at |
AT4G18280
|
[glycine-rich cell wall protein-related] |
0.556 |
0.017 |
| 60 |
251229_at |
AT3G62740
|
BGLU7, beta glucosidase 7 |
0.554 |
0.019 |
| 61 |
264513_at |
AT1G09420
|
G6PD4, glucose-6-phosphate dehydrogenase 4 |
0.551 |
0.450 |
| 62 |
257650_at |
AT3G16800
|
[Protein phosphatase 2C family protein] |
0.545 |
0.088 |
| 63 |
253182_at |
AT4G35190
|
LOG5, LONELY GUY 5 |
0.544 |
0.123 |
| 64 |
251346_at |
AT3G60980
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.543 |
0.117 |
| 65 |
247739_at |
AT5G59240
|
[Ribosomal protein S8e family protein] |
0.542 |
-0.027 |
| 66 |
248118_at |
AT5G55050
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.542 |
-0.111 |
| 67 |
257035_at |
AT3G19270
|
CYP707A4, cytochrome P450, family 707, subfamily A, polypeptide 4 |
0.538 |
0.608 |
| 68 |
258887_at |
AT3G05630
|
PDLZ2, PHOSPHOLIPASE D ZETA 2, PLDP2, phospholipase D P2, PLDZETA2, PHOSPHOLIPASE D ZETA 2 |
0.536 |
0.656 |
| 69 |
245879_at |
AT5G09420
|
AtmtOM64, ATTOC64-V, translocon at the outer membrane of chloroplasts 64-V, MTOM64, ARABIDOPSIS THALIANA TRANSLOCON AT THE OUTER MEMBRANE OF CHLOROPLASTS 64-V, OM64, outer membrane 64, TOC64-V, translocon at the outer membrane of chloroplasts 64-V |
0.528 |
0.190 |
| 70 |
258965_at |
AT3G10530
|
[Transducin/WD40 repeat-like superfamily protein] |
0.526 |
-0.401 |
| 71 |
252209_at |
AT3G50400
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.525 |
0.539 |
| 72 |
264211_at |
AT1G22770
|
FB, GI, GIGANTEA |
0.525 |
0.113 |
| 73 |
246519_at |
AT5G15780
|
[Pollen Ole e 1 allergen and extensin family protein] |
0.524 |
-0.200 |
| 74 |
262061_at |
AT1G80110
|
ATPP2-B11, phloem protein 2-B11, PP2-B11, phloem protein 2-B11 |
0.524 |
0.567 |
| 75 |
265154_at |
AT1G30960
|
[GTP-binding family protein] |
0.515 |
-0.168 |
| 76 |
257694_at |
AT3G12860
|
[NOP56-like pre RNA processing ribonucleoprotein] |
0.512 |
-0.028 |
| 77 |
247776_at |
AT5G58700
|
ATPLC4, PLC4, phosphatidylinositol-speciwc phospholipase C4 |
0.511 |
0.068 |
| 78 |
257237_at |
AT3G14890
|
[phosphoesterase] |
0.510 |
-0.144 |
| 79 |
252305_at |
AT3G49240
|
emb1796, embryo defective 1796 |
0.505 |
-0.411 |
| 80 |
254304_at |
AT4G22270
|
ATMRB1, ARABIDOPSIS MEMBRANE RELATED BIGGER1, MRB1, MEMBRANE RELATED BIGGER1 |
0.505 |
0.156 |
| 81 |
266510_at |
AT2G47990
|
EDA13, EMBRYO SAC DEVELOPMENT ARREST 13, EDA19, EMBRYO SAC DEVELOPMENT ARREST 19, SWA1, SLOW WALKER1 |
0.502 |
-0.168 |
| 82 |
261544_at |
AT1G63540
|
[hydroxyproline-rich glycoprotein family protein] |
0.502 |
0.069 |
| 83 |
252076_at |
AT3G51660
|
[Tautomerase/MIF superfamily protein] |
0.495 |
0.460 |
| 84 |
266209_at |
AT2G27550
|
ATC, centroradialis |
0.495 |
0.027 |
| 85 |
255278_at |
AT4G04940
|
[transducin family protein / WD-40 repeat family protein] |
0.495 |
-0.357 |
| 86 |
256014_at |
AT1G19200
|
unknown |
0.493 |
0.514 |
| 87 |
259133_at |
AT3G05400
|
[Major facilitator superfamily protein] |
0.492 |
0.677 |
| 88 |
251611_at |
AT3G57940
|
unknown |
