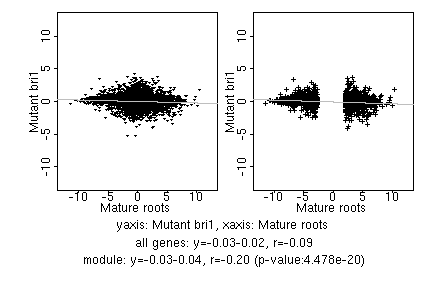
marker gene list
| Mature roots |
| Mutant bri1 |
| probeID | AGICode | Annotation | gene expression |
|
|---|---|---|---|---|
| 1 | 261577_at | AT1G01080 | 33 kDa ribonucleoprotein, chloroplast, putative / RNA-binding protein cp33, putative | -2.81 |
| 2 | 261581_at | AT1G01140 | CIPK9 (CBL-INTERACTING PROTEIN KINASE 9); kinase | -4.69 |
| 3 | 261572_at | AT1G01170 | ozone-responsive stress-related protein, putative | -2.83 |
| 4 | 261574_at | AT1G01190 | CYP78A8 (cytochrome P450, family 78, subfamily A, polypeptide 8); oxygen binding | 5.22 |
| 5 | 261053_at | AT1G01320 | tetratricopeptide repeat (TPR)-containing protein | -4.26 |
| 6 | 257490_x_at | AT1G01380 | ETC1 (ENHANCER OF TRY AND CPC 1); DNA binding / transcription factor | 2.22 |
| 7 | 259439_at | AT1G01480 | ACS2 (1-Amino-cyclopropane-1-carboxylate synthase 2) | 2.55 |
| 8 | 261586_at | AT1G01640 | speckle-type POZ protein-related | 3.91 |
| 9 | 261562_at | AT1G01750 | actin-depolymerizing factor, putative | 4.45 |
| 10 | 264180_at | AT1G02190 | CER1 protein, putative | -3.43 |
| 11 | 264146_at | AT1G02205 | CER1 (ECERIFERUM 1) | -6.32 |
| 12 | 264147_at | AT1G02205 | CER1 (ECERIFERUM 1) | -7.15 |
| 13 | 259443_at | AT1G02360 | chitinase, putative | 2.58 |
| 14 | 260916_at | AT1G02475 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G01883.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO40177.1); contains InterPro domain Streptomyces cyclase/dehydrase (InterPro:IPR005031) | -3.04 |
| 15 | 262121_at | AT1G02800 | ATCEL2 (Arabidopsis thaliana Cellulase 2); hydrolase, hydrolyzing O-glycosyl compounds | -3.29 |
| 16 | 262105_at | AT1G02810 | pectinesterase family protein | 3.80 |
| 17 | 262131_at | AT1G02900 | RALFL1 (RALF-LIKE 1) | 4.84 |
| 18 | 262120_at | AT1G02950 | ATGSTF4 (GLUTATHIONE S-TRANSFERASE 31); glutathione transferase | 2.32 |
| 19 | 263169_at | AT1G03010 | phototropic-responsive NPH3 family protein | 4.31 |
| 20 | 263114_at | AT1G03130 | PSAD-2 (photosystem I subunit D-2) | -6.09 |
| 21 | 264360_at | AT1G03310 | ATISA2/BE2/DBE1/ISA2 (DEBRANCHING ENZYME 1); alpha-amylase/ isoamylase | -2.56 |
| 22 | 264837_at | AT1G03600 | photosystem II family protein | -6.12 |
| 23 | 264839_at | AT1G03630 | POR C (PROTOCHLOROPHYLLIDE OXIDOREDUCTASE); NADPH dehydrogenase/ oxidoreductase/ protochlorophyllide reductase | -7.51 |
| 24 | 264845_at | AT1G03680 | ATHM1 (Arabidopsis thioredoxin M-type 1); thiol-disulfide exchange intermediate | -4.49 |
| 25 | 264842_at | AT1G03700 | integral membrane family protein | 3.72 |
| 26 | 265081_at | AT1G03840 | MGP (MAGPIE); nucleic acid binding / protein binding / transcription factor/ zinc ion binding | 4.70 |
| 27 | 265067_at | AT1G03850 | glutaredoxin family protein | 2.85 |
| 28 | 264323_at | AT1G04180 | flavin-containing monooxygenase family protein / FMO family protein | 4.71 |
| 29 | 263668_at | AT1G04350 | 2-oxoglutarate-dependent dioxygenase, putative | -3.66 |
| 30 | 263678_at | AT1G04420 | aldo/keto reductase family protein | -3.71 |
| 31 | 263658_at | AT1G04490 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G33360.2); similar to hypothetical protein [Vitis vinifera] (GB:CAN83005.1) | 3.45 |
| 32 | 261175_at | AT1G04800 | glycine-rich protein | -6.44 |
| 33 | 264584_at | AT1G05140 | membrane-associated zinc metalloprotease, putative | -2.81 |
| 34 | 264575_at | AT1G05190 | EMB2394 (EMBRYO DEFECTIVE 2394); structural constituent of ribosome | -3.97 |
| 35 | 264587_at | AT1G05200 | ATGLR3.4 (Arabidopsis thaliana glutamate receptor 3.4) | -2.87 |
| 36 | 264577_at | AT1G05260 | RCI3 (RARE COLD INDUCIBLE GENE 3); peroxidase | 5.58 |
| 37 | 263182_at | AT1G05575 | unknown protein | 3.51 |
| 38 | 260956_at | AT1G06040 | STO (SALT TOLERANCE); transcription factor/ zinc ion binding | -3.02 |
| 39 | 260783_at | AT1G06160 | ethylene-responsive factor, putative | 2.27 |
| 40 | 262626_at | AT1G06430 | FTSH8 (FtsH protease 8); ATP-dependent peptidase/ ATPase/ metallopeptidase/ zinc ion binding | -5.01 |
| 41 | 262629_at | AT1G06460 | ACD32.1 (ALPHA-CRYSTALLIN DOMAIN 31.2) | -4.87 |
| 42 | 262638_at | AT1G06650 | 2-oxoglutarate-dependent dioxygenase, putative | -2.87 |
| 43 | 262632_at | AT1G06680 | PSBP-1 (OXYGEN-EVOLVING ENHANCER PROTEIN 2); poly(U) binding | -7.36 |
| 44 | 262634_at | AT1G06690 | aldo/keto reductase family protein | -3.04 |
| 45 | 256049_at | AT1G07010 | calcineurin-like phosphoesterase family protein | -3.99 |
| 46 | 256060_at | AT1G07050 | CONSTANS-like protein-related | -3.61 |
| 47 | 256057_at | AT1G07180 | ATNDI1/NDA1 (ALTERNATIVE NAD(P)H DEHYDROGENASE 1); NADH dehydrogenase | -3.39 |
| 48 | 256053_at | AT1G07260 | UDP-glucoronosyl/UDP-glucosyl transferase family protein | 3.27 |
| 49 | 261075_at | AT1G07280 | binding | -3.38 |
| 50 | 261078_at | AT1G07320 | RPL4 (ribosomal protein L4); poly(U) binding / structural constituent of ribosome | -3.06 |
| 51 | 261084_at | AT1G07440 | tropinone reductase, putative / tropine dehydrogenase, putative | -4.12 |
| 52 | 261417_at | AT1G07700 | thioredoxin family protein | -3.70 |
| 53 | 261420_at | AT1G07720 | beta-ketoacyl-CoA synthase family protein | -3.11 |
| 54 | 260623_at | AT1G08090 | ATNRT2:1 (Arabidopsis thaliana high affinity nitrate transporter 2.1); nitrate transmembrane transporter | 2.44 |
| 55 | 261815_at | AT1G08320 | bZIP family transcription factor | 5.40 |
| 56 | 261746_at | AT1G08380 | PSAO (photosystem I subunit O) | -9.74 |
| 57 | 257481_at | AT1G08430 | ALMT1/ATALMT1 (AL-ACTIVATED MALATE TRANSPORTER 1); malate transmembrane transporter/ | 3.21 |
| 58 | 261745_at | AT1G08500 | plastocyanin-like domain-containing protein | 2.83 |
| 59 | 264781_at | AT1G08540 | SIGB (SIGMA FACTOR B); DNA binding / DNA-directed RNA polymerase/ transcription factor | -2.90 |
| 60 | 264799_at | AT1G08550 | NPQ1 (NON-PHOTOCHEMICAL QUENCHING 1) | -2.94 |
| 61 | 264794_at | AT1G08670 | epsin N-terminal homology (ENTH) domain-containing protein / clathrin assembly protein-related | 3.29 |
| 62 | 264643_at | AT1G08990 | PGSIP5 (PLANT GLYCOGENIN-LIKE STARCH INITIATION PROTEIN 5); transferase, transferring glycosyl groups | 4.74 |
| 63 | 264647_at | AT1G09090 | ATRBOHB (RESPIRATORY BURST OXIDASE HOMOLOG B); FAD binding / calcium ion binding / iron ion binding / oxidoreductase | 7.64 |
| 64 | 263709_at | AT1G09310 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G56580.1); similar to unknown [Populus trichocarpa] (GB:ABK94207.1); contains InterPro domain Protein of unknown function DUF538 (InterPro:IPR007493) | -8.70 |
| 65 | 263676_at | AT1G09340 | CRB; binding / catalytic/ coenzyme binding | -6.70 |
| 66 | 264510_at | AT1G09530 | PAP3/PIF3/POC1 (PHYTOCHROME INTERACTING FACTOR 3); DNA binding / protein binding / transcription factor/ transcription regulator | -2.76 |
| 67 | 264708_at | AT1G09740 | ethylene-responsive protein, putative | 3.52 |
| 68 | 264673_at | AT1G09795 | ATATP-PRT2 (ATP PHOSPHORIBOSYL TRANSFERASE 2); ATP phosphoribosyltransferase | -3.12 |
| 69 | 264661_at | AT1G09950 | transcription factor-related | 2.53 |
| 70 | 264525_at | AT1G10060 | ATBCAT-1; branched-chain-amino-acid transaminase/ catalytic | -3.02 |
| 71 | 264467_at | AT1G10140 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G58420.1); contains InterPro domain Uncharacterised conserved protein UCP031279 (InterPro:IPR016972) | 2.24 |
| 72 | 264435_at | AT1G10360 | ATGSTU18 (GLUTATHIONE S-TRANSFERASE 29); glutathione transferase | -3.31 |
| 73 | 264436_at | AT1G10370 | ATGSTU17/ERD9/GST30/GST30B (EARLY-RESPONSIVE TO DEHYDRATION 9); glutathione transferase | -3.94 |
| 74 | 264457_at | AT1G10400 | UDP-glycosyltransferase/ transferase, transferring glycosyl groups | 3.23 |
| 75 | 263211_at | AT1G10460 | GLP7 (GERMIN-LIKE PROTEIN 7); manganese ion binding / metal ion binding / nutrient reservoir | 2.49 |
| 76 | 263208_at | AT1G10480 | ZFP5 (ZINC FINGER PROTEIN 5); nucleic acid binding / transcription factor/ zinc ion binding | 3.01 |
| 77 | 260481_at | AT1G10960 | ATFD1 (FERREDOXIN 1); 2 iron, 2 sulfur cluster binding / electron carrier/ iron-sulfur cluster binding | -5.86 |
| 78 | 260475_at | AT1G11080 | SCPL31 (serine carboxypeptidase-like 31); serine carboxypeptidase | 3.28 |
| 79 | 262508_at | AT1G11300 | carbohydrate binding / kinase | -3.53 |
| 80 | 261873_at | AT1G11350 | S-locus lectin protein kinase family protein | -2.63 |
| 81 | 261826_at | AT1G11580 | ATPMEPCRA; pectinesterase | 3.63 |
| 82 | 262813_at | AT1G11670 | MATE efflux family protein | 2.68 |
| 83 | 264343_at | AT1G11850 | unknown protein | -7.47 |
| 84 | 264346_at | AT1G12010 | 1-aminocyclopropane-1-carboxylate oxidase, putative / ACC oxidase, putative | 5.91 |
| 85 | 264347_at | AT1G12040 | LRX1 (LEUCINE-RICH REPEAT/EXTENSIN 1); protein binding / structural constituent of cell wall | 2.64 |
| 86 | 264342_at | AT1G12080 | contains domain PTHR22683 (PTHR22683) | 2.65 |
| 87 | 260991_at | AT1G12160 | flavin-containing monooxygenase family protein / FMO family protein | 3.25 |
| 88 | 260968_at | AT1G12250 | thylakoid lumenal protein-related | -4.18 |
| 89 | 259528_at | AT1G12330 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G12900.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO16971.1) | -2.67 |
| 90 | 255937_at | AT1G12610 | DDF1 (DWARF AND DELAYED FLOWERING 1); DNA binding / transcription factor | 4.48 |
| 91 | 255934_at | AT1G12740 | CYP87A2 (cytochrome P450, family 87, subfamily A, polypeptide 2); oxygen binding | 4.29 |
| 92 | 261206_at | AT1G12800 | S1 RNA-binding domain-containing protein | -3.14 |
| 93 | 261196_at | AT1G12860 | basic helix-loop-helix (bHLH) family protein / F-box family protein | -2.52 |
| 94 | 261197_at | AT1G12900 | GAPA-2; glyceraldehyde-3-phosphate dehydrogenase | -10.20 |
| 95 | 259363_at | AT1G13270 | MAP1C (METHIONINE AMINOPEPTIDASE 1B); metalloexopeptidase | -2.79 |
| 96 | 259365_at | AT1G13300 | myb family transcription factor | 6.84 |
| 97 | 259388_at | AT1G13420 | sulfotransferase family protein | 7.18 |
| 98 | 256158_at | AT1G13590 | ATPSK1 (PHYTOSULFOKINE 1 PRECURSOR); growth factor | 2.92 |
| 99 | 256132_at | AT1G13610 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G30380.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO15341.1); contains InterPro domain Alpha/beta hydrolase fold-1 (InterPro:IPR000073) | 2.80 |
| 100 | 256096_at | AT1G13650 | similar to 18S pre-ribosomal assembly protein gar2-related [Arabidopsis thaliana] (TAIR:AT2G03810.4); similar to 18S pre-ribosomal assembly protein gar2-related [Arabidopsis thaliana] (TAIR:AT2G03810.2); similar to 18S pre-ribosomal assembly protein gar2-related [Arabidopsis thaliana] (TAIR:AT2G03810.3); similar to hypothetical protein Tc00.1047053503999.40 [Trypanosoma cruzi strain CL Brener] (GB:XP_813437.1) | -5.44 |
| 101 | 256099_at | AT1G13710 | CYP78A5 (cytochrome P450, family 78, subfamily A, polypeptide 5); oxygen binding | 2.59 |
| 102 | 259448_at | AT1G13790 | XH/XS domain-containing protein / XS zinc finger domain-containing protein | -2.99 |
| 103 | 262609_at | AT1G13930 | similar to nodulin-related [Arabidopsis thaliana] (TAIR:AT2G03440.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN63832.1) | -2.54 |
| 104 | 262648_at | AT1G14030 | ribulose-1,5 bisphosphate carboxylase oxygenase large subunit N-methyltransferase, putative | -3.63 |
| 105 | 262666_at | AT1G14080 | FUT6 (fucosyltransferase 6); fucosyltransferase/ transferase, transferring glycosyl groups | 5.85 |
| 106 | 262608_at | AT1G14120 | 2-oxoglutarate-dependent dioxygenase, putative | 5.12 |
| 107 | 262612_at | AT1G14150 | oxygen evolving enhancer 3 (PsbQ) family protein | -5.00 |
| 108 | 262656_at | AT1G14200 | zinc finger (C3HC4-type RING finger) family protein | 3.35 |
| 109 | 262658_at | AT1G14220 | ribonuclease T2 family protein | 4.63 |
| 110 | 262659_at | AT1G14240 | nucleoside phosphatase family protein / GDA1/CD39 family protein | 3.49 |
| 111 | 261483_at | AT1G14270 | CAAX amino terminal protease family protein | -3.50 |
| 112 | 261488_at | AT1G14345 | oxidoreductase | -4.51 |
| 113 | 261502_at | AT1G14440 | ATHB31 (ARABIDOPSIS THALIANA HOMEOBOX PROTEIN 31); transcription factor | -2.92 |
| 114 | 261476_at | AT1G14480 | protein binding | 3.99 |
| 115 | 262831_at | AT1G14730 | similar to ACYB-2 (Arabidopsis cytochrome b561 -2), carbon-monoxide oxygenase [Arabidopsis thaliana] (TAIR:AT4G25570.1); similar to ACYB-1 (Arabidopsis cytochrome b561 -1), carbon-monoxide oxygenase [Arabidopsis thaliana] (TAIR:AT5G38630.1); similar to cytochrome [Mesembryanthemum crystallinum] (GB:AAD11424.1); contains InterPro domain Cytochrome b561 / ferric reductase transmembrane (InterPro:IPR006593); contains InterPro domain Cytochrome b561; (InterPro:IPR004877) | 3.72 |
| 116 | 262838_at | AT1G14960 | major latex protein-related / MLP-related | 4.55 |
| 117 | 260744_at | AT1G15010 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G01300.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO42242.1) | 3.21 |
| 118 | 260741_at | AT1G15040 | glutamine amidotransferase-related | 5.79 |
| 119 | 262569_at | AT1G15180 | MATE efflux family protein | -3.44 |
| 120 | 262575_at | AT1G15210 | ATPDR7/PDR7 (PLEIOTROPIC DRUG RESISTANCE 7); ATPase, coupled to transmembrane movement of substances | 5.42 |
| 121 | 262598_at | AT1G15260 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G16070.1) | -3.98 |
| 122 | 262577_at | AT1G15290 | binding | -2.59 |
| 123 | 262603_at | AT1G15380 | lactoylglutathione lyase family protein / glyoxalase I family protein | 6.17 |
| 124 | 259491_at | AT1G15820 | LHCB6 (LIGHT HARVESTING COMPLEX PSII); chlorophyll binding | -8.48 |
| 125 | 259492_at | AT1G15830 | unknown protein | -2.87 |
| 126 | 261838_at | AT1G16030 | HSP70B (heat shock protein 70B); ATP binding | 2.33 |
| 127 | 261793_at | AT1G16080 | similar to unknown [Populus trichocarpa] (GB:ABK94042.1) | -2.56 |
| 128 | 262704_at | AT1G16530 | ASL9/LBD3 (LOB DOMAIN-CONTAINING PROTEIN 3) | 2.95 |
| 129 | 255764_at | AT1G16720 | HCF173 (HIGH CHLOROPHYLL FLUORESCENCE PHENOTYPE 173); binding / catalytic/ oxidoreductase/ transcription repressor | -3.01 |
| 130 | 255763_at | AT1G16730 | similar to unknown [Picea sitchensis] (GB:ABK21208.1) | -2.59 |
| 131 | 256115_at | AT1G16880 | uridylyltransferase-related | -2.60 |
| 132 | 262525_at | AT1G17060 | CYP72C1 (cytochrome P450, family 72, subfamily C, polypeptide 1); oxygen binding | 2.61 |
| 133 | 262536_at | AT1G17100 | SOUL heme-binding family protein | -5.80 |
| 134 | 262518_at | AT1G17170 | ATGSTU24 (ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE (CLASS TAU) 24); glutathione transferase | 2.56 |
| 135 | 262517_at | AT1G17180 | ATGSTU25 (Arabidopsis thaliana Glutathione S-transferase (class tau) 25); glutathione transferase | 2.95 |
| 136 | 262516_at | AT1G17190 | ATGSTU26 (Arabidopsis thaliana Glutathione S-transferase (class tau) 26); glutathione transferase | 2.83 |
| 137 | 262539_at | AT1G17200 | integral membrane family protein | -4.06 |
| 138 | 262483_at | AT1G17220 | FUG1 (FU-GAERI1); translation initiation factor | -3.82 |
| 139 | 260846_at | AT1G17300 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G17285.1) | 3.37 |
| 140 | 255889_at | AT1G17840 | ABCG11/COF1/DSO/WBC11 (DESPERADO); ATPase, coupled to transmembrane movement of substances | -3.52 |
| 141 | 255904_at | AT1G17860 | trypsin and protease inhibitor family protein / Kunitz family protein | 2.57 |
| 142 | 255903_at | AT1G17950 | MYB52 (myb domain protein 52); DNA binding / transcription factor | 2.71 |
| 143 | 256076_at | AT1G18060 | similar to unnamed protein product [Vitis vinifera] (GB:CAO68142.1) | -3.24 |
| 144 | 256128_at | AT1G18140 | LAC1 (Laccase 1); copper ion binding / oxidoreductase | 2.41 |
| 145 | 256130_at | AT1G18170 | immunophilin / FKBP-type peptidyl-prolyl cis-trans isomerase family protein | -2.58 |
| 146 | 261422_at | AT1G18730 | similar to unknown [Populus trichocarpa] (GB:ABK95263.1) | -9.42 |
| 147 | 261407_at | AT1G18810 | phytochrome kinase substrate-related | -4.57 |
| 148 | 261425_at | AT1G18880 | proton-dependent oligopeptide transport (POT) family protein | 3.60 |
| 149 | 259481_at | AT1G18970 | GLP4 (GERMIN-LIKE PROTEIN 4); manganese ion binding / metal ion binding / nutrient reservoir | 3.08 |
| 150 | 259478_at | AT1G18980 | germin-like protein, putative | 5.52 |
| 151 | 256015_at | AT1G19150 | LHCA6 (Photosystem I light harvesting complex gene 6); chlorophyll binding | -7.65 |
| 152 | 256039_at | AT1G19190 | hydrolase | 2.23 |
| 153 | 256009_at | AT1G19210 | AP2 domain-containing transcription factor, putative | 3.29 |
| 154 | 256012_at | AT1G19250 | FMO1 (FLAVIN-DEPENDENT MONOOXYGENASE 1); monooxygenase | 4.81 |
| 155 | 260668_at | AT1G19530 | similar to unnamed protein product [Vitis vinifera] (GB:CAO61141.1) | 8.17 |
| 156 | 261150_at | AT1G19640 | JMT (JASMONIC ACID CARBOXYL METHYLTRANSFERASE); jasmonate O-methyltransferase | -2.49 |
| 157 | 255786_at | AT1G19670 | ATCLH1 (CORONATINE-INDUCED PROTEIN 1) | -6.34 |
| 158 | 261141_at | AT1G19740 | ATP-dependent protease La (LON) domain-containing protein | -2.95 |
| 159 | 255814_at | AT1G19900 | glyoxal oxidase-related | 2.62 |
| 160 | 261218_at | AT1G20020 | ATLFNR2 (LEAF FNR 2); NADPH dehydrogenase/ oxidoreductase/ poly(U) binding | -7.97 |
| 161 | 261224_at | AT1G20160 | ATSBT5.2; subtilase | 3.14 |
| 162 | 255886_at | AT1G20340 | DRT112 (DNA-damage-repair/toleration protein 112); copper ion binding / electron carrier | -8.57 |
| 163 | 255941_at | AT1G20350 | ATTIM17-1 (Arabidopsis thaliana translocase inner membrane subunit 17-1); P-P-bond-hydrolysis-driven protein transmembrane transporter | -2.68 |
| 164 | 256081_at | AT1G20700 | WOX14 (WUSCHEL RELATED HOMEOBOX 14); DNA binding / transcription factor | 2.55 |
| 165 | 262805_at | AT1G20900 | ESC (ESCAROLA) | 2.49 |
| 166 | 261451_at | AT1G21060 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G76620.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO66316.1); contains InterPro domain Protein of unknown function DUF547 (InterPro:IPR006869) | -4.49 |
| 167 | 261459_at | AT1G21100 | O-methyltransferase, putative | 7.08 |
| 168 | 261453_at | AT1G21130 | O-methyltransferase, putative | -3.13 |
| 169 | 259561_at | AT1G21250 | WAK1 (CELL WALL-ASSOCIATED KINASE); kinase | -3.66 |
| 170 | 259553_x_at | AT1G21310 | ATEXT3 (EXTENSIN 3); structural constituent of cell wall | 3.54 |
| 171 | 260926_at | AT1G21360 | GLTP2 (GLYCOLIPID TRANSFER PROTEIN 2); glycolipid binding / glycolipid transporter | 4.29 |
| 172 | 260876_at | AT1G21460 | nodulin MtN3 family protein | -2.50 |
| 173 | 260877_at | AT1G21500 | similar to hypothetical protein OsI_030994 [Oryza sativa (indica cultivar-group)] (GB:EAZ09762.1); similar to Os09g0517000 [Oryza sativa (japonica cultivar-group)] (GB:NP_001063677.1); similar to unknown protein [Oryza sativa (japonica cultivar-group)] (GB:BAD33829.1) | -6.93 |
| 174 | 260921_at | AT1G21540 | AMP-binding protein, putative | -3.07 |
| 175 | 262503_at | AT1G21670 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G21680.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN73514.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO61906.1); contains InterPro domain WD40-like Beta Propeller (InterPro:IPR011659); contains InterPro domain Six-bladed beta-propeller, TolB-like (InterPro:IPR011042) | -2.62 |
| 176 | 260856_at | AT1G21910 | AP2 domain-containing transcription factor family protein | 2.88 |
| 177 | 255943_at | AT1G22370 | ATUGT85A5 (UDP-GLUCOSYL TRANSFERASE 85A5); transferase, transferring glycosyl groups | -3.14 |
| 178 | 261931_at | AT1G22430 | alcohol dehydrogenase, putative | -3.97 |
| 179 | 261930_at | AT1G22440 | alcohol dehydrogenase, putative | 5.28 |
| 180 | 261927_at | AT1G22500 | zinc finger (C3HC4-type RING finger) family protein | 5.82 |
| 181 | 261942_at | AT1G22590 | AGL87; transcription factor | -5.32 |
| 182 | 264201_at | AT1G22630 | heat shock protein binding / unfolded protein binding | -6.17 |
| 183 | 264195_at | AT1G22690 | gibberellin-responsive protein, putative | -2.96 |
| 184 | 264199_at | AT1G22700 | tetratricopeptide repeat (TPR)-containing protein | -2.50 |
| 185 | 264207_at | AT1G22750 | similar to unnamed protein product [Vitis vinifera] (GB:CAO67528.1); contains InterPro domain Protein of unknown function DUF1475 (InterPro:IPR009943) | -4.40 |
| 186 | 264773_at | AT1G22900 | similar to disease resistance-responsive family protein [Arabidopsis thaliana] (TAIR:AT5G42500.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO23438.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN61168.1); contains InterPro domain Plant disease resistance response protein (InterPro:IPR004265) | -3.26 |
| 187 | 264729_at | AT1G22990 | heavy-metal-associated domain-containing protein / copper chaperone (CCH)-related | 3.58 |
| 188 | 264901_at | AT1G23090 | AST91 (SULFATE TRANSPORTER 91); sulfate transmembrane transporter | -3.02 |
| 189 | 264887_at | AT1G23120 | major latex protein-related / MLP-related | 3.42 |
| 190 | 264899_at | AT1G23130 | Bet v I allergen family protein | -5.53 |
| 191 | 264892_at | AT1G23160 | auxin-responsive GH3 family protein | 3.72 |
| 192 | 262988_at | AT1G23310 | GGT1 (ALANINE-2-OXOGLUTARATE AMINOTRANSFERASE 1) | -3.72 |
| 193 | 263016_at | AT1G23410 | ubiquitin extension protein, putative / 40S ribosomal protein S27A (RPS27aA) | 2.85 |
| 194 | 265169_x_at | AT1G23720 | proline-rich extensin-like family protein | 3.20 |
| 195 | 265182_at | AT1G23740 | oxidoreductase, zinc-binding dehydrogenase family protein | -7.67 |
| 196 | 265131_at | AT1G23760 | JP630; polygalacturonase | 3.21 |
| 197 | 265188_at | AT1G23800 | ALDH2B7 (Aldehyde dehydrogenase 2B7); 3-chloroallyl aldehyde dehydrogenase/ aldehyde dehydrogenase (NAD) | 2.35 |
| 198 | 263032_at | AT1G23850 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G23840.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN74577.1) | 2.88 |
| 199 | 263034_at | AT1G24020 | MLP423 (MLP-LIKE PROTEIN 423) | -6.83 |
| 200 | 264861_at | AT1G24320 | alpha-glucosidase, putative | 3.68 |
| 201 | 265002_at | AT1G24400 | LHT2 (LYSINE HISTIDINE TRANSPORTER 2); amino acid transmembrane transporter | -2.92 |
| 202 | 265016_at | AT1G24420 | transferase family protein | 2.39 |
| 203 | 257405_at | AT1G24620 | polcalcin, putative / calcium-binding pollen allergen, putative | 3.57 |
| 204 | 245877_at | AT1G26220 | GCN5-related N-acetyltransferase (GNAT) family protein | -6.20 |
| 205 | 245876_at | AT1G26230 | chaperonin, putative | -3.12 |
| 206 | 261016_at | AT1G26560 | glycosyl hydrolase family 1 protein | -2.77 |
| 207 | 261012_at | AT1G26600 | CLE9 (CLAVATA3/ESR-RELATED 9); receptor binding | -4.23 |
| 208 | 263682_at | AT1G26860 | transposable element gene | 2.74 |
| 209 | 264988_at | AT1G27140 | ATGSTU14 (GLUTATHIONE S-TRANSFERASE 13); glutathione transferase | 4.37 |
| 210 | 264495_at | AT1G27380 | RIC2 (ROP-INTERACTIVE CRIB MOTIF-CONTAINING PROTEIN 2) | 2.35 |
| 211 | 264442_at | AT1G27480 | lecithin:cholesterol acyltransferase family protein / LACT family protein | -7.21 |
| 212 | 261648_at | AT1G27730 | STZ (SALT TOLERANCE ZINC FINGER); nucleic acid binding / transcription factor/ zinc ion binding | 2.38 |
| 213 | 259592_at | AT1G27950 | lipid transfer protein-related | -2.78 |
| 214 | 259596_at | AT1G28130 | GH3.17; indole-3-acetic acid amido synthetase | 3.07 |
| 215 | 261470_at | AT1G28370 | ATERF11/ERF11 (ERF domain protein 11); DNA binding / transcription factor/ transcription repressor | 2.40 |
| 216 | 261443_at | AT1G28480 | GRX480; thiol-disulfide exchange intermediate | 2.28 |
| 217 | 260898_at | AT1G29070 | ribosomal protein L34 family protein | -2.98 |
| 218 | 260883_at | AT1G29270 | similar to transcription regulator [Arabidopsis thaliana] (TAIR:AT2G40435.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN79056.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO48025.1) | 3.20 |
| 219 | 260882_at | AT1G29280 | WRKY65 (WRKY DNA-binding protein 65); transcription factor | 3.90 |
| 220 | 259789_at | AT1G29395 | COR414-TM1 (cold regulated 414 thylakoid membrane 1) | -3.32 |
| 221 | 257506_at | AT1G29440 | auxin-responsive family protein | -3.38 |
| 222 | 259784_at | AT1G29450 | auxin-responsive protein, putative | -4.88 |
| 223 | 259773_at | AT1G29500 | auxin-responsive protein, putative | -6.68 |
| 224 | 259783_at | AT1G29510 | SAUR68 (SMALL AUXIN UPREGULATED 68) | -4.69 |
| 225 | 259786_at | AT1G29660 | GDSL-motif lipase/hydrolase family protein | -3.60 |
| 226 | 259788_at | AT1G29670 | GDSL-motif lipase/hydrolase family protein | -8.06 |
| 227 | 259791_at | AT1G29700 | similar to unnamed protein product [Vitis vinifera] (GB:CAO42101.1); contains domain SSF56281 (SSF56281) | -2.68 |
| 228 | 246633_at | AT1G29720 | protein kinase family protein | -5.49 |
| 229 | 260025_at | AT1G30070 | SGS domain-containing protein | 2.47 |
| 230 | 256190_at | AT1G30100 | NCED5 (NINE-CIS-EPOXYCAROTENOID DIOXYGENASE 5) | -2.82 |
| 231 | 256306_at | AT1G30370 | lipase class 3 family protein | 4.26 |
| 232 | 256309_at | AT1G30380 | PSAK (PHOTOSYSTEM I SUBUNIT K) | -8.35 |
| 233 | 261806_at | AT1G30510 | ATRFNR2 (ROOT FNR 2); oxidoreductase | 2.77 |
| 234 | 261801_at | AT1G30520 | AAE14 (ACYL-ACTIVATING ENZYME 14); AMP binding | -3.79 |
| 235 | 261804_at | AT1G30530 | UDP-glucoronosyl/UDP-glucosyl transferase family protein | -3.52 |
| 236 | 263228_at | AT1G30700 | FAD-binding domain-containing protein | 4.72 |
| 237 | 263227_at | AT1G30750 | similar to Hypothetical protein CBG24759 [Caenorhabditis briggsae] (GB:CAE56916.1); contains InterPro domain Protein of unknown function DUF1720 (InterPro:IPR013182) | 3.80 |
| 238 | 264527_at | AT1G30760 | FAD-binding domain-containing protein | 5.36 |
| 239 | 264497_at | AT1G30840 | ATPUP4 (Arabidopsis thaliana purine permease 4); purine transmembrane transporter | 2.89 |
| 240 | 265102_at | AT1G30870 | cationic peroxidase, putative | 4.48 |
