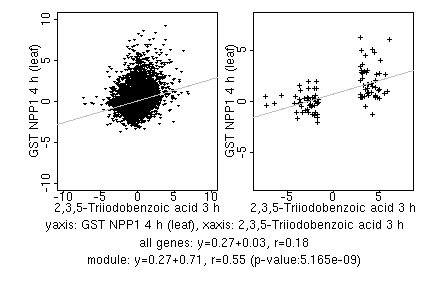
marker gene list
| 2,3,5-Triiodobenzoic acid3 h |
| GST NPP1 4 h (leaf) |
| probeID | AGICode | Annotation | gene expression |
|
|---|---|---|---|---|
| 1 | 263184_at | AT1G05560 | UGT1 (UDP-glucosyl transferase 75B1); UDP-glycosyltransferase/ transferase, transferring glycosyl groups | 4.92 |
| 2 | 263182_at | AT1G05575 | unknown protein | 3.88 |
| 3 | 264648_at | AT1G09080 | BIP3; ATP binding | 6.17 |
| 4 | 264347_at | AT1G12040 | LRX1 (LEUCINE-RICH REPEAT/EXTENSIN 1); protein binding / structural constituent of cell wall | -4.03 |
| 5 | 261763_at | AT1G15520 | ATPDR12/PDR12 (PLEIOTROPIC DRUG RESISTANCE 12); ATPase, coupled to transmembrane movement of substances | 4.31 |
| 6 | 261838_at | AT1G16030 | HSP70B (heat shock protein 70B); ATP binding | 4.70 |
| 7 | 262756_at | AT1G16370 | ATOCT6; carbohydrate transmembrane transporter/ sugar:hydrogen ion symporter | -1.55 |
| 8 | 262518_at | AT1G17170 | ATGSTU24 (ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE (CLASS TAU) 24); glutathione transferase | 4.67 |
| 9 | 262517_at | AT1G17180 | ATGSTU25 (Arabidopsis thaliana Glutathione S-transferase (class tau) 25); glutathione transferase | 5.05 |
| 10 | 255957_at | AT1G22160 | senescence-associated protein-related | -2.12 |
| 11 | 264770_at | AT1G23030 | armadillo/beta-catenin repeat family protein / U-box domain-containing protein | -1.80 |
| 12 | 265102_at | AT1G30870 | cationic peroxidase, putative | -3.74 |
| 13 | 260706_at | AT1G32350 | AOX1D (ALTERNATIVE OXIDASE 1D); alternative oxidase | 4.83 |
| 14 | 261157_at | AT1G34510 | peroxidase, putative | -6.25 |
| 15 | 256352_at | AT1G54970 | ATPRP1 (PROLINE-RICH PROTEIN 1); structural constituent of cell wall | -3.39 |
| 16 | 260344_at | AT1G69240 | hydrolase, alpha/beta fold family protein | -2.19 |
| 17 | 260405_at | AT1G69930 | ATGSTU11 (Arabidopsis thaliana Glutathione S-transferase (class tau) 11); glutathione transferase | 3.13 |
| 18 | 260248_at | AT1G74310 | ATHSP101 (HEAT SHOCK PROTEIN 101); ATP binding / ATPase | 5.09 |
| 19 | 259979_at | AT1G76600 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G21010.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN67638.1) | 3.79 |
| 20 | 260803_at | AT1G78340 | ATGSTU22 (Arabidopsis thaliana Glutathione S-transferase (class tau) 22); glutathione transferase | 3.55 |
| 21 | 261892_at | AT1G80840 | WRKY40 (WRKY DNA-binding protein 40); transcription factor | 3.31 |
| 22 | 263594_at | AT2G01880 | ATPAP7/PAP7 (purple acid phosphatase 7); acid phosphatase/ protein serine/threonine phosphatase | -1.62 |
| 23 | 264042_at | AT2G03760 | ST (steroid sulfotransferase); sulfotransferase | 4.26 |
| 24 | 265499_at | AT2G15480 | UGT73B5 (UDP-GLUCOSYL TRANSFERASE 73B5); UDP-glycosyltransferase/ transferase, transferring glycosyl groups | 4.56 |
| 25 | 266941_at | AT2G18980 | peroxidase, putative | -1.82 |
| 26 | 245076_at | AT2G23170 | GH3.3; indole-3-acetic acid amido synthetase | 5.07 |
| 27 | 264403_at | AT2G25150 | transferase family protein | -2.14 |
| 28 | 264404_at | AT2G25160 | CYP82F1 (cytochrome P450, family 82, subfamily F, polypeptide 1); oxygen binding | -2.68 |
| 29 | 266307_at | AT2G27000 | CYP705A8 (cytochrome P450, family 705, subfamily A, polypeptide 8); oxygen binding | -1.56 |
| 30 | 266296_at | AT2G29420 | ATGSTU7 (GLUTATHIONE S-TRANSFERASE 25); glutathione transferase | 3.84 |