0.491 |
-0.472 |
| 89 |
245998_at |
AT5G20830
|
ASUS1, atsus1, SUS1, sucrose synthase 1 |
0.486 |
0.413 |
| 90 |
263374_at |
AT2G20560
|
[DNAJ heat shock family protein] |
0.485 |
0.624 |
| 91 |
258166_at |
AT3G21540
|
[transducin family protein / WD-40 repeat family protein] |
0.481 |
-0.307 |
| 92 |
254085_at |
AT4G24960
|
ATHVA22D, ARABIDOPSIS THALIANA HVA22 HOMOLOGUE D, HVA22D, HVA22 homologue D |
0.478 |
0.015 |
| 93 |
257487_at |
AT1G71850
|
[Ubiquitin carboxyl-terminal hydrolase family protein] |
0.477 |
-0.263 |
| 94 |
258827_at |
AT3G07150
|
unknown |
0.473 |
0.132 |
| 95 |
263352_at |
AT2G22080
|
unknown |
0.471 |
-0.106 |
| 96 |
254818_at |
AT4G12470
|
AZI1, azelaic acid induced 1 |
0.467 |
0.008 |
| 97 |
260824_at |
AT1G06720
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.463 |
-0.540 |
| 98 |
259971_at |
AT1G76580
|
[Squamosa promoter-binding protein-like (SBP domain) transcription factor family protein] |
0.463 |
0.325 |
| 99 |
254090_at |
AT4G25010
|
AtSWEET14, SWEET14 |
0.457 |
0.222 |
| 100 |
257652_at |
AT3G16810
|
APUM24, pumilio 24, PUM24, pumilio 24 |
0.454 |
-0.322 |
| 101 |
250758_at |
AT5G06000
|
ATEIF3G2, ARABIDOPSIS THALIANA EUKARYOTIC TRANSLATION INITIATION FACTOR 3G2, EIF3G2, eukaryotic translation initiation factor 3G2 |
0.454 |
-0.139 |
| 102 |
262584_at |
AT1G15440
|
ATPWP2, PERIODIC TRYPTOPHAN PROTEIN 2, PWP2, periodic tryptophan protein 2 |
0.452 |
-0.219 |
| 103 |
249741_at |
AT5G24470
|
APRR5, pseudo-response regulator 5, PRR5, pseudo-response regulator 5 |
0.452 |
0.238 |
| 104 |
261377_at |
AT1G18850
|
unknown |
0.451 |
-0.198 |
| 105 |
248505_at |
AT5G50360
|
unknown |
0.451 |
0.303 |
| 106 |
267371_at |
AT2G44510
|
[CDK inhibitor P21 binding protein] |
0.450 |
-0.013 |
| 107 |
253219_at |
AT4G34990
|
AtMYB32, myb domain protein 32, MYB32, myb domain protein 32 |
0.446 |
0.072 |
| 108 |
251432_at |
AT3G59820
|
AtLETM1, LETM1, leucine zipper-EF-hand-containing transmembrane protein 1 |
0.446 |
-0.031 |
| 109 |
261168_at |
AT1G04945
|
[HIT-type Zinc finger family protein] |
0.445 |
-0.088 |
| 110 |
258505_at |
AT3G06530
|
[ARM repeat superfamily protein] |
0.443 |
-0.520 |
| 111 |
247612_at |
AT5G60730
|
[Anion-transporting ATPase] |
0.442 |
-0.050 |
| 112 |
260978_at |
AT1G53540
|
[HSP20-like chaperones superfamily protein] |
0.441 |
0.586 |
| 113 |
250418_at |
AT5G11240
|
[transducin family protein / WD-40 repeat family protein] |
0.441 |
-0.442 |
| 114 |
267309_at |
AT2G19385
|
[zinc ion binding] |
0.439 |
-0.244 |
| 115 |
255225_at |
AT4G05410
|
YAO, YAOZHE |
0.439 |
-0.185 |
| 116 |
256288_at |
AT3G12270
|
ATPRMT3, ARABIDOPSIS THALIANA PROTEIN ARGININE METHYLTRANSFERASE 3, PRMT3, protein arginine methyltransferase 3 |
0.438 |
-0.354 |
| 117 |
266934_at |
AT2G18900
|
[Transducin/WD40 repeat-like superfamily protein] |
0.434 |
-0.155 |
| 118 |
259503_at |
AT1G15870
|
[Mitochondrial glycoprotein family protein] |
0.434 |
0.017 |
| 119 |
251915_at |
AT3G53940
|