| 241 | 265155_at | AT1G30990 | major latex protein-related / MLP-related | 3.44 |
| 242 | 265160_at | AT1G31050 | transcription factor | 4.97 |
| 243 | 265159_at | AT1G31060 | unknown protein | 5.14 |
| 244 | 262557_at | AT1G31330 | PSAF (photosystem I subunit F) | -7.41 |
| 245 | 256489_at | AT1G31550 | GDSL-motif lipase, putative | -3.25 |
| 246 | 256497_at | AT1G31580 | ECS1 | -8.52 |
| 247 | 246601_at | AT1G31710 | copper amine oxidase, putative | 2.38 |
| 248 | 246268_at | AT1G31800 | CYP97A3/LUT5 (CYTOCHROME P450-TYPE MONOOXYGENASE 97A3); carotene beta-ring hydroxylase/ oxygen binding | -3.00 |
| 249 | 246313_at | AT1G31920 | pentatricopeptide (PPR) repeat-containing protein | -7.62 |
| 250 | 255751_at | AT1G31950 | terpene synthase/cyclase family protein | 4.61 |
| 251 | 255720_at | AT1G32060 | PRK (PHOSPHORIBULOKINASE); ATP binding / phosphoribulokinase/ protein binding | -7.49 |
| 252 | 255719_at | AT1G32080 | membrane protein, putative | -5.97 |
| 253 | 245795_at | AT1G32160 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G17800.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G17800.2); similar to unnamed protein product [Vitis vinifera] (GB:CAO63331.1); contains InterPro domain Protein of unknown function DUF760 (InterPro:IPR008479) | -2.55 |
| 254 | 245793_at | AT1G32220 | binding / catalytic/ coenzyme binding | -3.42 |
| 255 | 260693_at | AT1G32450 | proton-dependent oligopeptide transport (POT) family protein | 5.42 |
| 256 | 260704_at | AT1G32470 | glycine cleavage system H protein, mitochondrial, putative | -6.80 |
| 257 | 256469_at | AT1G32540 | LOL1 (LSD ONE LIKE 1) | -4.13 |
| 258 | 256468_at | AT1G32550 | ferredoxin family protein | -3.28 |
| 259 | 261191_at | AT1G32900 | starch synthase, putative | -4.71 |
| 260 | 261215_at | AT1G32970 | subtilase family protein | 4.47 |
| 261 | 261190_at | AT1G32990 | PRPL11 (PLASTID RIBOSOMAL PROTEIN L11); structural constituent of ribosome | -3.28 |
| 262 | 261567_at | AT1G33055 | unknown protein | 2.58 |
| 263 | 245765_at | AT1G33600 | leucine-rich repeat family protein | -4.19 |
| 264 | 261991_at | AT1G33700 | catalytic | 3.01 |
| 265 | 261985_at | AT1G33750 | terpene synthase/cyclase family protein | 6.47 |
| 266 | 261984_at | AT1G33760 | AP2 domain-containing transcription factor, putative | 3.80 |
| 267 | 261999_at | AT1G33800 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G09990.1); similar to unknown [Populus trichocarpa] (GB:ABK93991.1); contains InterPro domain Protein of unknown function DUF579, plant (InterPro:IPR006514) | 3.02 |
| 268 | 261981_at | AT1G33811 | GDSL-motif lipase/hydrolase family protein | -3.66 |
| 269 | 255982_at | AT1G34000 | OHP2 (ONE-HELIX PROTEIN 2) | -3.35 |
| 270 | 262566_at | AT1G34310 | ARF12 (AUXIN RESPONSE FACTOR 12); transcription factor | -5.34 |
| 271 | 262406_at | AT1G34670 | AtMYB93 (myb domain protein 93); DNA binding / transcription factor | 2.47 |
| 272 | 245755_at | AT1G35210 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G22470.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN82663.1) | 3.64 |
| 273 | 259548_at | AT1G35260 | MLP165 (MLP-LIKE PROTEIN 165) | 2.41 |
| 274 | 260106_at | AT1G35420 | dienelactone hydrolase family protein | -3.99 |
| 275 | 262029_at | AT1G35680 | 50S ribosomal protein L21, chloroplast / CL21 (RPL21) | -3.24 |
| 276 | 263194_at | AT1G36060 | AP2 domain-containing transcription factor, putative | 3.13 |
| 277 | 256499_at | AT1G36640 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G36622.1) | 3.39 |
| 278 | 256542_at | AT1G42550 | PMI1 (PLASTID MOVEMENT IMPAIRED1) | -5.21 |
| 279 | 259625_at | AT1G42970 | GAPB (GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE B SUBUNIT); glyceraldehyde-3-phosphate dehydrogenase | -8.18 |
| 280 | 255754_at | AT1G43040 | auxin-responsive protein, putative | 4.97 |
| 281 | 262721_at | AT1G43560 | ATY2 (Arabidopsis thioredoxin y2); thiol-disulfide exchange intermediate | -2.64 |
| 282 | 259460_at | AT1G44000 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G11911.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN81476.1) | -3.25 |
| 283 | 259454_at | AT1G44050 | DC1 domain-containing protein | 4.38 |
| 284 | 245737_at | AT1G44160 | DNAJ chaperone C-terminal domain-containing protein | 2.60 |
| 285 | 245242_at | AT1G44446 | CH1 (CHLOROPHYLL B BIOSYNTHESIS); chlorophyllide a oxygenase | -4.24 |
| 286 | 245213_at | AT1G44575 | NPQ4 (NONPHOTOCHEMICAL QUENCHING) | -6.61 |
| 287 | 261335_at | AT1G44800 | nodulin MtN21 family protein | 2.73 |
| 288 | 261327_at | AT1G44830 | AP2 domain-containing transcription factor TINY, putative | 3.19 |
| 289 | 261338_at | AT1G44920 | similar to unnamed protein product [Vitis vinifera] (GB:CAO61509.1) | -2.65 |
| 290 | 260941_at | AT1G44970 | peroxidase, putative | 2.33 |
| 291 | 245806_at | AT1G45474 | LHCA5 (Photosystem I light harvesting complex gene 5) | -5.87 |
| 292 | 262444_at | AT1G47480 | hydrolase | 3.71 |
| 293 | 262432_at | AT1G47530 | ripening-responsive protein, putative | -2.80 |
| 294 | 262235_at | AT1G48350 | ribosomal protein L18 family protein | -2.78 |
| 295 | 261298_at | AT1G48510 | cytochrome c oxidase assembly protein surfeit-related | 4.29 |
| 296 | 256137_at | AT1G48690 | auxin-responsive GH3 family protein | 4.94 |
| 297 | 260758_at | AT1G48930 | ATGH9C1 (ARABIDOPSIS THALIANA GLYCOSYL HYDROLASE 9C1); hydrolase, hydrolyzing O-glycosyl compounds | 2.44 |
| 298 | 260769_at | AT1G49010 | myb family transcription factor | -2.65 |
| 299 | 262446_at | AT1G49310 | unknown protein | 3.47 |
| 300 | 261606_at | AT1G49570 | peroxidase, putative | 3.81 |
| 301 | 261598_at | AT1G49750 | leucine-rich repeat family protein | -3.10 |
| 302 | 259813_at | AT1G49860 | ATGSTF14 (Arabidopsis thaliana Glutathione S-transferase (class phi) 14); glutathione transferase | 6.47 |
| 303 | 261638_at | AT1G49975 | similar to hypothetical protein [Vitis vinifera] (GB:CAN66219.1) | -4.58 |
| 304 | 261635_at | AT1G50020 | similar to unnamed protein product [Vitis vinifera] (GB:CAO49863.1) | -2.49 |
| 305 | 261691_at | AT1G50060 | pathogenesis-related protein, putative | 5.29 |
| 306 | 262473_at | AT1G50250 | FTSH1 (FtsH protease 1); ATP-dependent peptidase/ ATPase/ metallopeptidase | -5.91 |
| 307 | 261861_at | AT1G50450 | binding / catalytic | -3.55 |
| 308 | 261879_at | AT1G50520 | CYP705A27 (cytochrome P450, family 705, subfamily A, polypeptide 27); oxygen binding | 2.87 |
| 309 | 261878_at | AT1G50560 | CYP705A25 (cytochrome P450, family 705, subfamily A, polypeptide 25); oxygen binding | 3.72 |
| 310 | 261877_at | AT1G50580 | glycosyltransferase family protein | 5.84 |
| 311 | 256215_at | AT1G50900 | similar to hypothetical protein [Vitis vinifera] (GB:CAN65357.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO38995.1); contains InterPro domain Ankyrin (InterPro:IPR002110) | -3.14 |
| 312 | 245743_at | AT1G51080 | similar to unnamed protein product [Vitis vinifera] (GB:CAO41687.1) | -2.75 |
| 313 | 245747_at | AT1G51100 | similar to unnamed protein product [Vitis vinifera] (GB:CAO39038.1) | -2.89 |
| 314 | 245744_at | AT1G51110 | plastid-lipid associated protein PAP / fibrillin family protein | -7.49 |
| 315 | 245745_at | AT1G51110 | plastid-lipid associated protein PAP / fibrillin family protein | -6.81 |
| 316 | 265147_at | AT1G51380 | eukaryotic translation initiation factor 4A, putative / eIF-4A, putative | 2.27 |
| 317 | 265149_at | AT1G51400 | photosystem II 5 kD protein | -5.20 |
| 318 | 260490_at | AT1G51500 | CER5 (ECERIFERUM 5); ATPase, coupled to transmembrane movement of substances | -3.54 |
| 319 | 256168_at | AT1G51805 | leucine-rich repeat protein kinase, putative | -5.95 |
| 320 | 246375_at | AT1G51830 | ATP binding / kinase/ protein serine/threonine kinase | 3.60 |
| 321 | 246366_at | AT1G51850 | leucine-rich repeat protein kinase, putative | 6.19 |
| 322 | 246373_at | AT1G51860 | leucine-rich repeat protein kinase, putative | 4.20 |
| 323 | 265053_at | AT1G52000 | jacalin lectin family protein | -6.17 |
| 324 | 265048_at | AT1G52050 | jacalin lectin family protein | 4.28 |
| 325 | 265049_at | AT1G52060 | similar to jacalin lectin family protein [Arabidopsis thaliana] (TAIR:AT1G52070.1); similar to jasmonate inducible protein [Brassica napus] (GB:CAA72271.1); contains InterPro domain Mannose-binding lectin (InterPro:IPR001229) | 5.13 |
| 326 | 265050_at | AT1G52070 | jacalin lectin family protein | 6.64 |
| 327 | 259839_at | AT1G52190 | proton-dependent oligopeptide transport (POT) family protein | -2.90 |
| 328 | 259841_at | AT1G52200 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G18470.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO45876.1); contains InterPro domain Protein of unknown function Cys-rich (InterPro:IPR006461) | 2.74 |
| 329 | 259838_at | AT1G52220 | similar to PSAP/PSI-P/PTAC8/TMP14 (THYLAKOID MEMBRANE PHOSPHOPROTEIN OF 14 KDA) [Arabidopsis thaliana] (TAIR:AT2G46820.2); similar to PSAP/PSI-P/PTAC8/TMP14 (THYLAKOID MEMBRANE PHOSPHOPROTEIN OF 14 KDA), DNA binding [Arabidopsis thaliana] (TAIR:AT2G46820.1); similar to unknown [Populus trichocarpa] (GB:ABK95860.1) | -6.24 |
| 330 | 259840_at | AT1G52230 | PSAH-2/PSAH2/PSI-H (PHOTOSYSTEM I SUBUNIT H-2) | -8.95 |
| 331 | 259640_at | AT1G52400 | BGL1 (BETA-GLUCOSIDASE HOMOLOG 1); hydrolase, hydrolyzing O-glycosyl compounds | -3.56 |
| 332 | 262151_at | AT1G52510 | hydrolase, alpha/beta fold family protein | -6.56 |
| 333 | 262148_at | AT1G52560 | 26.5 kDa class I small heat shock protein-like (HSP26.5-P) | 5.45 |
| 334 | 262160_at | AT1G52590 | Identical to DCC family protein At1g52590, chloroplast precursor [Arabidopsis Thaliana] (GB:Q9SSR1;GB:Q8L9T1); similar to RNase H domain-containing protein [Arabidopsis thaliana] (TAIR:AT1G24090.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO62758.1); contains InterPro domain Putative thiol-disulphide oxidoreductase DCC (InterPro:IPR007263) | -2.48 |
| 335 | 260147_at | AT1G52790 | oxidoreductase, 2OG-Fe(II) oxygenase family protein | 5.10 |
| 336 | 260148_at | AT1G52800 | oxidoreductase, 2OG-Fe(II) oxygenase family protein | 2.94 |
| 337 | 260150_at | AT1G52820 | 2-oxoglutarate-dependent dioxygenase, putative | 5.52 |
| 338 | 260155_at | AT1G52870 | peroxisomal membrane protein-related | -4.08 |
| 339 | 261319_at | AT1G53090 | SPA4 (SPA1-RELATED 4); signal transducer | -2.54 |
| 340 | 260639_at | AT1G53180 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G15115.1) | 3.20 |
| 341 | 260978_at | AT1G53540 | 17.6 kDa class I small heat shock protein (HSP17.6C-CI) (AA 1-156) | 4.73 |
| 342 | 259965_at | AT1G53670 | MSRB1 (METHIONINE SULFOXIDE REDUCTASE B 1); peptide-methionine-(S)-S-oxide reductase | -3.15 |
| 343 | 259964_at | AT1G53680 | ATGSTU28 (Arabidopsis thaliana Glutathione S-transferase (class tau) 28); glutathione transferase | 3.37 |
| 344 | 262198_at | AT1G53830 | ATPME2 (Arabidopsis thaliana pectin methylesterase 2) | 4.29 |
| 345 | 263198_at | AT1G53990 | GLIP3 (GDSL-motif lipase 3); carboxylesterase/ lipase | 4.62 |
| 346 | 263161_at | AT1G54020 | myrosinase-associated protein, putative | -2.75 |
| 347 | 263174_at | AT1G54040 | ESP (EPITHIOSPECIFIER PROTEIN) | -7.03 |
| 348 | 263150_at | AT1G54050 | 17.4 kDa class III heat shock protein (HSP17.4-CIII) | 4.27 |
| 349 | 262954_at | AT1G54500 | rubredoxin family protein | -4.16 |
| 350 | 263005_at | AT1G54540 | similar to harpin-induced protein-related / HIN1-related / harpin-responsive protein-related [Arabidopsis thaliana] (TAIR:AT1G65690.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO62044.1); contains InterPro domain Harpin-induced 1 (InterPro:IPR010847) | 4.07 |
| 351 | 264185_at | AT1G54780 | thylakoid lumen 18.3 kDa protein | -7.68 |
| 352 | 264240_at | AT1G54820 | protein kinase family protein | -3.47 |
| 353 | 256349_at | AT1G54890 | late embryogenesis abundant protein-related / LEA protein-related | 3.50 |
| 354 | 259660_at | AT1G55260 | lipid binding | -3.94 |
| 355 | 259658_at | AT1G55370 | carbohydrate binding / catalytic | -3.22 |
| 356 | 265124_at | AT1G55430 | DC1 domain-containing protein | 4.25 |
| 357 | 265073_at | AT1G55480 | binding / protein binding | -4.42 |
| 358 | 265076_at | AT1G55490 | CPN60B (CHAPERONIN 60 BETA); ATP binding / protein binding / unfolded protein binding | -2.62 |
| 359 | 264545_at | AT1G55670 | PSAG | -10.52 |
| 360 | 256221_at | AT1G56300 | DNAJ heat shock N-terminal domain-containing protein | -2.62 |
| 361 | 256217_at | AT1G56320 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G49350.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO44880.1) | 2.23 |
| 362 | 259603_at | AT1G56500 | haloacid dehalogenase-like hydrolase family protein | -3.35 |
| 363 | 259633_at | AT1G56500 | haloacid dehalogenase-like hydrolase family protein | -2.89 |
| 364 | 245674_at | AT1G56680 | glycoside hydrolase family 19 protein | 3.96 |
| 365 | 246403_at | AT1G57590 | carboxylesterase | 2.26 |
| 366 | 246411_at | AT1G57770 | amine oxidase family | -4.42 |
| 367 | 256021_at | AT1G58270 | ZW9 | 3.29 |
| 368 | 256024_at | AT1G58340 | ZF14; transporter | 2.34 |
| 369 | 245220_at | AT1G59171 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G58936.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G58643.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G59312.1); contains InterPro domain Protein of unknown function DUF941 (InterPro:IPR009286) | 4.71 |
| 370 | 262072_at | AT1G59590 | ZCF37 | 2.72 |
| 371 | 262916_at | AT1G59700 | ATGSTU16 (Arabidopsis thaliana Glutathione S-transferase (class tau) 16); glutathione transferase | -3.73 |
| 372 | 262905_at | AT1G59730 | ATH7 (thioredoxin H-type 7); thiol-disulfide exchange intermediate | 3.37 |
| 373 | 262915_at | AT1G59940 | ARR3 (RESPONSE REGULATOR 3); transcription regulator/ two-component response regulator | 2.77 |
| 374 | 262913_at | AT1G59960 | aldo/keto reductase, putative | 2.25 |
| 375 | 263736_at | AT1G60000 | 29 kDa ribonucleoprotein, chloroplast, putative / RNA-binding protein cp29, putative | -5.13 |
| 376 | 264217_at | AT1G60190 | armadillo/beta-catenin repeat family protein / U-box domain-containing protein | 2.31 |
| 377 | 264920_at | AT1G60550 | naphthoate synthase, putative / dihydroxynaphthoic acid synthetase, putative / DHNA synthetase, putative | -4.14 |
| 378 | 264963_at | AT1G60600 | ABC4 (ABERRANT CHLOROPLAST DEVELOPMENT 4); 1,4-dihydroxy-2-naphthoate octaprenyltransferase/ prenyltransferase | -2.66 |
| 379 | 264941_at | AT1G60680 | AGD2 (ARF-GAP DOMAIN 2); aldo-keto reductase | 6.32 |
| 380 | 259727_at | AT1G60950 | FED A (FERREDOXIN 2); 2 iron, 2 sulfur cluster binding / electron carrier/ iron-sulfur cluster binding | -4.86 |
| 381 | 264886_at | AT1G61120 | terpene synthase/cyclase family protein | -2.89 |
| 382 | 264937_at | AT1G61130 | SCPL32; serine carboxypeptidase | 2.63 |
| 383 | 264758_at | AT1G61340 | F-box family protein | 3.69 |
| 384 | 265033_at | AT1G61520 | LHCA3 (Photosystem I light harvesting complex gene 3); chlorophyll binding | -9.24 |
| 385 | 265031_at | AT1G61590 | protein kinase, putative | 6.19 |
| 386 | 264400_at | AT1G61800 | GPT2 (glucose-6-phosphate/phosphate translocator 2); antiporter/ glucose-6-phosphate transmembrane transporter | -3.05 |
| 387 | 264280_at | AT1G61820 | BGLU46; hydrolase, hydrolyzing O-glycosyl compounds | 4.02 |
| 388 | 264310_at | AT1G62030 | DC1 domain-containing protein | -3.91 |
| 389 | 264734_at | AT1G62280 | SLAH1 (SLAC1 HOMOLOGUE 1); transporter | 3.59 |
| 390 | 265114_at | AT1G62440 | LRX2 (LEUCINE-RICH REPEAT/EXTENSIN 2); protein binding / structural constituent of cell wall | 2.23 |
| 391 | 262645_at | AT1G62750 | ATSCO1/ATSCO1/CPEF-G/SCO1 (SNOWY COTYLEDON1); translation elongation factor/ translation factor, nucleic acid binding | -4.85 |
| 392 | 262643_at | AT1G62770 | invertase/pectin methylesterase inhibitor family protein | 2.88 |
| 393 | 262693_at | AT1G62780 | similar to hypothetical protein [Vitis vinifera] (GB:CAN83165.1) | -4.45 |
| 394 | 261101_at | AT1G63030 | DDF2 (DWARF AND DELAYED FLOWERING 2); DNA binding / transcription factor | 3.88 |
| 395 | 260083_at | AT1G63220 | C2 domain-containing protein | 2.38 |
| 396 | 260086_at | AT1G63240 | unknown protein | -4.72 |
| 397 | 261551_at | AT1G63440 | HMA5 (HEAVY METAL ATPASE 5); ATPase, coupled to transmembrane movement of ions, phosphorylative mechanism | 2.34 |
| 398 | 261550_at | AT1G63450 | catalytic | 3.26 |
| 399 | 260241_at | AT1G63710 | CYP86A7 (cytochrome P450, family 86, subfamily A, polypeptide 7); oxygen binding | -3.33 |
| 400 | 260327_at | AT1G63840 | zinc finger (C3HC4-type RING finger) family protein | 2.30 |
| 401 | 260312_at | AT1G63880 | disease resistance protein (TIR-NBS-LRR class), putative | -3.28 |
| 402 | 262342_at | AT1G64150 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G13590.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO65879.1); contains InterPro domain Protein of unknown function UPF0016; (InterPro:IPR001727) | -2.89 |
| 403 | 262325_at | AT1G64160 | disease resistance-responsive family protein / dirigent family protein | 2.27 |
| 404 | 262336_at | AT1G64220 | TOM7-2 (TRANSLOCASE OF OUTER MEMBRANE 7 KDA SUBUNIT 2); P-P-bond-hydrolysis-driven protein transmembrane transporter | 2.47 |
| 405 | 259738_at | AT1G64355 | similar to unnamed protein product [Vitis vinifera] (GB:CAO65822.1) | -2.80 |
| 406 | 259766_at | AT1G64360 | unknown protein | -4.63 |
| 407 | 261958_at | AT1G64500 | glutaredoxin family protein | -5.92 |
| 408 | 261954_at | AT1G64510 | ribosomal protein S6 family protein | -3.71 |
| 409 | 261956_at | AT1G64590 | short-chain dehydrogenase/reductase (SDR) family protein | 3.33 |
| 410 | 261949_at | AT1G64670 | BDG1 (BODYGUARD1); hydrolase | -2.52 |
| 411 | 261948_at | AT1G64680 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G03055.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO62908.1) | -3.40 |
| 412 | 262870_at | AT1G64710 | alcohol dehydrogenase, putative | -3.84 |
| 413 | 262878_at | AT1G64770 | carbohydrate binding / catalytic | -3.05 |
| 414 | 262879_at | AT1G64860 | SIGA (SIGMA FACTOR A); DNA binding / DNA-directed RNA polymerase/ transcription factor | -2.61 |
| 415 | 262882_at | AT1G64900 | CYP89A2 (CYTOCHROME P450 89A2); oxygen binding | -2.78 |
| 416 | 262863_at | AT1G64910 | glycosyltransferase family protein | 2.33 |
| 417 | 262864_at | AT1G64920 | glycosyltransferase family protein | 3.29 |
| 418 | 262875_at | AT1G64970 | G-TMT (GAMMA-TOCOPHEROL METHYLTRANSFERASE) | -2.63 |
| 419 | 261907_at | AT1G65060 | 4CL3 (4-coumarate:CoA ligase 3); 4-coumarate-CoA ligase | -5.23 |
| 420 | 263142_at | AT1G65230 | similar to hypothetical protein [Vitis vinifera] (GB:CAN76393.1) | -5.29 |
| 421 | 264157_at | AT1G65310 | ATXTH17 (XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 17); hydrolase, acting on glycosyl bonds | 4.02 |
| 422 | 264163_at | AT1G65445 | transferase-related | -2.77 |
| 423 | 264636_at | AT1G65490 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G65486.1) | -4.82 |
| 424 | 264680_at | AT1G65510 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G65490.1) | 2.62 |
| 425 | 256515_at | AT1G66020 | terpene synthase/cyclase family protein | 4.57 |
| 426 | 256518_at | AT1G66080 | similar to hypothetical protein [Vitis vinifera] (GB:CAN77663.1); contains InterPro domain Protein of unknown function DUF775 (InterPro:IPR008493) | 3.23 |
| 427 | 256514_at | AT1G66130 | oxidoreductase N-terminal domain-containing protein | -3.11 |
| 428 | 256522_at | AT1G66160 | U-box domain-containing protein | 2.60 |
| 429 | 260135_at | AT1G66400 | calmodulin-related protein, putative | 2.77 |
| 430 | 256358_at | AT1G66470 | basic helix-loop-helix (bHLH) family protein | 2.95 |
| 431 | 256368_at | AT1G66800 | cinnamyl-alcohol dehydrogenase family / CAD family | 4.11 |
| 432 | 256379_at | AT1G66840 | Identical to Protein PLASTID MOVEMENT IMPAIRED 2 (PMI2) [Arabidopsis Thaliana] (GB:Q9C9N6); similar to PMI15 (plastid movement impaired 15) [Arabidopsis thaliana] (TAIR:AT5G38150.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN79831.1); contains InterPro domain Protein of unknown function DUF827, plant (InterPro:IPR008545) | -3.01 |
| 433 | 255856_at | AT1G66940 | protein kinase-related | -3.27 |
| 434 | 255852_at | AT1G66970 | glycerophosphoryl diester phosphodiesterase family protein | -4.39 |
| 435 | 255913_at | AT1G66980 | protein kinase family protein / glycerophosphoryl diester phosphodiesterase family protein | -4.71 |
| 436 | 255858_at | AT1G67030 | ZFP6 (ZINC FINGER PROTEIN 6); nucleic acid binding / transcription factor/ zinc ion binding | 3.45 |
| 437 | 264470_at | AT1G67110 | CYP735A2 (cytochrome P450, family 735, subfamily A, polypeptide 2); oxygen binding | 3.50 |
| 438 | 264998_at | AT1G67330 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G27930.1); similar to unknown [Populus trichocarpa] (GB:ABK93495.1); contains InterPro domain Protein of unknown function DUF579, plant (InterPro:IPR006514) | 4.25 |
| 439 | 245198_at | AT1G67700 | similar to unknown [Populus trichocarpa] (GB:ABK95743.1); contains domain PTHR11804:SF2 (PTHR11804:SF2); contains domain PTHR11804 (PTHR11804) | -3.06 |
| 440 | 245186_at | AT1G67710 | ARR11 (RESPONSE REGULATOR 11); transcription factor/ two-component response regulator | 4.16 |
| 441 | 245195_at | AT1G67740 | PSBY (photosystem II BY) | -7.34 |
| 442 | 245215_at | AT1G67830 | ATFXG1 (ALPHA-FUCOSIDASE 1); alpha-L-fucosidase/ carboxylesterase | -2.51 |
| 443 | 260014_at | AT1G68010 | HPR (HYDROXYPYRUVATE REDUCTASE); glycerate dehydrogenase/ poly(U) binding | -7.01 |
| 444 | 259991_at | AT1G68040 | S-adenosyl-L-methionine:carboxyl methyltransferase family protein | 2.72 |
| 445 | 260432_at | AT1G68150 | WRKY9 (WRKY DNA-binding protein 9); transcription factor | 2.77 |
| 446 | 260431_at | AT1G68190 | zinc finger (B-box type) family protein | -4.23 |
| 447 | 260484_at | AT1G68360 | zinc finger protein-related | 2.58 |
| 448 | 260266_at | AT1G68520 | zinc finger (B-box type) family protein | -5.13 |
| 449 | 260267_at | AT1G68530 | CUT1 (CUTICULAR 1); acyltransferase/ catalytic | -5.66 |
| 450 | 262283_at | AT1G68590 | plastid-specific 30S ribosomal protein 3, putative / PSRP-3, putative | -5.44 |
| 451 | 262232_at | AT1G68600 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G25480.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G17470.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO42118.1); contains InterPro domain Protein of unknown function UPF0005 (InterPro:IPR006214) | -3.77 |
| 452 | 260036_at | AT1G68830 | STN7 (STT7 HOMOLOG STN7); kinase/ protein kinase | -3.33 |
| 453 | 260035_at | AT1G68850 | peroxidase, putative | 3.67 |
| 454 | 260030_at | AT1G68880 | ATBZIP (ARABIDOPSIS THALIANA BASIC LEUCINE-ZIPPER 8); DNA binding / transcription factor | 4.14 |
| 455 | 259371_at | AT1G69080 | universal stress protein (USP) family protein | 2.40 |
| 456 | 260353_at | AT1G69230 | SP1L2 | 2.37 |
| 457 | 256302_at | AT1G69526 | UbiE/COQ5 methyltransferase family protein | 2.89 |
| 458 | 256299_at | AT1G69530 | ATEXPA1 (ARABIDOPSIS THALIANA EXPANSIN A1) | -3.04 |
| 459 | 256303_at | AT1G69550 | disease resistance protein (TIR-NBS class), putative | -2.65 |
| 460 | 260419_at | AT1G69730 | protein kinase family protein | -6.05 |
| 461 | 260403_at | AT1G69810 | WRKY36 (WRKY DNA-binding protein 36); transcription factor | 3.72 |
| 462 | 260410_at | AT1G69870 | proton-dependent oligopeptide transport (POT) family protein | -4.13 |
| 463 | 264341_at | AT1G70270 | unknown protein | -4.20 |
| 464 | 260366_at | AT1G70460 | protein kinase, putative | 2.51 |
| 465 | 260205_at | AT1G70700 | JAZ9/TIFY7 (JASMONATE-ZIM-DOMAIN PROTEIN 9) | -4.70 |
| 466 | 262288_at | AT1G70760 | inorganic carbon transport protein-related | -4.91 |
| 467 | 262309_at | AT1G70820 | phosphoglucomutase, putative / glucose phosphomutase, putative | -7.92 |
| 468 | 262312_at | AT1G70830 | MLP28 (MLP-LIKE PROTEIN 28) | -2.81 |
| 469 | 262260_at | AT1G70850 | MLP34 (MLP-LIKE PROTEIN 34) | 6.74 |
| 470 | 262301_at | AT1G70880 | Bet v I allergen family protein | 4.48 |
| 471 | 262315_at | AT1G70990 | proline-rich family protein | 2.69 |
| 472 | 262307_at | AT1G71000 | DNAJ heat shock N-terminal domain-containing protein | 2.68 |
| 473 | 259751_at | AT1G71030 | ATMYBL2 (Arabidopsis myb-like 2); DNA binding / transcription factor | -2.99 |
| 474 | 259935_at | AT1G71250 | GDSL-motif lipase/hydrolase family protein | -2.62 |
| 475 | 259952_at | AT1G71400 | disease resistance family protein / LRR family protein | 3.53 |
| 476 | 259943_at | AT1G71480 | nuclear transport factor 2 (NTF2) family protein | -3.02 |
| 477 | 259896_at | AT1G71500 | Rieske (2Fe-2S) domain-containing protein | -5.04 |
| 478 | 259947_at | AT1G71530 | protein kinase family protein | 3.54 |
| 479 | 261504_at | AT1G71692 | AGL12 (AGAMOUS-LIKE 12); transcription factor | 4.40 |
| 480 | 261507_at | AT1G71720 | S1 RNA-binding domain-containing protein | -2.90 |
| 481 | 256336_at | AT1G72030 | GCN5-related N-acetyltransferase (GNAT) family protein | -4.41 |
| 482 | 259854_at | AT1G72200 | zinc finger (C3HC4-type RING finger) family protein | 3.43 |
| 483 | 259828_at | AT1G72220 | zinc finger (C3HC4-type RING finger) family protein | 2.94 |
| 484 | 259850_at | AT1G72240 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G22470.1) | 2.81 |
| 485 | 259892_at | AT1G72610 | GLP1 (GERMIN-LIKE PROTEIN 1); manganese ion binding / metal ion binding / nutrient reservoir | -6.50 |
| 486 | 262368_at | AT1G73060 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G48790.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO49401.1) | -2.68 |
| 487 | 262377_at | AT1G73110 | ribulose bisphosphate carboxylase/oxygenase activase, putative / RuBisCO activase, putative | -3.40 |
| 488 | 262371_at | AT1G73160 | glycosyl transferase family 1 protein | 3.57 |
