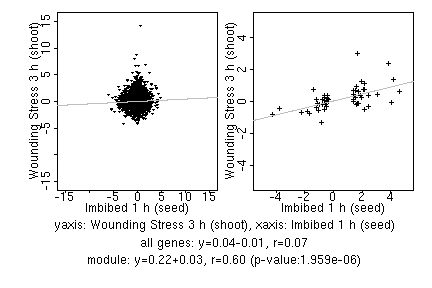
marker gene list
| Imbibed 1 h (seed) |
| Wounding Stress3 h (shoot) |
| probeID | AGICode | Annotation | gene expression |
|
|---|---|---|---|---|
| 1 | 264661_at | AT1G09950 | transcription factor-related | 1.48 |
| 2 | 246325_at | AT1G16540 | ABA3/ATABA3/LOS5/SIR3 (ABA DEFICIENT 3); Mo-molybdopterin cofactor sulfurase/ selenocysteine lyase | -0.32 |
| 3 | 260712_at | AT1G17550 | HAB2 (Homology to ABI2); protein serine/threonine phosphatase | -0.46 |
| 4 | 260924_at | AT1G21590 | protein kinase family protein | -0.30 |
| 5 | 245878_at | AT1G26190 | phosphoribulokinase/uridine kinase family protein | -0.27 |
| 6 | 261658_at | AT1G50040 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G19680.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO61535.1); contains InterPro domain Protein of unknown function DUF1005 (InterPro:IPR010410) | 1.71 |
| 7 | 259803_at | AT1G72150 | PATL1 (PATELLIN 1); transporter | 1.78 |
| 8 | 266203_at | AT2G02230 | ATPP2-B1 (Phloem protein 2-B1); carbohydrate binding | 1.46 |
| 9 | 263548_at | AT2G21660 | ATGRP7 (COLD, CIRCADIAN RHYTHM, AND RNA BINDING 2); RNA binding / double-stranded DNA binding / single-stranded DNA binding | -0.52 |
| 10 | 263790_at | AT2G24530 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G31440.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO43929.1) | -0.43 |
| 11 | 266843_at | AT2G26135 | zinc finger (C3HC4-type RING finger) family protein | -0.44 |
| 12 | 245096_at | AT2G40880 | FL3-27; cysteine protease inhibitor | 2.17 |
| 13 | 260496_at | AT2G41700 | ATPase, coupled to transmembrane movement of substances / amino acid transmembrane transporter | -0.55 |
| 14 | 260501_at | AT2G41770 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G57420.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO23590.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN82225.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO40788.1) | 1.98 |
| 15 | 245173_at | AT2G47520 | AP2 domain-containing transcription factor, putative | 2.51 |
| 16 | 259189_at | AT3G01700 | AGP11 (ARABINOGALACTAN PROTEIN 11) | -1.33 |
| 17 | 258979_at | AT3G09440 | heat shock cognate 70 kDa protein 3 (HSC70-3) (HSP70-3) | 1.82 |
| 18 | 257654_at | AT3G13310 | DNAJ heat shock N-terminal domain-containing protein | 3.17 |
| 19 | 257076_at | AT3G19680 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G50040.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO61535.1); contains InterPro domain Protein of unknown function DUF1005 (InterPro:IPR010410) | 4.14 |
| 20 | 258133_at | AT3G24500 | ATMBF1C/MBF1C (MULTIPROTEIN BRIDGING FACTOR 1C); DNA binding / transcription coactivator/ transcription factor | 1.47 |
| 21 | 256755_at | AT3G25600 | calmodulin, putative | 1.50 |
| 22 | 258073_at | AT3G25880 | auxin-resistance protein, putative | -3.69 |
| 23 | 256576_at | AT3G28210 | PMZ; zinc ion binding | 1.76 |
| 24 | 257111_x_at | AT3G30450 | transposable element gene | -4.18 |
| 25 | 252820_at | AT3G42640 | AHA8 (ARABIDOPSIS H(+)-ATPASE 8); ATPase | -0.92 |
| 26 | 252671_at | AT3G44190 | pyridine nucleotide-disulphide oxidoreductase family protein | 2.03 |
| 27 | 252679_at | AT3G44260 | CCR4-NOT transcription complex protein, putative | 1.46 |
