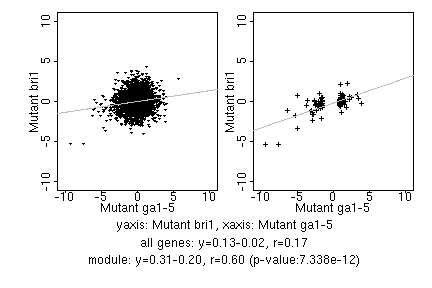
marker gene list
| Mutant ga1-5 |
| Mutant bri1 |
| probeID | AGICode | Annotation | gene expression |
|
|---|---|---|---|---|
| 1 | 261579_at | AT1G01050 | ATPPA1 (ARABIDOPSIS THALIANA PYROPHOSPHORYLASE 1); inorganic diphosphatase/ pyrophosphatase | 1.19 |
| 2 | 261057_at | AT1G01230 | ORMDL family protein | 1.25 |
| 3 | 261055_at | AT1G01300 | aspartyl protease family protein | 1.47 |
| 4 | 261049_at | AT1G01430 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G01080.1); similar to unknown protein Cr17 [Brassica napus] (GB:AAX51387.1); contains InterPro domain Protein of unknown function DUF231, plant (InterPro:IPR004253) | 1.08 |
| 5 | 261052_at | AT1G01440 | extra-large G-protein-related | 1.11 |
| 6 | 259438_at | AT1G01510 | AN (ANGUSTIFOLIA) | 1.36 |
| 7 | 259430_at | AT1G01610 | ATGPAT4/GPAT4 (GLYCEROL-3-PHOSPHATE ACYLTRANSFERASE 4); 1-acylglycerol-3-phosphate O-acyltransferase/ acyltransferase | 1.32 |
| 8 | 261560_at | AT1G01710 | acyl-CoA thioesterase family protein | 1.00 |
| 9 | 261562_at | AT1G01750 | actin-depolymerizing factor, putative | 1.68 |
| 10 | 261559_at | AT1G01780 | LIM domain-containing protein | 1.04 |
| 11 | 261621_at | AT1G01960 | EDA10 (embryo sac development arrest 10); guanyl-nucleotide exchange factor | 1.15 |
| 12 | 261624_at | AT1G02000 | GAE2 (UDP-D-GLUCURONATE 4-EPIMERASE 2); catalytic | 1.00 |
| 13 | 265044_at | AT1G03950 | VPS2.3 | -3.40 |
| 14 | 265194_at | AT1G05010 | EFE (ETHYLENE FORMING ENZYME) | -1.52 |
| 15 | 261826_at | AT1G11580 | ATPMEPCRA; pectinesterase | 1.05 |
| 16 | 262811_at | AT1G11700 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G61930.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO17934.1); contains InterPro domain Protein of unknown function DUF584 (InterPro:IPR007608) | -1.57 |
| 17 | 261768_at | AT1G15550 | GA4 (GA REQUIRING 4); gibberellin 3-beta-dioxygenase | 1.93 |
| 18 | 260656_at | AT1G19380 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G65650.1); similar to unknown [Vitis pseudoreticulata] (GB:ABC69762.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO61173.1); contains InterPro domain Protein of unknown function DUF1195 (InterPro:IPR010608) | -1.59 |
| 19 | 259561_at | AT1G21250 | WAK1 (CELL WALL-ASSOCIATED KINASE); kinase | -5.07 |
| 20 | 245642_at | AT1G25275 | similar to unnamed protein product [Vitis vinifera] (GB:CAO43403.1) | -1.30 |
| 21 | 259599_at | AT1G28110 | SCPL45; serine carboxypeptidase | 1.05 |
| 22 | 259632_at | AT1G56430 | nicotianamine synthase, putative | 1.20 |
| 23 | 264767_at | AT1G61380 | S-locus protein kinase, putative | -1.50 |
| 24 | 262644_at | AT1G62710 | BETA-VPE (vacuolar processing enzyme beta); cysteine-type endopeptidase | 1.17 |
| 25 | 261557_at | AT1G63640 | kinesin motor protein-related | -1.68 |
