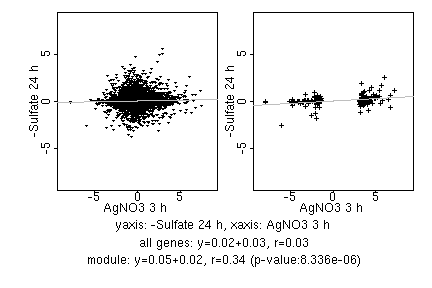
marker gene list
| AgNO3 3 h |
| -Sulfate 24 h |
| probeID | AGICode | Annotation | gene expression |
|
|---|---|---|---|---|
| 1 | 264574_at | AT1G05300 | ZIP5 (ZINC TRANSPORTER 5 PRECURSOR); cation transmembrane transporter | 3.35 |
| 2 | 263184_at | AT1G05560 | UGT1 (UDP-glucosyl transferase 75B1); UDP-glycosyltransferase/ transferase, transferring glycosyl groups | 3.69 |
| 3 | 263182_at | AT1G05575 | unknown protein | 3.35 |
| 4 | 261745_at | AT1G08500 | plastocyanin-like domain-containing protein | -1.42 |
| 5 | 264435_at | AT1G10360 | ATGSTU18 (GLUTATHIONE S-TRANSFERASE 29); glutathione transferase | -1.30 |
| 6 | 260969_at | AT1G12240 | ATBETAFRUCT4/VAC-INV (VACUOLAR INVERTASE); beta-fructofuranosidase/ hydrolase, hydrolyzing O-glycosyl compounds | -1.46 |
| 7 | 261488_at | AT1G14345 | oxidoreductase | -1.34 |
| 8 | 261474_at | AT1G14540 | anionic peroxidase, putative | 5.06 |
| 9 | 261475_at | AT1G14550 | anionic peroxidase, putative | 5.20 |
| 10 | 262598_at | AT1G15260 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G16070.1) | -1.90 |
| 11 | 261763_at | AT1G15520 | ATPDR12/PDR12 (PLEIOTROPIC DRUG RESISTANCE 12); ATPase, coupled to transmembrane movement of substances | 4.12 |
| 12 | 262518_at | AT1G17170 | ATGSTU24 (ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE (CLASS TAU) 24); glutathione transferase | 3.41 |
| 13 | 262517_at | AT1G17180 | ATGSTU25 (Arabidopsis thaliana Glutathione S-transferase (class tau) 25); glutathione transferase | 3.56 |
| 14 | 255753_at | AT1G18570 | MYB51 (MYB DOMAIN PROTEIN 51); DNA binding / transcription factor | 4.03 |
| 15 | 255940_at | AT1G20380 | prolyl oligopeptidase, putative / prolyl endopeptidase, putative / post-proline cleaving enzyme, putative | 3.34 |
| 16 | 260856_at | AT1G21910 | AP2 domain-containing transcription factor family protein | -1.59 |
| 17 | 261934_at | AT1G22400 | ATUGT85A1/UGT85A1 (UDP-GLUCOSYL TRANSFERASE 85A1); UDP-glycosyltransferase/ glucuronosyltransferase/ transferase, transferring glycosyl groups / transferase, transferring hexosyl groups | 3.22 |
| 18 | 264774_at | AT1G22890 | unknown protein | 4.32 |
| 19 | 264900_at | AT1G23080 | PIN7 (PIN-FORMED 7); auxin:hydrogen symporter/ transporter | -1.77 |
| 20 | 264866_at | AT1G24140 | matrixin family protein | 4.26 |
| 21 | 261648_at | AT1G27730 | STZ (SALT TOLERANCE ZINC FINGER); nucleic acid binding / transcription factor/ zinc ion binding | 3.25 |
| 22 | 259783_at | AT1G29510 | SAUR68 (SMALL AUXIN UPREGULATED 68) | -1.51 |
| 23 | 265102_at | AT1G30870 | cationic peroxidase, putative | -4.07 |
| 24 | 261999_at | AT1G33800 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G09990.1); similar to unknown [Populus trichocarpa] (GB:ABK93991.1); contains InterPro domain Protein of unknown function DUF579, plant (InterPro:IPR006514) | -1.97 |
| 25 | 256499_at | AT1G36640 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G36622.1) | 5.86 |
| 26 | 261607_at | AT1G49660 | ATCXE5 (ARABIDOPSIS THALIANA CARBOXYESTERASE 5); carboxylesterase | -1.63 |
| 27 | 265049_at | AT1G52060 | similar to jacalin lectin family protein [Arabidopsis thaliana] (TAIR:AT1G52070.1); similar to jasmonate inducible protein [Brassica napus] (GB:CAA72271.1); contains InterPro domain Mannose-binding lectin (InterPro:IPR001229) | -1.99 |
| 28 | 256352_at | AT1G54970 | ATPRP1 (PROLINE-RICH PROTEIN 1); structural constituent of cell wall | -3.48 |