| 28 | 246334_at | AT3G44850 | protein kinase-related | -0.72 |
| 29 | 252256_at | AT3G49430 | SRP34A (SER/ARG-RICH PROTEIN 34A); RNA binding | -0.42 |
| 30 | 252212_at | AT3G50310 | MAPKKK20 (Mitogen-activated protein kinase kinase kinase 20); kinase | 3.89 |
| 31 | 252000_at | AT3G52710 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G36220.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO15901.1) | -1.11 |
| 32 | 255807_at | AT4G10270 | wound-responsive family protein | 2.27 |
| 33 | 245585_at | AT4G14970 | similar to hypothetical protein [Vitis vinifera] (GB:CAN76433.1); similar to Os07g0154400 [Oryza sativa (japonica cultivar-group)] (GB:NP_001058915.1); similar to hypothetical protein OsI_024046 [Oryza sativa (indica cultivar-group)] (GB:EAZ02814.1) | -0.53 |
| 34 | 254605_at | AT4G18950 | ankyrin protein kinase, putative | 1.62 |
| 35 | 254458_at | AT4G21180 | DNAJ heat shock N-terminal domain-containing protein / sec63 domain-containing protein | -0.36 |
| 36 | 254120_at | AT4G24570 | mitochondrial substrate carrier family protein | 1.59 |
| 37 | 253915_at | AT4G27280 | calcium-binding EF hand family protein | 2.04 |
| 38 | 253832_at | AT4G27654 | unknown protein | 1.84 |
| 39 | 253669_at | AT4G30000 | dihydropterin pyrophosphokinase, putative / dihydropteroate synthase, putative / DHPS, putative | -1.10 |
| 40 | 253113_at | AT4G35985 | senescence/dehydration-associated protein-related | 4.69 |
| 41 | 250644_at | AT5G06750 | protein phosphatase 2C family protein / PP2C family protein | -0.66 |
| 42 | 246018_at | AT5G10695 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G57123.1); similar to unknown [Picea sitchensis] (GB:ABK22689.1) | 1.69 |
| 43 | 250358_at | AT5G11740 | AGP15 (ARABINOGALACTAN PROTEIN 15) | 2.27 |
| 44 | 249794_at | AT5G23530 | ATCXE18 (ARABIDOPSIS THALIANA CARBOXYESTERASE 18); carboxylesterase | 2.23 |
| 45 | 246765_at | AT5G27330 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G05130.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO68600.1); contains InterPro domain Prefoldin (InterPro:IPR009053) | -0.57 |
| 46 | 248981_at | AT5G45110 | NPR3 (NPR1-LIKE PROTEIN 3); protein binding | 2.53 |
| 47 | 247655_at | AT5G59820 | RHL41 (RESPONSIVE TO HIGH LIGHT 41); nucleic acid binding / transcription factor/ zinc ion binding | 4.28 |
| 48 | 245026_at | ATCG00140 | ATPase III subunit | -2.14 |
| 49 | 244995_at | ATCG00150 | Encodes a subunit of ATPase complex CF0, which is a proton channel that supplies the proton motive force to drive ATP synthesis by CF1 portion of the complex. | -1.71 |
| 50 | 244963_at | ATCG00570 | PSII cytochrome b559 | -1.06 |
| 51 | 244985_at | ATCG00810 | encodes a chloroplast ribosomal protein L22, a constituent of the large subunit of the ribosomal complex | -1.58 |
| 52 | 244938_at | ATCG01120 | encodes a chloroplast ribosomal protein S15, a constituent of the small subunit of the ribosomal complex | -0.75 |
| 53 | 257336_at | ATMG01410 | Identical to Uncharacterized mitochondrial protein AtMg01410 [Arabidopsis Thaliana] (GB:P92567;GB:Q1ZXV1); similar to hypothetical protein BrnapMp025 [Brassica napus] (GB:YP_717123.1); contains InterPro domain RNA-dependent RNA polymerase, mitoviral (InterPro:IPR008686) | -0.95 |