[Mitochondrial substrate carrier family protein] |
0.433 |
-0.001 |
| 120 |
252871_at |
AT4G40000
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.433 |
-0.088 |
| 121 |
259426_at |
AT1G01470
|
LEA14, LATE EMBRYOGENESIS ABUNDANT 14, LSR3, LIGHT STRESS-REGULATED 3 |
0.427 |
0.162 |
| 122 |
247319_at |
AT5G64050
|
ATERS, ERS, glutamate tRNA synthetase, OVA3, OVULE ABORTION 3 |
0.426 |
-0.136 |
| 123 |
249528_at |
AT5G38720
|
unknown |
0.426 |
-0.207 |
| 124 |
259516_at |
AT1G20450
|
ERD10, EARLY RESPONSIVE TO DEHYDRATION 10, LTI29, LOW TEMPERATURE INDUCED 29, LTI45, LOW TEMPERATURE INDUCED 45 |
0.425 |
0.056 |
| 125 |
261899_at |
AT1G80820
|
ATCCR2, CCR2, cinnamoyl coa reductase |
0.422 |
0.507 |
| 126 |
260331_at |
AT1G80270
|
PPR596, PENTATRICOPEPTIDE REPEAT 596 |
0.420 |
-0.490 |
| 127 |
247046_at |
AT5G66540
|
unknown |
0.419 |
-0.181 |
| 128 |
251631_at |
AT3G57430
|
OTP84, ORGANELLE TRANSCRIPT PROCESSING 84 |
0.419 |
0.017 |
| 129 |
261065_at |
AT1G07500
|
SMR5, SIAMESE-RELATED 5 |
0.417 |
0.627 |
| 130 |
247393_at |
AT5G63130
|
[Octicosapeptide/Phox/Bem1p family protein] |
0.413 |
0.528 |
| 131 |
250781_at |
AT5G05410
|
DREB2, DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN 2, DREB2A, DRE-binding protein 2A |
0.412 |
0.694 |
| 132 |
252872_at |
AT4G40010
|
SNRK2-7, SUCROSE NONFERMENTING 1-RELATED PROTEIN KINASE 2-7, SNRK2.7, SNF1-related protein kinase 2.7, SRK2F |
0.412 |
0.144 |
| 133 |
260585_at |
AT2G43650
|
EMB2777, EMBRYO DEFECTIVE 2777 |
0.410 |
-0.452 |
| 134 |
253496_at |
AT4G31870
|
ATGPX7, GLUTATHIONE PEROXIDASE 7, GPX7, glutathione peroxidase 7 |
0.408 |
-0.140 |
| 135 |
261651_at |
AT1G27760
|
ATSAT32, SALT-TOLERANCE 32, SAT32, SALT-TOLERANCE 32 |
0.407 |
0.288 |
| 136 |
247676_at |
AT5G59980
|
AtRPP30, GAF1, GAMETOPHYTE DEFECTIVE 1, RPP30 |
0.406 |
-0.110 |
| 137 |
248676_at |
AT5G48850
|
ATSDI1, SULPHUR DEFICIENCY-INDUCED 1 |
0.405 |
0.071 |
| 138 |
262113_at |
AT1G02820
|
AtLEA3, LEA3, late embryogenesis abundant 3 |
0.402 |
-0.244 |
| 139 |
266154_at |
AT2G12190
|
[Cytochrome P450 superfamily protein] |
0.400 |
0.021 |
| 140 |
256797_at |
AT3G18600
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.398 |
-0.535 |
| 141 |
265354_at |
AT2G16700
|
ADF5, actin depolymerizing factor 5, ATADF5 |
0.397 |
0.336 |
| 142 |
247378_at |
AT5G63120
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.395 |
0.213 |
| 143 |
261192_at |
AT1G32870
|
ANAC013, Arabidopsis NAC domain containing protein 13, ANAC13, NAC domain protein 13, NAC13, NAC domain protein 13 |
0.395 |
0.331 |
| 144 |
267576_at |
AT2G30640
|
MUG2, MUSTANG 2 |
0.395 |
0.341 |
| 145 |
262544_at |
AT1G15420
|
unknown |
0.394 |
-0.275 |
| 146 |
260410_at |
AT1G69870
|
AtNPF2.13, NPF2.13, NRT1/ PTR family 2.13, NRT1.7, nitrate transporter 1.7 |
0.393 |
0.420 |
| 147 |
253213_at |
AT4G34910
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.390 |
-0.115 |
| 148 |
251593_at |
AT3G57660
|
NRPA1, nuclear RNA polymerase A1 |
0.390 |
-0.328 |
| 149 |
259173_at |
AT3G03640
