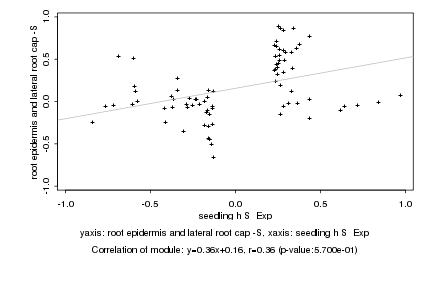
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
seedling h S Exp |
Log2 signal ratio
root epidermis and lateral root cap -S |
| 1 |
249866_at |
AT5G23010
|
IMS3, 2-ISOPROPYLMALATE SYNTHASE 3, MAM1, methylthioalkylmalate synthase 1 |
0.969 |
0.081 |
| 2 |
251524_at |
AT3G58990
|
IPMI1, isopropylmalate isomerase 1 |
0.837 |
-0.005 |
| 3 |
260745_at |
AT1G78370
|
ATGSTU20, glutathione S-transferase TAU 20, GSTU20, glutathione S-transferase TAU 20 |
0.716 |
-0.036 |
| 4 |
258851_at |
AT3G03190
|
ATGSTF11, glutathione S-transferase F11, ATGSTF6, ARABIDOPSIS GLUTATHIONE-S-TRANSFERASE 6, GSTF11, glutathione S-transferase F11 |
0.638 |
-0.055 |
| 5 |
255773_at |
AT1G18590
|
ATSOT17, SULFOTRANSFERASE 17, ATST5C, ARABIDOPSIS SULFOTRANSFERASE 5C, SOT17, sulfotransferase 17 |
0.618 |
-0.100 |
| 6 |
253975_at |
AT4G26600
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.435 |
0.773 |
| 7 |
254862_at |
AT4G12030
|
BASS5, BILE ACID:SODIUM SYMPORTER FAMILY PROTEIN 5, BAT5, bile acid transporter 5 |
0.435 |
-0.189 |
| 8 |
266395_at |
AT2G43100
|
ATLEUD1, IPMI2, isopropylmalate isomerase 2 |
0.433 |
0.027 |
| 9 |
255278_at |
AT4G04940
|
[transducin family protein / WD-40 repeat family protein] |
0.374 |
0.676 |
| 10 |
259264_at |
AT3G01260
|
[Galactose mutarotase-like superfamily protein] |
0.361 |
-0.013 |
| 11 |
250196_at |
AT5G14580
|
[polyribonucleotide nucleotidyltransferase, putative] |
0.358 |
0.635 |
| 12 |
266253_at |
AT2G27840
|
HD2D, histone deacetylase 2D, HDA13, HISTONE DEACETYLASE 13, HDT04, HDT4 |
0.341 |
0.866 |
| 13 |
257207_at |
AT3G14900
|
EMB3120, EMBRYO DEFECTIVE 3120 |
0.332 |
0.392 |
| 14 |
251106_at |
AT5G01500
|
TAAC, thylakoid ATP/ADP carrier |
0.328 |
0.124 |
| 15 |
245770_at |
AT1G30240
|
[FUNCTIONS IN: binding] |
0.324 |
0.585 |
| 16 |
265730_at |
AT2G32220
|
[Ribosomal L27e protein family] |
0.309 |
-0.021 |
| 17 |
261296_at |
AT1G48460
|
unknown |
0.289 |
0.584 |
| 18 |
266934_at |
AT2G18900
|
[Transducin/WD40 repeat-like superfamily protein] |
0.288 |
0.491 |
| 19 |
258166_at |
AT3G21540
|
[transducin family protein / WD-40 repeat family protein] |
0.280 |
0.614 |
| 20 |
251593_at |
AT3G57660
|
NRPA1, nuclear RNA polymerase A1 |
0.280 |
0.354 |
| 21 |
252625_at |
AT3G44750
|
ATHD2A, HD2A, HISTONE DEACETYLASE 2A, HDA3, histone deacetylase 3, HDT1 |
0.278 |
0.851 |
| 22 |
263714_at |
AT2G20610
|
ALF1, ABERRANT LATERAL ROOT FORMATION 1, HLS3, HOOKLESS 3, RTY, ROOTY, RTY1, ROOTY 1, SUR1, SUPERROOT 1 |
0.278 |
-0.048 |
| 23 |
248036_at |
AT5G55920
|
OLI2, OLIGOCELLULA 2 |
0.264 |
0.872 |
| 24 |
263477_at |
AT2G31790
|
[UDP-Glycosyltransferase superfamily protein] |
0.263 |
-0.146 |
| 25 |
245421_at |
AT4G17430
|