| 489 | 260101_at | AT1G73260 | trypsin and protease inhibitor family protein / Kunitz family protein | 5.35 |
| 490 | 260093_at | AT1G73270 | SCPL6 (serine carboxypeptidase-like 6); serine carboxypeptidase | 6.02 |
| 491 | 260092_at | AT1G73280 | SCPL3 (serine carboxypeptidase-like 3); serine carboxypeptidase | 2.90 |
| 492 | 260091_at | AT1G73290 | SCPL5 (serine carboxypeptidase-like 5); serine carboxypeptidase | 3.40 |
| 493 | 245736_at | AT1G73330 | ATDR4 (Arabidopsis thaliana drought-repressed 4) | 7.13 |
| 494 | 245728_at | AT1G73340 | oxygen binding | 4.36 |
| 495 | 245777_at | AT1G73540 | ATNUDT21 (Arabidopsis thaliana Nudix hydrolase homolog 21); hydrolase | 2.36 |
| 496 | 260077_at | AT1G73620 | thaumatin-like protein, putative / pathogenesis-related protein, putative | 2.78 |
| 497 | 260044_at | AT1G73655 | immunophilin / FKBP-type peptidyl-prolyl cis-trans isomerase family protein | -2.83 |
| 498 | 260067_at | AT1G73780 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | 4.55 |
| 499 | 260380_at | AT1G73870 | zinc finger (B-box type) family protein | -4.56 |
| 500 | 260392_at | AT1G74030 | enolase, putative | 2.38 |
| 501 | 260388_at | AT1G74070 | peptidyl-prolyl cis-trans isomerase cyclophilin-type family protein | -4.72 |
| 502 | 260248_at | AT1G74310 | ATHSP101 (HEAT SHOCK PROTEIN 101); ATP binding / ATPase | 5.09 |
| 503 | 260234_at | AT1G74460 | GDSL-motif lipase/hydrolase family protein | 4.08 |
| 504 | 260236_at | AT1G74470 | geranylgeranyl reductase | -6.24 |
| 505 | 260213_at | AT1G74490 | protein kinase, putative | 2.69 |
| 506 | 260230_at | AT1G74500 | bHLH family protein | 7.91 |
| 507 | 260225_at | AT1G74590 | ATGSTU10 (Arabidopsis thaliana Glutathione S-transferase (class tau) 10); glutathione transferase | 3.47 |
| 508 | 260226_at | AT1G74660 | MIF1 (MINI ZINC FINGER 1); DNA binding / transcription factor | 3.55 |
| 509 | 260221_at | AT1G74670 | gibberellin-responsive protein, putative | -6.52 |
| 510 | 262168_at | AT1G74730 | similar to unknown [Populus trichocarpa x Populus deltoides] (GB:ABK96654.1); contains InterPro domain Protein of unknown function DUF1118 (InterPro:IPR009500) | -5.33 |
| 511 | 262217_at | AT1G74770 | protein binding / zinc ion binding | 5.28 |
| 512 | 262218_at | AT1G74770 | protein binding / zinc ion binding | 3.32 |
| 513 | 262175_at | AT1G74880 | NDH-O (NAD(P)H:PLASTOQUINONE DEHYDROGENASE COMPLEX SUBUNIT O) | -3.67 |
| 514 | 262211_at | AT1G74930 | ORA47; DNA binding / transcription factor | 2.78 |
| 515 | 262172_at | AT1G74970 | RPS9 (RIBOSOMAL PROTEIN S9); structural constituent of ribosome | -3.37 |
| 516 | 259925_at | AT1G75040 | PR5 (PATHOGENESIS-RELATED GENE 5) | -3.35 |
| 517 | 259927_at | AT1G75100 | JAC1 (J-DOMAIN PROTEIN REQUIRED FOR CHLOROPLAST ACCUMULATION RESPONSE 1); heat shock protein binding | -2.83 |
| 518 | 256452_at | AT1G75240 | ATHB33 (ARABIDOPSIS THALIANA HOMEOBOX PROTEIN 33); DNA binding / transcription factor | -2.82 |
| 519 | 261119_at | AT1G75350 | EMB2184 (EMBRYO DEFECTIVE 2184); structural constituent of ribosome | -3.18 |
| 520 | 261118_at | AT1G75460 | ATP-dependent protease La (LON) domain-containing protein | -4.84 |
| 521 | 262970_at | AT1G75690 | chaperone protein dnaJ-related | -8.62 |
| 522 | 262978_at | AT1G75780 | TUB1 (tubulin beta-1 chain); structural molecule | 2.59 |
| 523 | 261751_at | AT1G76080 | ATCDSP32/CDSP32 (CHLOROPLASTIC DROUGHT-INDUCED STRESS PROTEIN OF 32 KD); thiol-disulfide exchange intermediate | -4.34 |
| 524 | 261727_at | AT1G76090 | SMT3 (S-adenosyl-methionine-sterol-C-methyltransferase 3); S-adenosylmethionine-dependent methyltransferase | 2.21 |
| 525 | 261769_at | AT1G76100 | plastocyanin | -8.44 |
| 526 | 261782_at | AT1G76110 | high mobility group (HMG1/2) family protein / ARID/BRIGHT DNA-binding domain-containing protein | -4.83 |
| 527 | 261777_at | AT1G76210 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G20520.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN69930.1); contains InterPro domain Protein of unknown function DUF241, plant (InterPro:IPR004320) | 5.35 |
| 528 | 261772_at | AT1G76240 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G17080.1); similar to Os06g0725500 [Oryza sativa (japonica cultivar-group)] (GB:NP_001058623.1); similar to hypothetical protein OsI_023643 [Oryza sativa (indica cultivar-group)] (GB:EAZ02411.1); contains InterPro domain Protein of unknown function DUF241, plant (InterPro:IPR004320) | 2.63 |
| 529 | 259981_at | AT1G76450 | oxygen-evolving complex-related | -3.95 |
| 530 | 259866_at | AT1G76640 | calmodulin-related protein, putative | 6.15 |
| 531 | 259879_at | AT1G76650 | CML38; calcium ion binding | 3.13 |
| 532 | 259880_at | AT1G76730 | 5-formyltetrahydrofolate cyclo-ligase family protein | -3.87 |
| 533 | 259878_at | AT1G76790 | O-methyltransferase family 2 protein | 3.13 |
| 534 | 264960_at | AT1G76930 | ATEXT4 (EXTENSIN 4) | 3.62 |
| 535 | 264959_at | AT1G77090 | thylakoid lumenal 29.8 kDa protein | -4.64 |
| 536 | 246390_at | AT1G77330 | 1-aminocyclopropane-1-carboxylate oxidase, putative / ACC oxidase, putative | 4.76 |
| 537 | 259707_at | AT1G77490 | TAPX; L-ascorbate peroxidase | -3.73 |
| 538 | 259729_at | AT1G77640 | AP2 domain-containing transcription factor, putative | 3.56 |
| 539 | 259680_at | AT1G77690 | amino acid permease, putative | 3.21 |
| 540 | 262133_at | AT1G78000 | SULTR1;2 (SULFATE TRANSPORTER 1;2); sulfate transmembrane transporter | 3.56 |
| 541 | 260059_at | AT1G78090 | ATTPPB (TREHALOSE-6-PHOSPHATE PHOSPHATASE) | 3.73 |
| 542 | 260081_at | AT1G78170 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G22250.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO61724.1) | -2.81 |
| 543 | 260803_at | AT1G78340 | ATGSTU22 (Arabidopsis thaliana Glutathione S-transferase (class tau) 22); glutathione transferase | 2.98 |
| 544 | 260745_at | AT1G78370 | ATGSTU20 (Arabidopsis thaliana Glutathione S-transferase (class tau) 20); glutathione transferase | -3.13 |
| 545 | 260804_at | AT1G78410 | VQ motif-containing protein | -2.78 |
| 546 | 263133_at | AT1G78450 | SOUL heme-binding family protein | -4.69 |
| 547 | 263126_at | AT1G78460 | SOUL heme-binding family protein | -2.93 |
| 548 | 263131_at | AT1G78630 | EMB1473 (EMBRYO DEFECTIVE 1473); structural constituent of ribosome | -3.35 |
| 549 | 264305_at | AT1G78815 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G16910.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN63025.1); contains InterPro domain Protein of unknown function DUF640 (InterPro:IPR006936) | 6.58 |
| 550 | 257428_at | AT1G78990 | transferase family protein | 5.13 |
| 551 | 264096_at | AT1G78995 | similar to unnamed protein product [Vitis vinifera] (GB:CAO41778.1) | -6.19 |
| 552 | 264092_at | AT1G79040 | PSBR (photosystem II subunit R) | -7.38 |
| 553 | 264145_at | AT1G79310 | ATMC7 (METACASPASE 7); caspase | 3.66 |
| 554 | 262945_at | AT1G79510 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G16320.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO64929.1); contains domain NTF2-like (SSF54427) | -3.07 |
| 555 | 261353_at | AT1G79600 | ABC1 family protein | -3.12 |
| 556 | 261345_at | AT1G79760 | DTA4 (DOWNSTREAM TARGET OF AGL15-4) | 4.48 |
| 557 | 260165_at | AT1G79850 | RPS17 (ribosomal protein S17); structural constituent of ribosome | -2.92 |
| 558 | 262041_at | AT1G80100 | AHP6 (ARABIDOPSIS HISTIDINE PHOSPHOTRANSFER PROTEIN 6); transferase, transferring phosphorus-containing groups | 2.70 |
| 559 | 262045_at | AT1G80240 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G25460.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO44183.1); contains InterPro domain Protein of unknown function DUF642 (InterPro:IPR006946) | 6.39 |
| 560 | 260298_at | AT1G80320 | oxidoreductase, 2OG-Fe(II) oxygenase family protein | 5.87 |
| 561 | 261899_at | AT1G80820 | CCR2 (CINNAMOYL COA REDUCTASE) | 3.62 |
| 562 | 261892_at | AT1G80840 | WRKY40 (WRKY DNA-binding protein 40); transcription factor | 2.77 |
| 563 | 262202_at | AT2G01110 | APG2 (ALBINO AND PALE GREEN 2) | -3.08 |
| 564 | 265742_at | AT2G01290 | ribose-5-phosphate isomerase | -2.78 |
| 565 | 265741_at | AT2G01320 | ABC transporter family protein | -3.18 |
| 566 | 266353_at | AT2G01520 | MLP328 (MLP-LIKE PROTEIN 328) | 7.30 |
| 567 | 266330_at | AT2G01530 | MLP329 (MLP-LIKE PROTEIN 329) | 7.19 |
| 568 | 266329_at | AT2G01590 | similar to unnamed protein product [Vitis vinifera] (GB:CAO42320.1) | -2.94 |
| 569 | 263597_at | AT2G01870 | similar to unnamed protein product [Vitis vinifera] (GB:CAO21902.1) | -2.91 |
| 570 | 263594_at | AT2G01880 | ATPAP7/PAP7 (purple acid phosphatase 7); acid phosphatase/ protein serine/threonine phosphatase | 3.71 |
| 571 | 263596_at | AT2G01900 | endonuclease/exonuclease/phosphatase family protein | 2.42 |
| 572 | 265250_at | AT2G01950 | BRL2 (BRI1-LIKE 2); ATP binding / protein serine/threonine kinase | 2.24 |
| 573 | 266141_at | AT2G02120 | LCR70/PDF2.1 (Low-molecular-weight cysteine-rich 70); protease inhibitor | 3.79 |
| 574 | 267217_at | AT2G02610 | DC1 domain-containing protein | 3.39 |
| 575 | 267240_at | AT2G02680 | DC1 domain-containing protein | 3.73 |
| 576 | 266708_at | AT2G03200 | aspartyl protease family protein | 2.25 |
| 577 | 266707_at | AT2G03310 | unknown protein | -5.66 |
| 578 | 265704_at | AT2G03420 | similar to expressed protein [Oryza sativa (japonica cultivar-group)] (GB:ABF94594.1); similar to hypothetical protein OsI_010237 [Oryza sativa (indica cultivar-group)] (GB:EAY89004.1) | -4.07 |
| 579 | 265699_at | AT2G03550 | hydrolase | -4.75 |
| 580 | 264029_at | AT2G03720 | MRH6 (morphogenesis of root hair 6) | 4.12 |
| 581 | 264037_at | AT2G03750 | sulfotransferase family protein | -5.19 |
| 582 | 263482_at | AT2G03980 | GDSL-motif lipase/hydrolase family protein | -3.45 |
| 583 | 263481_at | AT2G04025 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G13620.1) | 4.40 |
| 584 | 263410_at | AT2G04039 | similar to unnamed protein product [Vitis vinifera] (GB:CAO41070.1) | -6.01 |
| 585 | 263837_at | AT2G04500 | DC1 domain-containing protein | 2.28 |
| 586 | 263809_at | AT2G04570 | GDSL-motif lipase/hydrolase family protein | -5.56 |
| 587 | 263674_at | AT2G04790 | similar to unnamed protein product [Vitis vinifera] (GB:CAO23994.1) | -5.39 |
| 588 | 263673_at | AT2G04800 | unknown protein | 6.81 |
| 589 | 263046_at | AT2G05380 | GRP3S (GLYCINE-RICH PROTEIN 3 SHORT ISOFORM) | -4.21 |
| 590 | 265560_at | AT2G05520 | GRP-3 (GLYCINE-RICH PROTEIN 3) | -5.31 |
| 591 | 265569_at | AT2G05620 | PGR5 (PROTON GRADIENT REGULATION 5) | -5.49 |
| 592 | 265374_at | AT2G06520 | PSBX (photosystem II subunit X) | -6.38 |
| 593 | 265400_at | AT2G10940 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | -8.14 |
| 594 | 263276_at | AT2G14100 | CYP705A13 (cytochrome P450, family 705, subfamily A, polypeptide 13); oxygen binding | 3.80 |
| 595 | 263295_at | AT2G14210 | ANR1; DNA binding / transcription factor | 2.81 |
| 596 | 266611_at | AT2G14960 | GH3.1 | 2.56 |
| 597 | 265894_at | AT2G15050 | LTP; lipid binding | -6.38 |
| 598 | 265918_at | AT2G15090 | fatty acid elongase, putative | -3.25 |
| 599 | 265501_at | AT2G15490 | UGT73B4; UDP-glycosyltransferase/ transferase, transferring glycosyl groups | 2.50 |
| 600 | 265481_at | AT2G15960 | unknown protein | -2.91 |
| 601 | 263098_at | AT2G16005 | MD-2-related lipid recognition domain-containing protein / ML domain-containing protein | 4.85 |
| 602 | 263610_at | AT2G16230 | glycosyl hydrolase family 17 protein | 4.41 |
| 603 | 263556_at | AT2G16365 | F-box family protein | -2.68 |
| 604 | 265359_at | AT2G16720 | MYB7 (myb domain protein 7); DNA binding / transcription factor | 2.69 |
| 605 | 265405_at | AT2G16750 | protein kinase family protein | 2.46 |
| 606 | 266532_at | AT2G16890 | UDP-glucoronosyl/UDP-glucosyl transferase family protein | -3.92 |
| 607 | 266525_at | AT2G16970 | MEE15 (maternal effect embryo arrest 15); tetracycline transporter | 2.67 |
| 608 | 266526_at | AT2G16980 | tetracycline transporter | 3.40 |
| 609 | 263575_at | AT2G17070 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G17080.1); similar to CM0216.210.nc [Lotus japonicus] (GB:BAF98210.1); contains InterPro domain Protein of unknown function DUF241, plant (InterPro:IPR004320) | 3.78 |
| 610 | 263576_at | AT2G17080 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G17070.1); similar to CM0216.210.nc [Lotus japonicus] (GB:BAF98210.1); contains InterPro domain Protein of unknown function DUF241, plant (InterPro:IPR004320) | 4.99 |
| 611 | 264851_at | AT2G17290 | CPK6 (CALCIUM-DEPENDENT PROTEIN KINASE 6); calmodulin-dependent protein kinase/ kinase | 2.42 |
| 612 | 264908_at | AT2G17440 | leucine-rich repeat family protein | 2.56 |
| 613 | 257409_at | AT2G17470 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G25480.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G68600.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO42118.1); contains InterPro domain Protein of unknown function UPF0005 (InterPro:IPR006214) | -2.88 |
| 614 | 263073_at | AT2G17500 | auxin efflux carrier family protein | 2.23 |
| 615 | 264617_at | AT2G17660 | nitrate-responsive NOI protein, putative | 2.67 |
| 616 | 265819_at | AT2G17972 | similar to unnamed protein product [Vitis vinifera] (GB:CAO48225.1) | -3.50 |
| 617 | 265327_at | AT2G18210 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G36500.1); similar to hypothetical protein [Thellungiella halophila] (GB:ABB45855.1) | 5.80 |
| 618 | 265342_at | AT2G18300 | basic helix-loop-helix (bHLH) family protein | -4.74 |
| 619 | 265341_at | AT2G18360 | hydrolase, alpha/beta fold family protein | 2.26 |
| 620 | 265334_at | AT2G18370 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | 4.33 |
| 621 | 265329_at | AT2G18450 | SDH1-2 (Succinate dehydrogenase 1-2) | 3.52 |
| 622 | 266066_at | AT2G18800 | xyloglucan:xyloglucosyl transferase, putative / xyloglucan endotransglycosylase, putative / endo-xyloglucan transferase, putative | 6.52 |
| 623 | 266941_at | AT2G18980 | peroxidase, putative | 5.04 |
| 624 | 267439_at | AT2G19060 | GDSL-motif lipase/hydrolase family protein | 2.72 |
| 625 | 267488_at | AT2G19110 | HMA4 (Heavy metal ATPase 4); cadmium-transporting ATPase | 2.31 |
| 626 | 265948_at | AT2G19590 | ACO1 (ACC OXIDASE 1); 1-aminocyclopropane-1-carboxylate oxidase | 3.19 |
| 627 | 265939_at | AT2G19650 | DC1 domain-containing protein | -3.69 |
| 628 | 266688_at | AT2G19660 | DC1 domain-containing protein | 2.23 |
| 629 | 265588_at | AT2G19970 | pathogenesis-related protein, putative | 7.00 |
| 630 | 265586_at | AT2G19990 | PR-1-LIKE (PATHOGENESIS-RELATED PROTEIN-1-LIKE) | 3.27 |
| 631 | 265287_at | AT2G20260 | PSAE-2 (photosystem I subunit E-2); catalytic | -6.78 |
| 632 | 263373_at | AT2G20515 | similar to unnamed protein product [Vitis vinifera] (GB:CAO40634.1) | 3.19 |
| 633 | 263374_at | AT2G20560 | DNAJ heat shock family protein | 4.08 |
| 634 | 263715_at | AT2G20570 | GPRI1 (GOLDEN2-LIKE 1); transcription factor | -2.69 |
| 635 | 257396_at | AT2G20875 | EPF1 (EPIDERMAL PATTERNING FACTOR 1) | -3.75 |
| 636 | 265415_at | AT2G20890 | PSB29 (THYLAKOID FORMATION1) | -2.91 |
| 637 | 265439_at | AT2G21045 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G66170.2); similar to putative senescence-associated protein [Oryza sativa (japonica cultivar-group)] (GB:BAD07813.1); contains InterPro domain Rhodanese-like (InterPro:IPR001763) | 6.19 |
| 638 | 264010_at | AT2G21100 | disease resistance-responsive protein-related / dirigent protein-related | 3.00 |
| 639 | 264014_at | AT2G21210 | auxin-responsive protein, putative | -3.69 |
| 640 | 264016_at | AT2G21220 | auxin-responsive protein, putative | -4.23 |
| 641 | 263760_at | AT2G21280 | GC1 (GIANT CHLOROPLAST 1); binding / catalytic/ coenzyme binding | -7.00 |
| 642 | 263739_at | AT2G21320 | zinc finger (B-box type) family protein | -3.20 |
| 643 | 263761_at | AT2G21330 | fructose-bisphosphate aldolase, putative | -6.93 |
| 644 | 263773_at | AT2G21370 | xylulose kinase, putative | -4.35 |
| 645 | 263545_at | AT2G21560 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G39190.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN77202.1) | 2.73 |
| 646 | 257432_at | AT2G21850 | protein binding / zinc ion binding | 2.37 |
| 647 | 263880_at | AT2G21960 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G56180.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO71767.1) | -4.04 |
| 648 | 264005_at | AT2G22470 | AGP2 (ARABINOGALACTAN-PROTEIN 2) | 4.39 |
| 649 | 263998_at | AT2G22510 | hydroxyproline-rich glycoprotein family protein | 3.73 |
| 650 | 266456_at | AT2G22770 | NAI1; DNA binding / transcription factor | 2.82 |
| 651 | 266799_at | AT2G22860 | ATPSK2 (PHYTOSULFOKINE 2 PRECURSOR); growth factor | 2.70 |
| 652 | 266800_at | AT2G22880 | VQ motif-containing protein | 4.17 |
| 653 | 266827_at | AT2G22920 | SCPL12; serine carboxypeptidase | 3.85 |
| 654 | 266828_at | AT2G22930 | glycosyltransferase family protein | 4.59 |
| 655 | 267262_at | AT2G22990 | SNG1 (SINAPOYLGLUCOSE 1); serine carboxypeptidase | -4.75 |
| 656 | 267254_at | AT2G23030 | SNRK2-9/SNRK2.9 (SNF1-RELATED PROTEIN KINASE 2.9); kinase | 3.21 |
| 657 | 257353_at | AT2G23050 | phototropic-responsive NPH3 family protein | 5.57 |
| 658 | 245076_at | AT2G23170 | GH3.3; indole-3-acetic acid amido synthetase | -5.83 |
| 659 | 245075_at | AT2G23180 | CYP96A1 (cytochrome P450, family 96, subfamily A, polypeptide 1); oxygen binding | -4.47 |
| 660 | 267137_at | AT2G23410 | ACPT (ARABIDOPSIS CIS-PRENYLTRANSFERASE); dehydrodolichyl diphosphate synthase | 2.21 |
| 661 | 267121_at | AT2G23540 | GDSL-motif lipase/hydrolase family protein | 5.88 |
| 662 | 267287_at | AT2G23630 | SKS16 (SKU5 Similar 16); copper ion binding / pectinesterase | 3.04 |
| 663 | 267294_at | AT2G23670 | YCF37 (Arabidopsis homolog of Synechocystis YCF37) | -6.34 |
| 664 | 267298_at | AT2G23760 | BLH4 (SAWTOOTH 2); DNA binding / transcription factor | -2.85 |
| 665 | 266572_at | AT2G23840 | HNH endonuclease domain-containing protein | -3.67 |
| 666 | 266578_at | AT2G23910 | cinnamoyl-CoA reductase-related | -2.70 |
| 667 | 266575_at | AT2G24060 | translation initiation factor 3 (IF-3) family protein | -2.60 |
| 668 | 266570_at | AT2G24090 | ribosomal protein L35 family protein | -3.31 |
| 669 | 265995_at | AT2G24140 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G30830.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO66637.1); contains InterPro domain Protein of unknown function DUF593 (InterPro:IPR007656) | 3.41 |
| 670 | 265989_at | AT2G24260 | basic helix-loop-helix (bHLH) family protein | 2.76 |
| 671 | 265998_at | AT2G24270 | ALDH11A3 (Aldehyde dehydrogenase 11A3); 3-chloroallyl aldehyde dehydrogenase/ glyceraldehyde-3-phosphate dehydrogenase (NADP+) | -5.90 |
| 672 | 265688_at | AT2G24300 | calmodulin-binding protein | 3.39 |
| 673 | 265681_at | AT2G24370 | kinase | -2.57 |
| 674 | 265683_at | AT2G24400 | auxin-responsive protein, putative / small auxin up RNA (SAUR_D) | 4.93 |
| 675 | 263795_at | AT2G24610 | ATCNGC14 (cyclic nucleotide gated channel 14); calmodulin binding / cyclic nucleotide binding / ion channel | 2.61 |
| 676 | 263533_at | AT2G24820 | TIC55 (TRANSLOCON AT THE INNER ENVELOPE MEMBRANE OF CHLOROPLASTS 55); oxidoreductase | -4.60 |
| 677 | 263552_x_at | AT2G24980 | proline-rich extensin-like family protein | 2.84 |
| 678 | 264403_at | AT2G25150 | transferase family protein | 2.76 |
| 679 | 264404_at | AT2G25160 | CYP82F1 (cytochrome P450, family 82, subfamily F, polypeptide 1); oxygen binding | 4.16 |
| 680 | 265618_at | AT2G25460 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G04860.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN68625.1); contains InterPro domain C2 calcium-dependent membrane targeting (InterPro:IPR000008) | 2.93 |
| 681 | 265611_at | AT2G25510 | unknown protein | -5.99 |
| 682 | 265913_at | AT2G25625 | similar to unnamed protein product [Vitis vinifera] (GB:CAO44157.1) | -3.82 |
| 683 | 266649_at | AT2G25810 | TIP4;1 (tonoplast intrinsic protein 4;1); water channel | 4.38 |
| 684 | 266838_at | AT2G25980 | jacalin lectin family protein | 5.20 |
| 685 | 266841_at | AT2G26150 | ATHSFA2 (Arabidopsis thaliana heat shock transcription factor A2); DNA binding / transcription factor | 8.38 |
| 686 | 267377_at | AT2G26250 | FDH (FIDDLEHEAD); acyltransferase | -2.59 |
| 687 | 267372_at | AT2G26290 | ARSK1 (ROOT-SPECIFIC KINASE 1); kinase | 4.05 |
| 688 | 267376_at | AT2G26330 | ER (ERECTA) | -6.56 |
| 689 | 267379_at | AT2G26340 | similar to unnamed protein product [Vitis vinifera] (GB:CAO43885.1) | -2.54 |
| 690 | 245033_at | AT2G26380 | disease resistance protein-related / LRR protein-related | 2.59 |
| 691 | 245034_at | AT2G26390 | serpin, putative / serine protease inhibitor, putative | 2.51 |
| 692 | 245052_at | AT2G26440 | pectinesterase family protein | -2.77 |
| 693 | 245044_at | AT2G26500 | cytochrome b6f complex subunit (petM), putative | -8.53 |
| 694 | 245042_at | AT2G26540 | HEMD; uroporphyrinogen-III synthase | -2.51 |
| 695 | 245029_at | AT2G26580 | YAB5 (YABBY5); transcription factor | -4.21 |
| 696 | 267610_at | AT2G26650 | AKT1 (ARABIDOPSIS K TRANSPORTER 1); cyclic nucleotide binding / inward rectifier potassium channel | 5.54 |
| 697 | 266851_at | AT2G26820 | ATPP2-A3 (Phloem protein 2-A3); carbohydrate binding | 3.90 |
| 698 | 266857_at | AT2G26900 | bile acid:sodium symporter family protein | -2.61 |
| 699 | 266856_at | AT2G26910 | ATPDR4/PDR4 (PLEIOTROPIC DRUG RESISTANCE 4); ATPase, coupled to transmembrane movement of substances | -3.53 |
| 700 | 266307_at | AT2G27000 | CYP705A8 (cytochrome P450, family 705, subfamily A, polypeptide 8); oxygen binding | 5.96 |
| 701 | 266308_at | AT2G27010 | CYP705A9 (cytochrome P450, family 705, subfamily A, polypeptide 9); oxygen binding | 2.72 |
| 702 | 266316_at | AT2G27080 | harpin-induced protein-related / HIN1-related / harpin-responsive protein-related | 2.95 |
| 703 | 265628_at | AT2G27290 | similar to unnamed protein product [Vitis vinifera] (GB:CAO48682.1); contains InterPro domain Protein of unknown function DUF1279 (InterPro:IPR009688) | -2.56 |
| 704 | 265645_at | AT2G27370 | integral membrane family protein | 4.94 |
| 705 | 266209_at | AT2G27550 | ATC (ARABIDOPSIS THALIANA CENTRORADIALIS); phosphatidylethanolamine binding | 5.61 |
| 706 | 266207_at | AT2G27680 | aldo/keto reductase family protein | -2.99 |
| 707 | 266156_at | AT2G28110 | FRA8 (FRAGILE FIBER8); transferase | 3.05 |
| 708 | 265545_at | AT2G28250 | protein kinase family protein. Binds AtRop4. | 2.25 |
| 709 | 264057_at | AT2G28550 | RAP2.7/TOE1 (TARGET OF EAT1 1); DNA binding / transcription factor | -3.25 |
| 710 | 263442_at | AT2G28605 | Identical to PsbP-related thylakoid lumenal protein 3, chloroplast precursor [Arabidopsis Thaliana] (GB:Q8VY52;GB:P83050;GB:Q8S8B1); similar to unnamed protein product [Vitis vinifera] (GB:CAO71682.1); contains InterPro domain Mog1/PsbP/DUF1795, alpha/beta/alpha sandwich (InterPro:IPR016124); contains InterPro domain Mog1/PsbP, alpha/beta/alpha sandwich (InterPro:IPR016123) | -3.60 |
| 711 | 263443_at | AT2G28630 | beta-ketoacyl-CoA synthase family protein | -3.11 |
| 712 | 263437_at | AT2G28670 | disease resistance-responsive family protein / fibroin-related | 4.45 |
| 713 | 263412_at | AT2G28720 | histone H2B, putative | -2.52 |
| 714 | 266222_at | AT2G28780 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G09450.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO21693.1) | 7.60 |
| 715 | 266225_at | AT2G28900 | OEP16 (OUTER ENVELOPE PROTEIN 16); P-P-bond-hydrolysis-driven protein transmembrane transporter | -2.89 |
| 716 | 266784_at | AT2G28960 | leucine-rich repeat protein kinase, putative | 3.54 |
| 717 | 266785_at | AT2G28970 | leucine-rich repeat protein kinase, putative | 2.68 |
| 718 | 266783_at | AT2G29130 | LAC2 (laccase 2); copper ion binding / oxidoreductase | 5.68 |
| 719 | 266285_at | AT2G29180 | similar to 80C09_19 [Brassica rapa subsp. pekinensis] (GB:AAZ41830.1) | -3.81 |
| 720 | 266279_at | AT2G29290 | tropinone reductase, putative / tropine dehydrogenase, putative | -3.94 |
| 721 | 266277_at | AT2G29310 | tropinone reductase, putative / tropine dehydrogenase, putative | -3.98 |
| 722 | 266291_at | AT2G29320 | tropinone reductase, putative / tropine dehydrogenase, putative | -4.46 |
| 723 | 266276_at | AT2G29330 | TRI (TROPINONE REDUCTASE); oxidoreductase | 3.86 |
| 724 | 266265_at | AT2G29340 | short-chain dehydrogenase/reductase (SDR) family protein | -2.52 |
| 725 | 266292_at | AT2G29350 | SAG13 (Senescence-associated gene 13); oxidoreductase | -2.88 |
| 726 | 266293_at | AT2G29360 | tropinone reductase, putative / tropine dehydrogenase, putative | -6.09 |
| 727 | 266271_at | AT2G29440 | ATGSTU6 (GLUTATHIONE S-TRANSFERASE 24); glutathione transferase | 2.24 |
| 728 | 266299_at | AT2G29450 | ATGSTU5 (Arabidopsis thaliana Glutathione S-transferase (class tau) 5); glutathione transferase | -4.26 |
| 729 | 266269_at | AT2G29480 | ATGSTU2 (GLUTATHIONE S-TRANSFERASE 20); glutathione transferase | 2.79 |
| 730 | 266290_at | AT2G29490 | ATGSTU1 (GLUTATHIONE S-TRANSFERASE 19); glutathione transferase | 3.98 |
| 731 | 266294_at | AT2G29500 | 17.6 kDa class I small heat shock protein (HSP17.6B-CI) | 4.38 |
| 732 | 266673_at | AT2G29630 | thiamine biosynthesis family protein / thiC family protein | -5.01 |
| 733 | 266672_at | AT2G29650 | inorganic phosphate transporter, putative | -5.22 |
| 734 | 266617_at | AT2G29670 | binding | -2.48 |
| 735 | 266670_at | AT2G29740 | UDP-glucoronosyl/UDP-glucosyl transferase family protein | 2.66 |
| 736 | 266669_at | AT2G29750 | UDP-glucoronosyl/UDP-glucosyl transferase family protein | 7.03 |
| 737 | 267276_at | AT2G30130 | ASL5 (phosphoenolpyruvate carboxykinase1) | 6.28 |
| 738 | 267247_at | AT2G30170 | catalytic | -3.13 |
| 739 | 267307_at | AT2G30210 | LAC3 (laccase 3); copper ion binding / oxidoreductase | 4.97 |