| 26 | 260324_at | AT1G63970 | ISPF (Homolog of E. coli ispF (isoprenoids F)); 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase | -1.43 |
| 27 | 264636_at | AT1G65490 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G65486.1) | -2.34 |
| 28 | 264469_at | AT1G67100 | LBD40 (LOB DOMAIN-CONTAINING PROTEIN 40) | 2.34 |
| 29 | 264998_at | AT1G67330 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G27930.1); similar to unknown [Populus trichocarpa] (GB:ABK93495.1); contains InterPro domain Protein of unknown function DUF579, plant (InterPro:IPR006514) | 1.01 |
| 30 | 259831_at | AT1G69600 | ATHB29/ZFHD1 (ZINC FINGER HOMEODOMAIN 1); DNA binding / transcription activator/ transcription factor | 2.02 |
| 31 | 259752_at | AT1G71040 | LPR2 (LOW PHOSPHATE ROOT2); copper ion binding | -1.67 |
| 32 | 259854_at | AT1G72200 | zinc finger (C3HC4-type RING finger) family protein | 1.41 |
| 33 | 259918_at | AT1G72570 | DNA binding / transcription factor | 3.53 |
| 34 | 262381_at | AT1G72900 | disease resistance protein (TIR-NBS class), putative | -3.62 |
| 35 | 256332_at | AT1G76890 | GT2; transcription factor | -1.85 |
| 36 | 262050_at | AT1G80130 | binding | -2.82 |
| 37 | 261896_at | AT1G80670 | transducin family protein / WD-40 repeat family protein | 1.05 |
| 38 | 262206_at | AT2G01090 | ubiquinol-cytochrome C reductase complex 7.8 kDa protein, putative / mitochondrial hinge protein, putative | 1.05 |
| 39 | 266353_at | AT2G01520 | MLP328 (MLP-LIKE PROTEIN 328) | -1.50 |
| 40 | 266176_at | AT2G02250 | ATPP2-B2 (Phloem protein 2-B2); carbohydrate binding | 1.25 |
| 41 | 266070_at | AT2G18660 | EXLB3 (EXPANSIN-LIKE B3 PRECURSOR) | -9.28 |
| 42 | 245052_at | AT2G26440 | pectinesterase family protein | -1.58 |
| 43 | 245038_at | AT2G26560 | PLP2 (PHOSPHOLIPASE A 2A); nutrient reservoir | -4.76 |
| 44 | 265667_at | AT2G27470 | CCAAT-box binding transcription factor subunit HAP3-related | -1.57 |
| 45 | 266165_at | AT2G28190 | CSD2 (COPPER/ZINC SUPEROXIDE DISMUTASE 2); copper, zinc superoxide dismutase | 2.35 |
| 46 | 266644_at | AT2G29660 | zinc finger (C2H2 type) family protein | 1.08 |
| 47 | 266669_at | AT2G29750 | UDP-glucoronosyl/UDP-glucosyl transferase family protein | 1.39 |
| 48 | 267300_at | AT2G30140 | UDP-glucoronosyl/UDP-glucosyl transferase family protein | -1.55 |
| 49 | 267307_at | AT2G30210 | LAC3 (laccase 3); copper ion binding / oxidoreductase | 1.31 |
| 50 | 267546_at | AT2G32680 | disease resistance family protein | -1.88 |
| 51 | 263841_at | AT2G36870 | xyloglucan:xyloglucosyl transferase, putative / xyloglucan endotransglycosylase, putative / endo-xyloglucan transferase, putative | 1.02 |
| 52 | 265959_at | AT2G37240 | antioxidant/ oxidoreductase | -1.48 |
| 53 | 267035_at | AT2G38400 | AGT3 (ALANINE:GLYOXYLATE AMINOTRANSFERASE 3); alanine-glyoxylate transaminase | 1.02 |