|
BGLU25, beta glucosidase 25, GLUC |
0.390 |
0.290 |
| 150 |
251226_at |
AT3G62680
|
ATPRP3, ARABIDOPSIS THALIANA PROLINE-RICH PROTEIN 3, PRP3, proline-rich protein 3 |
0.390 |
0.107 |
| 151 |
254076_at |
AT4G25340
|
ATFKBP53, FKBP53, FK506 BINDING PROTEIN 53 |
0.390 |
-0.291 |
| 152 |
256497_at |
AT1G31580
|
CXC750, ECS1 |
-1.526 |
0.086 |
| 153 |
252462_at |
AT3G47250
|
unknown |
-1.490 |
0.221 |
| 154 |
256527_at |
AT1G66100
|
[Plant thionin] |
-1.430 |
-0.027 |
| 155 |
260221_at |
AT1G74670
|
GASA6, GA-stimulated Arabidopsis 6 |
-1.244 |
-1.425 |
| 156 |
255822_at |
AT2G40610
|
ATEXP8, ATEXPA8, expansin A8, ATHEXP ALPHA 1.11, EXP8, EXPA8, expansin A8 |
-1.157 |
-1.106 |
| 157 |
250942_at |
AT5G03350
|
[Legume lectin family protein] |
-1.144 |
0.037 |
| 158 |
253874_at |
AT4G27450
|
[Aluminium induced protein with YGL and LRDR motifs] |
-1.096 |
0.139 |
| 159 |
252648_at |
AT3G44630
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-1.088 |
0.184 |
| 160 |
267265_at |
AT2G22980
|
SCPL13, serine carboxypeptidase-like 13 |
-1.079 |
-0.768 |
| 161 |
265894_at |
AT2G15050
|
LTP, lipid transfer protein, LTP7, lipid transfer protein 7 |
-1.061 |
-0.909 |
| 162 |
265698_at |
AT2G32160
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-1.034 |
0.121 |
| 163 |
252703_at |
AT3G43740
|
[Leucine-rich repeat (LRR) family protein] |
-1.029 |
-0.204 |
| 164 |
260869_at |
AT1G43800
|
FTM1, FLORAL TRANSITION AT THE MERISTEM1, SAD6, STEAROYL-ACYL CARRIER PROTEIN Δ9-DESATURASE6 |
-1.010 |
0.040 |
| 165 |
264774_at |
AT1G22890
|
unknown |
-1.002 |
0.357 |
| 166 |
263023_at |
AT1G23960
|
unknown |
-0.967 |
0.155 |
| 167 |
252560_at |
AT3G46030
|
HTB11 |
-0.955 |
0.107 |
| 168 |
259384_at |
AT3G16450
|
JAL33, Jacalin-related lectin 33 |
-0.917 |
0.107 |
| 169 |
260312_at |
AT1G63880
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.905 |
0.005 |
| 170 |
245729_at |
AT1G73490
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.890 |
-0.020 |
| 171 |
254585_at |
AT4G19500
|
[nucleoside-triphosphatases] |
-0.881 |
-0.088 |
| 172 |
259629_at |
AT1G56510
|
ADR2, ACTIVATED DISEASE RESISTANCE 2, WRR4, WHITE RUST RESISTANCE 4 |
-0.875 |
0.081 |
| 173 |
252433_at |
AT3G47560
|
[alpha/beta-Hydrolases superfamily protein] |
-0.864 |
-0.446 |
| 174 |
256603_at |
AT3G28270
|
unknown |
-0.862 |
-0.293 |
| 175 |
246275_at |
AT4G36540
|
BEE2, BR enhanced expression 2 |
-0.845 |
-1.204 |
| 176 |
248839_at |
AT5G46690
|
bHLH071, beta HLH protein 71 |
-0.844 |
-0.636 |
| 177 |
249752_at |
AT5G24660
|
LSU2, RESPONSE TO LOW SULFUR 2 |
-0.826 |
-0.308 |
| 178 |
262010_at |
AT1G35612
|
[expressed protein] |
-0.815 |
-0.362 |
| 179 |
254553_at |
AT4G19530
|
[disease resistance protein (TIR-NBS-LRR class) family] |
-0.813 |
-0.721 |
| 180 |
248415_at |
AT5G51620
|
[Uncharacterised protein family (UPF0172)] |
-0.806 |
0.292 |
| 181 |
262705_at |
AT1G16260
|
[Wall-associated kinase family protein] |
-0.801 |
0.063 |
| 182 |
257964_at |
AT3G19850
|