| 740 | 255873_at | AT2G30340 | LBD13 (LOB DOMAIN-CONTAINING PROTEIN 13) | 5.57 |
| 741 | 267471_at | AT2G30390 | ferrochelatase II | -2.85 |
| 742 | 267516_at | AT2G30520 | RPT2 (ROOT PHOTOTROPISM 2); protein binding | -3.86 |
| 743 | 267517_at | AT2G30520 | RPT2 (ROOT PHOTOTROPISM 2); protein binding | -3.91 |
| 744 | 267497_at | AT2G30540 | glutaredoxin family protein | -4.45 |
| 745 | 267526_at | AT2G30570 | PSBW (PHOTOSYSTEM II REACTION CENTER W) | -7.03 |
| 746 | 267207_at | AT2G30840 | 2-oxoglutarate-dependent dioxygenase, putative | 3.49 |
| 747 | 267196_at | AT2G30950 | VAR2 (VARIEGATED 2); ATP-dependent peptidase/ ATPase/ metallopeptidase/ zinc ion binding | -4.02 |
| 748 | 267152_at | AT2G31040 | ATP synthase protein I -related | -2.90 |
| 749 | 266481_at | AT2G31070 | TCP10 (TCP DOMAIN PROTEIN 10); transcription factor | -3.48 |
| 750 | 266477_at | AT2G31085 | CLE6 (CLAVATA3/ESR-RELATED 6); receptor binding | 4.26 |
| 751 | 266469_at | AT2G31180 | AtMYB14/Myb14at (myb domain protein 14); DNA binding / transcription factor | 2.26 |
| 752 | 263252_at | AT2G31380 | STH (salt tolerance homologue); transcription factor/ zinc ion binding | -3.97 |
| 753 | 263470_at | AT2G31900 | XIF (Myosin-like protein XIF) | 2.33 |
| 754 | 265668_at | AT2G32020 | GCN5-related N-acetyltransferase (GNAT) family protein | 3.08 |
| 755 | 265675_at | AT2G32120 | HSP70T-2; ATP binding | 3.51 |
| 756 | 265723_at | AT2G32140 | transmembrane receptor | 2.92 |
| 757 | 266336_at | AT2G32270 | ZIP3 (ZINC TRANSPORTER 3 PRECURSOR); zinc ion transmembrane transporter | 2.56 |
| 758 | 266356_at | AT2G32300 | UCC1 (UCLACYANIN 1); copper ion binding | 4.04 |
| 759 | 267061_at | AT2G32480 | membrane-associated zinc metalloprotease, putative | -2.61 |
| 760 | 267119_at | AT2G32610 | ATCSLB01 (Cellulose synthase-like B1); transferase/ transferase, transferring glycosyl groups | 3.03 |
| 761 | 267549_at | AT2G32640 | similar to unnamed protein product [Vitis vinifera] (GB:CAO24043.1); contains domain G3DSA:3.50.50.60 (G3DSA:3.50.50.60); contains domain SSF51905 (SSF51905) | -2.64 |
| 762 | 267545_at | AT2G32690 | pseudogene, glycine-rich protein | -5.98 |
| 763 | 267645_at | AT2G32860 | glycosyl hydrolase family 1 protein | -3.32 |
| 764 | 245165_at | AT2G33180 | similar to unnamed protein product [Vitis vinifera] (GB:CAO41107.1) | -3.23 |
| 765 | 255793_at | AT2G33250 | similar to unnamed protein product [Vitis vinifera] (GB:CAO71510.1) | -5.20 |
| 766 | 255795_at | AT2G33380 | RD20 (RESPONSIVE TO DESSICATION 20); calcium ion binding | -4.28 |
| 767 | 255850_at | AT2G33450 | 50S ribosomal protein L28, chloroplast (CL28) | -2.68 |
| 768 | 267457_at | AT2G33790 | pollen Ole e 1 allergen and extensin family protein | 6.42 |
| 769 | 267435_at | AT2G33800 | ribosomal protein S5 family protein | -2.76 |
| 770 | 267459_at | AT2G33850 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G28400.1); similar to unknown [Brassica napus] (GB:AAC06020.1) | 2.35 |
| 771 | 256712_at | AT2G34020 | calcium ion binding | 4.20 |
| 772 | 256720_at | AT2G34140 | Dof-type zinc finger domain-containing protein | 3.62 |
| 773 | 267005_at | AT2G34460 | flavin reductase-related | -4.21 |
| 774 | 266995_at | AT2G34500 | CYP710A1 (cytochrome P450, family 710, subfamily A, polypeptide 1); C-22 sterol desaturase/ oxygen binding | 2.56 |
| 775 | 266900_at | AT2G34610 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G30190.1); similar to unnamed protein product [Tetraodon nigroviridis] (GB:CAG12037.1) | 4.27 |
| 776 | 266899_at | AT2G34620 | mitochondrial transcription termination factor-related / mTERF-related | -4.07 |
| 777 | 267425_at | AT2G34810 | FAD-binding domain-containing protein | -4.00 |
| 778 | 267427_at | AT2G34830 | WRKY35 (WRKY DNA-BINDING PROTEIN 35); transcription factor | 3.32 |
| 779 | 267430_at | AT2G34860 | EDA3 (embryo sac development arrest 3); heat shock protein binding / unfolded protein binding | -4.03 |
| 780 | 267409_at | AT2G34910 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G30850.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO15288.1) | 3.50 |
| 781 | 266548_at | AT2G35210 | AGD10/MEE28/RPA (MATERNAL EFFECT EMBRYO ARREST 28); DNA binding | 2.23 |
| 782 | 266551_at | AT2G35260 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G17840.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN74782.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO63413.1) | -6.26 |
| 783 | 266546_at | AT2G35270 | DNA-binding protein-related | 4.94 |
| 784 | 266636_at | AT2G35370 | GDCH (Glycine decarboxylase complex H) | -7.32 |
| 785 | 266625_at | AT2G35380 | peroxidase 20 (PER20) (P20) | 2.88 |
| 786 | 266642_at | AT2G35410 | 33 kDa ribonucleoprotein, chloroplast, putative / RNA-binding protein cp33, putative | -2.98 |
| 787 | 266638_at | AT2G35490 | plastid-lipid associated protein PAP, putative | -3.29 |
| 788 | 266608_at | AT2G35500 | shikimate kinase-related | -2.75 |
| 789 | 263947_at | AT2G35820 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G35810.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO69007.1); contains domain G3DSA:2.60.120.480 (G3DSA:2.60.120.480) | -2.76 |
| 790 | 263935_at | AT2G35930 | U-box domain-containing protein | 3.39 |
| 791 | 263951_at | AT2G35960 | NHL12 (NDR1/HIN1-like 12) | -3.18 |
| 792 | 263963_at | AT2G36080 | DNA-binding protein, putative | -3.22 |
| 793 | 263284_at | AT2G36100 | integral membrane family protein | 5.14 |
| 794 | 263285_at | AT2G36120 | pseudogene, glycine-rich protein | 5.24 |
| 795 | 263287_at | AT2G36145 | similar to unnamed protein product [Vitis vinifera] (GB:CAO15876.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN74693.1) | -7.44 |
| 796 | 263931_at | AT2G36220 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G52710.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO15901.1) | 2.59 |
| 797 | 265204_at | AT2G36650 | similar to CHUP1 (CHLOROPLAST UNUSUAL POSITIONING 1) [Arabidopsis thaliana] (TAIR:AT3G25690.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN72920.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO62954.1) | 5.28 |
| 798 | 265198_at | AT2G36760 | UGT73C2 (UDP-GLUCOSYL TRANSFERASE 73C2); UDP-glycosyltransferase/ transferase, transferring glycosyl groups / transferase, transferring hexosyl groups | -2.62 |
| 799 | 263846_at | AT2G36990 | SIGF (RNA POLYMERASE SIGMA-70 FACTOR); DNA binding / DNA-directed RNA polymerase/ transcription factor | -3.76 |
| 800 | 265471_at | AT2G37130 | peroxidase 21 (PER21) (P21) (PRXR5) | 4.55 |
| 801 | 265966_at | AT2G37220 | 29 kDa ribonucleoprotein, chloroplast, putative / RNA-binding protein cp29, putative | -2.68 |
| 802 | 265959_at | AT2G37240 | antioxidant/ oxidoreductase | -3.04 |
| 803 | 266010_at | AT2G37430 | zinc finger (C2H2 type) family protein (ZAT11) | 3.40 |
| 804 | 266011_at | AT2G37440 | endonuclease/exonuclease/phosphatase family protein | 5.46 |
| 805 | 267158_at | AT2G37640 | ATEXPA3 (ARABIDOPSIS THALIANA EXPANSIN A3) | -2.65 |
| 806 | 267172_at | AT2G37660 | binding / catalytic/ coenzyme binding | -4.85 |
| 807 | 267178_at | AT2G37750 | unknown protein | 4.48 |
| 808 | 266097_at | AT2G37970 | SOUL-1; binding | -4.19 |
| 809 | 267088_at | AT2G38140 | PSRP4 (PLASTID-SPECIFIC RIBOSOMAL PROTEIN 4); structural constituent of ribosome | -2.60 |
| 810 | 267093_at | AT2G38170 | CAX1 (CATION EXCHANGER 1); calcium ion transmembrane transporter/ calcium:hydrogen antiporter | -2.71 |
| 811 | 267027_at | AT2G38330 | MATE efflux family protein | -2.55 |
| 812 | 266421_at | AT2G38540 | LP1 (nonspecific lipid transfer protein 1) | -6.11 |
| 813 | 266419_at | AT2G38760 | ANNAT3 (ANNEXIN ARABIDOPSIS 3); calcium ion binding / calcium-dependent phospholipid binding | 2.24 |
| 814 | 266172_at | AT2G39010 | PIP2;6/PIP2E (plasma membrane intrinsic protein 2;6); water channel | -4.12 |
| 815 | 266191_at | AT2G39040 | peroxidase, putative | 4.64 |
| 816 | 266196_at | AT2G39110 | protein kinase, putative | 4.14 |
| 817 | 266992_at | AT2G39200 | MLO12 (MILDEW RESISTANCE LOCUS O 12); calmodulin binding | 2.54 |
| 818 | 266988_at | AT2G39310 | jacalin lectin family protein | 4.95 |
| 819 | 266974_at | AT2G39370 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G37380.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO41570.1) | 3.96 |
| 820 | 266976_at | AT2G39410 | hydrolase, alpha/beta fold family protein | 2.41 |
| 821 | 266978_at | AT2G39430 | disease resistance-responsive protein-related / dirigent protein-related | 2.73 |
| 822 | 266979_at | AT2G39470 | PPL2 (PSBP-LIKE PROTEIN 2); calcium ion binding | -4.92 |
| 823 | 266967_at | AT2G39530 | integral membrane protein, putative | 5.80 |
| 824 | 267623_at | AT2G39650 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G14620.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO69213.1); contains InterPro domain Protein of unknown function DUF506, plant (InterPro:IPR006502) | 2.54 |
| 825 | 245061_at | AT2G39730 | RCA (RUBISCO ACTIVASE) | -7.15 |
| 826 | 245088_at | AT2G39850 | subtilase | -4.28 |
| 827 | 267361_at | AT2G39920 | acid phosphatase class B family protein | -3.89 |
| 828 | 265722_at | AT2G40100 | LHCB4.3 (LIGHT HARVESTING COMPLEX PSII); chlorophyll binding | -5.24 |
| 829 | 255823_at | AT2G40470 | LBD15 (LOB DOMAIN-CONTAINING PROTEIN 15) | 2.55 |
| 830 | 255826_at | AT2G40490 | HEME2; uroporphyrinogen decarboxylase | -3.00 |
| 831 | 255822_at | AT2G40610 | ATEXPA8 (ARABIDOPSIS THALIANA EXPANSIN A8) | -3.14 |
| 832 | 266078_at | AT2G40670 | ARR16 (response regulator 16); transcription regulator/ two-component response regulator | -3.59 |
| 833 | 245096_at | AT2G40880 | FL3-27; cysteine protease inhibitor | -3.67 |
| 834 | 267077_at | AT2G40970 | myb family transcription factor | 3.06 |
| 835 | 267076_at | AT2G41090 | calmodulin-like calcium-binding protein, 22 kDa (CaBP-22) | -4.91 |
| 836 | 267101_at | AT2G41480 | peroxidase | 4.20 |
| 837 | 245119_at | AT2G41640 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G57380.1); similar to glycosyltransferase [Medicago truncatula] (GB:CAI30145.1); contains InterPro domain Protein of unknown function DUF563 (InterPro:IPR007657) | 2.55 |
| 838 | 245113_at | AT2G41660 | MIZ1 (MIZU-KUSSEI 1) | 3.23 |
| 839 | 260555_at | AT2G41780 | unknown protein | 2.85 |
| 840 | 260553_at | AT2G41800 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G41810.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO23583.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN80832.1); contains InterPro domain Protein of unknown function DUF642 (InterPro:IPR006946); contains InterPro domain Galactose-binding like (InterPro:IPR008979) | 5.76 |
| 841 | 260495_at | AT2G41810 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G41800.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO23583.1); contains InterPro domain Protein of unknown function DUF642 (InterPro:IPR006946); contains InterPro domain Galactose-binding like (InterPro:IPR008979) | 3.74 |
| 842 | 267535_at | AT2G41940 | ZFP8 (ZINC FINGER PROTEIN 8); nucleic acid binding / transcription factor/ zinc ion binding | -2.54 |
| 843 | 267630_at | AT2G42130 | Identical to Probable plastid-lipid-associated protein 13, chloroplast precursor (PAP13) [Arabidopsis Thaliana] (GB:Q8S9M1;GB:O48521;GB:Q84X37;GB:Q84X38;GB:Q84X39;GB:Q8GY49); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G58010.1); similar to unknown [Populus trichocarpa] (GB:ABK96151.1) | -3.10 |
| 844 | 267635_at | AT2G42220 | rhodanese-like domain-containing protein | -6.98 |
| 845 | 267626_at | AT2G42250 | CYP712A1 (cytochrome P450, family 712, subfamily A, polypeptide 1); oxygen binding | 4.06 |
| 846 | 265852_at | AT2G42350 | zinc finger (C3HC4-type RING finger) family protein | 3.23 |
| 847 | 265877_at | AT2G42380 | bZIP transcription factor family protein | -3.37 |
| 848 | 263495_at | AT2G42530 | COR15B | -3.79 |
| 849 | 263497_at | AT2G42540 | COR15A (COLD-REGULATED 15A) | -6.97 |
| 850 | 263496_at | AT2G42570 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G31110.2); similar to unknown [Pisum sativum] (GB:ABA29158.1); contains InterPro domain Protein of unknown function DUF231, plant (InterPro:IPR004253) | 2.58 |
| 851 | 263498_at | AT2G42610 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G07090.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN66524.1); contains InterPro domain Protein of unknown function DUF640 (InterPro:IPR006936) | 3.08 |
| 852 | 263987_at | AT2G42690 | lipase, putative | -2.65 |
| 853 | 263972_at | AT2G42760 | similar to unnamed protein product [Vitis vinifera] (GB:CAO69913.1) | 2.41 |
| 854 | 263970_at | AT2G42850 | CYP718 (cytochrome P450, family 718); oxygen binding | 3.50 |
| 855 | 265248_at | AT2G43010 | PIF4 (PHYTOCHROME INTERACTING FACTOR 4); DNA binding / transcription factor | -2.60 |
| 856 | 265247_at | AT2G43030 | ribosomal protein L3 family protein | -2.82 |
| 857 | 265246_at | AT2G43050 | ATPMEPCRD; pectinesterase | 2.59 |
| 858 | 257373_at | AT2G43140 | DNA binding / transcription factor | 5.16 |
| 859 | 260535_at | AT2G43390 | unknown protein | 3.13 |
| 860 | 260551_at | AT2G43510 | ATTI1 (ARABIDOPSIS THALIANA TRYPSIN INHIBITOR PROTEIN 1) | -3.39 |
| 861 | 260549_at | AT2G43535 | trypsin inhibitor, putative | 3.63 |
| 862 | 260547_at | AT2G43550 | trypsin inhibitor, putative | -2.53 |
| 863 | 260542_at | AT2G43560 | immunophilin / FKBP-type peptidyl-prolyl cis-trans isomerase family protein | -4.18 |
| 864 | 260558_at | AT2G43600 | glycoside hydrolase family 19 protein | 5.02 |
| 865 | 260557_at | AT2G43610 | glycoside hydrolase family 19 protein | 5.14 |
| 866 | 260563_at | AT2G43840 | UDP-glucoronosyl/UDP-glucosyl transferase family protein | 2.67 |
| 867 | 267222_at | AT2G43880 | polygalacturonase, putative / pectinase, putative | 3.15 |
| 868 | 267228_at | AT2G43890 | polygalacturonase, putative / pectinase, putative | 4.21 |
| 869 | 267226_at | AT2G44010 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G59880.1) | 5.73 |
| 870 | 267191_at | AT2G44110 | MLO15 (MILDEW RESISTANCE LOCUS O 15); calmodulin binding | 3.38 |
| 871 | 267384_at | AT2G44370 | DC1 domain-containing protein | 2.61 |
| 872 | 267385_at | AT2G44380 | DC1 domain-containing protein | 4.62 |
| 873 | 267388_at | AT2G44450 | glycosyl hydrolase family 1 protein | 2.29 |
| 874 | 267391_at | AT2G44480 | glycosyl hydrolase family 1 protein | 2.49 |
| 875 | 266884_at | AT2G44790 | UCC2 (UCLACYANIN 2); copper ion binding | 5.68 |
| 876 | 266883_at | AT2G44810 | DAD1 (DEFECTIVE ANTHER DEHISCENCE 1); triacylglycerol lipase | 2.59 |
| 877 | 266821_at | AT2G44840 | ATERF13/EREBP (ETHYLENE-RESPONSIVE ELEMENT BINDING FACTOR 13); DNA binding / transcription factor | 4.80 |
| 878 | 266813_at | AT2G44920 | thylakoid lumenal 15 kDa protein, chloroplast | -4.44 |
| 879 | 266125_at | AT2G45050 | zinc finger (GATA type) family protein | 2.64 |
| 880 | 266123_at | AT2G45180 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | -3.63 |
| 881 | 245138_at | AT2G45190 | AFO (ABNORMAL FLORAL ORGANS); transcription factor | -5.34 |
| 882 | 245136_at | AT2G45210 | auxin-responsive protein-related | 5.97 |
| 883 | 245148_at | AT2G45220 | pectinesterase family protein | 2.38 |
| 884 | 245141_at | AT2G45400 | BEN1; oxidoreductase, acting on CH-OH group of donors | 2.46 |
| 885 | 245139_at | AT2G45430 | DNA-binding protein-related | 3.94 |
| 886 | 267502_at | AT2G45550 | CYP76C4 (cytochrome P450, family 76, subfamily C, polypeptide 4); oxygen binding | 3.16 |
| 887 | 267559_at | AT2G45570 | CYP76C2 (cytochrome P450, family 76, subfamily C, polypeptide 2); oxygen binding | -2.93 |
| 888 | 266925_at | AT2G45740 | PEX11D | -3.71 |
| 889 | 266920_at | AT2G45750 | dehydration-responsive family protein | 2.86 |
| 890 | 266918_at | AT2G45800 | LIM domain-containing protein | -3.02 |
| 891 | 266917_at | AT2G45830 | DTA2 (DOWNSTREAM TARGET OF AGL15 2) | 2.30 |
| 892 | 266929_at | AT2G45850 | DNA-binding family protein | -2.99 |
| 893 | 266913_at | AT2G45890 | ATROPGEF4/ROPGEF4 (KINASE PARTNER PROTEIN-LIKE); Rho guanyl-nucleotide exchange factor/ | 3.16 |
| 894 | 266581_at | AT2G46140 | late embryogenesis abundant protein, putative / LEA protein, putative | 2.61 |
| 895 | 266590_at | AT2G46240 | BAG6 (ARABIDOPSIS THALIANA BCL-2-ASSOCIATED ATHANOGENE 6); calmodulin binding / protein binding | 4.22 |
| 896 | 263779_at | AT2G46340 | SPA1 (SUPPRESSOR OF PHYA-105 1); signal transducer | -6.94 |
| 897 | 263780_at | AT2G46340 | SPA1 (SUPPRESSOR OF PHYA-105 1); signal transducer | -3.68 |
| 898 | 263783_at | AT2G46400 | WRKY46 (WRKY DNA-binding protein 46); transcription factor | 2.34 |
| 899 | 263787_at | AT2G46420 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G61700.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO39750.1); contains InterPro domain Conserved hypothetical protein CHP01589, plant (InterPro:IPR006476) | -3.85 |
| 900 | 266711_at | AT2G46740 | FAD-binding domain-containing protein | 3.45 |
| 901 | 266713_at | AT2G46760 | FAD-binding domain-containing protein | 2.73 |
| 902 | 266716_at | AT2G46820 | PSAP/PSI-P/PTAC8/TMP14 (THYLAKOID MEMBRANE PHOSPHOPROTEIN OF 14 KDA); DNA binding | -5.07 |
| 903 | 266760_at | AT2G46870 | NGA1 (NGATHA1); transcription factor | -3.59 |
| 904 | 266757_at | AT2G46940 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G62070.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN75643.1) | 3.17 |
| 905 | 266752_at | AT2G47000 | MDR4/PGP4 (P-GLYCOPROTEIN4); ATPase, coupled to transmembrane movement of substances / xenobiotic-transporting ATPase | 3.93 |
| 906 | 266737_at | AT2G47140 | short-chain dehydrogenase/reductase (SDR) family protein | 2.92 |
| 907 | 260582_at | AT2G47200 | unknown protein | 3.46 |
| 908 | 260531_at | AT2G47240 | long-chain-fatty-acid--CoA ligase family protein / long-chain acyl-CoA synthetase family protein | -5.32 |
| 909 | 260527_at | AT2G47270 | transcription factor/ transcription regulator | 4.17 |
| 910 | 260529_at | AT2G47400 | CP12-1 (CP12 domain-containing protein 1) | -4.07 |
| 911 | 245123_at | AT2G47450 | CAO (CHAOS); chromatin binding | -4.08 |
| 912 | 245172_at | AT2G47540 | pollen Ole e 1 allergen and extensin family protein | 2.66 |
| 913 | 245151_at | AT2G47550 | pectinesterase family protein | 3.06 |
| 914 | 245171_at | AT2G47560 | zinc finger (C3HC4-type RING finger) family protein | 2.31 |
| 915 | 245150_at | AT2G47590 | PHR2 (PHOTOLYASE/BLUE-LIGHT RECEPTOR 2) | -2.62 |
| 916 | 266488_at | AT2G47670 | invertase/pectin methylesterase inhibitor family protein | 2.36 |
| 917 | 266507_at | AT2G47860 | phototropic-responsive NPH3 family protein | 4.11 |
| 918 | 266483_at | AT2G47910 | CRR6 (CHLORORESPIRATORY REDUCTION 6) | -4.68 |
| 919 | 262349_at | AT2G48130 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | 3.77 |
| 920 | 262317_at | AT2G48140 | EDA4 (embryo sac development arrest 4); lipid binding | 4.44 |
| 921 | 259275_at | AT3G01060 | similar to unnamed protein product [Vitis vinifera] (GB:CAO15045.1); similar to unknown [Brassica rapa] (GB:ABC41272.1) | -4.00 |
| 922 | 259276_at | AT3G01190 | peroxidase 27 (PER27) (P27) (PRXR7) | 9.19 |
| 923 | 259274_at | AT3G01220 | ATHB20 (ARABIDOPSIS THALIANA HOMEOBOX PROTEIN 20); DNA binding / transcription factor | 4.48 |
| 924 | 259264_at | AT3G01260 | aldose 1-epimerase family protein | 5.92 |
| 925 | 258956_at | AT3G01440 | oxygen evolving enhancer 3 (PsbQ) family protein | -4.29 |
| 926 | 259193_at | AT3G01480 | CYP38 (CYCLOPHILIN 38); peptidyl-prolyl cis-trans isomerase | -4.29 |
| 927 | 259163_at | AT3G01490 | protein kinase, putative | -5.57 |
| 928 | 259161_at | AT3G01500 | CA1 (CARBONIC ANHYDRASE 1); carbonate dehydratase/ zinc ion binding | -9.35 |
| 929 | 259185_at | AT3G01550 | triose phosphate/phosphate translocator, putative | -4.03 |
| 930 | 259179_at | AT3G01660 | methyltransferase | -3.32 |
| 931 | 259129_at | AT3G02150 | PTF1 (PLASTID TRANSCRIPTION FACTOR 1); transcription factor | -2.65 |
| 932 | 259120_at | AT3G02240 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G02242.1) | 3.22 |
| 933 | 258497_at | AT3G02380 | COL2 (CONSTANS-LIKE 2); transcription factor/ zinc ion binding | -3.78 |
| 934 | 258495_at | AT3G02690 | integral membrane family protein | -3.00 |
| 935 | 258607_at | AT3G02730 | ATF1/TRXF1 (THIOREDOXIN F-TYPE 1); thiol-disulfide exchange intermediate | -4.96 |
| 936 | 258621_at | AT3G02830 | ZFN1 (ZINC FINGER PROTEIN 1); nucleic acid binding | -3.18 |
| 937 | 258606_at | AT3G02840 | immediate-early fungal elicitor family protein | 3.25 |
| 938 | 258629_at | AT3G02850 | SKOR (stelar K+ outward rectifier); cyclic nucleotide binding / outward rectifier potassium channel | 4.75 |
| 939 | 259169_at | AT3G03520 | phosphoesterase family protein | 3.53 |
| 940 | 259197_at | AT3G03670 | peroxidase, putative | 2.91 |
| 941 | 259331_at | AT3G03840 | auxin-responsive protein, putative | -3.56 |
| 942 | 258805_at | AT3G04010 | glycosyl hydrolase family 17 protein | 2.34 |
| 943 | 258537_at | AT3G04210 | disease resistance protein (TIR-NBS class), putative | -5.62 |
| 944 | 258573_at | AT3G04260 | PTAC3 (PLASTID TRANSCRIPTIONALLY ACTIVE3); DNA binding | -2.52 |
| 945 | 258589_at | AT3G04290 | ATLTL1/LTL1 (LI-TOLERANT LIPASE 1); carboxylesterase | -6.53 |
| 946 | 258800_at | AT3G04550 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G28500.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO65032.1) | -3.45 |
| 947 | 258795_at | AT3G04570 | DNA-binding protein-related | 6.94 |
| 948 | 258804_at | AT3G04760 | pentatricopeptide (PPR) repeat-containing protein | -5.13 |
| 949 | 259351_at | AT3G05150 | sugar transporter family protein | 5.10 |
| 950 | 259308_at | AT3G05180 | GDSL-motif lipase/hydrolase family protein | -3.37 |
| 951 | 258732_at | AT3G05820 | beta-fructofuranosidase, putative / invertase, putative / saccharase, putative / beta-fructosidase, putative | -3.67 |
| 952 | 258751_at | AT3G05890 | RCI2B (RARE-COLD-INDUCIBLE 2B) | 3.79 |
| 953 | 258745_at | AT3G05920 | heavy-metal-associated domain-containing protein | 6.96 |
| 954 | 258746_at | AT3G05950 | germin-like protein, putative | 6.14 |
| 955 | 258905_at | AT3G06390 | integral membrane family protein | 6.21 |
| 956 | 258902_at | AT3G06483 | PDK (PYRUVATE DEHYDROGENASE KINASE); ATP binding / pyruvate dehydrogenase (acetyl-transferring) kinase | -2.62 |
| 957 | 258540_at | AT3G06990 | DC1 domain-containing protein | 2.59 |
| 958 | 258541_at | AT3G07000 | DC1 domain-containing protein | 2.33 |
| 959 | 259251_at | AT3G07600 | heavy-metal-associated domain-containing protein | 3.29 |
| 960 | 259226_at | AT3G07700 | ABC1 family protein | -2.62 |
| 961 | 258643_at | AT3G08010 | ATAB2; RNA binding | -3.23 |
| 962 | 258674_at | AT3G08740 | elongation factor P (EF-P) family protein | -2.66 |
| 963 | 258675_at | AT3G08770 | LTP6 (Lipid transfer protein 6); lipid binding | -2.68 |
| 964 | 258982_at | AT3G08870 | lectin protein kinase, putative | -3.31 |
| 965 | 258989_at | AT3G08920 | rhodanese-like domain-containing protein | -3.23 |
| 966 | 258993_at | AT3G08940 | LHCB4.2 (LIGHT HARVESTING COMPLEX PSII); chlorophyll binding | -6.43 |
| 967 | 259207_at | AT3G09050 | similar to unknown [Populus trichocarpa x Populus deltoides] (GB:ABK96465.1) | -3.08 |
| 968 | 259036_at | AT3G09220 | LAC7 (laccase 7); copper ion binding / oxidoreductase | 2.81 |
| 969 | 259009_at | AT3G09260 | PYK10 (phosphate starvation-response 3.1); hydrolase, hydrolyzing O-glycosyl compounds | 6.65 |
| 970 | 259040_at | AT3G09270 | ATGSTU8 (Arabidopsis thaliana Glutathione S-transferase (class tau) 8); glutathione transferase | 3.19 |
| 971 | 259028_at | AT3G09290 | TAC1 (TELOMERASE ACTIVATOR1); nucleic acid binding / transcription factor/ zinc ion binding | 4.55 |
| 972 | 259008_at | AT3G09390 | MT2A (METALLOTHIONEIN 2A) | -2.56 |
| 973 | 258708_at | AT3G09580 | amine oxidase family protein | -4.90 |
| 974 | 258941_at | AT3G09940 | ATMDAR3/MDHAR (MONODEHYDROASCORBATE REDUCTASE); monodehydroascorbate reductase (NADH) | 4.47 |
| 975 | 258929_at | AT3G10060 | immunophilin, putative / FKBP-type peptidyl-prolyl cis-trans isomerase, putative | -3.35 |
| 976 | 258932_at | AT3G10150 | ATPAP16/PAP16 (purple acid phosphatase 16); acid phosphatase/ protein serine/threonine phosphatase | -3.81 |
| 977 | 259150_at | AT3G10320 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G41640.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO23616.1); contains InterPro domain Protein of unknown function DUF563 (InterPro:IPR007657) | 2.50 |
| 978 | 258962_at | AT3G10570 | CYP77A6 (cytochrome P450, family 77, subfamily A, polypeptide 6); oxygen binding | -4.31 |
| 979 | 258945_at | AT3G10660 | CPK2 (CALMODULIN-DOMAIN PROTEIN KINASE CDPK ISOFORM 2); calmodulin-dependent protein kinase/ kinase | 2.69 |
| 980 | 257533_at | AT3G10840 | hydrolase, alpha/beta fold family protein | -3.15 |
| 981 | 256442_at | AT3G10930 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G05300.1) | 3.13 |
| 982 | 256427_at | AT3G11090 | LBD21 (LOB DOMAIN-CONTAINING PROTEIN 21) | -4.18 |
| 983 | 256252_at | AT3G11340 | UDP-glucoronosyl/UDP-glucosyl transferase family protein | 4.19 |
| 984 | 259260_at | AT3G11370 | DC1 domain-containing protein | 2.76 |
| 985 | 259286_at | AT3G11480 | BSMT1; S-adenosylmethionine-dependent methyltransferase | -2.55 |
| 986 | 259291_at | AT3G11550 | integral membrane family protein | 6.88 |
| 987 | 256273_at | AT3G12090 | TET6 (TETRASPANIN6) | 2.25 |
| 988 | 256243_at | AT3G12500 | ATHCHIB (BASIC CHITINASE); chitinase | 5.11 |
| 989 | 256283_at | AT3G12540 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G39690.1); similar to At3g12540-like protein [Boechera stricta] (GB:ABB89771.1); contains InterPro domain Protein of unknown function DUF547 (InterPro:IPR006869) | 2.83 |
| 990 | 256245_at | AT3G12580 | HSP70 (heat shock protein 70); ATP binding | 4.33 |
