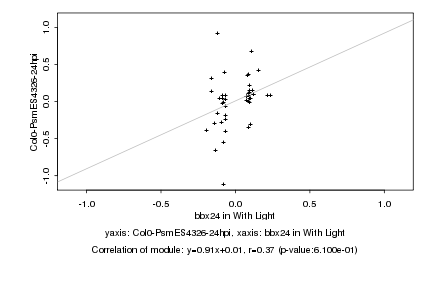
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
bbx24 in With Light |
Log2 signal ratio
Col0-PsmES4326-24hpi |
| 1 |
260973_at |
AT1G53490
|
HEI10, homolog of human HEI10 ( Enhancer of cell Invasion No.10) |
0.234 |
0.087 |
| 2 |
266578_at |
AT2G23910
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.213 |
0.091 |
| 3 |
247519_at |
AT5G61430
|
ANAC100, NAC domain containing protein 100, ATNAC5, NAC100, NAC domain containing protein 100 |
0.150 |
0.428 |
| 4 |
261215_at |
AT1G32970
|
[Subtilisin-like serine endopeptidase family protein] |
0.120 |
0.108 |
| 5 |
254993_at |
AT4G10730
|
[Protein kinase superfamily protein] |
0.117 |
0.106 |
| 6 |
257736_at |
AT3G27410
|
unknown |
0.111 |
0.153 |
| 7 |
252684_at |
AT3G44400
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.105 |
0.681 |
| 8 |
256913_at |
AT3G23870
|
unknown |
0.097 |
0.042 |
| 9 |
266873_at |
AT2G44740
|
CYCP4;1, cyclin p4;1 |
0.097 |
-0.309 |
| 10 |
255090_at |
AT4G09360
|
[NB-ARC domain-containing disease resistance protein] |
0.092 |
0.053 |
| 11 |
256762_at |
AT3G25655
|
IDL1, inflorescence deficient in abscission (IDA)-like 1 |
0.092 |
0.156 |
| 12 |
254781_at |
AT4G12840
|
unknown |
0.092 |
0.226 |
| 13 |
250629_at |
AT5G07390
|
ATRBOHA, respiratory burst oxidase homolog A, RBOHA, respiratory burst oxidase homolog A |
0.091 |
0.122 |
| 14 |
247752_at |
AT5G59060
|
unknown |
0.088 |
-0.010 |
| 15 |
263700_at |
AT1G31150
|
[FUNCTIONS IN: sequence-specific DNA binding transcription factor activity] |
0.087 |
-0.348 |
| 16 |
255481_at |
AT4G02460
|
AtPMS1, PMS1, POSTMEIOTIC SEGREGATION 1 |
0.086 |
0.010 |
| 17 |
250168_at |
AT5G15320
|
unknown |
0.084 |
0.088 |
| 18 |
255751_at |
AT1G31950
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
0.082 |
0.112 |
| 19 |
257058_at |
AT3G15352
|
ATCOX17, ARABIDOPSIS THALIANA CYTOCHROME C OXIDASE 17, COX17, cytochrome c oxidase 17 |
0.082 |
0.375 |
| 20 |
267364_at |
AT2G40080
|
ELF4, EARLY FLOWERING 4 |
0.077 |
0.355 |
| 21 |
256097_at |
AT1G13670
|
unknown |
0.074 |
0.074 |
| 22 |
257353_at |
AT2G23050
|
MEL4, MAB4/ENP/NPY1-LIKE 4, NPY4, NAKED PINS IN YUC MUTANTS 4 |
0.073 |
0.078 |
| 23 |
253663_at |
AT4G30160
|
ATVLN4, VLN4, villin 4 |
0.069 |
0.025 |
| 24 |
250180_at |
AT5G14450
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.201 |
-0.383 |
| 25 |
262838_at |
AT1G14960
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.164 |
0.141 |
| 26 |
248657_at |
AT5G48570
|
ATFKBP65, FKBP65, ROF2 |
-0.161 |
0.320 |
| 27 |
247717_at |
AT5G59320
|
LTP3, lipid transfer protein 3 |
-0.146 |
-0.284 |
| 28 |
260041_at |
AT1G68780
|
[RNI-like superfamily protein] |
-0.139 |
-0.649 |
| 29 |
264527_at |
AT1G30760
|
[FAD-binding Berberine family protein] |
-0.125 |
-0.151 |
| 30 |
266821_at |
AT2G44840
|
ATERF13, ETHYLENE-RESPONSIVE ELEMENT BINDING FACTOR 13, EREBP, ERF13, ethylene-responsive element binding factor 13 |
-0.123 |
0.924 |
| 31 |
262332_at |
AT1G64030
|
ATSRP3, SRP3, serpin 3 |
-0.110 |
0.042 |
| 32 |
257744_at |
AT3G29230
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.094 |
-0.273 |
| 33 |
255637_at |
AT4G00750
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.089 |
-0.024 |
| 34 |
263417_at |
AT2G17180
|
DAZ1, DUO1-ACTIVATED ZINC FINGER 1 |
-0.089 |
0.044 |
| 35 |
247524_at |
AT5G61440
|
ACHT5, atypical CYS HIS rich thioredoxin 5 |
-0.088 |
0.090 |
| 36 |
246439_at |
AT5G17600
|
[RING/U-box superfamily protein] |
-0.085 |
-0.543 |
| 37 |
255822_at |
AT2G40610
|
ATEXP8, ATEXPA8, expansin A8, ATHEXP ALPHA 1.11, EXP8, EXPA8, expansin A8 |
-0.083 |
-1.106 |
| 38 |
254758_at |
AT4G13260
|
AtYUC2, YUC2, YUCCA2 |
-0.082 |
-0.002 |
| 39 |
259037_at |
AT3G09350
|
Fes1A, Fes1A |
-0.079 |
0.398 |
| 40 |
255798_at |
AT2G33255
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.072 |
-0.182 |
| 41 |
263790_at |
AT2G24530
|
unknown |
-0.072 |
0.082 |
| 42 |
266682_at |
AT2G19780
|
[Leucine-rich repeat (LRR) family protein] |
-0.071 |
-0.235 |
| 43 |
261493_at |
AT1G14410
|
ATWHY1, A. THALIANA WHIRLY 1, PTAC1, WHY1, WHIRLY 1 |
-0.069 |
-0.064 |
| 44 |
256741_at |
AT3G29375
|
[XH domain-containing protein] |
-0.068 |
-0.395 |
| 45 |
257582_at |
AT1G50720
|
[Stigma-specific Stig1 family protein] |
-0.068 |
0.033 |