[Phototropic-responsive NPH3 family protein] |
-0.789 |
-0.949 |
| 183 |
250063_at |
AT5G17880
|
CSA1, constitutive shade-avoidance1 |
-0.772 |
0.085 |
| 184 |
252485_at |
AT3G46530
|
RPP13, RECOGNITION OF PERONOSPORA PARASITICA 13 |
-0.772 |
0.159 |
| 185 |
257272_at |
AT3G28130
|
UMAMIT44, Usually multiple acids move in and out Transporters 44 |
-0.768 |
-0.592 |
| 186 |
259839_at |
AT1G52190
|
AtNPF1.2, NPF1.2, NRT1/ PTR family 1.2, NRT1.11, nitrate transporter 1.11 |
-0.767 |
-0.886 |
| 187 |
266279_at |
AT2G29290
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.760 |
-0.970 |
| 188 |
259529_at |
AT1G12400
|
[Nucleotide excision repair, TFIIH, subunit TTDA] |
-0.758 |
-0.019 |
| 189 |
245637_at |
AT1G25230
|
[Calcineurin-like metallo-phosphoesterase superfamily protein] |
-0.754 |
-0.335 |
| 190 |
262458_at |
AT1G11280
|
[S-locus lectin protein kinase family protein] |
-0.738 |
-0.298 |
| 191 |
257714_at |
AT3G27360
|
H3.1, histone 3.1 |
-0.722 |
-0.312 |
| 192 |
257254_at |
AT3G21950
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.721 |
-0.267 |
| 193 |
256299_at |
AT1G69530
|
AT-EXP1, ATEXP1, ATEXPA1, expansin A1, ATHEXP ALPHA 1.2, EXP1, EXPANSIN 1, EXPA1, expansin A1 |
-0.719 |
-0.700 |
| 194 |
254870_at |
AT4G11900
|
[S-locus lectin protein kinase family protein] |
-0.712 |
0.105 |
| 195 |
252168_at |
AT3G50440
|
ATMES10, ARABIDOPSIS THALIANA METHYL ESTERASE 10, MES10, methyl esterase 10 |
-0.705 |
-0.165 |
| 196 |
249850_at |
AT5G23240
|
DJC76, DNA J protein C76 |
-0.697 |
0.552 |
| 197 |
250142_at |
AT5G14650
|
[Pectin lyase-like superfamily protein] |
-0.697 |
0.198 |
| 198 |
258618_at |
AT3G02885
|
GASA5, GAST1 protein homolog 5 |
-0.690 |
-0.173 |
| 199 |
254032_at |
AT4G25940
|
[ENTH/ANTH/VHS superfamily protein] |
-0.687 |
0.228 |
| 200 |
257057_at |
AT3G15310
|
unknown |
-0.686 |
-1.019 |
| 201 |
245451_at |
AT4G16890
|
BAL, BALL, SNC1, SUPPRESSOR OF NPR1-1, CONSTITUTIVE 1 |
-0.683 |
0.171 |
| 202 |
254378_at |
AT4G21810
|
DER2.1, DERLIN-2.1 |
-0.680 |
0.079 |
| 203 |
252250_at |
AT3G49790
|
[Carbohydrate-binding protein] |
-0.670 |
0.216 |
| 204 |
262286_at |
AT1G68585
|
unknown |
-0.665 |
-0.415 |
| 205 |
255484_at |
AT4G02540
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.660 |
0.007 |
| 206 |
257615_at |
AT3G26510
|
[Octicosapeptide/Phox/Bem1p family protein] |
-0.656 |
-0.029 |
| 207 |
263150_at |
AT1G54050
|
[HSP20-like chaperones superfamily protein] |
-0.647 |
-0.349 |
| 208 |
264238_at |
AT1G54740
|
unknown |
-0.639 |
-0.349 |
| 209 |
264482_at |
AT1G77210
|
AtSTP14, sugar transport protein 14, STP14, sugar transport protein 14 |
-0.634 |
-0.022 |
| 210 |
265297_at |
AT2G14080
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.627 |
0.159 |
| 211 |
250433_at |
AT5G10400
|
H3.1, histone 3.1 |
-0.623 |
0.039 |
| 212 |
263674_at |
AT2G04790
|
unknown |
-0.620 |
-0.815 |
| 213 |
267349_at |
AT2G40010
|
[Ribosomal protein L10 family protein] |
-0.620 |
-0.207 |
| 214 |
249694_at |
AT5G35790
|