| 29 | 262085_at | AT1G56060 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G32190.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO68639.1); contains domain PD188784 (PD188784) | 3.89 |
| 30 | 246405_at | AT1G57630 | disease resistance protein (TIR class), putative | 7.35 |
| 31 | 264746_at | AT1G62300 | WRKY6 (WRKY DNA-binding protein 6); transcription factor | 3.40 |
| 32 | 261099_at | AT1G62980 | ATEXPA18 (ARABIDOPSIS THALIANA EXPANSIN A18) | -2.93 |
| 33 | 262325_at | AT1G64160 | disease resistance-responsive family protein / dirigent family protein | 6.13 |
| 34 | 264632_at | AT1G65640 | DEGP4 (DEGP PROTEASE 4); serine-type peptidase/ trypsin | -1.31 |
| 35 | 262930_at | AT1G65690 | harpin-induced protein-related / HIN1-related / harpin-responsive protein-related | 3.20 |
| 36 | 255858_at | AT1G67030 | ZFP6 (ZINC FINGER PROTEIN 6); nucleic acid binding / transcription factor/ zinc ion binding | -1.54 |
| 37 | 245193_at | AT1G67810 | Fe-S metabolism associated domain-containing protein | 3.52 |
| 38 | 260406_at | AT1G69920 | ATGSTU12 (Arabidopsis thaliana Glutathione S-transferase (class tau) 12); glutathione transferase | 6.79 |
| 39 | 260405_at | AT1G69930 | ATGSTU11 (Arabidopsis thaliana Glutathione S-transferase (class tau) 11); glutathione transferase | 4.34 |
| 40 | 260239_at | AT1G74360 | leucine-rich repeat transmembrane protein kinase, putative | 3.63 |
| 41 | 264096_at | AT1G78995 | similar to unnamed protein product [Vitis vinifera] (GB:CAO41778.1) | -1.76 |
| 42 | 263403_at | AT2G04040 | ATDTX1; antiporter/ multidrug efflux pump/ multidrug transporter/ transporter | 3.40 |
| 43 | 263513_at | AT2G12400 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G25270.1); similar to hypothetical protein OsJ_008445 [Oryza sativa (japonica cultivar-group)] (GB:EAZ24962.1) | -1.61 |
| 44 | 265499_at | AT2G15480 | UGT73B5 (UDP-GLUCOSYL TRANSFERASE 73B5); UDP-glycosyltransferase/ transferase, transferring glycosyl groups | 3.98 |
| 45 | 266941_at | AT2G18980 | peroxidase, putative | -1.93 |
| 46 | 266800_at | AT2G22880 | VQ motif-containing protein | 5.57 |
| 47 | 267260_at | AT2G23130 | AGP17 (ARABINOGALACTAN PROTEIN 17) | -1.69 |
| 48 | 245082_at | AT2G23270 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G37290.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN62855.1) | 4.86 |
| 49 | 267128_at | AT2G23620 | esterase, putative | -3.12 |
| 50 | 263800_at | AT2G24600 | ankyrin repeat family protein | 3.50 |
| 51 | 263539_at | AT2G24850 | TAT3 (TYROSINE AMINOTRANSFERASE 3); transaminase | 4.58 |
| 52 | 263552_x_at | AT2G24980 | proline-rich extensin-like family protein | -3.51 |
| 53 | 264403_at | AT2G25150 | transferase family protein | -1.95 |
| 54 | 266658_at | AT2G25735 | unknown protein | 3.74 |
| 55 | 245033_at | AT2G26380 | disease resistance protein-related / LRR protein-related | 3.46 |
| 56 | 245041_at | AT2G26530 | AR781 | 3.32 |
| 57 | 264078_at | AT2G28470 | BGAL8 (BETA-GALACTOSIDASE 8); beta-galactosidase | -1.62 |
| 58 | 267300_at | AT2G30140 | UDP-glucoronosyl/UDP-glucosyl transferase family protein | 3.74 |
| 59 | 267567_at | AT2G30770 | CYP71A13 (CYTOCHROME P450, FAMILY 71, SUBFAMILY A, POLYPEPTIDE 13); indoleacetaldoxime dehydratase/ oxygen binding | 4.12 |
| 60 | 263942_at | AT2G35860 | FLA16 (FASCICLIN-LIKE ARABINOGALACTAN PROTEIN 16 PRECURSOR) | -1.45 |
| 61 | 266167_at | AT2G38860 | YLS5 (yellow-leaf-specific gene 5) | 3.60 |