| 991 | 257699_at | AT3G12780 | PGK1 (PHOSPHOGLYCERATE KINASE 1); phosphoglycerate kinase | -3.07 |
| 992 | 257690_at | AT3G12830 | auxin-responsive family protein | 2.43 |
| 993 | 257856_at | AT3G12930 | similar to unknown [Populus trichocarpa] (GB:ABK94112.1); contains InterPro domain Iojap-related protein (InterPro:IPR004394) | -2.85 |
| 994 | 257190_at | AT3G13120 | 30S ribosomal protein S10, chloroplast, putative | -2.94 |
| 995 | 256955_at | AT3G13390 | SKS11 (SKU5 Similar 11); copper ion binding / oxidoreductase | -3.61 |
| 996 | 256647_at | AT3G13610 | oxidoreductase, 2OG-Fe(II) oxygenase family protein | 5.21 |
| 997 | 256781_at | AT3G13650 | disease resistance response | 2.84 |
| 998 | 256774_at | AT3G13760 | DC1 domain-containing protein | 7.85 |
| 999 | 256999_at | AT3G14200 | DNAJ heat shock N-terminal domain-containing protein | 2.50 |
| 1000 | 257008_at | AT3G14210 | ESM1 (EPITHIOSPECIFIER MODIFIER 1); carboxylesterase | -5.64 |
| 1001 | 258362_at | AT3G14280 | similar to hypothetical protein [Vitis vinifera] (GB:CAN62135.1) | 3.36 |
| 1002 | 258063_at | AT3G14620 | CYP72A8 (cytochrome P450, family 72, subfamily A, polypeptide 8); oxygen binding | -2.79 |
| 1003 | 258064_at | AT3G14680 | CYP72A14 (cytochrome P450, family 72, subfamily A, polypeptide 14); oxygen binding | 7.53 |
| 1004 | 256599_at | AT3G14760 | similar to unnamed protein product [Vitis vinifera] (GB:CAO39487.1) | -3.51 |
| 1005 | 257217_at | AT3G14940 | ATPPC3 (PHOSPHOENOLPYRUVATE CARBOXYLASE 3); phosphoenolpyruvate carboxylase | 3.16 |
| 1006 | 257267_at | AT3G15030 | TCP4 (TCP FAMILY TRANSCRIPTION FACTOR 4); transcription factor | -3.18 |
| 1007 | 256856_at | AT3G15110 | similar to unnamed protein product [Vitis vinifera] (GB:CAO39343.1) | -3.07 |
| 1008 | 256855_at | AT3G15190 | chloroplast 30S ribosomal protein S20, putative | -2.79 |
| 1009 | 257079_at | AT3G15240 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G53900.2); similar to unnamed protein product [Vitis vinifera] (GB:CAO39314.1); contains domain gb def: At3g15240 (PTHR13902:SF3); contains domain SERINE/THREONINE-PROTEIN KINASE WNK (WITH NO LYSINE)-RELATED (PTHR13902) | 6.31 |
| 1010 | 257080_at | AT3G15240 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G53900.2); similar to unnamed protein product [Vitis vinifera] (GB:CAO39314.1); contains domain gb def: At3g15240 (PTHR13902:SF3); contains domain SERINE/THREONINE-PROTEIN KINASE WNK (WITH NO LYSINE)-RELATED (PTHR13902) | 6.77 |
| 1011 | 257051_at | AT3G15270 | SPL5 (SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 5); DNA binding / transcription factor | -4.07 |
| 1012 | 258397_at | AT3G15357 | unknown protein | 3.66 |
| 1013 | 258398_at | AT3G15360 | ATHM4 (Arabidopsis thioredoxin M-type 4); thiol-disulfide exchange intermediate | -3.42 |
| 1014 | 258388_at | AT3G15370 | ATEXPA12 (ARABIDOPSIS THALIANA EXPANSIN 12) | 3.18 |
| 1015 | 258386_at | AT3G15520 | peptidyl-prolyl cis-trans isomerase TLP38, chloroplast / thylakoid lumen PPIase of 38 kDa / cyclophilin / rotamase | -3.02 |
| 1016 | 257294_at | AT3G15570 | phototropic-responsive NPH3 family protein | -6.59 |
| 1017 | 258223_at | AT3G15840 | PIFI (POST-ILLUMINATION CHLOROPHYLL FLUORESCENCE INCREASE) | -7.38 |
| 1018 | 258250_at | AT3G15850 | FAD5 (FATTY ACID DESATURASE 5); oxidoreductase | -3.19 |
| 1019 | 257800_at | AT3G15900 | similar to unknown [Populus trichocarpa] (GB:ABK95820.1) | -2.83 |
| 1020 | 257798_at | AT3G15950 | (TSA1-LIKE); protein binding | 5.53 |
| 1021 | 258333_at | AT3G16000 | MFP1 (MAR BINDING FILAMENT-LIKE PROTEIN 1) | -3.36 |
| 1022 | 258336_at | AT3G16050 | A37 (PYRIDOXINE BIOSYNTHESIS 1.2); protein heterodimerization | 3.62 |
| 1023 | 258285_at | AT3G16140 | PSAH-1 (photosystem I subunit H-1) | -7.54 |
| 1024 | 258055_at | AT3G16250 | ferredoxin-related | -6.79 |
| 1025 | 259380_at | AT3G16340 | ATPDR1/PDR1 (PLEIOTROPIC DRUG RESISTANCE 1); ATPase, coupled to transmembrane movement of substances | 2.61 |
| 1026 | 259375_at | AT3G16370 | GDSL-motif lipase/hydrolase family protein | -3.81 |
| 1027 | 259328_at | AT3G16440 | ATMLP-300B (MYROSINASE-BINDING PROTEIN-LIKE PROTEIN-300B) | 4.91 |
| 1028 | 259384_at | AT3G16450 | jacalin lectin family protein | 5.85 |
| 1029 | 259327_at | AT3G16460 | jacalin lectin family protein | 6.04 |
| 1030 | 257205_at | AT3G16520 | UDP-glucoronosyl/UDP-glucosyl transferase family protein | -3.29 |
| 1031 | 257206_at | AT3G16530 | legume lectin family protein | -2.89 |
| 1032 | 258421_at | AT3G16690 | nodulin MtN3 family protein | 2.86 |
| 1033 | 257932_at | AT3G17040 | HCF107 (HIGH CHLOROPHYLL FLUORESCENT 107); binding | -4.94 |
| 1034 | 257874_at | AT3G17110 | pseudogene, glycine-rich protein | 3.96 |
| 1035 | 257295_at | AT3G17420 | GPK1 (Glyoxysomal protein kinase 1); kinase | 2.27 |
| 1036 | 258349_at | AT3G17609 | HYH (HY5-HOMOLOG); DNA binding / transcription factor | -3.48 |
| 1037 | 258377_at | AT3G17690 | ATCNGC19 (cyclic nucleotide gated channel 19); calmodulin binding / cyclic nucleotide binding / ion channel | 2.81 |
| 1038 | 258161_at | AT3G17930 | similar to unknown [Populus trichocarpa] (GB:ABK96142.1) | -3.32 |
| 1039 | 258156_at | AT3G18050 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G28100.1); similar to unknown [Populus trichocarpa] (GB:ABK95654.1) | -2.74 |
| 1040 | 258145_at | AT3G18200 | nodulin MtN21 family protein | 5.83 |
| 1041 | 257066_at | AT3G18280 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | 3.06 |
| 1042 | 257717_at | AT3G18390 | EMB1865 (EMBRYO DEFECTIVE 1865) | -2.83 |
| 1043 | 256655_at | AT3G18890 | binding / catalytic/ coenzyme binding | -3.20 |
| 1044 | 258017_at | AT3G19370 | similar to transport protein-related [Arabidopsis thaliana] (TAIR:AT2G23360.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO47839.1); contains InterPro domain Protein of unknown function DUF869, plant (InterPro:IPR008587) | 2.34 |
| 1045 | 258005_at | AT3G19390 | cysteine proteinase, putative / thiol protease, putative | 5.50 |
| 1046 | 258008_at | AT3G19430 | late embryogenesis abundant protein-related / LEA protein-related | 4.56 |
| 1047 | 258025_at | AT3G19480 | D-3-phosphoglycerate dehydrogenase, putative / 3-PGDH, putative | -5.66 |
| 1048 | 256569_at | AT3G19550 | similar to hypothetical protein [Vitis vinifera] (GB:CAN60715.1) | -3.10 |
| 1049 | 257022_at | AT3G19580 | AZF2 (ARABIDOPSIS ZINC-FINGER PROTEIN 2); nucleic acid binding / transcription factor/ zinc ion binding | 2.43 |
| 1050 | 257017_at | AT3G19620 | glycosyl hydrolase family 3 protein | -3.08 |
| 1051 | 257966_at | AT3G19800 | similar to unnamed protein product [Vitis vinifera] (GB:CAO15360.1) | -6.05 |
| 1052 | 257964_at | AT3G19850 | phototropic-responsive NPH3 family protein | -2.92 |
| 1053 | 257142_at | AT3G20090 | CYP705A18 (cytochrome P450, family 705, subfamily A, polypeptide 18); oxygen binding | 2.22 |
| 1054 | 257113_at | AT3G20130 | CYP705A22 (cytochrome P450, family 705, subfamily A, polypeptide 22); oxygen binding | 2.47 |
| 1055 | 257117_at | AT3G20160 | geranylgeranyl pyrophosphate synthase, putative / GGPP synthetase, putative / farnesyltranstransferase, putative | 3.15 |
| 1056 | 257670_at | AT3G20340 | Expression of the gene is downregulated in the presence of paraquat, an inducer of photoxidative stress. | 2.73 |
| 1057 | 257673_at | AT3G20370 | meprin and TRAF homology domain-containing protein / MATH domain-containing protein | 5.50 |
| 1058 | 257671_at | AT3G20450 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G11905.1); similar to unknown [Populus trichocarpa] (GB:ABK95443.1); similar to Os05g0272900 [Oryza sativa (japonica cultivar-group)] (GB:NP_001055067.1); contains InterPro domain B-cell receptor-associated 31-like; (InterPro:IPR008417) | 2.45 |
| 1059 | 257974_at | AT3G20820 | leucine-rich repeat family protein | -4.02 |
| 1060 | 257986_at | AT3G20865 | AGP40 (ARABINOGALACTAN-PROTEIN 40) | -3.45 |
| 1061 | 256803_at | AT3G20960 | CYP705A33 (cytochrome P450, family 705, subfamily A, polypeptide 33); oxygen binding | 2.31 |
| 1062 | 256979_at | AT3G21055 | PSBTN (photosystem II subunit T) | -8.99 |
| 1063 | 258047_at | AT3G21240 | 4CL2 (4-coumarate:CoA ligase 2); 4-coumarate-CoA ligase | 2.48 |
| 1064 | 256811_at | AT3G21340 | leucine-rich repeat protein kinase, putative | 3.28 |
| 1065 | 258167_at | AT3G21560 | UGT84A2; UDP-glycosyltransferase/ sinapate 1-glucosyltransferase | -3.99 |
| 1066 | 258181_at | AT3G21670 | nitrate transporter (NTP3) | -4.98 |
| 1067 | 258178_at | AT3G21680 | unknown protein | 4.21 |
| 1068 | 257946_at | AT3G21710 | unknown protein | 3.70 |
| 1069 | 257949_at | AT3G21750 | UDP-glucoronosyl/UDP-glucosyl transferase family protein | -2.54 |
| 1070 | 257952_at | AT3G21770 | peroxidase 30 (PER30) (P30) (PRXR9) | 4.69 |
| 1071 | 256792_at | AT3G22150 | pentatricopeptide (PPR) repeat-containing protein | -2.88 |
| 1072 | 256796_at | AT3G22210 | unknown protein | -5.59 |
| 1073 | 256766_at | AT3G22231 | PCC1 (PATHOGEN AND CIRCADIAN CONTROLLED 1) | -4.84 |
| 1074 | 258456_at | AT3G22420 | WNK2 (WITH NO K 2); kinase | -3.87 |
| 1075 | 256935_at | AT3G22570 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | 5.95 |
| 1076 | 256933_at | AT3G22600 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | 3.14 |
| 1077 | 256937_at | AT3G22620 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | 4.95 |
| 1078 | 258325_at | AT3G22830 | AT-HSFA6B (Arabidopsis thaliana heat shock transcription factor A6B); DNA binding / transcription factor | 2.71 |
| 1079 | 258321_at | AT3G22840 | ELIP1 (EARLY LIGHT-INDUCABLE PROTEIN); chlorophyll binding | -2.89 |
| 1080 | 256839_at | AT3G22930 | calmodulin, putative | 2.69 |
| 1081 | 257761_at | AT3G23090 | similar to WDL1 (WVD2-LIKE 1) [Arabidopsis thaliana] (TAIR:AT3G04630.2); similar to WDL1 (WVD2-LIKE 1) [Arabidopsis thaliana] (TAIR:AT3G04630.3); similar to seed specific protein Bn15D14A [Brassica napus] (GB:AAP37969.1); contains InterPro domain Targeting for Xklp2 (InterPro:IPR009675) | 3.54 |
| 1082 | 257924_at | AT3G23190 | lesion inducing protein-related | 4.09 |
| 1083 | 257919_at | AT3G23250 | AtMYB15/AtY19/MYB15 (myb domain protein 15); DNA binding / transcription factor | 5.53 |
| 1084 | 258293_at | AT3G23430 | PHO1 (PHOSPHATE 1) | 4.66 |
| 1085 | 258103_at | AT3G23630 | ATIPT7 (Arabidopsis thaliana isopentenyltransferase 7); transferase, transferring alkyl or aryl (other than methyl) groups | 3.13 |
| 1086 | 257172_at | AT3G23700 | S1 RNA-binding domain-containing protein | -3.81 |
| 1087 | 257171_at | AT3G23760 | similar to transferase, transferring glycosyl groups [Arabidopsis thaliana] (TAIR:AT4G14100.1); similar to unknown [Populus trichocarpa] (GB:ABK94230.1) | -6.16 |
| 1088 | 257197_at | AT3G23800 | selenium-binding family protein | 5.93 |
| 1089 | 257247_at | AT3G24140 | FMA (FAMA); DNA binding / transcription activator/ transcription factor | -2.62 |
| 1090 | 257244_at | AT3G24240 | leucine-rich repeat transmembrane protein kinase, putative | 4.18 |
| 1091 | 257163_at | AT3G24310 | MYB305 (myb domain protein 305); DNA binding / transcription factor | 3.43 |
| 1092 | 257168_at | AT3G24430 | HCF101 (HIGH-CHLOROPHYLL-FLUORESCENCE 101); ATP binding | -3.44 |
| 1093 | 256621_at | AT3G24450 | copper-binding family protein | 2.26 |
| 1094 | 258133_at | AT3G24500 | ATMBF1C/MBF1C (MULTIPROTEIN BRIDGING FACTOR 1C); DNA binding / transcription coactivator/ transcription factor | 4.37 |
| 1095 | 257595_at | AT3G24750 | unknown protein | 4.20 |
| 1096 | 257823_at | AT3G25190 | nodulin, putative | 6.89 |
| 1097 | 257840_at | AT3G25250 | AGC2-1 (OXIDATIVE SIGNAL-INDUCIBLE1); kinase | 3.92 |
| 1098 | 257909_at | AT3G25480 | rhodanese-like domain-containing protein | -2.67 |
| 1099 | 256754_at | AT3G25690 | CHUP1 (CHLOROPLAST UNUSUAL POSITIONING 1) | -3.27 |
| 1100 | 257644_at | AT3G25780 | AOC3 (ALLENE OXIDE CYCLASE 3) | 3.52 |
| 1101 | 257645_at | AT3G25790 | myb family transcription factor | 2.66 |
| 1102 | 258076_at | AT3G25920 | RPL15 (ribosomal protein L15) | -3.37 |
| 1103 | 258080_at | AT3G25930 | universal stress protein (USP) family protein | 6.29 |
| 1104 | 258087_at | AT3G26060 | ATPRX Q; antioxidant/ peroxiredoxin | -5.77 |
| 1105 | 257552_at | AT3G26130 | glycosyl hydrolase family 5 protein / cellulase family protein | 2.45 |
| 1106 | 257636_at | AT3G26200 | CYP71B22 (cytochrome P450, family 71, subfamily B, polypeptide 22); oxygen binding | -3.18 |
| 1107 | 257635_at | AT3G26280 | CYP71B4 (cytochrome P450, family 71, subfamily B, polypeptide 4); oxygen binding | -3.35 |
| 1108 | 257628_at | AT3G26290 | CYP71B26 (cytochrome P450, family 71, subfamily B, polypeptide 26); oxygen binding | -2.65 |
| 1109 | 256874_at | AT3G26320 | CYP71B36 (cytochrome P450, family 71, subfamily B, polypeptide 36); oxygen binding | -3.36 |
| 1110 | 256875_at | AT3G26330 | CYP71B37 (cytochrome P450, family 71, subfamily B, polypeptide 37); oxygen binding | 2.84 |
| 1111 | 256878_at | AT3G26460 | major latex protein-related / MLP-related | 5.32 |
| 1112 | 256877_at | AT3G26470 | similar to ADR1-L1 (ADR1-LIKE 1), ATP binding / protein binding [Arabidopsis thaliana] (TAIR:AT4G33300.2); similar to ADR1-L1 (ADR1-LIKE 1), ATP binding / protein binding [Arabidopsis thaliana] (TAIR:AT4G33300.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO61278.1); contains InterPro domain Disease resistance, plant (InterPro:IPR014011) | 3.63 |
| 1113 | 257617_at | AT3G26550 | DC1 domain-containing protein | 2.87 |
| 1114 | 257311_at | AT3G26570 | PHT2;1 (phosphate transporter 2;1) | -4.62 |
| 1115 | 257611_at | AT3G26580 | binding | -2.82 |
| 1116 | 257613_at | AT3G26610 | polygalacturonase, putative / pectinase, putative | 2.90 |
| 1117 | 257807_at | AT3G26650 | GAPA (GLYCERALDEHYDE 3-PHOSPHATE DEHYDROGENASE A SUBUNIT); glyceraldehyde-3-phosphate dehydrogenase | -8.01 |
| 1118 | 257831_at | AT3G26710 | similar to unnamed protein product [Vitis vinifera] (GB:CAO49107.1) | -3.00 |
| 1119 | 257832_at | AT3G26740 | CCL (CCR-LIKE) | -4.24 |
| 1120 | 258257_at | AT3G26770 | short-chain dehydrogenase/reductase (SDR) family protein | 3.81 |
| 1121 | 257810_at | AT3G27070 | TOM20-1 (TRANSLOCASE OUTER MEMBRANE 20-1) | 2.38 |
| 1122 | 256750_at | AT3G27150 | kelch repeat-containing F-box family protein | 4.31 |
| 1123 | 256753_at | AT3G27160 | GHS1 (GLUCOSE HYPERSENSITIVE 1); structural constituent of ribosome | -3.17 |
| 1124 | 256751_at | AT3G27170 | CLC-B (chloride channel protein B); anion channel/ voltage-gated chloride channel | 2.34 |
| 1125 | 257154_at | AT3G27210 | Identical to Uncharacterized protein At3g27210 (Y-2) [Arabidopsis Thaliana] (GB:Q9LK32); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G40860.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO14583.1) | 2.21 |
| 1126 | 258028_at | AT3G27473 | DC1 domain-containing protein | 2.22 |
| 1127 | 258239_at | AT3G27690 | LHCB2:4 (Photosystem II light harvesting complex gene 2.3); chlorophyll binding | -5.31 |
| 1128 | 257222_at | AT3G27925 | DEGP1 (DEGP PROTEASE 1); serine-type peptidase | -3.31 |
| 1129 | 257300_at | AT3G28080 | nodulin MtN21 family protein | -3.43 |
| 1130 | 256576_at | AT3G28210 | PMZ; zinc ion binding | 2.46 |
| 1131 | 256577_at | AT3G28220 | meprin and TRAF homology domain-containing protein / MATH domain-containing protein | -5.69 |
| 1132 | 256603_at | AT3G28270 | Identical to UPF0496 protein At3g28270 [Arabidopsis Thaliana] (GB:Q9LHD9;GB:Q8H7D0;GB:Q8H7E9;GB:Q944H3); similar to AT14A [Arabidopsis thaliana] (TAIR:AT3G28290.1); similar to AT14A [Arabidopsis thaliana] (TAIR:AT3G28300.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO22906.1); contains InterPro domain Protein of unknown function DUF677 (InterPro:IPR007749) | -6.48 |
| 1133 | 256633_at | AT3G28340 | GATL10 (Galacturonosyltransferase-like 10); polygalacturonate 4-alpha-galacturonosyltransferase/ transferase, transferring hexosyl groups | 2.46 |
| 1134 | 257903_at | AT3G28460 | similar to hypothetical protein [Vitis vinifera] (GB:CAN74970.1); contains InterPro domain Protein of unknown function methylase putative (InterPro:IPR016065) | -2.51 |
| 1135 | 256593_at | AT3G28510 | AAA-type ATPase family protein | 3.36 |
| 1136 | 256596_at | AT3G28540 | AAA-type ATPase family protein | -4.19 |
| 1137 | 257565_at | AT3G28620 | zinc finger (C3HC4-type RING finger) family protein | 2.34 |
| 1138 | 256585_at | AT3G28760 | similar to unnamed protein product [Vitis vinifera] (GB:CAO14940.1); contains InterPro domain 3-dehydroquinate synthase, prokaryotic-type (InterPro:IPR002812) | -2.70 |
| 1139 | 256622_at | AT3G28920 | ATHB34 (ARABIDOPSIS THALIANA HOMEOBOX PROTEIN 34); DNA binding / transcription factor | -5.18 |
| 1140 | 258003_at | AT3G29030 | ATEXPA5 (ARABIDOPSIS THALIANA EXPANSIN A5) | -5.42 |
| 1141 | 258059_at | AT3G29035 | ANAC059/ATNAC3 (Arabidopsis NAC domain containing protein 59); protein heterodimerization/ transcription factor | 2.43 |
| 1142 | 258192_at | AT3G29110 | terpene synthase/cyclase family protein | 2.40 |
| 1143 | 257774_at | AT3G29250 | oxidoreductase | 7.98 |
| 1144 | 256736_at | AT3G29410 | terpene synthase/cyclase family protein | 7.05 |
| 1145 | 256738_at | AT3G29430 | geranylgeranyl pyrophosphate synthase, putative / GGPP synthetase, putative / farnesyltranstransferase, putative | 7.36 |
| 1146 | 256563_at | AT3G29780 | RALFL27 (RALF-LIKE 27) | 5.29 |
| 1147 | 256703_at | AT3G30260 | MADS-box protein (AGL79) | 5.84 |
| 1148 | 256710_at | AT3G30350 | unknown protein | 4.03 |
| 1149 | 256940_at | AT3G30720 | unknown protein | -3.13 |
| 1150 | 256684_at | AT3G32040 | geranylgeranyl pyrophosphate synthase, putative / GGPP synthetase, putative / farnesyltranstransferase, putative | 4.01 |
| 1151 | 252746_at | AT3G43190 | SUS4; UDP-glycosyltransferase/ sucrose synthase/ transferase, transferring glycosyl groups | 2.81 |
| 1152 | 252711_at | AT3G43720 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | -2.95 |
| 1153 | 252688_at | AT3G44020 | thylakoid lumenal P17.1 protein | -3.90 |
| 1154 | 252679_at | AT3G44260 | CCR4-NOT transcription complex protein, putative | 2.72 |
| 1155 | 252677_at | AT3G44320 | NIT3 (NITRILASE 3) | 3.06 |
| 1156 | 252661_at | AT3G44450 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G52740.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO70571.1) | -3.12 |
| 1157 | 252636_at | AT3G44510 | similar to esterase/lipase/thioesterase family protein [Arabidopsis thaliana] (TAIR:AT1G08310.1); similar to esterase/lipase/thioesterase family protein [Arabidopsis thaliana] (TAIR:AT1G08310.2); similar to hypothetical protein [Vitis vinifera] (GB:CAN60741.1); contains domain SSF53474 (SSF53474) | 5.21 |
| 1158 | 252638_at | AT3G44540 | oxidoreductase, acting on the CH-CH group of donors | 3.19 |
| 1159 | 252648_at | AT3G44630 | disease resistance protein RPP1-WsB-like (TIR-NBS-LRR class), putative | -2.79 |
| 1160 | 246335_at | AT3G44880 | ACD1 (ACCELERATED CELL DEATH 1) | -4.53 |
| 1161 | 246339_at | AT3G44890 | RPL9 (ribosomal protein L9); structural constituent of ribosome | -3.09 |
| 1162 | 252607_at | AT3G44990 | XTR8 (xyloglucan:xyloglucosyl transferase 8); hydrolase, acting on glycosyl bonds | 3.91 |
| 1163 | 252604_at | AT3G45060 | ATNRT2.6 (Arabidopsis thaliana high affinity nitrate transporter 2.6); nitrate transmembrane transporter | 3.35 |
| 1164 | 252618_at | AT3G45140 | LOX2 (LIPOXYGENASE 2) | -8.02 |
| 1165 | 252612_at | AT3G45160 | contains domain PROKAR_LIPOPROTEIN (PS51257) | 3.02 |
| 1166 | 252568_at | AT3G45410 | lectin protein kinase family protein | 2.28 |
| 1167 | 252536_at | AT3G45700 | proton-dependent oligopeptide transport (POT) family protein | 4.45 |
| 1168 | 252537_at | AT3G45710 | proton-dependent oligopeptide transport (POT) family protein | 6.48 |
| 1169 | 252543_at | AT3G45780 | PHOT1 (phototropin 1); kinase | -2.92 |
| 1170 | 252557_at | AT3G45960 | ATEXLA3 (ARABIDOPSIS THALIANA EXPANSIN-LIKE A3) | 5.13 |
| 1171 | 252534_at | AT3G46130 | MYB111 (myb domain protein 111); DNA binding / transcription factor | 2.42 |
| 1172 | 252505_at | AT3G46170 | short-chain dehydrogenase/reductase (SDR) family protein | 2.41 |
| 1173 | 252515_at | AT3G46230 | ATHSP17.4 (Arabidopsis thaliana heat shock protein 17.4) | 5.13 |
| 1174 | 252511_at | AT3G46280 | protein kinase-related | 4.14 |
| 1175 | 252530_at | AT3G46500 | oxidoreductase, 2OG-Fe(II) oxygenase family protein | 4.37 |
| 1176 | 252485_at | AT3G46530 | RPP13 (RECOGNITION OF PERONOSPORA PARASITICA 13); ATP binding | -6.80 |
| 1177 | 252484_at | AT3G46690 | UDP-glucoronosyl/UDP-glucosyl transferase family protein | 3.64 |
| 1178 | 252488_at | AT3G46700 | UDP-glycosyltransferase/ transferase, transferring glycosyl groups | 3.52 |
| 1179 | 252490_at | AT3G46720 | UDP-glucoronosyl/UDP-glucosyl transferase family protein | 2.23 |
| 1180 | 252441_at | AT3G46780 | PTAC16 (PLASTID TRANSCRIPTIONALLY ACTIVE18); binding / catalytic | -9.48 |
| 1181 | 252468_at | AT3G46970 | ATPHS2/PHS2 (ALPHA-GLUCAN PHOSPHORYLASE 2); phosphorylase/ transferase, transferring glycosyl groups | -2.79 |
| 1182 | 252447_at | AT3G47040 | glycosyl hydrolase family 3 protein | 2.56 |
| 1183 | 252463_at | AT3G47070 | similar to unknown [Populus trichocarpa] (GB:ABK95428.1) | -6.62 |
| 1184 | 252458_at | AT3G47210 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G47250.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G47250.3); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G47250.2); similar to unnamed protein product [Vitis vinifera] (GB:CAO15956.1); contains InterPro domain Protein of unknown function DUF247, plant (InterPro:IPR004158) | 4.35 |
| 1185 | 252462_at | AT3G47250 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G47200.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G47200.2); similar to unnamed protein product [Vitis vinifera] (GB:CAO15956.1); contains InterPro domain Protein of unknown function DUF247, plant (InterPro:IPR004158) | -4.28 |
| 1186 | 252412_at | AT3G47295 | unknown protein | -4.18 |
| 1187 | 252411_at | AT3G47430 | PEX11B | -3.87 |
| 1188 | 252430_at | AT3G47470 | LHCA4 (Photosystem I light harvesting complex gene 4); chlorophyll binding | -6.15 |
| 1189 | 252409_at | AT3G47650 | bundle-sheath defective protein 2 family / bsd2 family | -4.79 |
| 1190 | 252391_at | AT3G47860 | apolipoprotein D-related | -3.69 |
| 1191 | 252353_at | AT3G48200 | similar to hypothetical protein OsI_020499 [Oryza sativa (indica cultivar-group)] (GB:EAY99266.1); similar to hypothetical protein OsJ_018975 [Oryza sativa (japonica cultivar-group)] (GB:EAZ35492.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO64300.1) | -4.00 |
| 1192 | 252366_at | AT3G48420 | haloacid dehalogenase-like hydrolase family protein | -6.40 |
| 1193 | 252368_at | AT3G48520 | CYP94B3 (cytochrome P450, family 94, subfamily B, polypeptide 3); oxygen binding | 2.44 |
| 1194 | 252315_at | AT3G48690 | ATCXE12 (ARABIDOPSIS THALIANA CARBOXYESTERASE 12); carboxylesterase | -3.53 |
| 1195 | 252317_at | AT3G48720 | transferase family protein | -3.17 |
| 1196 | 252318_at | AT3G48730 | GSA2 (GLUTAMATE-1-SEMIALDEHYDE 2,1-AMINOMUTASE 2); glutamate-1-semialdehyde 2,1-aminomutase | -2.94 |
| 1197 | 252327_at | AT3G48740 | nodulin MtN3 family protein | -2.90 |
| 1198 | 252302_at | AT3G49190 | condensation domain-containing protein | 2.82 |
| 1199 | 252280_at | AT3G49260 | IQD21 (IQ-DOMAIN 21, IQ-domain 21); calmodulin binding | -3.47 |
| 1200 | 252265_at | AT3G49620 | DIN11 (DARK INDUCIBLE 11); oxidoreductase | 4.03 |
| 1201 | 252248_at | AT3G49750 | leucine-rich repeat family protein | 2.21 |
| 1202 | 252222_at | AT3G49845 | contains InterPro domain XYPPX repeat (InterPro:IPR006031) | 5.51 |
| 1203 | 252224_at | AT3G49860 | ATARLA1B (ADP-ribosylation factor-like A1B); GTP binding | 4.33 |
| 1204 | 252227_at | AT3G49900 | BTB/POZ domain-containing protein | -2.64 |
| 1205 | 252240_at | AT3G50010 | DC1 domain-containing protein | 2.43 |
| 1206 | 252195_at | AT3G50190 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G50140.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G50130.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO71911.1); contains InterPro domain Protein of unknown function DUF247, plant (InterPro:IPR004158) | 5.19 |
| 1207 | 252211_at | AT3G50220 | nucleic acid binding / pancreatic ribonuclease | 2.37 |
| 1208 | 252199_at | AT3G50270 | transferase family protein | -3.41 |
| 1209 | 252202_at | AT3G50300 | transferase family protein | 4.56 |
| 1210 | 252168_at | AT3G50440 | hydrolase | -4.99 |
| 1211 | 252172_at | AT3G50640 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G66800.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN61148.1) | 3.46 |
| 1212 | 252181_at | AT3G50685 | similar to unnamed protein product [Vitis vinifera] (GB:CAO22868.1) | -3.12 |
| 1213 | 252130_at | AT3G50820 | PSBO-2/PSBO2 (PHOTOSYSTEM II SUBUNIT O-2); oxygen evolving/ poly(U) binding | -9.34 |
| 1214 | 252129_at | AT3G50890 | ATHB28 (ARABIDOPSIS THALIANA HOMEOBOX PROTEIN 28); DNA binding / transcription factor | -3.53 |
| 1215 | 252131_at | AT3G50930 | AAA-type ATPase family protein | 3.08 |
| 1216 | 252123_at | AT3G51240 | F3H (TRANSPARENT TESTA 6); naringenin 3-dioxygenase | -4.67 |
| 1217 | 252098_at | AT3G51330 | aspartyl protease family protein | 3.77 |
| 1218 | 252092_at | AT3G51420 | strictosidine synthase family protein | -6.23 |
| 1219 | 252116_at | AT3G51510 | similar to unnamed protein product [Vitis vinifera] (GB:CAO14460.1) | -5.38 |
| 1220 | 252070_at | AT3G51680 | short-chain dehydrogenase/reductase (SDR) family protein | 4.55 |
| 1221 | 246308_at | AT3G51820 | ATG4/CHLG/G4 (CHLOROPHYLL SYNTHASE); chlorophyll synthetase | -5.70 |
| 1222 | 246310_at | AT3G51895 | SULTR3;1 (SULFATE TRANSPORTER 1); sulfate transmembrane transporter | -3.80 |
| 1223 | 252081_at | AT3G51910 | AT-HSFA7A (Arabidopsis thaliana heat shock transcription factor A7A); DNA binding / transcription factor | 5.39 |
