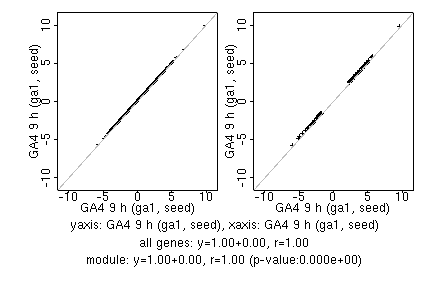marker gene list
| GA4 9 h (ga1, seed) |
| GA4 9 h (ga1, seed) |
| probeID | AGICode | Annotation | gene expression |
|
|---|---|---|---|---|
| 1 | 260914_at | AT1G02640 | BXL2 (BETA-XYLOSIDASE 2); hydrolase, hydrolyzing O-glycosyl compounds | 4.50 |
| 2 | 262105_at | AT1G02810 | pectinesterase family protein | 3.12 |
| 3 | 265084_at | AT1G03790 | zinc finger (CCCH-type) family protein | -1.75 |
| 4 | 263184_at | AT1G05560 | UGT1 (UDP-glucosyl transferase 75B1); UDP-glycosyltransferase/ transferase, transferring glycosyl groups | -2.00 |
| 5 | 261077_at | AT1G07430 | protein phosphatase 2C, putative / PP2C, putative | -1.61 |
| 6 | 264804_at | AT1G08590 | CLAVATA1 receptor kinase (CLV1) | 2.87 |
| 7 | 264777_at | AT1G08630 | THA1 (THREONINE ALDOLASE 1); aldehyde-lyase/ threonine aldolase | -1.88 |
| 8 | 264653_at | AT1G08980 | ATAMI1 (AMIDASE-LIKE PROTEIN 1); amidase | -1.80 |
| 9 | 262826_at | AT1G13080 | CYP71B2 (CYTOCHROME P450 71B2); oxygen binding | 2.91 |
| 10 | 262828_at | AT1G14950 | major latex protein-related / MLP-related | -1.79 |
| 11 | 255900_at | AT1G17830 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G73210.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO45300.1); contains InterPro domain Protein of unknown function DUF789 (InterPro:IPR008507) | -1.99 |
| 12 | 259541_at | AT1G20650 | protein kinase | -1.82 |
| 13 | 262505_at | AT1G21680 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G21670.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN73514.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO61906.1); similar to hypothetical protein OsJ_012725 [Oryza sativa (japonica cultivar-group)] (GB:EAZ29242.1); contains InterPro domain WD40-like Beta Propeller (InterPro:IPR011659); contains InterPro domain Six-bladed beta-propeller, TolB-like (InterPro:IPR011042) | -2.18 |
| 14 | 245642_at | AT1G25275 | similar to unnamed protein product [Vitis vinifera] (GB:CAO43403.1) | -2.59 |
| 15 | 245688_at | AT1G28290 | AGP31 (ARABINOGALACTAN-PROTEIN 31); structural constituent of cell wall | 2.89 |
| 16 | 261866_at | AT1G50420 | SCL3 (SCARECROW-LIKE 3); transcription factor | -3.84 |
| 17 | 245629_at | AT1G56580 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G09310.1); similar to unknown [Populus trichocarpa] (GB:ABK94967.1); contains InterPro domain Protein of unknown function DUF538 (InterPro:IPR007493) | 2.86 |
| 18 | 264999_at | AT1G67310 | calmodulin binding / transcription regulator | -1.67 |
| 19 | 256299_at | AT1G69530 | ATEXPA1 (ARABIDOPSIS THALIANA EXPANSIN A1) | 4.48 |
| 20 | 264313_at | AT1G70410 | carbonic anhydrase, putative / carbonate dehydratase, putative | 4.83 |
| 21 | 262310_at | AT1G70840 | MLP31 (MLP-LIKE PROTEIN 31) | -4.96 |
| 22 | 260387_at | AT1G74100 | sulfotransferase family protein | 3.70 |
| 23 | 260221_at | AT1G74670 | gibberellin-responsive protein, putative | 5.29 |
| 24 | 262967_at | AT1G75730 | similar to hypothetical protein [Vitis vinifera] (GB:CAN68413.1) | -1.95 |
| 25 | 262049_at | AT1G80180 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G15400.2); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G15400.3); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G15400.1); similar to hypothetical protein MtrDRAFT_AC148340g12v2 [Medicago truncatula] (GB:ABD28396.1) | -1.72 |
| 26 | 263325_at | AT2G04240 | XERICO; protein binding / zinc ion binding | -3.21 |