[O-fucosyltransferase family protein] |
0.261 |
0.200 |
| 26 |
246457_at |
AT5G16750
|
TOZ, TORMOZEMBRYO DEFECTIVE |
0.255 |
0.617 |
| 27 |
258009_at |
AT3G19440
|
[Pseudouridine synthase family protein] |
0.254 |
0.552 |
| 28 |
255225_at |
AT4G05410
|
YAO, YAOZHE |
0.254 |
0.487 |
| 29 |
249426_at |
AT5G39840
|
[ATP-dependent RNA helicase, mitochondrial, putative] |
0.251 |
0.895 |
| 30 |
253252_at |
AT4G34740
|
ASE2, GLN phosphoribosyl pyrophosphate amidotransferase 2, ATASE2, GLN phosphoribosyl pyrophosphate amidotransferase 2, ATPURF2, CIA1, CHLOROPLAST IMPORT APPARATUS 1, DOV1, DIFFERENTIAL DEVELOPMENT OF VASCULAR ASSOCIATED CELLS |
0.250 |
0.458 |
| 31 |
254455_at |
AT4G21140
|
unknown |
0.244 |
0.320 |
| 32 |
245494_at |
AT4G16390
|
SVR7, suppressor of variegation 7 |
0.243 |
0.408 |
| 33 |
265961_at |
AT2G37400
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.241 |
0.720 |
| 34 |
254080_at |
AT4G25630
|
ATFIB2, FIB2, fibrillarin 2 |
0.239 |
0.661 |
| 35 |
262766_at |
AT1G13160
|
[ARM repeat superfamily protein] |
0.236 |
0.443 |
| 36 |
247040_at |
AT5G67150
|
[HXXXD-type acyl-transferase family protein] |
0.235 |
0.390 |
| 37 |
256065_at |
AT1G07070
|
[Ribosomal protein L35Ae family protein] |
0.232 |
0.536 |
| 38 |
262573_at |
AT1G15390
|
ATDEF1, PDF1A, peptide deformylase 1A |
0.232 |
0.245 |
| 39 |
253566_at |
AT4G31210
|
[DNA topoisomerase, type IA, core] |
0.231 |
0.538 |
| 40 |
253598_at |
AT4G30800
|
[Nucleic acid-binding, OB-fold-like protein] |
0.227 |
0.672 |
| 41 |
252252_at |
AT3G49180
|
RID3, ROOT INITIATION DEFECTIVE 3 |
0.225 |
0.377 |
| 42 |
254430_at |
AT4G20820
|
[FAD-binding Berberine family protein] |
-0.842 |
-0.245 |
| 43 |
258983_at |
AT3G08860
|
PYD4, PYRIMIDINE 4 |
-0.770 |
-0.055 |
| 44 |
259133_at |
AT3G05400
|
[Major facilitator superfamily protein] |
-0.718 |
-0.042 |
| 45 |
252269_at |
AT3G49580
|
LSU1, RESPONSE TO LOW SULFUR 1 |
-0.694 |
0.538 |
| 46 |
263168_at |
AT1G03020
|
[Thioredoxin superfamily protein] |
-0.608 |
-0.027 |
| 47 |
256244_at |
AT3G12520
|
SULTR4;2, sulfate transporter 4;2 |
-0.600 |
0.520 |
| 48 |
254343_at |
AT4G21990
|
APR3, APS reductase 3, ATAPR3, PRH-26, PRH26, PAPS REDUCTASE HOMOLOG 26 |
-0.598 |
0.184 |
| 49 |
261177_at |
AT1G04770
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.590 |
0.120 |
| 50 |
250025_at |
AT5G18290
|
SIP1;2, SIP1B, SMALL AND BASIC INTRINSIC PROTEIN 1B |
-0.580 |
0.009 |
| 51 |
251130_at |
AT5G01180
|
AtNPF8.2, ATPTR5, ARABIDOPSIS THALIANA PEPTIDE TRANSPORTER 5, NPF8.2, NRT1/ PTR family 8.2, PTR5, peptide transporter 5 |
-0.419 |
-0.082 |
| 52 |
262134_at |
AT1G77990
|
AST56, SULTR2;2, SULPHATE TRANSPORTER 2;2 |
-0.414 |
-0.243 |
| 53 |
262133_at |
AT1G78000
|
SEL1, SELENATE RESISTANT 1, SULTR1;2, sulfate transporter 1;2 |