| 54 | 266984_at | AT2G39570 | ACT domain-containing protein | 1.09 |
| 55 | 267337_at | AT2G39980 | transferase family protein | -1.60 |
| 56 | 267076_at | AT2G41090 | calmodulin-like calcium-binding protein, 22 kDa (CaBP-22) | -2.96 |
| 57 | 266884_at | AT2G44790 | UCC2 (UCLACYANIN 2); copper ion binding | 1.22 |
| 58 | 245139_at | AT2G45430 | DNA-binding protein-related | 1.17 |
| 59 | 259353_at | AT3G05190 | aminotransferase class IV family protein | 1.79 |
| 60 | 258939_at | AT3G10020 | similar to Os12g0147200 [Oryza sativa (japonica cultivar-group)] (GB:NP_001066153.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO15981.1); similar to Os11g0149200 [Oryza sativa (japonica cultivar-group)] (GB:NP_001065754.1) | 1.13 |
| 61 | 257654_at | AT3G13310 | DNAJ heat shock N-terminal domain-containing protein | 1.28 |
| 62 | 257206_at | AT3G16530 | legume lectin family protein | -2.70 |
| 63 | 256922_at | AT3G19010 | oxidoreductase, 2OG-Fe(II) oxygenase family protein | -2.10 |
| 64 | 257143_at | AT3G20110 | CYP705A20 (cytochrome P450, family 705, subfamily A, polypeptide 20); oxygen binding | 1.02 |
| 65 | 257264_at | AT3G22060 | receptor protein kinase-related | -2.20 |
| 66 | 257623_at | AT3G26210 | CYP71B23 (cytochrome P450, family 71, subfamily B, polypeptide 23); oxygen binding | -2.69 |
| 67 | 258277_at | AT3G26830 | PAD3 (PHYTOALEXIN DEFICIENT 3); oxygen binding | -2.02 |
| 68 | 252345_at | AT3G48640 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G66670.2); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G66670.1) | -2.78 |
| 69 | 252037_at | AT3G51920 | CAM9 (CALMODULIN 9); calcium ion binding | -1.36 |
| 70 | 256681_at | AT3G52340 | SPP2; sucrose-phosphatase | -1.38 |
| 71 | 252010_at | AT3G52740 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G44450.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO70571.1) | 1.32 |
| 72 | 251887_at | AT3G54170 | ATFIP37 (ARABIDOPSIS THALIANA FKBP12 INTERACTING PROTEIN 37) | -1.37 |
| 73 | 251654_at | AT3G57140 | SDP1-LIKE (SDP1-LIKE); GTP binding | 1.38 |
| 74 | 255116_at | AT4G08850 | leucine-rich repeat family protein / protein kinase family protein | -1.89 |
| 75 | 255070_at | AT4G09020 | ATISA3/ISA3 (ISOAMYLASE 3); alpha-amylase | -1.89 |
| 76 | 255016_at | AT4G10120 | ATSPS4F; sucrose-phosphate synthase/ transferase, transferring glycosyl groups | -1.81 |
| 77 | 245325_at | AT4G14130 | XTR7 (XYLOGLUCAN ENDOTRANSGLYCOSYLASE 7); hydrolase, acting on glycosyl bonds | 1.12 |
| 78 | 245293_at | AT4G16660 | heat shock protein 70, putative / HSP70, putative | -1.66 |
| 79 | 245346_at | AT4G17090 | CT-BMY (BETA-AMYLASE 3, BETA-AMYLASE 8); beta-amylase | -1.58 |
| 80 | 254452_at | AT4G21100 | DDB1B (DAMAGED DNA BINDING PROTEIN 1 B) | 4.05 |
| 81 | 254333_at | AT4G22753 | SMO1-3 (STEROL 4-ALPHA METHYL OXIDASE); catalytic | -1.28 |