| 1224 | 252032_at | AT3G52150 | RNA recognition motif (RRM)-containing protein | -4.19 |
| 1225 | 256676_at | AT3G52180 | ATPTPKIS1/DSP4/SEX4 (STARCH-EXCESS 4); polysaccharide binding / protein tyrosine/serine/threonine phosphatase | -3.31 |
| 1226 | 256678_at | AT3G52380 | CP33 (PIGMENT DEFECTIVE 322); RNA binding | -3.31 |
| 1227 | 252045_at | AT3G52450 | U-box domain-containing protein | 4.03 |
| 1228 | 252011_at | AT3G52720 | carbonic anhydrase family protein | -5.54 |
| 1229 | 252010_at | AT3G52740 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G44450.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO70571.1) | -5.49 |
| 1230 | 252017_at | AT3G52970 | CYP76G1 (cytochrome P450, family 76, subfamily G, polypeptide 1); oxygen binding | 3.31 |
| 1231 | 251969_at | AT3G53130 | LUT1 (LUTEIN DEFICIENT 1); oxygen binding | -3.42 |
| 1232 | 251987_at | AT3G53280 | CYP71B5 (CYTOCHROME P450 71B5); oxygen binding | -5.77 |
| 1233 | 251941_at | AT3G53470 | similar to unnamed protein product [Vitis vinifera] (GB:CAO18045.1) | -2.69 |
| 1234 | 251942_at | AT3G53480 | ATPDR9/PDR9 (PLEIOTROPIC DRUG RESISTANCE 9); ATPase, coupled to transmembrane movement of substances | 5.05 |
| 1235 | 251906_at | AT3G53720 | ATCHX20 (CATION/H+ EXCHANGER 20); monovalent cation:proton antiporter | -4.06 |
| 1236 | 251929_at | AT3G53920 | SIGC (RNA polymerase sigma subunit C); DNA binding / DNA-directed RNA polymerase/ transcription factor | -2.48 |
| 1237 | 251928_at | AT3G53980 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | 5.79 |
| 1238 | 251918_at | AT3G54040 | photoassimilate-responsive protein-related | 2.79 |
| 1239 | 251885_at | AT3G54050 | fructose-1,6-bisphosphatase, putative / D-fructose-1,6-bisphosphate 1-phosphohydrolase, putative / FBPase, putative | -7.39 |
| 1240 | 251935_at | AT3G54090 | pfkB-type carbohydrate kinase family protein | -2.55 |
| 1241 | 251883_at | AT3G54210 | ribosomal protein L17 family protein | -2.98 |
| 1242 | 251886_at | AT3G54260 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G42570.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO70127.1); contains InterPro domain Protein of unknown function DUF231, plant (InterPro:IPR004253) | 3.48 |
| 1243 | 251878_at | AT3G54310 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G38430.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO70793.1) | 2.36 |
| 1244 | 251857_at | AT3G54770 | RNA recognition motif (RRM)-containing protein | 6.62 |
| 1245 | 251814_at | AT3G54890 | LHCA1; chlorophyll binding | -7.32 |
| 1246 | 251824_at | AT3G55090 | ATPase, coupled to transmembrane movement of substances | 3.18 |
| 1247 | 251826_at | AT3G55110 | ABC transporter family protein | -2.86 |
| 1248 | 251827_at | AT3G55120 | A11/CFI/TT5 (TRANSPARENT TESTA 5); chalcone isomerase | -3.43 |
| 1249 | 251785_at | AT3G55130 | ATWBC19 (WHITE-BROWN COMPLEX HOMOLOG 19); ATPase, coupled to transmembrane movement of substances | -4.69 |
| 1250 | 251784_at | AT3G55330 | PPL1 (PSBP-LIKE PROTEIN 1); calcium ion binding | -5.58 |
| 1251 | 251759_at | AT3G55630 | ATDFD (A. THALIANA DHFS-FPGS HOMOLOG D); tetrahydrofolylpolyglutamate synthase | -3.88 |
| 1252 | 251762_at | AT3G55800 | SBPASE (SEDOHEPTULOSE-BISPHOSPHATASE); phosphoric ester hydrolase/ sedoheptulose-bisphosphatase | -7.47 |
| 1253 | 251770_at | AT3G55970 | oxidoreductase, 2OG-Fe(II) oxygenase family protein | -2.73 |
| 1254 | 251713_at | AT3G56080 | dehydration-responsive protein-related | -2.58 |
| 1255 | 251723_at | AT3G56230 | speckle-type POZ protein-related | 2.38 |
| 1256 | 251725_at | AT3G56260 | unknown protein | -2.88 |
| 1257 | 251727_at | AT3G56290 | similar to hypothetical protein [Vitis vinifera] (GB:CAN75527.1) | -4.11 |
| 1258 | 251701_at | AT3G56650 | thylakoid lumenal 20 kDa protein | -2.94 |
| 1259 | 246344_at | AT3G56730 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G62050.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO17351.1); contains InterPro domain Protein of unknown function DUF537 (InterPro:IPR007491) | 2.26 |
| 1260 | 246294_at | AT3G56910 | PSRP5 (PLASTID-SPECIFIC 50S RIBOSOMAL PROTEIN 5) | -2.61 |
| 1261 | 251664_at | AT3G56940 | AT103 (DICARBOXYLATE DIIRON 1) | -6.96 |
| 1262 | 251668_at | AT3G57010 | strictosidine synthase family protein | 4.49 |
| 1263 | 251658_at | AT3G57020 | strictosidine synthase family protein | -3.35 |
| 1264 | 251669_at | AT3G57180 | GTP binding | -4.23 |
| 1265 | 251670_at | AT3G57190 | peptide chain release factor, putative | -2.81 |
| 1266 | 251625_at | AT3G57260 | BGL2 (PATHOGENESIS-RELATED PROTEIN 2); glucan 1,3-beta-glucosidase/ hydrolase, hydrolyzing O-glycosyl compounds | -3.38 |
| 1267 | 251647_at | AT3G57770 | protein kinase, putative | -3.37 |
| 1268 | 251612_at | AT3G57950 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G42180.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN83225.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO23380.1) | 4.15 |
| 1269 | 251531_at | AT3G58550 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | 3.81 |
| 1270 | 251553_at | AT3G58710 | WRKY69 (WRKY DNA-binding protein 69); transcription factor | 2.35 |
| 1271 | 251545_at | AT3G58810 | MTPA2; efflux transmembrane transporter/ zinc ion transmembrane transporter | 2.72 |
| 1272 | 251497_at | AT3G59060 | PIL6 (PHYTOCHROME-INTERACTING FACTOR 5); DNA binding / transcription factor | -7.14 |
| 1273 | 251466_at | AT3G59340 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G59310.1); similar to Os05g0299500 [Oryza sativa (japonica cultivar-group)] (GB:NP_001055130.1); similar to hypothetical protein OsI_018658 [Oryza sativa (indica cultivar-group)] (GB:EAY97425.1); contains InterPro domain Protein of unknown function DUF914, eukaryotic (InterPro:IPR009262) | 3.06 |
| 1274 | 251517_at | AT3G59370 | contains domain PTHR22683 (PTHR22683) | 5.64 |
| 1275 | 251519_at | AT3G59400 | GUN4 (Genomes uncoupled 4) | -10.62 |
| 1276 | 251478_at | AT3G59690 | IQD13 (IQ-domain 13); calmodulin binding | 2.53 |
| 1277 | 251434_at | AT3G59850 | polygalacturonase, putative / pectinase, putative | 4.57 |
| 1278 | 251443_at | AT3G59940 | kelch repeat-containing F-box family protein | -2.76 |
| 1279 | 251400_at | AT3G60420 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G60450.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO70569.1); contains InterPro domain Phosphoglycerate mutase (InterPro:IPR013078); contains InterPro domain PRIB5 (InterPro:IPR012398) | -3.34 |
| 1280 | 251423_at | AT3G60550 | CYCP3;2 (cyclin p3;2); cyclin-dependent protein kinase | 3.91 |
| 1281 | 251380_at | AT3G60700 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G70040.1); contains InterPro domain Protein of unknown function DUF1163 (InterPro:IPR009544) | 2.65 |
| 1282 | 251353_at | AT3G61080 | fructosamine kinase family protein | -3.52 |
| 1283 | 251336_at | AT3G61190 | BAP1 (BON ASSOCIATION PROTEIN 1) | 4.20 |
| 1284 | 251309_at | AT3G61220 | short-chain dehydrogenase/reductase (SDR) family protein | -4.12 |
| 1285 | 251364_at | AT3G61300 | C2 domain-containing protein | 2.72 |
| 1286 | 251315_at | AT3G61410 | similar to protein kinase family protein / U-box domain-containing protein [Arabidopsis thaliana] (TAIR:AT2G45910.1); similar to Protein kinase; U box [Medicago truncatula] (GB:ABD32822.1); contains domain Adenine nucleotide alpha hydrolases-like (SSF52402) | 3.42 |
| 1287 | 251330_at | AT3G61550 | zinc finger (C3HC4-type RING finger) family protein | -2.60 |
| 1288 | 251274_at | AT3G61700 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G46420.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO39750.1); contains InterPro domain Conserved hypothetical protein CHP01589, plant (InterPro:IPR006476) | -4.90 |
| 1289 | 251243_at | AT3G61870 | similar to unknown [Populus trichocarpa] (GB:ABK95718.1) | -5.24 |
| 1290 | 251304_at | AT3G61990 | O-methyltransferase family 3 protein | 4.09 |
| 1291 | 251305_at | AT3G62030 | ROC4 (ROTAMASE CYP 4); peptidyl-prolyl cis-trans isomerase | -3.17 |
| 1292 | 251298_at | AT3G62040 | hydrolase | 5.35 |
| 1293 | 251259_at | AT3G62260 | protein phosphatase 2C, putative / PP2C, putative | 3.19 |
| 1294 | 251255_at | AT3G62280 | carboxylesterase | 3.17 |
| 1295 | 251218_at | AT3G62410 | CP12-2 | -6.78 |
| 1296 | 251221_at | AT3G62550 | universal stress protein (USP) family protein | -5.55 |
| 1297 | 251226_at | AT3G62680 | PRP3 (PROLINE-RICH PROTEIN 3); structural constituent of cell wall | 5.59 |
| 1298 | 251231_at | AT3G62760 | ATGSTF13 (Arabidopsis thaliana Glutathione S-transferase (class phi) 13); glutathione transferase | 4.79 |
| 1299 | 251181_at | AT3G62820 | invertase/pectin methylesterase inhibitor family protein | -2.72 |
| 1300 | 251193_at | AT3G62910 | APG3 (ALBINO AND PALE GREEN); translation release factor | -3.63 |
| 1301 | 251157_at | AT3G63140 | mRNA-binding protein, putative | -7.13 |
| 1302 | 251155_at | AT3G63160 | unknown protein | -8.91 |
| 1303 | 251172_at | AT3G63190 | RRF (RIBOSOME RECYCLING FACTOR, CHLOROPLAST PRECURSOR) | -2.54 |
| 1304 | 251166_at | AT3G63350 | AT-HSFA7B (Arabidopsis thaliana heat shock transcription factor A7B); DNA binding / transcription factor | 5.71 |
| 1305 | 251167_at | AT3G63360 | defensin-related | 3.08 |
| 1306 | 251118_at | AT3G63410 | APG1 (ALBINO OR PALE GREEN MUTANT 1); methyltransferase | -2.79 |
| 1307 | 251116_at | AT3G63470 | SCPL40 (serine carboxypeptidase-like 40); serine carboxypeptidase | 2.35 |
| 1308 | 251120_at | AT3G63490 | ribosomal protein L1 family protein | -3.84 |
| 1309 | 251146_at | AT3G63520 | CCD1 (CAROTENOID CLEAVAGE DIOXYGENASE 1) | -2.75 |
| 1310 | 255694_at | AT4G00050 | UNE10 (unfertilized embryo sac 10); DNA binding / transcription factor | -2.78 |
| 1311 | 255695_at | AT4G00080 | UNE11 (unfertilized embryo sac 11); pectinesterase inhibitor | 3.79 |
| 1312 | 255709_at | AT4G00180 | YAB3 (YABBY3); transcription factor | -3.55 |
| 1313 | 255690_at | AT4G00360 | CYP86A2 (ABERRANT INDUCTION OF TYPE THREE GENES 1); oxygen binding | -2.50 |
| 1314 | 255691_at | AT4G00370 | ANTR2 (anion transporter 2); organic anion transmembrane transporter | -2.88 |
| 1315 | 255632_at | AT4G00680 | actin-depolymerizing factor, putative | 5.13 |
| 1316 | 255637_at | AT4G00750 | dehydration-responsive family protein | -3.44 |
| 1317 | 255659_at | AT4G00895 | ATP synthase delta chain-related | -4.60 |
| 1318 | 255648_at | AT4G00910 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G08440.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO14409.1); contains InterPro domain Protein of unknown function UPF0005 (InterPro:IPR006214) | 3.33 |
| 1319 | 255601_at | AT4G01030 | pentatricopeptide (PPR) repeat-containing protein | -2.85 |
| 1320 | 255572_at | AT4G01050 | hydroxyproline-rich glycoprotein family protein | -5.45 |
| 1321 | 255607_at | AT4G01130 | acetylesterase, putative | -3.10 |
| 1322 | 255567_at | AT4G01150 | Identical to Uncharacterized protein At4g01150, chloroplast precursor [Arabidopsis Thaliana] (GB:O04616;GB:Q38835); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G38100.1); similar to unknown [Populus trichocarpa] (GB:ABK95729.1) | -5.04 |
| 1323 | 255623_at | AT4G01310 | ribosomal protein L5 family protein | -2.86 |
| 1324 | 255621_at | AT4G01390 | meprin and TRAF homology domain-containing protein / MATH domain-containing protein | 3.78 |
| 1325 | 255578_at | AT4G01450 | nodulin MtN21 family protein | 2.45 |
| 1326 | 255579_at | AT4G01460 | basic helix-loop-helix (bHLH) family protein | -3.21 |
| 1327 | 255591_at | AT4G01630 | ATEXPA17 (ARABIDOPSIS THALIANA EXPANSIN A17) | 2.60 |
| 1328 | 255528_at | AT4G02090 | similar to unnamed protein product [Vitis vinifera] (GB:CAO40099.1) | 5.86 |
| 1329 | 255516_at | AT4G02270 | pollen Ole e 1 allergen and extensin family protein | 2.48 |
| 1330 | 255517_at | AT4G02290 | ATGH9B13 (ARABIDOPSIS THALIANA GLYCOSYL HYDROLASE 9B13); hydrolase, hydrolyzing O-glycosyl compounds | 2.58 |
| 1331 | 255503_at | AT4G02420 | lectin protein kinase, putative | -3.87 |
| 1332 | 255440_at | AT4G02530 | chloroplast thylakoid lumen protein | -3.50 |
| 1333 | 255457_at | AT4G02770 | PSAD-1 (photosystem I subunit D-1) | -8.73 |
| 1334 | 255447_at | AT4G02790 | GTP-binding family protein | -3.04 |
| 1335 | 255450_at | AT4G02850 | phenazine biosynthesis PhzC/PhzF family protein | 2.69 |
| 1336 | 255436_at | AT4G03150 | similar to unnamed protein product [Vitis vinifera] (GB:CAO22424.1) | -3.87 |
| 1337 | 255435_at | AT4G03280 | PETC (PHOTOSYNTHETIC ELECTRON TRANSFER C) | -7.35 |
| 1338 | 255403_at | AT4G03400 | DFL2 (DWARF IN LIGHT 2) | -2.69 |
| 1339 | 255385_at | AT4G03610 | phosphonate metabolism protein-related | 2.76 |
| 1340 | 255331_at | AT4G04330 | similar to unnamed protein product [Vitis vinifera] (GB:CAO69665.1) | -3.94 |
| 1341 | 255340_at | AT4G04490 | protein kinase family protein | -3.76 |
| 1342 | 255290_at | AT4G04640 | ATPC1 (ATP synthase gamma chain 1) | -5.26 |
| 1343 | 255302_at | AT4G04830 | methionine sulfoxide reductase domain-containing protein / SeIR domain-containing protein | 2.41 |
| 1344 | 255298_at | AT4G04840 | methionine sulfoxide reductase domain-containing protein / SeIR domain-containing protein | -3.19 |
| 1345 | 255304_at | AT4G04850 | KEA3 (K+ efflux antiporter 3); potassium:hydrogen antiporter | -2.63 |
| 1346 | 255277_at | AT4G04890 | PDF2 (PROTODERMAL FACTOR2); DNA binding / transcription factor | -3.36 |
| 1347 | 255252_at | AT4G04990 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G61260.1); contains InterPro domain Protein of unknown function DUF761, plant (InterPro:IPR008480) | 2.90 |
| 1348 | 255248_at | AT4G05180 | PSBQ/PSBQ-2/PSII-Q (PHOTOSYSTEM II SUBUNIT Q-2); calcium ion binding | -6.96 |
| 1349 | 255266_at | AT4G05200 | protein kinase family protein | 3.43 |
| 1350 | 255160_at | AT4G07820 | pathogenesis-related protein, putative | 4.55 |
| 1351 | 255177_at | AT4G08040 | ACS11 (1-Amino-cyclopropane-1-carboxylate synthase 11); 1-aminocyclopropane-1-carboxylate synthase | 3.25 |
| 1352 | 255127_at | AT4G08300 | nodulin MtN21 family protein | 4.15 |
| 1353 | 255110_at | AT4G08770 | peroxidase, putative | 4.83 |
| 1354 | 255111_at | AT4G08780 | peroxidase, putative | 3.43 |
| 1355 | 255078_at | AT4G09010 | APX4 (ASCORBATE PEROXIDASE 4); peroxidase | -5.42 |
| 1356 | 255074_at | AT4G09100 | zinc finger (C3HC4-type RING finger) family protein | 2.31 |
| 1357 | 255088_at | AT4G09350 | DNAJ heat shock N-terminal domain-containing protein | -4.10 |
| 1358 | 255046_at | AT4G09650 | ATP synthase delta chain, chloroplast, putative / H(+)-transporting two-sector ATPase, delta (OSCP) subunit, putative | -5.52 |
| 1359 | 254998_at | AT4G09760 | choline kinase, putative | -3.21 |
| 1360 | 255025_at | AT4G09900 | hydrolase, alpha/beta fold family protein | -6.07 |
| 1361 | 255008_at | AT4G10060 | catalytic | -4.05 |
| 1362 | 255016_at | AT4G10120 | ATSPS4F; sucrose-phosphate synthase/ transferase, transferring glycosyl groups | -5.65 |
| 1363 | 255809_at | AT4G10300 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G04300.1); similar to unknown [Populus trichocarpa] (GB:ABK95374.1); contains InterPro domain RmlC-like jelly roll fold (InterPro:IPR014710); contains InterPro domain Protein of unknown function DUF861, cupin-3 (InterPro:IPR008579); contains InterPro domain Cupin, RmlC-type (InterPro:IPR011051) | -3.27 |
| 1364 | 255812_at | AT4G10310 | HKT1 (HIGH-AFFINITY K+ TRANSPORTER 1); sodium ion transmembrane transporter | 5.19 |
| 1365 | 254970_at | AT4G10340 | LHCB5 (LIGHT HARVESTING COMPLEX OF PHOTOSYSTEM II 5); chlorophyll binding | -7.49 |
| 1366 | 254971_at | AT4G10380 | NIP5;1/NLM6/NLM8 (NOD26-like intrinsic protein 5;1); boron transporter/ water channel | 3.96 |
| 1367 | 254911_at | AT4G11100 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G03060.1) | -5.24 |
| 1368 | 254910_at | AT4G11175 | translation initiation factor IF-1, chloroplast, putative | -2.58 |
| 1369 | 254907_at | AT4G11190 | disease resistance-responsive family protein / dirigent family protein | 7.07 |
| 1370 | 254909_at | AT4G11210 | disease resistance-responsive family protein / dirigent family protein | 4.25 |
| 1371 | 254926_at | AT4G11280 | ACS6 (1-AMINOCYCLOPROPANE-1-CARBOXYLIC ACID (ACC) SYNTHASE 6) | 3.24 |
| 1372 | 254914_at | AT4G11290 | peroxidase, putative | 4.93 |
| 1373 | 254889_at | AT4G11650 | ATOSM34 (OSMOTIN 34) | 6.96 |
| 1374 | 254886_at | AT4G11760 | LCR17 (Low-molecular-weight cysteine-rich 17) | -2.48 |
| 1375 | 254888_at | AT4G11780 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G23020.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN60407.1) | 3.73 |
| 1376 | 254896_at | AT4G11880 | AGL14 (AGAMOUS-LIKE 14); DNA binding / transcription factor | 7.12 |
| 1377 | 254853_at | AT4G12080 | DNA-binding family protein | 4.79 |
| 1378 | 254839_at | AT4G12400 | stress-inducible protein, putative | 4.21 |
| 1379 | 254828_at | AT4G12550 | AIR1 (Auxin-Induced in Root cultures 1); lipid binding | 9.20 |
| 1380 | 254790_at | AT4G12800 | PSAL (photosystem I subunit L) | -8.30 |
| 1381 | 254783_at | AT4G12830 | hydrolase, alpha/beta fold family protein | -2.87 |
| 1382 | 254794_at | AT4G12970 | similar to unnamed protein product [Vitis vinifera] (GB:CAO17947.1) | -7.42 |
| 1383 | 254804_at | AT4G13010 | oxidoreductase, zinc-binding dehydrogenase family protein | -3.04 |
| 1384 | 254797_at | AT4G13030 | similar to unnamed protein product [Vitis vinifera] (GB:CAO18001.1); contains domain SSF52540 (SSF52540) | 2.37 |
| 1385 | 254759_at | AT4G13180 | short-chain dehydrogenase/reductase (SDR) family protein | 2.65 |
| 1386 | 254764_at | AT4G13250 | short-chain dehydrogenase/reductase (SDR) family protein | -4.27 |
| 1387 | 254772_at | AT4G13390 | proline-rich extensin-like family protein | 2.37 |
| 1388 | 254715_at | AT4G13550 | lipase class 3 family protein | -3.46 |
| 1389 | 254716_at | AT4G13560 | UNE15 (unfertilized embryo sac 15) | -4.79 |
| 1390 | 254718_at | AT4G13580 | disease resistance-responsive family protein | 7.76 |
| 1391 | 254726_at | AT4G13660 | pinoresinol-lariciresinol reductase, putative | 5.56 |
| 1392 | 254727_at | AT4G13670 | PTAC5 (PLASTID TRANSCRIPTIONALLY ACTIVE5); heat shock protein binding / unfolded protein binding | -3.03 |
| 1393 | 254737_at | AT4G13840 | transferase family protein | -3.56 |
| 1394 | 245386_at | AT4G14010 | RALFL32 (RALF-LIKE 32) | 2.86 |
| 1395 | 245267_at | AT4G14060 | major latex protein-related / MLP-related | 6.34 |
| 1396 | 245621_at | AT4G14070 | AAE15 (ACYL-ACTIVATING ENZYME 15); long-chain-fatty-acid-[acyl-carrier-protein] ligase | -2.65 |
| 1397 | 245325_at | AT4G14130 | XTR7 (XYLOGLUCAN ENDOTRANSGLYCOSYLASE 7); hydrolase, acting on glycosyl bonds | 3.65 |
| 1398 | 245265_at | AT4G14400 | ACD6 (ACCELERATED CELL DEATH 6); protein binding | -2.88 |
| 1399 | 245328_at | AT4G14465 | DNA-binding protein-related | 3.13 |
| 1400 | 245592_at | AT4G14540 | CCAAT-box binding transcription factor subunit B (NF-YB) (HAP3 ) (AHAP3) family | -5.59 |
| 1401 | 245567_at | AT4G14630 | GLP9 (GERMIN-LIKE PROTEIN 9); manganese ion binding / metal ion binding / nutrient reservoir | 4.37 |
| 1402 | 245306_at | AT4G14690 | ELIP2 (EARLY LIGHT-INDUCIBLE PROTEIN 2); chlorophyll binding | -2.77 |
| 1403 | 245396_at | AT4G14870 | P-P-bond-hydrolysis-driven protein transmembrane transporter | -2.68 |
| 1404 | 245347_at | AT4G14890 | ferredoxin family protein | -3.64 |
| 1405 | 245586_at | AT4G14980 | DC1 domain-containing protein | 4.42 |
| 1406 | 245275_at | AT4G15210 | ATBETA-AMY (BETA-AMYLASE); beta-amylase | -5.27 |
| 1407 | 245550_at | AT4G15330 | CYP705A1 (cytochrome P450, family 705, subfamily A, polypeptide 1); oxygen binding | 3.63 |
| 1408 | 245258_at | AT4G15340 | ATPEN1 (Arabidopsis thaliana pentacyclic triterpene synthase 1); catalytic/ lyase | 5.61 |
| 1409 | 245554_at | AT4G15380 | CYP705A4 (cytochrome P450, family 705, subfamily A, polypeptide 4); oxygen binding | 2.69 |
| 1410 | 245555_at | AT4G15390 | transferase family protein | 10.12 |
| 1411 | 245556_at | AT4G15400 | transferase family protein | 6.33 |
| 1412 | 245253_at | AT4G15440 | HPL1 (HYDROPEROXIDE LYASE 1); heme binding / iron ion binding / monooxygenase | -4.04 |
| 1413 | 245352_at | AT4G15490 | UGT84A3; UDP-glycosyltransferase/ sinapate 1-glucosyltransferase/ transferase, transferring glycosyl groups | -3.72 |
| 1414 | 245528_at | AT4G15530 | PPDK (PYRUVATE ORTHOPHOSPHATE DIKINASE); kinase/ pyruvate, phosphate dikinase | -3.30 |
| 1415 | 245317_at | AT4G15610 | integral membrane family protein | 3.06 |
| 1416 | 245501_at | AT4G15620 | integral membrane family protein | -6.23 |
| 1417 | 245304_at | AT4G15630 | integral membrane family protein | -2.71 |
| 1418 | 245523_at | AT4G15910 | ATDI21 (Arabidopsis thaliana drought-induced 21) | 2.67 |
| 1419 | 245353_at | AT4G16000 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G15990.1) | 2.53 |
| 1420 | 245319_at | AT4G16146 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G69510.2); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G69510.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G69510.3); similar to unnamed protein product [Vitis vinifera] (GB:CAO44882.1); contains InterPro domain Lg106-like (InterPro:IPR012482) | -3.04 |
| 1421 | 245393_at | AT4G16260 | glycosyl hydrolase family 17 protein | 5.76 |
| 1422 | 245388_at | AT4G16410 | similar to hypothetical protein [Vitis vinifera] (GB:CAN73304.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO39941.1); contains InterPro domain Protein of unknown function DUF751 (InterPro:IPR008470) | -5.39 |
| 1423 | 245336_at | AT4G16515 | unknown protein | -3.02 |
| 1424 | 245349_at | AT4G16690 | esterase/lipase/thioesterase family protein | -2.78 |
| 1425 | 245448_at | AT4G16860 | RPP4 (RECOGNITION OF PERONOSPORA PARASITICA 4) | -5.23 |
| 1426 | 245450_at | AT4G16880 | disease resistance protein-related | -4.55 |
| 1427 | 245456_at | AT4G16950 | RPP5 (RECOGNITION OF PERONOSPORA PARASITICA 5) | -2.52 |
| 1428 | 245318_at | AT4G16980 | arabinogalactan-protein family | -4.40 |
| 1429 | 245460_at | AT4G16990 | disease resistance protein (TIR-NBS class), putative | -4.00 |
| 1430 | 245346_at | AT4G17090 | CT-BMY (BETA-AMYLASE 3, BETA-AMYLASE 8); beta-amylase | -3.18 |
| 1431 | 245305_at | AT4G17215 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G47635.1); similar to hypothetical protein 40.t00006 [Brassica oleracea] (GB:ABD65131.1) | 2.48 |
| 1432 | 245410_at | AT4G17220 | ATMAP70-5 (microtubule-associated proteins 70-5); microtubule binding | 3.81 |
| 1433 | 245264_at | AT4G17245 | zinc finger (C3HC4-type RING finger) family protein | -4.52 |
| 1434 | 245412_at | AT4G17280 | auxin-responsive family protein | 3.22 |
| 1435 | 245399_at | AT4G17340 | DELTA-TIP2/TIP2;2 (tonoplast intrinsic protein 2;2); water channel | 3.01 |
| 1436 | 245250_at | AT4G17490 | ATERF6 (ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 6); DNA binding / transcription factor | 3.21 |
| 1437 | 245357_at | AT4G17560 | ribosomal protein L19 family protein | -2.62 |
| 1438 | 245354_at | AT4G17600 | LIL3:1; transcription factor | -6.92 |
| 1439 | 245263_at | AT4G17740 | C-terminal processing protease, putative | -2.68 |
| 1440 | 245382_at | AT4G17800 | DNA-binding protein-related | 4.92 |
| 1441 | 254691_at | AT4G17840 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G35260.1); similar to hypothetical protein 40.t00061 [Brassica oleracea] (GB:ABD65174.1) | -2.74 |
| 1442 | 254663_at | AT4G18290 | KAT2 (K+ ATPase 2); cyclic nucleotide binding / inward rectifier potassium channel | -3.47 |
| 1443 | 254665_at | AT4G18340 | glycosyl hydrolase family 17 protein | 2.29 |
| 1444 | 254630_at | AT4G18360 | (S)-2-hydroxy-acid oxidase, peroxisomal, putative / glycolate oxidase, putative / short chain alpha-hydroxy acid oxidase, putative | 2.96 |
| 1445 | 254623_at | AT4G18480 | CHLI1 (CHLORINA 42); magnesium chelatase | -4.48 |
| 1446 | 254644_at | AT4G18510 | CLE2 (CLAVATA3/ESR-RELATED); receptor binding | 6.70 |
| 1447 | 254648_at | AT4G18550 | lipase class 3 family protein | 3.22 |
| 1448 | 254631_at | AT4G18610 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G42610.2); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G42610.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN72178.1); similar to unknown [Populus trichocarpa] (GB:ABK93578.1); contains InterPro domain Protein of unknown function DUF640 (InterPro:IPR006936) | 3.66 |
| 1449 | 254638_at | AT4G18740 | transcription termination factor | -2.94 |
| 1450 | 254642_at | AT4G18810 | binding / catalytic/ transcription repressor | -6.50 |
| 1451 | 254609_at | AT4G18970 | GDSL-motif lipase/hydrolase family protein | -5.23 |
| 1452 | 254606_at | AT4G19030 | NLM1 (NOD26-like intrinsic protein 1;1); water channel | 4.72 |
| 1453 | 254612_at | AT4G19100 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G52780.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN79943.1) | -3.27 |
| 1454 | 254564_at | AT4G19170 | NCED4 (NINE-CIS-EPOXYCAROTENOID DIOXYGENASE 4) | -3.92 |
| 1455 | 254550_at | AT4G19690 | IRT1 (IRON-REGULATED TRANSPORTER 1); cadmium ion transmembrane transporter/ iron ion transmembrane transporter/ manganese ion transmembrane transporter/ zinc ion transmembrane transporter | 4.25 |
| 1456 | 254524_at | AT4G20000 | VQ motif-containing protein | 4.85 |
| 1457 | 254500_at | AT4G20110 | vacuolar sorting receptor, putative | 2.76 |
| 1458 | 254506_at | AT4G20140 | leucine-rich repeat transmembrane protein kinase, putative | 2.69 |
| 1459 | 254510_at | AT4G20210 | terpene synthase/cyclase family protein | 6.60 |
| 1460 | 254512_at | AT4G20230 | terpene synthase/cyclase family protein | 4.28 |
| 1461 | 254480_at | AT4G20360 | AtRABE1b/AtRab8D (Arabidopsis Rab GTPase homolog E1b); translation elongation factor | -2.64 |
| 1462 | 254474_at | AT4G20390 | integral membrane family protein | 3.81 |