| 62 | 265260_at | AT2G43000 | ANAC042 (Arabidopsis NAC domain containing protein 42); transcription factor | 3.50 |
| 63 | 258815_at | AT3G04000 | short-chain dehydrogenase/reductase (SDR) family protein | 3.40 |
| 64 | 258468_at | AT3G06070 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G19190.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO71112.1) | -1.59 |
| 65 | 258528_at | AT3G06770 | glycoside hydrolase family 28 protein / polygalacturonase (pectinase) family protein | -1.31 |
| 66 | 258832_at | AT3G07070 | protein kinase family protein | -3.50 |
| 67 | 259033_at | AT3G09410 | pectinacetylesterase family protein | 4.45 |
| 68 | 258941_at | AT3G09940 | ATMDAR3/MDHAR (MONODEHYDROASCORBATE REDUCTASE); monodehydroascorbate reductase (NADH) | 3.49 |
| 69 | 258765_at | AT3G10710 | pectinesterase family protein | -7.84 |
| 70 | 256283_at | AT3G12540 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G39690.1); similar to At3g12540-like protein [Boechera stricta] (GB:ABB89771.1); contains InterPro domain Protein of unknown function DUF547 (InterPro:IPR006869) | -4.83 |
| 71 | 258355_at | AT3G14330 | pentatricopeptide (PPR) repeat-containing protein | -1.44 |
| 72 | 257557_at | AT3G14490 | terpene synthase/cyclase family protein | -2.11 |
| 73 | 257217_at | AT3G14940 | ATPPC3 (PHOSPHOENOLPYRUVATE CARBOXYLASE 3); phosphoenolpyruvate carboxylase | -2.06 |
| 74 | 258145_at | AT3G18200 | nodulin MtN21 family protein | -4.63 |
| 75 | 257061_at | AT3G18250 | contains domain PROKAR_LIPOPROTEIN (PS51257) | 4.54 |
| 76 | 257540_at | AT3G21520 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G21550.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO71058.1); contains InterPro domain Protein of unknown function DUF679 (InterPro:IPR007770) | 4.66 |
| 77 | 257919_at | AT3G23250 | AtMYB15/AtY19/MYB15 (myb domain protein 15); DNA binding / transcription factor | 3.98 |
| 78 | 258100_at | AT3G23550 | MATE efflux family protein | 5.69 |
| 79 | 257644_at | AT3G25780 | AOC3 (ALLENE OXIDE CYCLASE 3) | 3.55 |
| 80 | 258277_at | AT3G26830 | PAD3 (PHYTOALEXIN DEFICIENT 3); oxygen binding | 6.54 |
| 81 | 252502_at | AT3G46900 | COPT2 (Copper transporter 2); copper ion transmembrane transporter | -2.31 |
| 82 | 252364_at | AT3G48450 | nitrate-responsive NOI protein, putative | 3.48 |
| 83 | 252238_at | AT3G49960 | peroxidase, putative | -7.90 |
| 84 | 252202_at | AT3G50300 | transferase family protein | -1.49 |
| 85 | 251884_at | AT3G54150 | embryo-abundant protein-related | 4.36 |
| 86 | 251895_at | AT3G54420 | ATEP3 (Arabidopsis thaliana chitinase class IV); chitinase | 3.20 |
| 87 | 251843_x_at | AT3G54590 | ATHRGP1; structural constituent of cell wall | -2.42 |
| 88 | 251575_at | AT3G58120 | bZIP transcription factor family protein | -2.76 |
| 89 | 251524_at | AT3G58990 | aconitase C-terminal domain-containing protein | -1.84 |
| 90 | 251226_at | AT3G62680 | PRP3 (PROLINE-RICH PROTEIN 3); structural constituent of cell wall | -4.42 |
| 91 | 251176_at | AT3G63380 | calcium-transporting ATPase, plasma membrane-type, putative / Ca(2+)-ATPase, putative (ACA12) | 4.01 |
| 92 | 255694_at | AT4G00050 | UNE10 (unfertilized embryo sac 10); DNA binding / transcription factor | -1.94 |
| 93 | 255587_at | AT4G01480 | ATPPA5 (ARABIDOPSIS THALIANA PYROPHOSPHORYLASE 5); inorganic diphosphatase/ pyrophosphatase | -1.33 |
| 94 | 255595_at | AT4G01700 | chitinase, putative | 3.24 |
| 95 | 255543_at | AT4G01870 | tolB protein-related | 3.49 |