| 82 | 254256_at | AT4G23180 | CRK10 (CYSTEINE-RICH RLK10); kinase | -2.52 |
| 83 | 254200_at | AT4G24110 | similar to unnamed protein product [Vitis vinifera] (GB:CAO62924.1) | 1.28 |
| 84 | 253648_at | AT4G29940 | PRHA (PATHOGENESIS RELATED HOMEODOMAIN PROTEIN A); transcription factor | -4.79 |
| 85 | 253414_at | AT4G33050 | EDA39 (embryo sac development arrest 39); calmodulin binding | -2.62 |
| 86 | 253039_at | AT4G37760 | SQE3 (SQUALENE EPOXIDASE 3); oxidoreductase | 1.40 |
| 87 | 251054_at | AT5G01540 | lectin protein kinase, putative | -1.54 |
| 88 | 250844_at | AT5G04470 | SIM (SIAMESE) | 1.16 |
| 89 | 250214_at | AT5G13870 | EXGT-A4 (ENDOXYLOGLUCAN TRANSFERASE A4); hydrolase, acting on glycosyl bonds | 2.69 |
| 90 | 249756_at | AT5G24313 | unknown protein | 1.16 |
| 91 | 249353_at | AT5G40420 | OLEO2 (OLEOSIN 2) | 3.62 |
| 92 | 249051_at | AT5G44390 | FAD-binding domain-containing protein | -2.50 |
| 93 | 248725_at | AT5G47980 | transferase family protein | 1.22 |
| 94 | 248686_at | AT5G48540 | 33 kDa secretory protein-related | -1.36 |
| 95 | 248427_at | AT5G51750 | ATSBT1.3; subtilase | 1.08 |
| 96 | 248327_at | AT5G52750 | heavy-metal-associated domain-containing protein | -6.14 |
| 97 | 248330_at | AT5G52810 | ornithine cyclodeaminase/mu-crystallin family protein | -1.54 |
| 98 | 248271_at | AT5G53420 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G27900.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G27900.2); similar to Os05g0595300 [Oryza sativa (japonica cultivar-group)] (GB:NP_001056514.1); similar to hypothetical protein OsI_020507 [Oryza sativa (indica cultivar-group)] (GB:EAY99274.1); contains InterPro domain CCT (InterPro:IPR010402) | -1.46 |
| 99 | 248276_at | AT5G53550 | YSL3 (YELLOW STRIPE LIKE 3); oligopeptide transporter | -1.92 |
| 100 | 248186_at | AT5G53880 | similar to unknown [Populus trichocarpa] (GB:ABK92577.1) | -2.01 |
| 101 | 248197_at | AT5G54190 | PORA (Protochlorophyllide reductase A); oxidoreductase/ protochlorophyllide reductase | 1.74 |
| 102 | 247573_at | AT5G61160 | AACT1 (ANTHOCYANIN 5-AROMATIC ACYLTRANSFERASE 1); transferase | -7.47 |
| 103 | 247314_at | AT5G64000 | SAL2; 3'(2'),5'-bisphosphate nucleotidase/ inositol or phosphatidylinositol phosphatase | -2.35 |
| 104 | 247091_at | AT5G66390 | peroxidase 72 (PER72) (P72) (PRXR8) | 1.22 |
| 105 | 247074_at | AT5G66590 | allergen V5/Tpx-1-related family protein | 1.53 |
| 106 | 247030_at | AT5G67210 | nucleic acid binding / pancreatic ribonuclease | 1.29 |
| 107 | 244982_at | ATCG00780 | encodes a chloroplast ribosomal protein L14, a constituent of the large subunit of the ribosomal complex | -2.16 |
| 108 | 244984_at | ATCG00800 | encodes a chloroplast ribosomal protein S3, a constituent of the small subunit of the ribosomal complex | -1.47 |