| 1463 | 254447_at | AT4G20860 | FAD-binding domain-containing protein | 2.32 |
| 1464 | 254460_at | AT4G21210 | phosphoprotein phosphatase/ protein kinase | -6.02 |
| 1465 | 254398_at | AT4G21280 | PSBQ/PSBQ-1/PSBQA; calcium ion binding | -7.52 |
| 1466 | 254416_at | AT4G21380 | ARK3 (Arabidopsis Receptor Kinase 3); kinase | -2.79 |
| 1467 | 254408_at | AT4G21390 | B120; protein kinase/ sugar binding | 2.35 |
| 1468 | 254392_at | AT4G21600 | ENDO5 (ENDONUCLEASE 5); T/G mismatch-specific endonuclease/ endonuclease/ nucleic acid binding / single-stranded DNA specific endodeoxyribonuclease | 4.35 |
| 1469 | 254370_at | AT4G21750 | ATML1 (MERISTEM LAYER 1); DNA binding / transcription factor | -2.93 |
| 1470 | 254387_at | AT4G21850 | methionine sulfoxide reductase domain-containing protein / SeIR domain-containing protein | 4.28 |
| 1471 | 254384_at | AT4G21870 | 26.5 kDa class P-related heat shock protein (HSP26.5-P) | -4.48 |
| 1472 | 254305_at | AT4G22200 | AKT2 (Arabidopsis K+ transporter 2); cyclic nucleotide binding / inward rectifier potassium channel | -3.21 |
| 1473 | 254361_at | AT4G22212 | Encodes a defensin-like (DEFL) family protein. | 8.11 |
| 1474 | 254326_at | AT4G22610 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | 3.16 |
| 1475 | 254333_at | AT4G22753 | SMO1-3 (STEROL 4-ALPHA METHYL OXIDASE); catalytic | -2.87 |
| 1476 | 254298_at | AT4G22890 | PGR5-LIKE A | -4.08 |
| 1477 | 254286_at | AT4G22950 | AGL19 (AGAMOUS-LIKE 19); transcription factor | 3.01 |
| 1478 | 254247_at | AT4G23260 | protein kinase | -2.63 |
| 1479 | 254264_at | AT4G23510 | disease resistance protein (TIR class), putative | 3.24 |
| 1480 | 254232_at | AT4G23600 | CORI3 (CORONATINE INDUCED 1, JASMONIC ACID RESPONSIVE 2); transaminase | -3.86 |
| 1481 | 254226_at | AT4G23690 | disease resistance-responsive family protein / dirigent family protein | 4.01 |
| 1482 | 254215_at | AT4G23700 | ATCHX17 (CATION/H+ EXCHANGER 17); monovalent cation:proton antiporter | 3.36 |
| 1483 | 254221_at | AT4G23820 | glycoside hydrolase family 28 protein / polygalacturonase (pectinase) family protein | -3.19 |
| 1484 | 254187_at | AT4G23890 | similar to unnamed protein product [Vitis vinifera] (GB:CAO69542.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN73381.1) | -5.07 |
| 1485 | 254174_at | AT4G24120 | YSL1 (YELLOW STRIPE LIKE 1); oligopeptide transporter | -3.06 |
| 1486 | 254203_at | AT4G24150 | AtGRF8 (GROWTH-REGULATING FACTOR 8) | 2.54 |
| 1487 | 254206_at | AT4G24180 | similar to thaumatin, putative [Arabidopsis thaliana] (TAIR:AT4G38660.2); similar to unnamed protein product [Vitis vinifera] (GB:CAO48203.1); contains InterPro domain Thaumatin, pathogenesis-related (InterPro:IPR001938) | 2.99 |
| 1488 | 254122_at | AT4G24510 | CER2 (ECERIFERUM 2); transferase | -7.87 |
| 1489 | 254130_at | AT4G24540 | AGL24 (AGAMOUS-LIKE 24); protein binding / protein heterodimerization/ protein homodimerization/ transcription factor | -3.54 |
| 1490 | 254120_at | AT4G24570 | mitochondrial substrate carrier family protein | 2.32 |
| 1491 | 254132_at | AT4G24660 | ATHB22/MEE68 (ARABIDOPSIS THALIANA HOMEOBOX PROTEIN 22); DNA binding / transcription factor | -2.93 |
| 1492 | 254145_at | AT4G24700 | unknown protein | -6.10 |
| 1493 | 254126_at | AT4G24770 | RBP31 (31-KDA RNA BINDING PROTEIN); RNA binding / poly(U) binding | -2.71 |
| 1494 | 254137_at | AT4G24930 | thylakoid lumenal 17.9 kDa protein, chloroplast | -2.91 |
| 1495 | 254102_at | AT4G25050 | ACP4 (ACYL CARRIER PROTEIN 4) | -5.73 |
| 1496 | 254105_at | AT4G25080 | CHLM (MAGNESIUM-PROTOPORPHYRIN IX METHYLTRANSFERASE); magnesium protoporphyrin IX methyltransferase | -3.02 |
| 1497 | 254092_at | AT4G25090 | respiratory burst oxidase, putative / NADPH oxidase, putative | 5.07 |
| 1498 | 254059_at | AT4G25200 | ATHSP23.6-MITO (MITOCHONDRION-LOCALIZED SMALL HEAT SHOCK PROTEIN 23.6) | 5.76 |
| 1499 | 254056_at | AT4G25250 | invertase/pectin methylesterase inhibitor family protein | 7.23 |
| 1500 | 254069_at | AT4G25434 | ATNUDT10 (ARABIDOPSIS THALIANA NUDIX HYDROLASE HOMOLOG 10); catalytic | 2.87 |
| 1501 | 254075_at | AT4G25470 | CBF2 (FREEZING TOLERANCE QTL 4); DNA binding / transcription activator/ transcription factor | 3.86 |
| 1502 | 254074_at | AT4G25490 | CBF1 (C-REPEAT/DRE BINDING FACTOR 1); DNA binding / transcription activator/ transcription factor | 3.66 |
| 1503 | 254021_at | AT4G25650 | ACD1-LIKE; electron carrier | -2.81 |
| 1504 | 254020_at | AT4G25700 | BETA-OHASE 1 (BETA-HYDROXYLASE 1); beta-carotene hydroxylase | -3.17 |
| 1505 | 254042_at | AT4G25810 | XTR6 (XYLOGLUCAN ENDOTRANSGLYCOSYLASE 6); hydrolase, acting on glycosyl bonds | 2.69 |
| 1506 | 254044_at | AT4G25820 | XTR9 (XYLOGLUCAN ENDOTRANSGLYCOSYLASE 9); hydrolase, acting on glycosyl bonds | 2.71 |
| 1507 | 254041_at | AT4G25830 | integral membrane family protein | -3.21 |
| 1508 | 254038_at | AT4G25910 | NFU3 (NFU domain protein 3) | -2.91 |
| 1509 | 254034_at | AT4G25960 | PGP2 (P-GLYCOPROTEIN 2); ATPase, coupled to transmembrane movement of substances | -4.81 |
| 1510 | 253998_at | AT4G26010 | peroxidase, putative | 4.92 |
| 1511 | 253985_at | AT4G26220 | caffeoyl-CoA 3-O-methyltransferase, putative | 4.67 |
| 1512 | 253957_at | AT4G26320 | AGP13 (ARABINOGALACTAN PROTEIN 13) | 7.20 |
| 1513 | 254011_at | AT4G26370 | antitermination NusB domain-containing protein | -3.35 |
| 1514 | 253963_at | AT4G26470 | calcium ion binding | 2.71 |
| 1515 | 253966_at | AT4G26520 | fructose-bisphosphate aldolase, cytoplasmic | -5.42 |
| 1516 | 253971_at | AT4G26530 | fructose-bisphosphate aldolase, putative | -7.79 |
| 1517 | 253956_at | AT4G26700 | ATFIM1 (Arabidopsis thaliana fimbrin 1); actin binding | -2.97 |
| 1518 | 253922_at | AT4G26850 | VTC2 (VITAMIN C DEFECTIVE 2) | -4.34 |
| 1519 | 253943_at | AT4G27030 | small conjugating protein ligase | -5.68 |
| 1520 | 253910_at | AT4G27290 | S-locus protein kinase, putative | 3.92 |
| 1521 | 253871_at | AT4G27440 | PORB (PROTOCHLOROPHYLLIDE OXIDOREDUCTASE B); oxidoreductase/ protochlorophyllide reductase | -6.75 |
| 1522 | 253858_at | AT4G27600 | pfkB-type carbohydrate kinase family protein | -2.79 |
| 1523 | 253830_at | AT4G27652 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G27657.1) | 2.99 |
| 1524 | 253832_at | AT4G27654 | unknown protein | 3.70 |
| 1525 | 253859_at | AT4G27657 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G27652.1) | 2.56 |
| 1526 | 253860_at | AT4G27700 | rhodanese-like domain-containing protein | -5.12 |
| 1527 | 253834_at | AT4G27800 | protein phosphatase 2C PPH1 / PP2C PPH1 (PPH1) | -2.76 |
| 1528 | 253825_at | AT4G28025 | similar to unnamed protein product [Vitis vinifera] (GB:CAO66511.1); contains domain Chlorophyll a-b binding protein (SSF103511) | -2.67 |
| 1529 | 253823_at | AT4G28030 | GCN5-related N-acetyltransferase (GNAT) family protein | -2.82 |
| 1530 | 253849_at | AT4G28080 | binding | -5.36 |
| 1531 | 253822_at | AT4G28410 | aminotransferase-related | 6.95 |
| 1532 | 253786_at | AT4G28650 | leucine-rich repeat transmembrane protein kinase, putative | 3.49 |
| 1533 | 253790_at | AT4G28660 | PSB28 (PHOTOSYSTEM II REACTION CENTER PSB28 PROTEIN) | -5.95 |
| 1534 | 253765_at | AT4G28740 | similar to LPA1 (LOW PSII ACCUMULATION1), binding [Arabidopsis thaliana] (TAIR:AT1G02910.1); similar to OSJNBa0043L24.5 [Oryza sativa (japonica cultivar-group)] (GB:CAE04717.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO40509.1) | -5.41 |
| 1535 | 253738_at | AT4G28750 | PSAE-1 (PSA E1 KNOCKOUT); catalytic | -9.07 |
| 1536 | 253741_at | AT4G28890 | protein binding / ubiquitin-protein ligase/ zinc ion binding | 4.53 |
| 1537 | 253743_at | AT4G28940 | catalytic | 2.24 |
| 1538 | 253758_at | AT4G29060 | EMB2726 (EMBRYO DEFECTIVE 2726); translation elongation factor | -4.03 |
| 1539 | 253710_at | AT4G29230 | ANAC075 (Arabidopsis NAC domain containing protein 75); transcription factor | 2.25 |
| 1540 | 253720_at | AT4G29270 | acid phosphatase class B family protein | 4.53 |
| 1541 | 253688_at | AT4G29590 | methyltransferase | -2.58 |
| 1542 | 253684_at | AT4G29690 | type I phosphodiesterase/nucleotide pyrophosphatase family protein | 4.20 |
| 1543 | 253643_at | AT4G29780 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G12010.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO43835.1); contains domain PTHR22930 (PTHR22930) | 2.44 |
| 1544 | 253699_at | AT4G29800 | PLA IVD/PLP8 (Patatin-like protein 8) | 3.15 |
| 1545 | 253667_at | AT4G30170 | peroxidase, putative | 8.52 |
| 1546 | 253628_at | AT4G30280 | ATXTH18/XTH18 (XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 18); hydrolase, acting on glycosyl bonds | 4.21 |
| 1547 | 253608_at | AT4G30290 | ATXTH19 (XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 19); hydrolase, acting on glycosyl bonds | 5.03 |
| 1548 | 253618_at | AT4G30420 | nodulin MtN21 family protein | 2.40 |
| 1549 | 253632_at | AT4G30430 | TET9 (TETRASPANIN9) | 2.37 |
| 1550 | 253629_at | AT4G30450 | glycine-rich protein | 3.98 |
| 1551 | 253619_at | AT4G30460 | glycine-rich protein | 3.25 |
| 1552 | 253626_at | AT4G30640 | F-box family protein (FBL19) | 2.22 |
| 1553 | 253582_at | AT4G30670 | contains domain PROKAR_LIPOPROTEIN (PS51257) | 2.60 |
| 1554 | 253595_at | AT4G30830 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G24140.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO66637.1); contains InterPro domain Protein of unknown function DUF593 (InterPro:IPR007656) | 2.61 |
| 1555 | 253547_at | AT4G30950 | FAD6 (FATTY ACID DESATURASE 6); omega-6 fatty acid desaturase | -2.88 |
| 1556 | 253556_at | AT4G31100 | wall-associated kinase, putative | 6.61 |
| 1557 | 253515_at | AT4G31320 | auxin-responsive protein, putative / small auxin up RNA (SAUR_C) | 5.01 |
| 1558 | 253527_at | AT4G31470 | pathogenesis-related protein, putative | 3.89 |
| 1559 | 253510_at | AT4G31730 | GDU1 (GLUTAMINE DUMPER 1) | 3.21 |
| 1560 | 253454_at | AT4G31875 | unknown protein | 3.29 |
| 1561 | 253483_at | AT4G31910 | transferase family protein | 4.00 |
| 1562 | 253502_at | AT4G31940 | CYP82C4 (cytochrome P450, family 82, subfamily C, polypeptide 4); oxygen binding | 4.99 |
| 1563 | 253420_at | AT4G32260 | ATP synthase family | -4.09 |
| 1564 | 253391_at | AT4G32590 | ferredoxin-related | -2.48 |
| 1565 | 253392_at | AT4G32650 | ATKC1 (ARABIDOPSIS THALIANA K+ RECTIFYING CHANNEL 1); cyclic nucleotide binding / inward rectifier potassium channel | 3.49 |
| 1566 | 253394_at | AT4G32770 | VTE1 (VITAMIN E DEFICIENT 1) | -2.75 |
| 1567 | 253400_at | AT4G32860 | similar to unknown [Populus trichocarpa x Populus deltoides] (GB:ABK96753.1) | 5.49 |
| 1568 | 253411_at | AT4G32980 | ATH1 (ARABIDOPSIS THALIANA HOMEOBOX GENE 1); transcription factor | -2.49 |
| 1569 | 253387_at | AT4G33010 | ATGLDP1 (ARABIDOPSIS THALIANA GLYCINE DECARBOXYLASE P-PROTEIN 1); glycine dehydrogenase (decarboxylating) | -2.72 |
| 1570 | 253416_at | AT4G33070 | pyruvate decarboxylase, putative | 2.69 |
| 1571 | 253372_at | AT4G33220 | pectinesterase family protein | -2.94 |
| 1572 | 253332_at | AT4G33420 | peroxidase, putative | 4.64 |
| 1573 | 253337_at | AT4G33470 | HDA14 (histone deacetylase 14); histone deacetylase | -2.48 |
| 1574 | 253305_at | AT4G33666 | unknown protein | -3.25 |
| 1575 | 253301_at | AT4G33720 | pathogenesis-related protein, putative | 10.53 |
| 1576 | 253353_at | AT4G33730 | pathogenesis-related protein, putative | 2.85 |
| 1577 | 253283_at | AT4G34090 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G23370.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO47838.1) | -3.38 |
| 1578 | 253272_at | AT4G34190 | SEP1 (STRESS ENHANCED PROTEIN 1) | -4.14 |
| 1579 | 253259_at | AT4G34410 | AP2 domain-containing transcription factor, putative | 4.01 |
| 1580 | 253246_at | AT4G34600 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G16385.1) | 4.37 |
| 1581 | 253251_at | AT4G34730 | ribosome-binding factor A family protein | -3.13 |
| 1582 | 253207_at | AT4G34770 | auxin-responsive family protein | -4.45 |
| 1583 | 253208_at | AT4G34830 | binding | -4.18 |
| 1584 | 253209_at | AT4G34830 | binding | -6.16 |
| 1585 | 253211_at | AT4G34880 | amidase family protein | 2.27 |
| 1586 | 253172_at | AT4G35060 | heavy-metal-associated domain-containing protein / copper chaperone (CCH)-related | 2.25 |
| 1587 | 253197_at | AT4G35250 | vestitone reductase-related | -5.05 |
| 1588 | 253191_at | AT4G35350 | XCP1 (XYLEM CYSTEINE PEPTIDASE 1); cysteine-type peptidase | 2.28 |
| 1589 | 253193_at | AT4G35380 | guanine nucleotide exchange family protein | 2.79 |
| 1590 | 246216_at | AT4G36380 | ROT3 (ROTUNDIFOLIA 3); oxygen binding / steroid hydroxylase | 2.35 |
| 1591 | 246228_at | AT4G36430 | peroxidase, putative | 5.05 |
| 1592 | 246236_at | AT4G36470 | S-adenosyl-L-methionine:carboxyl methyltransferase family protein | 2.32 |
| 1593 | 246199_at | AT4G36530 | hydrolase, alpha/beta fold family protein | -3.23 |
| 1594 | 246275_at | AT4G36540 | BEE2 (BR ENHANCED EXPRESSION 2); DNA binding / transcription factor | -3.38 |
| 1595 | 246197_at | AT4G37010 | caltractin, putative / centrin, putative | 4.73 |
| 1596 | 246229_at | AT4G37160 | SKS15 (SKU5 Similar 15); copper ion binding | 4.80 |
| 1597 | 246226_at | AT4G37200 | HCF164 (High chlorophyll fluorescence 164); thiol-disulfide exchange intermediate | -2.67 |
| 1598 | 253052_at | AT4G37310 | CYP81H1 (cytochrome P450, family 81, subfamily H, polypeptide 1); oxygen binding | -4.02 |
| 1599 | 253098_at | AT4G37340 | CYP81D3 (cytochrome P450, family 81, subfamily D, polypeptide 3); oxygen binding | 3.56 |
| 1600 | 253073_at | AT4G37410 | CYP81F4 (cytochrome P450, family 81, subfamily F, polypeptide 4); oxygen binding | 5.60 |
| 1601 | 253053_at | AT4G37470 | hydrolase, alpha/beta fold family protein | -3.40 |
| 1602 | 253048_at | AT4G37560 | formamidase, putative / formamide amidohydrolase, putative | -3.19 |
| 1603 | 253064_at | AT4G37730 | ATBZIP7 (ARABIDOPSIS THALIANA BASIC LEUCINE-ZIPPER 7); DNA binding / transcription factor | 2.48 |
| 1604 | 253039_at | AT4G37760 | SQE3 (SQUALENE EPOXIDASE 3); oxidoreductase | -4.58 |
| 1605 | 253009_at | AT4G37930 | SHM1 (SERINE HYDROXYMETHYLTRANSFERASE 1); glycine hydroxymethyltransferase/ poly(U) binding | -4.95 |
| 1606 | 253014_at | AT4G37940 | AGL21 (AGAMOUS-LIKE 21); transcription factor | 6.17 |
| 1607 | 252983_at | AT4G37980 | ELI3-1 (ELICITOR-ACTIVATED GENE 3); binding / catalytic/ oxidoreductase/ zinc ion binding | -7.51 |
| 1608 | 253024_at | AT4G38080 | hydroxyproline-rich glycoprotein family protein | 7.43 |
| 1609 | 252988_at | AT4G38410 | dehydrin, putative | 3.00 |
| 1610 | 252993_at | AT4G38540 | monooxygenase, putative (MO2) | 2.77 |
| 1611 | 252950_at | AT4G38690 | 1-phosphatidylinositol phosphodiesterase-related | -2.75 |
| 1612 | 252971_at | AT4G38770 | PRP4 (PROLINE-RICH PROTEIN 4) | -4.08 |
| 1613 | 252972_at | AT4G38840 | auxin-responsive protein, putative | -4.38 |
| 1614 | 252965_at | AT4G38860 | auxin-responsive protein, putative | -5.56 |
| 1615 | 252929_at | AT4G38970 | fructose-bisphosphate aldolase, putative | -7.78 |
| 1616 | 252943_at | AT4G39330 | mannitol dehydrogenase, putative | -3.70 |
| 1617 | 252882_at | AT4G39675 | unknown protein | 9.63 |
| 1618 | 252853_at | AT4G39710 | immunophilin, putative / FKBP-type peptidyl-prolyl cis-trans isomerase, putative | -3.95 |
| 1619 | 252876_at | AT4G39970 | haloacid dehalogenase-like hydrolase family protein | -3.03 |
| 1620 | 251142_at | AT5G01015 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G65295.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN82885.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO45347.1) | -3.27 |
| 1621 | 251082_at | AT5G01530 | chlorophyll A-B binding protein CP29 (LHCB4) | -5.79 |
| 1622 | 251072_at | AT5G01740 | similar to SAG20 (WOUND-INDUCED PROTEIN 12) [Arabidopsis thaliana] (TAIR:AT3G10985.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO48816.1); contains InterPro domain Wound-induced protein, Wun1 (InterPro:IPR009798) | 2.61 |
| 1623 | 251061_at | AT5G01830 | armadillo/beta-catenin repeat family protein / U-box domain-containing protein | 4.47 |
| 1624 | 251065_at | AT5G01870 | lipid transfer protein, putative | 4.61 |
| 1625 | 251068_at | AT5G01920 | STN8 (state transition 8); kinase | -3.34 |
| 1626 | 251032_at | AT5G02030 | LSN (LARSON); DNA binding / transcription factor | -2.84 |
| 1627 | 251031_at | AT5G02120 | OHP (ONE HELIX PROTEIN) | -7.57 |
| 1628 | 251021_at | AT5G02140 | thaumatin-like protein, putative | 2.81 |
| 1629 | 251036_at | AT5G02160 | unknown protein | -5.43 |
| 1630 | 251038_at | AT5G02240 | binding / catalytic/ coenzyme binding | -2.72 |
| 1631 | 251044_at | AT5G02350 | DC1 domain-containing protein | 2.31 |
| 1632 | 251012_at | AT5G02580 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G55240.1); similar to hypothetical protein OsI_019565 [Oryza sativa (indica cultivar-group)] (GB:EAY98332.1); similar to Os05g0462000 [Oryza sativa (japonica cultivar-group)] (GB:NP_001055765.1); contains InterPro domain Conserved hypothetical protein CHP01589, plant (InterPro:IPR006476) | 2.79 |
| 1633 | 251009_at | AT5G02640 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G46300.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN66779.1) | 2.46 |
| 1634 | 251008_at | AT5G02710 | similar to unnamed protein product [Vitis vinifera] (GB:CAO18074.1); contains InterPro domain Protein of unknown function UPF0153 (InterPro:IPR005358) | -3.97 |
| 1635 | 250983_at | AT5G02780 | In2-1 protein, putative | 3.24 |
| 1636 | 250967_at | AT5G02790 | In2-1 protein, putative | -2.99 |
| 1637 | 250960_at | AT5G02940 | similar to phosphotransferase-related [Arabidopsis thaliana] (TAIR:AT5G43745.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO71282.1); similar to hypothetical protein OsI_009890 [Oryza sativa (indica cultivar-group)] (GB:EAY88657.1); similar to Unknown protein [Oryza sativa (japonica cultivar-group)] (GB:AAN06856.1); contains InterPro domain Protein of unknown function DUF1012 (InterPro:IPR010420) | -5.50 |
| 1638 | 250936_at | AT5G03120 | similar to hypothetical protein [Vitis vinifera] (GB:CAN68657.1) | -5.74 |
| 1639 | 250942_at | AT5G03350 | legume lectin family protein | -7.13 |
| 1640 | 250952_at | AT5G03570 | iron-responsive transporter-related | 3.17 |
| 1641 | 250906_at | AT5G03650 | SBE2.2 (STARCH BRANCHING ENZYME 2.2); 1,4-alpha-glucan branching enzyme | -2.99 |
| 1642 | 250869_at | AT5G03840 | TFL1 (TERMINAL FLOWER 1); phosphatidylethanolamine binding | 2.43 |
| 1643 | 250884_at | AT5G03940 | FFC (FIFTY-FOUR CHLOROPLAST HOMOLOGUE); 7S RNA binding / GTP binding / mRNA binding | -3.00 |
| 1644 | 250874_at | AT5G04010 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G03920.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO23344.1); contains domain SSF81383 (SSF81383) | 2.24 |
| 1645 | 245689_at | AT5G04120 | phosphoglycerate/bisphosphoglycerate mutase family protein | 6.68 |
| 1646 | 245701_at | AT5G04140 | GLS1/GLU1/GLUS (FERREDOXIN-DEPENDENT GLUTAMATE SYNTHASE 1); glutamate synthase (ferredoxin) | -6.63 |
| 1647 | 245692_at | AT5G04150 | BHLH101; DNA binding / transcription factor | 2.73 |
| 1648 | 245711_at | AT5G04340 | C2H2 (ZINC FINGER OF ARABIDOPSIS THALIANA 6); nucleic acid binding / transcription factor/ zinc ion binding | 2.58 |
| 1649 | 245713_at | AT5G04370 | NAMT1; S-adenosylmethionine-dependent methyltransferase | 4.54 |
| 1650 | 250842_at | AT5G04490 | VTE5 (VITAMIN E PATHWAY GENE5); phosphatidate cytidylyltransferase/ phytol kinase | -2.82 |
| 1651 | 250891_at | AT5G04530 | beta-ketoacyl-CoA synthase family protein | -3.68 |
| 1652 | 250855_at | AT5G04730 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G04700.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO46503.1); contains InterPro domain Ankyrin (InterPro:IPR002110) | 2.33 |
| 1653 | 250812_at | AT5G04900 | short-chain dehydrogenase/reductase (SDR) family protein | -3.87 |
| 1654 | 250801_at | AT5G04960 | pectinesterase family protein | 2.21 |
| 1655 | 250802_at | AT5G04970 | pectinesterase, putative | 6.16 |
| 1656 | 250820_at | AT5G05160 | leucine-rich repeat transmembrane protein kinase, putative | 2.63 |
| 1657 | 250824_at | AT5G05200 | ABC1 family protein | -2.52 |
| 1658 | 250794_at | AT5G05270 | chalcone-flavanone isomerase family protein | -4.84 |
| 1659 | 250771_at | AT5G05400 | disease resistance protein (CC-NBS-LRR class), putative | 2.23 |
| 1660 | 250781_at | AT5G05410 | DREB2A (DRE-BINDING PROTEIN 2A); DNA binding / transcription activator/ transcription factor | 4.27 |
| 1661 | 250778_at | AT5G05500 | pollen Ole e 1 allergen and extensin family protein | 2.69 |
| 1662 | 250739_at | AT5G05740 | ATEGY2; metalloendopeptidase | -3.81 |
| 1663 | 250741_at | AT5G05790 | myb family transcription factor | 2.51 |
| 1664 | 250746_at | AT5G05880 | UDP-glucoronosyl/UDP-glucosyl transferase family protein | 5.88 |
| 1665 | 250751_at | AT5G05890 | UDP-glucoronosyl/UDP-glucosyl transferase family protein | 2.67 |
| 1666 | 250764_at | AT5G05960 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | 4.60 |
| 1667 | 250717_at | AT5G06200 | integral membrane family protein | 2.25 |
| 1668 | 250733_at | AT5G06290 | 2-cys peroxiredoxin, chloroplast, putative | -2.83 |
| 1669 | 250690_at | AT5G06530 | ABC transporter family protein | -2.85 |
| 1670 | 250680_at | AT5G06570 | hydrolase | 2.54 |
| 1671 | 250702_at | AT5G06730 | peroxidase, putative | 4.90 |
| 1672 | 250695_at | AT5G06740 | lectin protein kinase family protein | 5.12 |
| 1673 | 250697_at | AT5G06800 | myb family transcription factor | 3.61 |
| 1674 | 250669_at | AT5G06870 | PGIP2 (POLYGALACTURONASE INHIBITING PROTEIN 2); protein binding | -4.01 |
| 1675 | 250651_at | AT5G06900 | CYP93D1 (cytochrome P450, family 93, subfamily D, polypeptide 1); oxygen binding | 4.41 |
| 1676 | 250668_at | AT5G07020 | proline-rich family protein | -3.66 |
| 1677 | 250642_at | AT5G07180 | ERL2 (ERECTA-LIKE 2); kinase | -3.43 |
| 1678 | 250629_at | AT5G07390 | ATRBOHA (RESPIRATORY BURST OXIDASE HOMOLOG A); FAD binding / calcium ion binding / iron ion binding / oxidoreductase | 3.20 |
| 1679 | 250596_at | AT5G07780 | formin homology 2 domain-containing protein / FH2 domain-containing protein | 2.27 |
| 1680 | 250558_at | AT5G07990 | TT7 (TRANSPARENT TESTA 7); flavonoid 3'-monooxygenase/ oxygen binding | -2.99 |
| 1681 | 250565_at | AT5G08000 | E13L3 (GLUCAN ENDO-1,3-BETA-GLUCOSIDASE-LIKE PROTEIN 3) | -3.18 |
| 1682 | 250563_at | AT5G08050 | similar to unnamed protein product [Vitis vinifera] (GB:CAO62462.1); similar to unknown [Populus trichocarpa] (GB:ABK95457.1); contains InterPro domain Protein of unknown function DUF1118 (InterPro:IPR009500); contains InterPro domain Uncharacterised conserved protein UCP022207 (InterPro:IPR016801) | -5.98 |
| 1683 | 250575_at | AT5G08240 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G23160.1) | 3.15 |
| 1684 | 246033_at | AT5G08280 | HEMC (HYDROXYMETHYLBILANE SYNTHASE); hydroxymethylbilane synthase | -2.56 |
| 1685 | 246007_at | AT5G08410 | FTRA2 (ferredoxin/thioredoxin reductase subunit A (variable subunit) 2); ferredoxin:thioredoxin reductase | -2.61 |
| 1686 | 250533_at | AT5G08640 | FLS (FLAVONOL SYNTHASE) | -5.29 |
| 1687 | 250531_at | AT5G08650 | GTP-binding protein LepA, putative | -2.95 |
| 1688 | 245889_at | AT5G09480 | hydroxyproline-rich glycoprotein family protein | 5.12 |
| 1689 | 250500_at | AT5G09530 | hydroxyproline-rich glycoprotein family protein | 6.02 |
| 1690 | 250498_at | AT5G09660 | PMDH2 (PEROXISOMAL NAD-MALATE DEHYDROGENASE 2); malate dehydrogenase | -5.65 |
| 1691 | 250455_at | AT5G09980 | PROPEP4 (Elicitor peptide 4 precursor) | 2.64 |
| 1692 | 250467_at | AT5G10100 | trehalose-6-phosphate phosphatase, putative | 2.64 |
| 1693 | 250469_at | AT5G10130 | pollen Ole e 1 allergen and extensin family protein | 5.87 |
| 1694 | 250472_at | AT5G10210 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G65030.1); similar to 80A08_18 [Brassica rapa subsp. pekinensis] (GB:AAZ67603.1); contains InterPro domain C2 calcium-dependent membrane targeting (InterPro:IPR000008) | 5.84 |
| 1695 | 250437_at | AT5G10430 | AGP4 (ARABINOGALACTAN-PROTEIN 4) | 3.81 |
| 1696 | 250443_at | AT5G10520 | RBK1 (ROP BINDING PROTEIN KINASES 1); kinase | 2.90 |
| 1697 | 250425_at | AT5G10530 | lectin protein kinase, putative | 2.52 |
| 1698 | 250438_at | AT5G10580 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G31330.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN79714.1); contains InterPro domain Protein of unknown function DUF599 (InterPro:IPR006747) | 5.13 |
| 1699 | 246019_at | AT5G10690 | pentatricopeptide (PPR) repeat-containing protein / CBS domain-containing protein | -2.90 |
| 1700 | 246018_at | AT5G10695 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G57123.1); similar to unknown [Picea sitchensis] (GB:ABK22689.1) | 2.26 |
| 1701 | 246016_at | AT5G10720 | AHK5 (CYTOKININ INDEPENDENT 2) | 5.48 |
| 1702 | 250445_at | AT5G10760 | aspartyl protease family protein | -3.16 |
| 1703 | 250449_at | AT5G10830 | embryo-abundant protein-related | 2.82 |
| 1704 | 245901_at | AT5G11060 | KNAT4 (KNOTTED1-LIKE HOMEOBOX GENE 4); transcription factor | -2.93 |
| 1705 | 250302_at | AT5G11920 | ATCWINV6 (6-&1-FRUCTAN EXOHYDROLASE); hydrolase, hydrolyzing O-glycosyl compounds / inulinase/ levanase | 3.41 |
| 1706 | 250296_at | AT5G12020 | HSP17.6II (17.6 KDA CLASS II HEAT SHOCK PROTEIN) | 4.31 |
| 1707 | 250351_at | AT5G12030 | AT-HSP17.6A (Arabidopsis thaliana heat shock protein 17.6A) | 5.53 |
| 1708 | 250323_at | AT5G12880 | proline-rich family protein | 3.15 |
| 1709 | 245976_at | AT5G13080 | WRKY75 (WRKY DNA-BINDING PROTEIN 75); transcription factor | 3.21 |
| 1710 | 245979_at | AT5G13150 | ATEXO70C1 (exocyst subunit EXO70 family protein C1); protein binding | 3.28 |
| 1711 | 250286_at | AT5G13320 | PBS3 (AVRPPHB SUSCEPTIBLE 3) | -3.17 |
