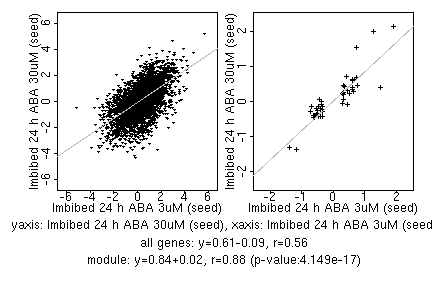
marker gene list
| Imbibed 24 h ABA 3uM (seed) |
| Imbibed 24 h ABA 30uM (seed) |
| probeID | AGICode | Annotation | gene expression |
|
|---|---|---|---|---|
| 1 | 263164_at | AT1G03070 | glutamate binding / | 0.81 |
| 2 | 264460_at | AT1G10170 | ATNFXL1; transcription factor | 0.34 |
| 3 | 245637_at | AT1G25230 | purple acid phosphatase family protein | -1.13 |
| 4 | 256425_at | AT1G33560 | ADR1 (ACTIVATED DISEASE RESISTANCE 1) | 0.78 |
| 5 | 263193_at | AT1G36050 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G22200.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO61738.1); contains InterPro domain Protein of unknown function DUF1692 (InterPro:IPR012936) | -0.32 |
| 6 | 245242_at | AT1G44446 | CH1 (CHLOROPHYLL B BIOSYNTHESIS); chlorophyllide a oxygenase | 0.47 |
| 7 | 262234_at | AT1G48270 | GCR1 (G-PROTEIN-COUPLED RECEPTOR 1) | -0.57 |
| 8 | 261865_at | AT1G50430 | DWF5 (DWARF 5); sterol delta7 reductase | -0.49 |
| 9 | 261906_at | AT1G65080 | OXA1 family protein | 0.53 |
| 10 | 260367_at | AT1G69760 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G26920.1); similar to hypothetical protein [Ricinus communis] (GB:CAH56540.1) | -0.37 |
| 11 | 260411_at | AT1G69890 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G27100.1); similar to unknown [Populus trichocarpa] (GB:ABK94560.1); contains InterPro domain Protein of unknown function DUF569 (InterPro:IPR007679); contains InterPro domain Actin-crosslinking proteins (InterPro:IPR008999) | 0.44 |
| 12 | 262290_at | AT1G70985 | hydroxyproline-rich glycoprotein family protein | 1.29 |
| 13 | 265702_at | AT2G03450 | ATPAP9/PAP9 (purple acid phosphatase 9); acid phosphatase/ protein serine/threonine phosphatase | 0.66 |
| 14 | 264032_at | AT2G03800 | GEK1 (GEKO1) | -0.54 |
| 15 | 266574_at | AT2G23890 | 5' nucleotidase family protein | -0.32 |
| 16 | 267373_at | AT2G26280 | CID7; ATP binding / damaged DNA binding | -0.34 |
| 17 | 267614_at | AT2G26710 | BAS1/CYP734A1 (PHYB ACTIVATION TAGGED SUPPRESSOR 1); oxygen binding / steroid hydroxylase | -0.59 |
| 18 | 264081_at | AT2G28540 | nucleic acid binding / nucleotide binding / protein binding / zinc ion binding | 0.63 |
| 19 | 267180_at | AT2G37570 | SLT1 (SODIUM- AND LITHIUM-TOLERANT 1) | 0.35 |
| 20 | 258933_at | AT3G09980 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G03660.1); similar to unknown [Populus trichocarpa] (GB:ABK95581.1); similar to unknown [Populus trichocarpa] (GB:ABK93998.1); contains InterPro domain Protein of unknown function DUF662 (InterPro:IPR007033) | -0.45 |
| 21 | 257547_at | AT3G13000 | transcription factor | 0.63 |
| 22 | 258435_at | AT3G16740 | F-box family protein | 0.69 |
| 23 | 252482_at | AT3G46670 | UDP-glucoronosyl/UDP-glucosyl transferase family protein | 0.39 |
| 24 | 252398_at | AT3G47990 | zinc finger (C3HC4-type RING finger) family protein | -0.35 |
| 25 | 252347_at | AT3G48130 | ribosomal protein L13 homolog | -1.34 |