| 27 | 264001_at | AT2G22420 | peroxidase 17 (PER17) (P17) | 3.51 |
| 28 | 265276_at | AT2G28400 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G60680.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO21845.1); contains InterPro domain Protein of unknown function DUF584 (InterPro:IPR007608) | -3.05 |
| 29 | 267461_at | AT2G33830 | dormancy/auxin associated family protein | -3.03 |
| 30 | 267092_at | AT2G38120 | AUX1 (AUXIN RESISTANT 1); amino acid transmembrane transporter/ transporter | 2.67 |
| 31 | 266167_at | AT2G38860 | YLS5 (yellow-leaf-specific gene 5) | 2.94 |
| 32 | 255822_at | AT2G40610 | ATEXPA8 (ARABIDOPSIS THALIANA EXPANSIN A8) | 3.72 |
| 33 | 265266_at | AT2G42890 | AML2; RNA binding | -1.59 |
| 34 | 245136_at | AT2G45210 | auxin-responsive protein-related | -5.80 |
| 35 | 266326_at | AT2G46650 | B5 #1 (cytochrome b5 family protein #1); heme binding / transition metal ion binding | 4.43 |
| 36 | 258947_at | AT3G01830 | calmodulin-related protein, putative | -4.32 |
| 37 | 259302_at | AT3G05120 | ATGID1A/GID1A (GA INSENSITIVE DWARF1A); hydrolase | -2.12 |
| 38 | 259018_at | AT3G07390 | AIR12 (Auxin-Induced in Root cultures 12); extracellular matrix structural constituent | 3.27 |
| 39 | 259009_at | AT3G09260 | PYK10 (phosphate starvation-response 3.1); hydrolase, hydrolyzing O-glycosyl compounds | 3.34 |
| 40 | 258764_at | AT3G10720 | pectinesterase, putative | 5.67 |
| 41 | 256275_at | AT3G12110 | ACT11 (ACTIN-11); structural constituent of cytoskeleton | 2.54 |
| 42 | 258402_at | AT3G15450 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G27450.1); similar to unknown [Populus trichocarpa] (GB:ABK93866.1); contains domain PTHR11772 (PTHR11772); contains domain G3DSA:3.60.20.10 (G3DSA:3.60.20.10); contains domain SSF56235 (SSF56235) | -1.63 |
| 43 | 258252_at | AT3G15720 | glycoside hydrolase family 28 protein / polygalacturonase (pectinase) family protein | 5.80 |
| 44 | 258263_at | AT3G15780 | unknown protein | -2.30 |
| 45 | 258338_at | AT3G16150 | L-asparaginase, putative / L-asparagine amidohydrolase, putative | 3.92 |
| 46 | 256814_at | AT3G21370 | glycosyl hydrolase family 1 protein | -3.61 |
| 47 | 252711_at | AT3G43720 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | 3.67 |
| 48 | 252607_at | AT3G44990 | XTR8 (xyloglucan:xyloglucosyl transferase 8); hydrolase, acting on glycosyl bonds | 5.01 |
| 49 | 252606_at | AT3G45010 | SCPL48 (serine carboxypeptidase-like 48); serine carboxypeptidase | 4.33 |
| 50 | 252058_at | AT3G52470 | harpin-induced family protein / HIN1 family protein / harpin-responsive family protein | 2.51 |
| 51 | 251443_at | AT3G59940 | kelch repeat-containing F-box family protein | -2.07 |
| 52 | 251342_at | AT3G60690 | auxin-responsive family protein | -2.12 |
| 53 | 255578_at | AT4G01450 | nodulin MtN21 family protein | 3.74 |
| 54 | 255471_at | AT4G03050 | AOP3 (2-oxoglutarate?dependent dioxygenase 3); oxidoreductase, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors | -2.81 |
| 55 | 255433_at | AT4G03210 | XTH9 (XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 9); hydrolase, acting on glycosyl bonds | 2.48 |
| 56 | 255310_at | AT4G04955 | ATALN (ARABIDOPSIS ALLANTOINASE); allantoinase/ hydrolase | -1.81 |
| 57 | 254791_at | AT4G12910 | SCPL20 (serine carboxypeptidase-like 20); serine carboxypeptidase | 3.91 |
| 58 | 254419_at | AT4G21490 | NDB3; NADH dehydrogenase | 5.05 |
| 59 | 254110_at | AT4G25260 | invertase/pectin methylesterase inhibitor family protein | 2.51 |