G6PD1, glucose-6-phosphate dehydrogenase 1 |
-0.610 |
-0.621 |
| 215 |
255786_at |
AT1G19670
|
ATCLH1, chlorophyllase 1, ATHCOR1, CORONATINE-INDUCED PROTEIN 1, CLH1, chlorophyllase 1, CORI1, CORONATINE-INDUCED PROTEIN 1 |
-0.609 |
0.170 |
| 216 |
250008_at |
AT5G18630
|
[alpha/beta-Hydrolases superfamily protein] |
-0.608 |
-0.010 |
| 217 |
265342_at |
AT2G18300
|
HBI1, HOMOLOG OF BEE2 INTERACTING WITH IBH 1 |
-0.608 |
-1.045 |
| 218 |
264379_at |
AT2G25200
|
unknown |
-0.607 |
-0.140 |
| 219 |
261459_at |
AT1G21100
|
IGMT1, indole glucosinolate O-methyltransferase 1 |
-0.605 |
0.441 |
| 220 |
264507_at |
AT1G09415
|
NIMIN-3, NIM1-interacting 3 |
-0.604 |
-0.029 |
| 221 |
252081_at |
AT3G51910
|
AT-HSFA7A, ARABIDOPSIS THALIANA HEAT SHOCK TRANSCRIPTION FACTOR A7A, HSFA7A, heat shock transcription factor A7A |
-0.598 |
0.063 |
| 222 |
252534_at |
AT3G46130
|
ATMYB48, myb domain protein 48, ATMYB48-1, ATMYB48-2, ATMYB48-3, MYB48, myb domain protein 48 |
-0.595 |
-0.603 |
| 223 |
255913_at |
AT1G66980
|
GDPDL2, Glycerophosphodiester phosphodiesterase (GDPD) like 2, SNC4, suppressor of npr1-1 constitutive 4 |
-0.589 |
0.057 |
| 224 |
249144_at |
AT5G43270
|
SPL2, squamosa promoter binding protein-like 2 |
-0.584 |
-0.192 |
| 225 |
256914_at |
AT3G23880
|
[F-box and associated interaction domains-containing protein] |
-0.581 |
-0.581 |
| 226 |
264658_at |
AT1G09910
|
[Rhamnogalacturonate lyase family protein] |
-0.581 |
0.232 |
| 227 |
261873_at |
AT1G11350
|
CBRLK1, CALMODULIN-BINDING RECEPTOR-LIKE PROTEIN KINASE, RKS2, SD1-13, S-domain-1 13 |
-0.572 |
-0.628 |
| 228 |
257966_at |
AT3G19800
|
unknown |
-0.569 |
-0.223 |
| 229 |
264041_at |
AT2G03710
|
AGL3, AGAMOUS-like 3, SEP4, SEPALLATA 4 |
-0.568 |
-0.092 |
| 230 |
261453_at |
AT1G21130
|
IGMT4, indole glucosinolate O-methyltransferase 4 |
-0.567 |
0.129 |
| 231 |
249331_at |
AT5G40950
|
RPL27, ribosomal protein large subunit 27 |
-0.565 |
-0.319 |
| 232 |
264697_at |
AT1G70210
|
ATCYCD1;1, CYCD1;1, CYCLIN D1;1 |
-0.562 |
-0.309 |
| 233 |
258275_at |
AT3G15760
|
unknown |
-0.561 |
0.471 |
| 234 |
255008_at |
AT4G10060
|
[Beta-glucosidase, GBA2 type family protein] |
-0.559 |
-0.070 |
| 235 |
267359_at |
AT2G40020
|
[Nucleolar histone methyltransferase-related protein] |
-0.557 |
0.006 |
| 236 |
254481_at |
AT4G20480
|
[Putative endonuclease or glycosyl hydrolase] |
-0.550 |
0.094 |
| 237 |
251720_at |
AT3G56160
|
[Sodium Bile acid symporter family] |
-0.543 |
-0.491 |
| 238 |
255591_at |
AT4G01630
|
ATEXP17, EXPANSIN 17, ATEXPA17, expansin A17, ATHEXP ALPHA 1.13, EXPA17, expansin A17 |
-0.542 |
-0.048 |
| 239 |
248079_at |
AT5G55790
|
unknown |
-0.541 |
-0.112 |
| 240 |
262669_at |
AT1G62850
|
[Class I peptide chain release factor] |
-0.540 |
0.120 |
| 241 |
252972_at |
AT4G38840
|
SAUR14, SMALL AUXIN UPREGULATED RNA 14 |
-0.540 |
-0.891 |
| 242 |
266265_at |
AT2G29340
|
[NAD-dependent epimerase/dehydratase family protein] |
-0.538 |
-0.145 |
| 243 |
264101_at |
AT1G79000
|