| 31 | 266290_at | AT2G29490 | ATGSTU1 (GLUTATHIONE S-TRANSFERASE 19); glutathione transferase | 3.92 |
| 32 | 266294_at | AT2G29500 | 17.6 kDa class I small heat shock protein (HSP17.6B-CI) | 4.72 |
| 33 | 266674_at | AT2G29620 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G07330.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN81231.1) | -2.60 |
| 34 | 267028_at | AT2G38470 | WRKY33 (WRKY DNA-binding protein 33); transcription factor | 3.06 |
| 35 | 267008_at | AT2G39350 | ABC transporter family protein | 3.25 |
| 36 | 260522_x_at | AT2G41730 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G24640.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO14635.1) | 4.71 |
| 37 | 266590_at | AT2G46240 | BAG6 (ARABIDOPSIS THALIANA BCL-2-ASSOCIATED ATHANOGENE 6); calmodulin binding / protein binding | 3.09 |
| 38 | 266752_at | AT2G47000 | MDR4/PGP4 (P-GLYCOPROTEIN4); ATPase, coupled to transmembrane movement of substances / xenobiotic-transporting ATPase | 3.40 |
| 39 | 245173_at | AT2G47520 | AP2 domain-containing transcription factor, putative | 5.95 |
| 40 | 259351_at | AT3G05150 | sugar transporter family protein | -2.63 |
| 41 | 259106_at | AT3G05490 | RALFL22 (RALF-LIKE 22) | -1.65 |
| 42 | 258832_at | AT3G07070 | protein kinase family protein | -3.59 |
| 43 | 258765_at | AT3G10710 | pectinesterase family protein | -7.29 |
| 44 | 256283_at | AT3G12540 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G39690.1); similar to At3g12540-like protein [Boechera stricta] (GB:ABB89771.1); contains InterPro domain Protein of unknown function DUF547 (InterPro:IPR006869) | -4.08 |
| 45 | 258452_at | AT3G22370 | AOX1A (alternative oxidase 1A); alternative oxidase | 4.33 |
| 46 | 257645_at | AT3G25790 | myb family transcription factor | -1.68 |
| 47 | 256980_at | AT3G26932 | DRB3 (DSRNA-BINDING PROTEIN 3); double-stranded RNA binding | -1.79 |
| 48 | 256589_at | AT3G28740 | cytochrome P450 family protein | 5.60 |
| 49 | 252238_at | AT3G49960 | peroxidase, putative | -7.13 |
| 50 | 252131_at | AT3G50930 | AAA-type ATPase family protein | 3.53 |
| 51 | 251824_at | AT3G55090 | ATPase, coupled to transmembrane movement of substances | 4.69 |
| 52 | 251226_at | AT3G62680 | PRP3 (PROLINE-RICH PROTEIN 3); structural constituent of cell wall | -3.45 |
| 53 | 251174_at | AT3G63200 | PLA IIIB/PLP9 (Patatin-like protein 9); nutrient reservoir | -1.65 |
| 54 | 255516_at | AT4G02270 | pollen Ole e 1 allergen and extensin family protein | -2.32 |
| 55 | 254907_at | AT4G11190 | disease resistance-responsive family protein / dirigent family protein | -1.59 |
| 56 | 254889_at | AT4G11650 | ATOSM34 (OSMOTIN 34) | -2.58 |
| 57 | 254759_at | AT4G13180 | short-chain dehydrogenase/reductase (SDR) family protein | 3.28 |
| 58 | 254718_at | AT4G13580 | disease resistance-responsive family protein | -3.03 |
| 59 | 245399_at | AT4G17340 | DELTA-TIP2/TIP2;2 (tonoplast intrinsic protein 2;2); water channel | -2.48 |
| 60 | 254534_at | AT4G19680 | IRT2 (iron-responsive transporter 2); iron ion transmembrane transporter/ zinc ion transmembrane transporter | -3.51 |
| 61 | 254318_at | AT4G22530 | embryo-abundant protein-related | 3.84 |
| 62 | 254044_at | AT4G25820 | XTR9 (XYLOGLUCAN ENDOTRANSGLYCOSYLASE 9); hydrolase, acting on glycosyl bonds | -2.42 |
| 63 | 253998_at | AT4G26010 | peroxidase, putative | -2.64 |
| 64 | 253796_at | AT4G28460 | unknown protein | 3.63 |
| 65 | 253734_at | AT4G29180 | leucine-rich repeat protein kinase, putative | -1.91 |