| 60 | 254065_at | AT4G25420 | GA5 (GA REQUIRING 5); gibberellin 20-oxidase/ gibberellin 3-beta-dioxygenase | -2.15 |
| 61 | 253946_at | AT4G26790 | GDSL-motif lipase/hydrolase family protein | 4.67 |
| 62 | 253776_at | AT4G28390 | AAC3 (ADP/ATP CARRIER 3); ATP:ADP antiporter/ binding | -1.62 |
| 63 | 253708_at | AT4G29210 | GGT3/GGT4 (GAMMA-GLUTAMYL TRANSPEPTIDASE 3); gamma-glutamyltransferase/ glutathione gamma-glutamylcysteinyltransferase | 2.82 |
| 64 | 253608_at | AT4G30290 | ATXTH19 (XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 19); hydrolase, acting on glycosyl bonds | 3.25 |
| 65 | 253534_at | AT4G31500 | CYP83B1 (CYTOCHROME P450 MONOOXYGENASE 83B1); oxygen binding | 4.22 |
| 66 | 253322_at | AT4G33980 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G42900.2); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G42900.3); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G42900.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN64989.1) | -2.53 |
| 67 | 253218_at | AT4G34980 | SLP2 (subtilisin-like serine protease 2); subtilase | 2.96 |
| 68 | 253104_at | AT4G36010 | pathogenesis-related thaumatin family protein | -1.79 |
| 69 | 246250_at | AT4G36880 | CP1 (CYSTEINE PROTEINASE1); cysteine-type peptidase | 4.96 |
| 70 | 252870_at | AT4G39940 | AKN2 (APS-KINASE 2); ATP binding / kinase/ transferase, transferring phosphorus-containing groups | 3.22 |
| 71 | 250780_at | AT5G05290 | ATEXPA2 (ARABIDOPSIS THALIANA EXPANSIN A2) | 9.79 |
| 72 | 250611_at | AT5G07200 | YAP169 (GIBBERELLIN 20 OXIDASE 3); gibberellin 20-oxidase | -3.16 |
| 73 | 250261_at | AT5G13400 | proton-dependent oligopeptide transport (POT) family protein | 2.54 |
| 74 | 250214_at | AT5G13870 | EXGT-A4 (ENDOXYLOGLUCAN TRANSFERASE A4); hydrolase, acting on glycosyl bonds | 3.44 |
| 75 | 249937_at | AT5G22470 | NAD+ ADP-ribosyltransferase | -1.73 |
| 76 | 249769_at | AT5G24120 | SIGE (RNA polymerase sigma subunit E); DNA binding / DNA-directed RNA polymerase/ sigma factor/ transcription factor | -3.42 |
| 77 | 249688_at | AT5G36160 | aminotransferase-related | -2.91 |
| 78 | 249376_at | AT5G40645 | nitrate-responsive NOI protein, putative | 3.98 |
| 79 | 249045_at | AT5G44380 | FAD-binding domain-containing protein | -2.14 |
| 80 | 248912_at | AT5G45670 | GDSL-motif lipase/hydrolase family protein | 3.46 |
| 81 | 248794_at | AT5G47220 | ATERF-2/ATERF2/ERF2 (ETHYLENE RESPONSE FACTOR 2); DNA binding / transcription activator/ transcription factor | -4.72 |
| 82 | 248756_at | AT5G47560 | ATSDAT/ATTDT (TONOPLAST DICARBOXYLATE TRANSPORTER); malate transmembrane transporter/ sodium:dicarboxylate symporter | -1.59 |
| 83 | 248759_at | AT5G47610 | zinc finger (C3HC4-type RING finger) family protein | -1.87 |
| 84 | 248371_at | AT5G51810 | AT2353/ATGA20OX2/GA20OX2 (GIBBERELLIN 20 OXIDASE 2); gibberellin 20-oxidase | -2.55 |
| 85 | 248193_at | AT5G54080 | HGO (HOMOGENTISATE 1,2-DIOXYGENASE); homogentisate 1,2-dioxygenase | -1.80 |
| 86 | 247949_at | AT5G57220 | CYP81F2 (cytochrome P450, family 81, subfamily F, polypeptide 2); oxygen binding | 3.46 |
| 87 | 247543_at | AT5G61600 | ethylene-responsive element-binding family protein | -1.61 |
| 88 | 247212_at | AT5G65040 | senescence-associated protein-related | -1.99 |
| 89 | 247097_at | AT5G66460 | (1-4)-beta-mannan endohydrolase, putative | 3.12 |
| 90 | 247026_at | AT5G67080 | MAPKKK19 (Mitogen-activated protein kinase kinase kinase 19); kinase | 3.64 |