-0.378 |
0.069 |
| 54 |
248713_at |
AT5G48180
|
AtNSP5, NSP5, nitrile specifier protein 5 |
-0.373 |
-0.060 |
| 55 |
262229_at |
AT1G68620
|
[alpha/beta-Hydrolases superfamily protein] |
-0.369 |
0.033 |
| 56 |
249869_at |
AT5G23050
|
AAE17, acyl-activating enzyme 17 |
-0.346 |
0.132 |
| 57 |
264745_at |
AT1G62180
|
5'adenylylphosphosulfate reductase 2 |
-0.343 |
0.275 |
| 58 |
253525_at |
AT4G31330
|
unknown |
-0.311 |
-0.347 |
| 59 |
252712_at |
AT3G43800
|
ATGSTU27, glutathione S-transferase tau 27, GSTU27, glutathione S-transferase tau 27 |
-0.291 |
-0.034 |
| 60 |
252677_at |
AT3G44320
|
AtNIT3, NITRILASE 3, NIT3, nitrilase 3 |
-0.288 |
-0.065 |
| 61 |
253203_at |
AT4G34710
|
ADC2, arginine decarboxylase 2, ATADC2, SPE2 |
-0.271 |
0.037 |
| 62 |
251060_at |
AT5G01820
|
ATCIPK14, ATSR1, serine/threonine protein kinase 1, CIPK14, CBL-INTERACTING PROTEIN KINASE 14, PKS24, SOS2-like protein kinase 24, SnRK3.15, SNF1-RELATED PROTEIN KINASE 3.15, SR1, serine/threonine protein kinase 1 |
-0.258 |
-0.037 |
| 63 |
249850_at |
AT5G23240
|
DJC76, DNA J protein C76 |
-0.239 |
0.030 |
| 64 |
249917_at |
AT5G22460
|
[alpha/beta-Hydrolases superfamily protein] |
-0.235 |
0.032 |
| 65 |
254836_at |
AT4G12330
|
CYP706A7, cytochrome P450, family 706, subfamily A, polypeptide 7 |
-0.217 |
-0.028 |
| 66 |
245689_at |
AT5G04120
|
[Phosphoglycerate mutase family protein] |
-0.186 |
-0.283 |
| 67 |
254926_at |
AT4G11280
|
ACS6, 1-aminocyclopropane-1-carboxylic acid (acc) synthase 6, ATACS6 |
-0.183 |
0.006 |
| 68 |
256633_at |
AT3G28340
|
GATL10, galacturonosyltransferase-like 10, GolS8, galactinol synthase 8 |
-0.171 |
-0.120 |
| 69 |
249533_at |
AT5G38790
|
unknown |
-0.168 |
-0.095 |
| 70 |
261292_at |
AT1G36940
|
unknown |
-0.167 |
0.056 |
| 71 |
256453_at |
AT1G75270
|
DHAR2, dehydroascorbate reductase 2 |
-0.162 |
0.142 |
| 72 |
253454_at |
AT4G31875
|
unknown |
-0.160 |
-0.429 |
| 73 |
255740_at |
AT1G25390
|
[Protein kinase superfamily protein] |
-0.160 |
-0.293 |
| 74 |
253990_at |
AT4G26160
|
ACHT1, atypical CYS HIS rich thioredoxin 1 |
-0.158 |
-0.438 |
| 75 |
246396_at |
AT1G58180
|
ATBCA6, A. THALIANA BETA CARBONIC ANHYDRASE 6, BCA6, beta carbonic anhydrase 6 |
-0.158 |
-0.448 |
| 76 |
250054_at |
AT5G17860
|
CAX7, calcium exchanger 7 |
-0.154 |
-0.148 |
| 77 |
262887_at |
AT1G14780
|
[MAC/Perforin domain-containing protein] |
-0.143 |
-0.507 |
| 78 |
265443_at |
AT2G20750
|
ATEXPB1, expansin B1, ATHEXP BETA 1.5, EXPB1, expansin B1 |
-0.139 |
-0.076 |
| 79 |
263287_at |
AT2G36145
|
unknown |
-0.139 |
-0.049 |
| 80 |
249010_at |
AT5G44580
|
unknown |
-0.138 |
-0.270 |
| 81 |
251315_at |
AT3G61410
|
unknown |
-0.135 |
-0.653 |
| 82 |
257923_at |
AT3G23160
|
unknown |
-0.133 |
0.123 |