| 66 | 253720_at | AT4G29270 | acid phosphatase class B family protein | -1.74 |
| 67 | 253667_at | AT4G30170 | peroxidase, putative | -1.97 |
| 68 | 253569_at | AT4G31250 | leucine-rich repeat transmembrane protein kinase, putative | -1.53 |
| 69 | 253483_at | AT4G31910 | transferase family protein | -2.92 |
| 70 | 253353_at | AT4G33730 | pathogenesis-related protein, putative | -5.49 |
| 71 | 246203_at | AT4G36610 | hydrolase, alpha/beta fold family protein | 3.16 |
| 72 | 253044_at | AT4G37290 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G23270.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN62855.1) | 5.25 |
| 73 | 253046_at | AT4G37370 | CYP81D8 (cytochrome P450, family 81, subfamily D, polypeptide 8); oxygen binding | 5.74 |
| 74 | 252997_at | AT4G38400 | ATEXLA2 (ARABIDOPSIS THALIANA EXPANSIN-LIKE A2) | -3.60 |
| 75 | 250801_at | AT5G04960 | pectinesterase family protein | -3.21 |
| 76 | 250781_at | AT5G05410 | DREB2A (DRE-BINDING PROTEIN 2A); DNA binding / transcription activator/ transcription factor | 3.03 |
| 77 | 250778_at | AT5G05500 | pollen Ole e 1 allergen and extensin family protein | -2.67 |
| 78 | 250682_x_at | AT5G06630 | proline-rich extensin-like family protein | -2.21 |
| 79 | 250683_x_at | AT5G06640 | proline-rich extensin-like family protein | -2.22 |
| 80 | 250252_at | AT5G13750 | ZIFL1 (ZINC INDUCED FACILITATOR-LIKE 1); tetracycline:hydrogen antiporter/ transporter | 4.39 |
| 81 | 246584_at | AT5G14730 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G01513.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO64332.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN65788.1) | 3.60 |
| 82 | 249848_at | AT5G23220 | NIC3 (NICOTINAMIDASE 3); catalytic/ nicotinamidase | -1.96 |
| 83 | 246858_at | AT5G25930 | leucine-rich repeat family protein / protein kinase family protein | 3.29 |
| 84 | 246652_at | AT5G35190 | proline-rich extensin-like family protein | -2.67 |
| 85 | 249494_at | AT5G39050 | transferase family protein | 3.06 |
| 86 | 248434_at | AT5G51440 | 23.5 kDa mitochondrial small heat shock protein (HSP23.5-M) | 3.92 |
| 87 | 248381_at | AT5G51830 | pfkB-type carbohydrate kinase family protein | 3.72 |
| 88 | 248203_at | AT5G54230 | MYB49 (myb domain protein 49); transcription factor | -2.39 |
| 89 | 247949_at | AT5G57220 | CYP81F2 (cytochrome P450, family 81, subfamily F, polypeptide 2); oxygen binding | 3.04 |
| 90 | 247871_at | AT5G57530 | xyloglucan:xyloglucosyl transferase, putative / xyloglucan endotransglycosylase, putative / endo-xyloglucan transferase, putative | -3.47 |
| 91 | 247691_at | AT5G59720 | HSP18.2 (HEAT SHOCK PROTEIN 18.2) | 5.12 |
| 92 | 247573_at | AT5G61160 | AACT1 (ANTHOCYANIN 5-AROMATIC ACYLTRANSFERASE 1); transferase | 3.10 |
| 93 | 247435_at | AT5G62480 | ATGSTU9 (GLUTATHIONE S-TRANSFERASE TAU 9); glutathione transferase | 3.15 |
| 94 | 247351_at | AT5G63790 | ANAC102 (Arabidopsis NAC domain containing protein 102); transcription factor | 3.84 |
| 95 | 247283_at | AT5G64250 | 2-nitropropane dioxygenase family / NPD family | 3.44 |
| 96 | 247293_at | AT5G64510 | similar to unnamed protein product [Vitis vinifera] (GB:CAO49799.1) | 4.21 |
| 97 | 247170_at | AT5G65530 | protein kinase, putative | -3.92 |
| 98 | 247059_at | AT5G66690 | UGT72E2; UDP-glycosyltransferase/ coniferyl-alcohol glucosyltransferase/ transferase, transferring glycosyl groups | -1.94 |