| 26 | 251940_at | AT3G53450 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G37210.1); similar to Conserved hypothetical protein 730 [Medicago truncatula] (GB:ABD32794.1); contains InterPro domain Conserved hypothetical protein CHP00730 (InterPro:IPR005269) | -0.56 |
| 27 | 251561_at | AT3G57870 | AHUS5 (SUMO CONJUGATION ENZYME 1); ubiquitin-protein ligase | -0.30 |
| 28 | 251522_at | AT3G59430 | similar to hypothetical protein [Vitis vinifera] (GB:CAN70075.1) | 0.32 |
| 29 | 255593_at | AT4G01650 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G08720.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO40206.1); contains InterPro domain Streptomyces cyclase/dehydrase (InterPro:IPR005031) | -0.41 |
| 30 | 255327_at | AT4G04320 | malonyl-CoA decarboxylase family protein | 0.36 |
| 31 | 255055_at | AT4G09810 | transporter-related | -0.49 |
| 32 | 255009_at | AT4G10070 | KH domain-containing protein | 0.33 |
| 33 | 254424_at | AT4G21510 | F-box family protein | -0.45 |
| 34 | 254083_at | AT4G24920 | protein transport protein SEC61 gamma subunit, putative | -0.43 |
| 35 | 253869_at | AT4G27510 | similar to hypothetical protein [Vitis vinifera] (GB:CAN77561.1) | -0.47 |
| 36 | 253724_at | AT4G29285 | LCR24 (Low-molecular-weight cysteine-rich 24) | 1.51 |
| 37 | 253156_at | AT4G35730 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G34220.2); similar to unnamed protein product [Vitis vinifera] (GB:CAO23719.1); contains InterPro domain Protein of unknown function DUF292, eukaryotic (InterPro:IPR005061) | 0.64 |
| 38 | 252976_s_at | AT4G38550 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G20950.1); contains InterPro domain Phospholipase-like, arabidopsis (InterPro:IPR007942) | 0.47 |
| 39 | 252860_at | AT4G39790 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G27090.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO49895.1); contains InterPro domain Protein of unknown function DUF632 (InterPro:IPR006867); contains InterPro domain Protein of unknown function DUF630 (InterPro:IPR006868) | 0.65 |
| 40 | 251144_at | AT5G01210 | transferase family protein | 0.77 |
| 41 | 250967_at | AT5G02790 | In2-1 protein, putative | -0.60 |
| 42 | 245693_at | AT5G04260 | thioredoxin family protein | -0.66 |
| 43 | 250761_at | AT5G06050 | dehydration-responsive protein-related | -0.30 |
| 44 | 250544_at | AT5G08080 | SYP132 (syntaxin 132); SNAP receptor | -0.46 |
| 45 | 246085_at | AT5G20540 | ATBRXL4/BRX-LIKE4 (BREVIS RADIX-LIKE 4) | 0.47 |
| 46 | 245988_at | AT5G20610 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G26160.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN67684.1) | 0.36 |
| 47 | 246613_at | AT5G35360 | CAC2 (acetyl co-enzyme A carboxylase biotin carboxylase subunit) | -0.38 |
| 48 | 248213_at | AT5G53660 | AtGRF7 (GROWTH-REGULATING FACTOR 7) | 0.65 |
| 49 | 247509_at | AT5G62020 | AT-HSFB2A (Arabidopsis thaliana heat shock transcription factor B2A); DNA binding / transcription factor | -0.70 |
| 50 | 244973_at | ATCG00690 | Encodes photosystem II 5 kD protein subunit PSII-T. This is a plastid-encoded gene (PsbTc) which also has a nuclear-encoded paralog (PsbTn). | 1.95 |