ATHAC1, ARABIDOPSIS HISTONE ACETYLTRANSFERASE OF THE CBP FAMILY 1, ATHPCAT2, ARABIDOPSIS THALIANA P300/CBP ACETYLTRANSFERASE-RELATED PROTEIN 2, HAC1, histone acetyltransferase of the CBP family 1, PCAT2, P300/CBP ACETYLTRANSFERASE-RELATED PROTEIN 2 |
-0.536 |
0.034 |
| 244 |
247284_at |
AT5G64410
|
ATOPT4, ARABIDOPSIS THALIANA OLIGOPEPTIDE TRANSPORTER 4, OPT4, oligopeptide transporter 4 |
-0.535 |
-0.346 |
| 245 |
265149_at |
AT1G51400
|
[Photosystem II 5 kD protein] |
-0.534 |
-0.256 |
| 246 |
250217_at |
AT5G14120
|
[Major facilitator superfamily protein] |
-0.529 |
0.126 |
| 247 |
253061_at |
AT4G37610
|
BT5, BTB and TAZ domain protein 5 |
-0.527 |
0.227 |
| 248 |
252529_at |
AT3G46490
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.525 |
-0.197 |
| 249 |
251952_at |
AT3G53650
|
[Histone superfamily protein] |
-0.522 |
0.638 |
| 250 |
260070_at |
AT1G73830
|
BEE3, BR enhanced expression 3 |
-0.514 |
-0.087 |
| 251 |
263106_at |
AT2G05160
|
[CCCH-type zinc fingerfamily protein with RNA-binding domain] |
-0.510 |
-0.507 |
| 252 |
255285_at |
AT4G04630
|
unknown |
-0.510 |
-0.324 |
| 253 |
263179_at |
AT1G05710
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.509 |
0.484 |
| 254 |
252199_at |
AT3G50270
|
[HXXXD-type acyl-transferase family protein] |
-0.504 |
-0.270 |
| 255 |
256303_at |
AT1G69550
|
[disease resistance protein (TIR-NBS-LRR class)] |
-0.502 |
0.245 |
| 256 |
260167_at |
AT1G71970
|
unknown |
-0.501 |
-0.180 |
| 257 |
266277_at |
AT2G29310
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.500 |
-0.656 |
| 258 |
260317_at |
AT1G63800
|
UBC5, ubiquitin-conjugating enzyme 5 |
-0.498 |
-0.071 |
| 259 |
264191_at |
AT1G54730
|
[Major facilitator superfamily protein] |
-0.498 |
-0.391 |
| 260 |
259671_at |
AT1G52290
|
AtPERK15, proline-rich extensin-like receptor kinase 15, PERK15, proline-rich extensin-like receptor kinase 15 |
-0.497 |
0.075 |
| 261 |
253597_at |
AT4G30690
|
[Translation initiation factor 3 protein] |
-0.496 |
-0.294 |
| 262 |
260770_at |
AT1G49200
|
[RING/U-box superfamily protein] |
-0.496 |
-0.406 |
| 263 |
246478_at |
AT5G15980
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.485 |
-0.181 |
| 264 |
266460_at |
AT2G47930
|
AGP26, arabinogalactan protein 26, ATAGP26, ARABIDOPSIS THALIANA ARABINOGALACTAN PROTEIN 26 |
-0.483 |
-0.680 |
| 265 |
262884_at |
AT1G64720
|
CP5 |
-0.473 |
-0.199 |
| 266 |
266790_at |
AT2G28950
|
ATEXP6, ARABIDOPSIS THALIANA TEXPANSIN 6, ATEXPA6, expansin A6, ATHEXP ALPHA 1.8, EXPA6, expansin A6 |
-0.473 |
-1.003 |
| 267 |
249265_at |
AT5G41700
|
ATUBC8, ARABIDOPSIS THALIANA UBIQUITIN CONJUGATING ENZYME 8, UBC8, ubiquitin conjugating enzyme 8 |
-0.470 |
-0.028 |
| 268 |
261611_at |
AT1G49730
|
[Protein kinase superfamily protein] |
-0.467 |
-0.474 |
| 269 |
249073_at |
AT5G44020
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
-0.455 |
-0.826 |
| 270 |
247072_at |
AT5G66490
|
unknown |
-0.453 |
-0.221 |
| 271 |
251919_at |
AT3G53800
|
Fes1B, Fes1B |
-0.453 |