| 96 | 255516_at | AT4G02270 | pollen Ole e 1 allergen and extensin family protein | -3.17 |
| 97 | 255500_at | AT4G02390 | APP (ARABIDOPSIS POLY(ADP-RIBOSE) POLYMERASE); NAD+ ADP-ribosyltransferase | -1.59 |
| 98 | 255298_at | AT4G04840 | methionine sulfoxide reductase domain-containing protein / SeIR domain-containing protein | -2.41 |
| 99 | 255140_x_at | AT4G08410 | proline-rich extensin-like family protein | -3.28 |
| 100 | 255110_at | AT4G08770 | peroxidase, putative | 3.39 |
| 101 | 255111_at | AT4G08780 | peroxidase, putative | 3.44 |
| 102 | 255005_at | AT4G09990 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G33800.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO16316.1); contains InterPro domain Protein of unknown function DUF579, plant (InterPro:IPR006514) | -1.82 |
| 103 | 254926_at | AT4G11280 | ACS6 (1-AMINOCYCLOPROPANE-1-CARBOXYLIC ACID (ACC) SYNTHASE 6) | 4.22 |
| 104 | 245325_at | AT4G14130 | XTR7 (XYLOGLUCAN ENDOTRANSGLYCOSYLASE 7); hydrolase, acting on glycosyl bonds | -1.59 |
| 105 | 245399_at | AT4G17340 | DELTA-TIP2/TIP2;2 (tonoplast intrinsic protein 2;2); water channel | -3.40 |
| 106 | 245250_at | AT4G17490 | ATERF6 (ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 6); DNA binding / transcription factor | 4.08 |
| 107 | 254432_at | AT4G20830 | FAD-binding domain-containing protein | 3.24 |
| 108 | 254318_at | AT4G22530 | embryo-abundant protein-related | 4.13 |
| 109 | 254255_at | AT4G23220 | protein kinase family protein | 3.76 |
| 110 | 254215_at | AT4G23700 | ATCHX17 (CATION/H+ EXCHANGER 17); monovalent cation:proton antiporter | 3.72 |
| 111 | 254025_at | AT4G25790 | allergen V5/Tpx-1-related family protein | -2.77 |
| 112 | 254044_at | AT4G25820 | XTR9 (XYLOGLUCAN ENDOTRANSGLYCOSYLASE 9); hydrolase, acting on glycosyl bonds | -3.44 |
| 113 | 253998_at | AT4G26010 | peroxidase, putative | -3.91 |
| 114 | 253957_at | AT4G26320 | AGP13 (ARABINOGALACTAN PROTEIN 13) | -2.78 |
| 115 | 253827_at | AT4G28085 | unknown protein | 4.91 |
| 116 | 253763_at | AT4G28850 | xyloglucan:xyloglucosyl transferase, putative / xyloglucan endotransglycosylase, putative / endo-xyloglucan transferase, putative | -6.02 |
| 117 | 253732_at | AT4G29140 | MATE efflux protein-related | -4.68 |
| 118 | 253734_at | AT4G29180 | leucine-rich repeat protein kinase, putative | -1.90 |
| 119 | 253720_at | AT4G29270 | acid phosphatase class B family protein | -1.31 |
| 120 | 253667_at | AT4G30170 | peroxidase, putative | -2.14 |
| 121 | 253579_at | AT4G30610 | BRS1 (BRI1 SUPPRESSOR 1) | -1.74 |
| 122 | 253582_at | AT4G30670 | contains domain PROKAR_LIPOPROTEIN (PS51257) | -2.80 |
| 123 | 253255_at | AT4G34760 | auxin-responsive family protein | -2.01 |
| 124 | 253173_at | AT4G35110 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G16900.1); contains InterPro domain Phospholipase-like, arabidopsis (InterPro:IPR007942) | 3.66 |
| 125 | 246275_at | AT4G36540 | BEE2 (BR ENHANCED EXPRESSION 2); DNA binding / transcription factor | -1.55 |
| 126 | 253044_at | AT4G37290 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G23270.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN62855.1) | 5.59 |
| 127 | 253046_at | AT4G37370 | CYP81D8 (cytochrome P450, family 81, subfamily D, polypeptide 8); oxygen binding | 3.23 |
| 128 | 252908_at | AT4G39670 | glycolipid binding / glycolipid transporter | 3.29 |
| 129 | 252833_at | AT4G40090 | AGP3 (ARABINOGALACTAN-PROTEIN 3) | -3.92 |