-0.445 |
| 272 |
254795_at |
AT4G12990
|
unknown |
-0.453 |
0.121 |
| 273 |
256225_at |
AT1G56220
|
[Dormancy/auxin associated family protein] |
-0.451 |
-0.231 |
| 274 |
247742_at |
AT5G58980
|
[Neutral/alkaline non-lysosomal ceramidase] |
-0.449 |
0.119 |
| 275 |
251746_at |
AT3G56060
|
[Glucose-methanol-choline (GMC) oxidoreductase family protein] |
-0.448 |
-0.310 |
| 276 |
250265_at |
AT5G12900
|
unknown |
-0.448 |
-0.440 |
| 277 |
246759_at |
AT5G27950
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.444 |
0.038 |
| 278 |
252444_at |
AT3G46980
|
PHT4;3, phosphate transporter 4;3 |
-0.443 |
0.018 |
| 279 |
256503_at |
AT1G75250
|
ATRL6, RAD-like 6, RL6, RAD-like 6, RSM3, RADIALIS-LIKE SANT/MYB 3 |
-0.441 |
-0.481 |
| 280 |
266707_at |
AT2G03310
|
unknown |
-0.440 |
-0.417 |
| 281 |
257071_at |
AT3G28180
|
ATCSLC04, Cellulose-synthase-like C4, ATCSLC4, CSLC04, CSLC04, Cellulose-synthase-like C4, CSLC4, CELLULOSE-SYNTHASE LIKE C4 |
-0.440 |
0.018 |
| 282 |
250649_at |
AT5G06690
|
WCRKC1, WCRKC thioredoxin 1 |
-0.435 |
-0.309 |
| 283 |
265530_at |
AT2G06050
|
AtOPR3, DDE1, DELAYED DEHISCENCE 1, OPR3, oxophytodienoate-reductase 3 |
-0.433 |
0.416 |
| 284 |
253302_at |
AT4G33660
|
unknown |
-0.430 |
-0.231 |
| 285 |
250016_at |
AT5G18100
|
CSD3, copper/zinc superoxide dismutase 3 |
-0.429 |
-0.251 |
| 286 |
248301_at |
AT5G53150
|
[DNAJ heat shock N-terminal domain-containing protein] |
-0.426 |
0.095 |
| 287 |
254561_at |
AT4G19160
|
unknown |
-0.424 |
-0.085 |
| 288 |
254333_at |
AT4G22753
|
ATSMO1-3, SMO1-3, sterol 4-alpha methyl oxidase 1-3 |
-0.422 |
-0.244 |
| 289 |
259398_at |
AT1G17700
|
PRA1.F1, prenylated RAB acceptor 1.F1 |
-0.419 |
-0.232 |
| 290 |
257476_at |
AT1G80960
|
[F-box and Leucine Rich Repeat domains containing protein] |
-0.418 |
0.284 |
| 291 |
259476_at |
AT1G19000
|
[Homeodomain-like superfamily protein] |
-0.417 |
-0.142 |
| 292 |
245325_at |
AT4G14130
|
XTH15, xyloglucan endotransglucosylase/hydrolase 15, XTR7, xyloglucan endotransglycosylase 7 |
-0.417 |
0.041 |
| 293 |
253141_at |
AT4G35440
|
ATCLC-E, CLC-E, chloride channel E, CLCE, CHLORIDE CHANNEL E |
-0.416 |
-0.506 |
| 294 |
259207_at |
AT3G09050
|
unknown |
-0.414 |
-0.639 |
| 295 |
258617_at |
AT3G03000
|
[EF hand calcium-binding protein family] |
-0.414 |
0.268 |
| 296 |
263202_at |
AT1G05630
|
5PTASE13, inositol-polyphosphate 5-phosphatase 13, AT5PTASE13, Arabidopsis thaliana inositol-polyphosphate 5-phosphatase 13 |
-0.413 |
0.122 |
| 297 |
265175_at |
AT1G23480
|
ATCSLA03, cellulose synthase-like A3, ATCSLA3, CSLA03, CSLA03, cellulose synthase-like A3, CSLA3, CELLULOSE SYNTHASE-LIKE A3 |
-0.413 |
-0.551 |
| 298 |
264963_at |
AT1G60600
|
ABC4, ABERRANT CHLOROPLAST DEVELOPMENT 4 |
-0.411 |
-0.610 |
| 299 |
256021_at |
AT1G58270
|
ZW9 |
-0.411 |
-0.107 |
| 300 |
250017_at |
AT5G18140
|
DJC69, DNA J protein A69 |
-0.410 |
-0.166 |
| 301 |
249337_at |
AT5G41080
|
AtGDPD2, GDPD2, glycerophosphodiester phosphodiesterase 2 |
-0.410 |
0.746 |