| 130 | 251017_at | AT5G02760 | protein phosphatase 2C family protein / PP2C family protein | -3.10 |
| 131 | 250983_at | AT5G02780 | In2-1 protein, putative | 3.63 |
| 132 | 250801_at | AT5G04960 | pectinesterase family protein | -3.97 |
| 133 | 250778_at | AT5G05500 | pollen Ole e 1 allergen and extensin family protein | -3.44 |
| 134 | 250738_at | AT5G05730 | ASA1 (ANTHRANILATE SYNTHASE ALPHA SUBUNIT 1); anthranilate synthase | 3.33 |
| 135 | 250682_x_at | AT5G06630 | proline-rich extensin-like family protein | -3.10 |
| 136 | 250683_x_at | AT5G06640 | proline-rich extensin-like family protein | -2.94 |
| 137 | 250695_at | AT5G06740 | lectin protein kinase family protein | 4.45 |
| 138 | 250670_at | AT5G06860 | PGIP1 (POLYGALACTURONASE INHIBITING PROTEIN 1); protein binding | 3.51 |
| 139 | 246487_at | AT5G16030 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G02500.1); similar to hypothetical protein [Cleome spinosa] (GB:ABD96956.1) | -1.88 |
| 140 | 250054_at | AT5G17860 | CAX7 (CALCIUM EXCHANGER 7); calcium:sodium antiporter/ cation:cation antiporter | 3.21 |
| 141 | 249983_at | AT5G18470 | curculin-like (mannose-binding) lectin family protein | 3.92 |
| 142 | 246831_at | AT5G26340 | MSS1 (SUGAR TRANSPORT PROTEIN 13); carbohydrate transmembrane transporter/ hexose:hydrogen symporter/ high-affinity hydrogen:glucose symporter/ sugar:hydrogen ion symporter | 3.86 |
| 143 | 246652_at | AT5G35190 | proline-rich extensin-like family protein | -3.75 |
| 144 | 249459_at | AT5G39580 | peroxidase, putative | 4.64 |
| 145 | 249417_at | AT5G39670 | calcium-binding EF hand family protein | 3.75 |
| 146 | 248681_at | AT5G48900 | pectate lyase family protein | -1.51 |
| 147 | 248652_at | AT5G49270 | COBL9/MRH4/SHV2 (COBRA-LIKE 9, SHAVEN 2); carbohydrate binding | -1.86 |
| 148 | 248381_at | AT5G51830 | pfkB-type carbohydrate kinase family protein | 4.09 |
| 149 | 248327_at | AT5G52750 | heavy-metal-associated domain-containing protein | 3.60 |
| 150 | 248252_at | AT5G53250 | AGP22/ATAGP22 (ARABINOGALACTAN PROTEINS 22) | -4.54 |
| 151 | 247940_at | AT5G57190 | PSD2 (PHOSPHATIDYLSERINE DECARBOXYLASE 2); phosphatidylserine decarboxylase | 3.64 |
| 152 | 247949_at | AT5G57220 | CYP81F2 (cytochrome P450, family 81, subfamily F, polypeptide 2); oxygen binding | 5.76 |
| 153 | 247871_at | AT5G57530 | xyloglucan:xyloglucosyl transferase, putative / xyloglucan endotransglycosylase, putative / endo-xyloglucan transferase, putative | -4.46 |
| 154 | 247914_at | AT5G57540 | xyloglucan:xyloglucosyl transferase, putative / xyloglucan endotransglycosylase, putative / endo-xyloglucan transferase, putative | -3.98 |
| 155 | 247878_at | AT5G57760 | unknown protein | -2.10 |
| 156 | 247678_at | AT5G59520 | ZIP2 (ZINC TRANSPORTER 2 PRECURSOR); transferase, transferring glycosyl groups / zinc ion transmembrane transporter | -1.46 |
| 157 | 247573_at | AT5G61160 | AACT1 (ANTHOCYANIN 5-AROMATIC ACYLTRANSFERASE 1); transferase | 5.34 |
| 158 | 247536_at | AT5G61650 | CYCP4;2 (CYCLIN P4;2); cyclin-dependent protein kinase | -2.41 |
| 159 | 247215_at | AT5G64905 | PROPEP3 (Elicitor peptide 3 precursor) | 4.17 |
| 160 | 247145_at | AT5G65600 | legume lectin family protein / protein kinase family protein | 6.80 |
| 161 | 247162_at | AT5G65730 | xyloglucan:xyloglucosyl transferase, putative / xyloglucan endotransglycosylase, putative / endo-xyloglucan transferase, putative | -3.12 |
| 162 | 246991_at | AT5G67400 | peroxidase 73 (PER73) (P73) (PRXR11) | -4.99 |