| 1712 | 245852_at | AT5G13510 | ribosomal protein L10 family protein | -2.78 |
| 1713 | 250243_at | AT5G13630 | GUN5 (GENOMES UNCOUPLED 5) | -8.22 |
| 1714 | 250256_at | AT5G13650 | elongation factor family protein | -2.63 |
| 1715 | 250247_at | AT5G13720 | structural constituent of ribosome | -3.03 |
| 1716 | 250255_at | AT5G13730 | SIG4 (SIGMA FACTOR 4); DNA binding / DNA-directed RNA polymerase/ transcription factor | -4.84 |
| 1717 | 250257_at | AT5G13770 | pentatricopeptide (PPR) repeat-containing protein | -6.16 |
| 1718 | 250214_at | AT5G13870 | EXGT-A4 (ENDOXYLOGLUCAN TRANSFERASE A4); hydrolase, acting on glycosyl bonds | 2.67 |
| 1719 | 250207_at | AT5G13930 | ATCHS/CHS/TT4 (CHALCONE SYNTHASE); naringenin-chalcone synthase | -8.29 |
| 1720 | 250208_at | AT5G14000 | ANAC084 (Arabidopsis NAC domain containing protein 84); transcription factor | 2.94 |
| 1721 | 250205_at | AT5G14020 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G73390.2); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G73390.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G73390.3); similar to unnamed protein product [Vitis vinifera] (GB:CAO49073.1); contains InterPro domain BRO1 (InterPro:IPR004328) | 3.56 |
| 1722 | 250223_at | AT5G14070 | ROXY2; thiol-disulfide exchange intermediate | 2.67 |
| 1723 | 250224_at | AT5G14150 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G11420.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN70048.1); contains InterPro domain Protein of unknown function DUF642 (InterPro:IPR006946) | 2.66 |
| 1724 | 250190_at | AT5G14320 | 30S ribosomal protein S13, chloroplast (CS13) | -3.24 |
| 1725 | 250187_at | AT5G14370 | similar to CIL [Arabidopsis thaliana] (TAIR:AT4G25990.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO14646.1); contains InterPro domain CCT (InterPro:IPR010402) | -2.81 |
| 1726 | 250189_at | AT5G14410 | unknown protein | -2.53 |
| 1727 | 250142_at | AT5G14650 | polygalacturonase, putative / pectinase, putative | 2.38 |
| 1728 | 250146_at | AT5G14660 | PDF1B (PEPTIDE DEFORMYLASE 1B); peptide deformylase | -2.65 |
| 1729 | 250145_at | AT5G14690 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G01516.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO14758.1) | 2.86 |
| 1730 | 246596_at | AT5G14740 | CA2 (BETA CARBONIC ANHYDRASE 2); carbonate dehydratase/ zinc ion binding | -6.70 |
| 1731 | 246585_at | AT5G14750 | ATMYB66/WER/WER1 (WEREWOLF 1); DNA binding / protein binding / transcription factor/ transcription regulator | 5.78 |
| 1732 | 246548_at | AT5G14910 | heavy-metal-associated domain-containing protein | -3.19 |
| 1733 | 250153_at | AT5G15130 | WRKY72 (WRKY DNA-binding protein 72); transcription factor | 4.04 |
| 1734 | 250108_at | AT5G15150 | ATHB-3 (ARABIDOPSIS THALIANA HOMEOBOX 3); DNA binding / transcription factor | 2.83 |
| 1735 | 250157_at | AT5G15180 | peroxidase, putative | 4.98 |
| 1736 | 250160_at | AT5G15210 | ATHB30/ZFHD3 (ZINC FINGER HOMEODOMAIN 3); DNA binding / transcription factor | -2.57 |
| 1737 | 246530_at | AT5G15725 | unknown protein | 2.85 |
| 1738 | 246519_at | AT5G15780 | pollen Ole e 1 allergen and extensin family protein | -2.97 |
| 1739 | 246522_at | AT5G15830 | ATBZIP3 (ARABIDOPSIS THALIANA BASIC LEUCINE-ZIPPER 3); DNA binding / transcription factor | 3.44 |
| 1740 | 246523_at | AT5G15850 | COL1 (CONSTANS-LIKE 1); transcription factor/ zinc ion binding | -6.24 |
| 1741 | 246534_at | AT5G15890 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G15900.1); similar to hypothetical protein [Cleome spinosa] (GB:ABD96865.1); contains InterPro domain Protein of unknown function DUF231, plant (InterPro:IPR004253) | 4.77 |
| 1742 | 246488_at | AT5G16010 | 3-oxo-5-alpha-steroid 4-dehydrogenase family protein / steroid 5-alpha-reductase family protein | -3.34 |
| 1743 | 246487_at | AT5G16030 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G02500.1); similar to hypothetical protein [Cleome spinosa] (GB:ABD96956.1) | -3.91 |
| 1744 | 250133_at | AT5G16400 | ATF2/TRXF2 (THIOREDOXIN F2); thiol-disulfide exchange intermediate | -4.07 |
| 1745 | 250103_at | AT5G16600 | MYB43 (myb domain protein 43); DNA binding / transcription factor | 2.53 |
| 1746 | 246417_at | AT5G16990 | NADP-dependent oxidoreductase, putative | -2.75 |
| 1747 | 246468_at | AT5G17050 | UDP-glucoronosyl/UDP-glucosyl transferase family protein | -2.81 |
| 1748 | 250073_at | AT5G17170 | ENH1 (ENHANCER OF SOS3-1); metal ion binding | -5.57 |
| 1749 | 250095_at | AT5G17230 | PSY (PHYTOENE SYNTHASE); geranylgeranyl-diphosphate geranylgeranyltransferase | -3.90 |
| 1750 | 250090_at | AT5G17330 | GAD (Glutamate decarboxylase 1); calmodulin binding | 6.25 |
| 1751 | 250098_at | AT5G17350 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G03280.1); similar to unknown [Populus trichocarpa] (GB:ABK95625.1) | 4.08 |
| 1752 | 250075_at | AT5G17670 | hydrolase, acting on ester bonds | -3.59 |
| 1753 | 250059_at | AT5G17820 | peroxidase 57 (PER57) (P57) (PRXR10) | 8.19 |
| 1754 | 250054_at | AT5G17860 | CAX7 (CALCIUM EXCHANGER 7); calcium:sodium antiporter/ cation:cation antiporter | -3.02 |
| 1755 | 250012_x_at | AT5G18060 | auxin-responsive protein, putative | -3.51 |
| 1756 | 249996_at | AT5G18600 | glutaredoxin family protein | -5.71 |
| 1757 | 250006_at | AT5G18660 | DVR (PALE-GREEN AND CHLOROPHYLL B REDUCED 2); 3,8-divinyl protochlorophyllide a 8-vinyl reductase | -4.94 |
| 1758 | 249972_at | AT5G19040 | ATIPT5 (Arabidopsis thaliana isopentenyltransferase 5); transferase, transferring alkyl or aryl (other than methyl) groups | 4.47 |
| 1759 | 249971_at | AT5G19110 | extracellular dermal glycoprotein-related / EDGP-related | 3.84 |
| 1760 | 249922_at | AT5G19140 | auxin/aluminum-responsive protein, putative | -2.64 |
| 1761 | 249927_at | AT5G19220 | ADG2 (ADPG PYROPHOSPHORYLASE 2); glucose-1-phosphate adenylyltransferase | -5.49 |
| 1762 | 246063_at | AT5G19340 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G05980.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN75979.1) | 2.21 |
| 1763 | 246067_at | AT5G19410 | ABC transporter family protein | 2.83 |
| 1764 | 245944_at | AT5G19520 | mechanosensitive ion channel domain-containing protein / MS ion channel domain-containing protein | 4.70 |
| 1765 | 245946_at | AT5G19580 | glyoxal oxidase-related | -4.22 |
| 1766 | 245936_at | AT5G19850 | hydrolase, alpha/beta fold family protein | -2.64 |
| 1767 | 246158_at | AT5G19855 | similar to unnamed protein product [Vitis vinifera] (GB:CAO21907.1) | -3.04 |
| 1768 | 246149_at | AT5G19890 | peroxidase, putative | 4.25 |
| 1769 | 246154_at | AT5G19940 | plastid-lipid associated protein PAP-related / fibrillin-related | -4.64 |
| 1770 | 246142_at | AT5G19970 | similar to unnamed protein product [Vitis vinifera] (GB:CAO65601.1) | 2.66 |
| 1771 | 246069_at | AT5G20220 | zinc knuckle (CCHC-type) family protein | -2.53 |
| 1772 | 246098_at | AT5G20400 | oxidoreductase, 2OG-Fe(II) oxygenase family protein | 3.34 |
| 1773 | 246082_at | AT5G20480 | EFR (EF-TU RECEPTOR); ATP binding / kinase/ protein serine/threonine kinase | -3.54 |
| 1774 | 246093_at | AT5G20550 | oxidoreductase, 2OG-Fe(II) oxygenase family protein | 6.18 |
| 1775 | 246004_at | AT5G20630 | GLP3 (GERMIN-LIKE PROTEIN 3); manganese ion binding / metal ion binding / nutrient reservoir | -7.32 |
| 1776 | 246002_at | AT5G20740 | invertase/pectin methylesterase inhibitor family protein | -5.81 |
| 1777 | 246001_at | AT5G20790 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G43110.1) | -3.19 |
| 1778 | 246159_at | AT5G20935 | similar to unnamed protein product [Vitis vinifera] (GB:CAO67410.1) | -6.47 |
| 1779 | 246133_at | AT5G20960 | AAO1 (ALDEHYDE OXIDASE 1) | 3.84 |
| 1780 | 246021_at | AT5G21100 | L-ascorbate oxidase, putative | -4.29 |
| 1781 | 249928_at | AT5G22250 | CCR4-NOT transcription complex protein, putative | 2.85 |
| 1782 | 249917_at | AT5G22460 | esterase/lipase/thioesterase family protein | -3.75 |
| 1783 | 249896_at | AT5G22530 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G22520.1) | 2.47 |
| 1784 | 249897_at | AT5G22550 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G22560.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN75021.1); contains InterPro domain Protein of unknown function DUF247, plant (InterPro:IPR004158) | 3.18 |
| 1785 | 249894_at | AT5G22580 | Identical to Uncharacterized protein At5g22580 [Arabidopsis Thaliana] (GB:Q9FK81); similar to stable protein 1-related [Arabidopsis thaliana] (TAIR:AT3G17210.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN68720.1); contains InterPro domain Dimeric alpha-beta barrel (InterPro:IPR011008); contains InterPro domain Stress responsive alpha-beta barrel (InterPro:IPR013097) | -2.96 |
| 1786 | 249867_at | AT5G23020 | IMS2/MAM-L/MAM3 (METHYLTHIOALKYMALATE SYNTHASE-LIKE); 2-isopropylmalate synthase/ methylthioalkylmalate synthase | 5.15 |
| 1787 | 249876_at | AT5G23060 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G59780.1); similar to extracellular Ca2+ sensing receptor [Glycine max] (GB:ABY57763.1); contains InterPro domain Rhodanese-like (InterPro:IPR001763) | -5.68 |
| 1788 | 249875_at | AT5G23120 | HCF136 (High chlorophyll fluorescence 136) | -4.23 |
| 1789 | 249881_at | AT5G23190 | CYP86B1 (cytochrome P450, family 86, subfamily B, polypeptide 1); oxygen binding | 2.47 |
| 1790 | 249848_at | AT5G23220 | NIC3 (NICOTINAMIDASE 3); catalytic/ nicotinamidase | 3.35 |
| 1791 | 249850_at | AT5G23240 | DNAJ heat shock N-terminal domain-containing protein | -4.38 |
| 1792 | 249812_at | AT5G23830 | MD-2-related lipid recognition domain-containing protein / ML domain-containing protein | 7.10 |
| 1793 | 249814_at | AT5G23840 | MD-2-related lipid recognition domain-containing protein / ML domain-containing protein | 7.42 |
| 1794 | 249765_at | AT5G24030 | SLAH3 (SLAC1 HOMOLOGUE 3); transporter | 2.41 |
| 1795 | 249766_at | AT5G24070 | peroxidase family protein | 4.48 |
| 1796 | 249768_at | AT5G24100 | leucine-rich repeat transmembrane protein kinase, putative | 2.44 |
| 1797 | 249769_at | AT5G24120 | SIGE (RNA polymerase sigma subunit E); DNA binding / DNA-directed RNA polymerase/ sigma factor/ transcription factor | -2.90 |
| 1798 | 249774_at | AT5G24150 | SQP1 (Squalene monooxygenase 1) | -3.97 |
| 1799 | 249788_at | AT5G24330 | ATXR6 (Arabidopsis thaliana Trithorax- related protein 6); DNA binding | 3.56 |
| 1800 | 249729_at | AT5G24410 | glucosamine/galactosamine-6-phosphate isomerase-related | 5.47 |
| 1801 | 249732_at | AT5G24420 | glucosamine/galactosamine-6-phosphate isomerase-related | -3.45 |
| 1802 | 249750_at | AT5G24570 | unknown protein | -3.42 |
| 1803 | 246966_at | AT5G24850 | CRY3 (CRYPTOCHROME 3); DNA binding / DNA photolyase/ FMN binding | -2.79 |
| 1804 | 246977_at | AT5G24930 | zinc finger (B-box type) family protein | -8.22 |
| 1805 | 246933_at | AT5G25160 | ZFP3 (ZINC FINGER PROTEIN 3); nucleic acid binding / transcription factor/ zinc ion binding | 3.99 |
| 1806 | 246908_at | AT5G25610 | RD22 (RESPONSIVE TO DESSICATION 22) | -5.51 |
| 1807 | 246901_at | AT5G25630 | pentatricopeptide (PPR) repeat-containing protein | -3.44 |
| 1808 | 246902_at | AT5G25640 | similar to rhomboid family protein [Arabidopsis thaliana] (TAIR:AT5G25752.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO44068.1); contains InterPro domain Peptidase S54, rhomboid; (InterPro:IPR002610) | 2.77 |
| 1809 | 246912_at | AT5G25820 | exostosin family protein | 3.88 |
| 1810 | 246825_at | AT5G26260 | meprin and TRAF homology domain-containing protein / MATH domain-containing protein | 9.77 |
| 1811 | 246855_at | AT5G26280 | meprin and TRAF homology domain-containing protein / MATH domain-containing protein | 7.02 |
| 1812 | 246826_at | AT5G26310 | UGT72E3; UDP-glycosyltransferase/ coniferyl-alcohol glucosyltransferase/ transferase, transferring glycosyl groups | 5.28 |
| 1813 | 246844_at | AT5G26660 | ATMYB4 (myb domain protein 4); transcription repressor | 2.29 |
| 1814 | 246792_at | AT5G27290 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G54680.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G54680.2); similar to unnamed protein product [Vitis vinifera] (GB:CAO68604.1) | -3.48 |
| 1815 | 246736_at | AT5G27560 | similar to unnamed protein product [Vitis vinifera] (GB:CAO17622.1) | -2.77 |
| 1816 | 246761_at | AT5G27980 | seed maturation family protein | -2.73 |
| 1817 | 246703_at | AT5G28080 | WNK9 (Arabidopsis WNK kinase 9); kinase | 2.91 |
| 1818 | 245952_at | AT5G28500 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G04550.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO65032.1) | -5.03 |
| 1819 | 255945_at | AT5G28610 | similar to glycine-rich protein [Arabidopsis thaliana] (TAIR:AT5G28630.1) | 5.00 |
| 1820 | 246108_at | AT5G28630 | glycine-rich protein | 3.63 |
| 1821 | 246673_at | AT5G30510 | RPS1 (ribosomal protein S1); RNA binding | -3.11 |
| 1822 | 246651_at | AT5G35170 | adenylate kinase family protein | -3.62 |
| 1823 | 246652_at | AT5G35190 | proline-rich extensin-like family protein | 5.19 |
| 1824 | 249727_at | AT5G35490 | unknown protein | -2.81 |
| 1825 | 249705_at | AT5G35580 | kinase | 6.63 |
| 1826 | 249694_at | AT5G35790 | G6PD1 (GLUCOSE-6-PHOSPHATE DEHYDROGENASE 1); glucose-6-phosphate dehydrogenase | -5.85 |
| 1827 | 249675_at | AT5G35940 | jacalin lectin family protein | 4.67 |
| 1828 | 249677_at | AT5G35970 | DNA-binding protein, putative | -2.97 |
| 1829 | 246616_at | AT5G36260 | aspartyl protease family protein | 2.60 |
| 1830 | 249645_at | AT5G36910 | THI2.2 (THIONIN 2.2); toxin receptor binding | -3.18 |
| 1831 | 249610_at | AT5G37360 | similar to unnamed protein product [Vitis vinifera] (GB:CAO38751.1) | -2.96 |
| 1832 | 249617_at | AT5G37450 | leucine-rich repeat transmembrane protein kinase, putative | 2.85 |
| 1833 | 249626_at | AT5G37540 | aspartyl protease family protein | 2.32 |
| 1834 | 249575_at | AT5G37670 | 15.7 kDa class I-related small heat shock protein-like (HSP15.7-CI) | 2.95 |
| 1835 | 249576_at | AT5G37690 | GDSL-motif lipase/hydrolase family protein | 2.91 |
| 1836 | 249599_at | AT5G37990 | S-adenosylmethionine-dependent methyltransferase | 6.13 |
| 1837 | 249567_at | AT5G38020 | S-adenosyl-L-methionine:carboxyl methyltransferase family protein | 3.57 |
| 1838 | 249545_at | AT5G38030 | MATE efflux family protein | 4.05 |
| 1839 | 249557_at | AT5G38280 | PR5K (PR5-like receptor kinase); kinase/ transmembrane receptor protein serine/threonine kinase | 2.89 |
| 1840 | 249562_at | AT5G38350 | disease resistance protein (NBS-LRR class), putative | 2.23 |
| 1841 | 249510_at | AT5G38510 | rhomboid family protein | -2.59 |
| 1842 | 249524_at | AT5G38520 | hydrolase, alpha/beta fold family protein | -5.59 |
| 1843 | 249522_at | AT5G38700 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G02170.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO40081.1) | 5.23 |
| 1844 | 249533_at | AT5G38790 | unknown protein | 3.33 |
| 1845 | 249482_at | AT5G38980 | unknown protein | -3.67 |
| 1846 | 249494_at | AT5G39050 | transferase family protein | -5.22 |
| 1847 | 249472_at | AT5G39210 | CRR7 (CHLORORESPIRATORY REDUCTION 7) | -4.97 |
| 1848 | 249417_at | AT5G39670 | calcium-binding EF hand family protein | 2.73 |
| 1849 | 249421_at | AT5G39830 | DEG8/DEGP8 (DEGP PROTEASE 8); peptidase/ serine-type peptidase/ trypsin | -2.48 |
| 1850 | 249429_at | AT5G39880 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G28750.1) | -3.39 |
| 1851 | 249435_at | AT5G39970 | catalytic | 4.12 |
| 1852 | 249439_at | AT5G40020 | pathogenesis-related thaumatin family protein | 2.41 |
| 1853 | 249410_at | AT5G40380 | protein kinase family protein | -6.00 |
| 1854 | 249355_at | AT5G40500 | similar to unnamed protein product [Vitis vinifera] (GB:CAO14700.1) | -2.66 |
| 1855 | 249358_at | AT5G40510 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G27570.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO14698.1); contains InterPro domain Thioredoxin-like fold (InterPro:IPR012336); contains InterPro domain Thioredoxin fold (InterPro:IPR012335); contains InterPro domain Sucraseferredoxin-like (InterPro:IPR009737) | 3.43 |
| 1856 | 249367_at | AT5G40630 | ubiquitin family protein | 2.33 |
| 1857 | 249375_at | AT5G40730 | AGP24 (ARABINOGALACTAN PROTEIN 24) | 3.54 |
| 1858 | 249331_at | AT5G40950 | RPL27; structural constituent of ribosome | -3.02 |
| 1859 | 249289_at | AT5G41040 | transferase family protein | 2.91 |
| 1860 | 249288_at | AT5G41050 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G26960.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO49052.1) | -2.67 |
| 1861 | 249230_at | AT5G42070 | similar to unnamed protein product [Vitis vinifera] (GB:CAO69519.1) | -3.47 |
| 1862 | 249227_at | AT5G42180 | peroxidase 64 (PER64) (P64) (PRXR4) | 4.91 |
| 1863 | 249244_at | AT5G42270 | VAR1 (VARIEGATED 1); ATP-dependent peptidase/ ATPase/ metallopeptidase | -3.92 |
| 1864 | 249247_at | AT5G42310 | pentatricopeptide (PPR) repeat-containing protein | -2.77 |
| 1865 | 249197_at | AT5G42380 | CML37/CML39; calcium ion binding | 4.88 |
| 1866 | 249202_at | AT5G42580 | CYP705A12 (cytochrome P450, family 705, subfamily A, polypeptide 12); oxygen binding | 3.29 |
| 1867 | 249203_at | AT5G42590 | CYP71A16 (cytochrome P450, family 71, subfamily A, polypeptide 16); oxygen binding | 4.91 |
| 1868 | 249205_at | AT5G42600 | MRN (MARNERAL SYNTHASE); catalytic | 6.15 |
| 1869 | 249206_at | AT5G42630 | ATS/KAN4 (ABERRANT TESTA SHAPE); DNA binding / transcription factor | 3.29 |
| 1870 | 249211_at | AT5G42680 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G39610.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO61286.1); contains InterPro domain Protein of unknown function DUF617, plant (InterPro:IPR006460) | -2.50 |
| 1871 | 249191_at | AT5G42760 | similar to unknown [Populus trichocarpa] (GB:ABK95091.1); contains InterPro domain O-methyltransferase, N-terminal (InterPro:IPR003455); contains InterPro domain Conserved hypothetical protein CHP00027, methylltransferase (InterPro:IPR011610) | -2.56 |
| 1872 | 249174_at | AT5G42900 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G33980.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN75496.1) | -3.17 |
| 1873 | 249185_at | AT5G43030 | DC1 domain-containing protein | 5.59 |
| 1874 | 249186_at | AT5G43040 | DC1 domain-containing protein | 2.36 |
| 1875 | 249136_at | AT5G43180 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G10580.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN66486.1); contains InterPro domain Protein of unknown function DUF599 (InterPro:IPR006747) | 5.06 |
| 1876 | 249128_at | AT5G43440 | 2-oxoglutarate-dependent dioxygenase, putative | -3.61 |
| 1877 | 249125_at | AT5G43450 | 2-oxoglutarate-dependent dioxygenase, putative | -4.17 |
| 1878 | 249097_at | AT5G43520 | DC1 domain-containing protein | 5.38 |
| 1879 | 249101_at | AT5G43580 | serine-type endopeptidase inhibitor | 4.35 |
| 1880 | 249108_at | AT5G43690 | sulfotransferase family protein | 2.38 |
| 1881 | 249120_at | AT5G43750 | similar to unnamed protein product [Vitis vinifera] (GB:CAO71280.1) | -3.61 |
| 1882 | 249094_at | AT5G43890 | SUPER1/YUCCA5 (SUPPRESSOR OF ER1); monooxygenase | 3.51 |
| 1883 | 249034_at | AT5G44160 | NUC (NUTCRACKER); nucleic acid binding / transcription factor/ zinc ion binding | 4.27 |
| 1884 | 249035_at | AT5G44190 | GLK2 (GOLDEN2-LIKE 2); DNA binding / transcription factor | -2.92 |
| 1885 | 249042_at | AT5G44350 | ethylene-responsive nuclear protein -related | 3.55 |
| 1886 | 249045_at | AT5G44380 | FAD-binding domain-containing protein | 5.35 |
| 1887 | 249055_at | AT5G44460 | calcium-binding protein, putative | 2.60 |
| 1888 | 249057_at | AT5G44480 | DUR (DEFECTIVE UGE IN ROOT); catalytic | 3.05 |
| 1889 | 249061_at | AT5G44550 | integral membrane family protein | 3.46 |
| 1890 | 249010_at | AT5G44580 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G44582.1) | -2.56 |
| 1891 | 249009_at | AT5G44610 | MAP18 (MICROTUBULE-ASSOCIATED PROTEIN 18); microtubule binding | 4.18 |
| 1892 | 249007_at | AT5G44650 | similar to unnamed protein product [Vitis vinifera] (GB:CAO44475.1) | -3.26 |
| 1893 | 249008_at | AT5G44680 | methyladenine glycosylase family protein | -3.03 |
| 1894 | 249027_at | AT5G44790 | RAN1 (RESPONSIVE-TO-ANTAGONIST1); ATPase, coupled to transmembrane movement of ions, phosphorylative mechanism | 2.30 |
| 1895 | 249029_at | AT5G44870 | disease resistance protein (TIR-NBS-LRR class), putative | -4.24 |
| 1896 | 249033_at | AT5G44920 | Toll-Interleukin-Resistance (TIR) domain-containing protein | 4.43 |
| 1897 | 248978_at | AT5G45070 | ATPP2-A8 (Phloem protein 2-A8); transmembrane receptor | 3.55 |
| 1898 | 248979_at | AT5G45080 | ATPP2-A6 (Phloem protein 2-A6); transmembrane receptor | 2.59 |
| 1899 | 248990_at | AT5G45210 | disease resistance protein (TIR-NBS-LRR class), putative | 5.29 |
| 1900 | 248964_at | AT5G45340 | CYP707A3 (cytochrome P450, family 707, subfamily A, polypeptide 3); oxygen binding | 2.53 |
| 1901 | 248961_at | AT5G45650 | subtilase family protein | -3.17 |
| 1902 | 248962_at | AT5G45680 | FK506-binding protein 1 (FKBP13) | -4.91 |
| 1903 | 248920_at | AT5G45930 | CHLI2; magnesium chelatase | -6.35 |
| 1904 | 248921_at | AT5G45950 | GDSL-motif lipase/hydrolase family protein | -7.89 |
| 1905 | 248886_at | AT5G46110 | APE2 (ACCLIMATION OF PHOTOSYNTHESIS TO ENVIRONMENT); antiporter/ triose-phosphate transmembrane transporter | -5.76 |
| 1906 | 248888_at | AT5G46240 | KAT1 (K+ ATPASE 1); cyclic nucleotide binding / inward rectifier potassium channel | -6.01 |
| 1907 | 248854_at | AT5G46580 | pentatricopeptide (PPR) repeat-containing protein | -2.94 |
| 1908 | 248828_at | AT5G47110 | lil3 protein, putative | -4.99 |
| 1909 | 248798_at | AT5G47190 | ribosomal protein L19 family protein | -2.71 |
| 1910 | 248799_at | AT5G47230 | ERF5 (ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 5); DNA binding / transcription activator/ transcription factor | 2.90 |
| 1911 | 248784_at | AT5G47380 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G66600.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G66600.3); similar to hypothetical protein 24.t00022 [Brassica oleracea] (GB:ABD64944.1); contains InterPro domain Protein of unknown function DUF547 (InterPro:IPR006869) | -4.63 |
| 1912 | 248790_at | AT5G47450 | AtTIP2;3 (Arabidopsis thaliana tonoplast intrinsic protein 2;3); water channel | 6.03 |
| 1913 | 248759_at | AT5G47610 | zinc finger (C3HC4-type RING finger) family protein | -3.38 |
| 1914 | 248723_at | AT5G47950 | transferase family protein | 3.14 |
| 1915 | 248725_at | AT5G47980 | transferase family protein | 4.42 |
| 1916 | 248727_at | AT5G47990 | CYP705A5 (cytochrome P450, family 705, subfamily A, polypeptide 5); oxygen binding | 5.02 |
| 1917 | 248728_at | AT5G48000 | CYP708A2 (cytochrome P450, family 708, subfamily A, polypeptide 2); oxygen binding | 7.37 |
| 1918 | 248729_at | AT5G48010 | pentacyclic triterpene synthase, putative | 5.93 |
| 1919 | 248732_at | AT5G48070 | ATXTH20 (XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 20); hydrolase, acting on glycosyl bonds | 2.99 |
| 1920 | 248736_at | AT5G48110 | terpene synthase/cyclase family protein | 7.18 |
| 1921 | 248687_at | AT5G48300 | ADG1 (ADP GLUCOSE PYROPHOSPHORYLASE SMALL SUBUNIT 1); glucose-1-phosphate adenylyltransferase | -3.25 |
| 1922 | 248703_at | AT5G48430 | aspartic-type endopeptidase/ pepsin A | 4.11 |
| 1923 | 248683_at | AT5G48490 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | -3.93 |
| 1924 | 248708_at | AT5G48560 | basic helix-loop-helix (bHLH) family protein | 3.64 |
| 1925 | 248657_at | AT5G48570 | peptidyl-prolyl cis-trans isomerase, putative / FK506-binding protein, putative | 3.47 |
| 1926 | 248624_at | AT5G48790 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G73060.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO49997.1) | -2.58 |
| 1927 | 248625_at | AT5G48880 | KAT5/PKT1/PKT2 (PEROXISOMAL 3-KETO-ACYL-COA THIOLASE 1, PEROXISOMAL 3-KETO-ACYL-COA THIOLASE 2); acetyl-CoA C-acyltransferase | -4.78 |
| 1928 | 248626_at | AT5G48940 | leucine-rich repeat transmembrane protein kinase, putative | 3.27 |
| 1929 | 248621_at | AT5G49350 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G56320.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO44880.1); contains domain PTHR10499 (PTHR10499) | 3.68 |
| 1930 | 248569_at | AT5G49770 | leucine-rich repeat transmembrane protein kinase, putative | 3.75 |
| 1931 | 248577_at | AT5G49870 | jacalin lectin family protein | 3.21 |
| 1932 | 248537_at | AT5G50100 | similar to PBng143 [Vigna radiata] (GB:BAB82450.1); contains InterPro domain Thioredoxin-like fold (InterPro:IPR012336); contains InterPro domain Putative thiol-disulphide oxidoreductase DCC (InterPro:IPR007263) | -5.83 |
| 1933 | 248535_at | AT5G50120 | transducin family protein / WD-40 repeat family protein | 2.53 |
| 1934 | 248540_at | AT5G50160 | ATFRO8/FRO8 (FERRIC REDUCTION OXIDASE 8); ferric-chelate reductase/ oxidoreductase | -2.99 |
| 1935 | 248509_at | AT5G50335 | unknown protein | -5.74 |
| 1936 | 248460_at | AT5G50915 | basic helix-loop-helix (bHLH) family protein | -2.92 |
| 1937 | 248491_at | AT5G51010 | rubredoxin family protein | -2.95 |
| 1938 | 248486_at | AT5G51060 | RHD2 (ROOT HAIR DEFECTIVE 2) | 3.15 |
| 1939 | 248449_at | AT5G51110 | similar to dehydratase family [Arabidopsis thaliana] (TAIR:AT1G29810.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO62291.1); contains InterPro domain Transcriptional coactivator/pterin dehydratase (InterPro:IPR001533) | -4.62 |
| 1940 | 248448_at | AT5G51190 | AP2 domain-containing transcription factor, putative | 3.07 |
| 1941 | 248434_at | AT5G51440 | 23.5 kDa mitochondrial small heat shock protein (HSP23.5-M) | 4.29 |
| 1942 | 248406_at | AT5G51490 | pectinesterase family protein | 3.71 |
| 1943 | 248407_at | AT5G51500 | pectinesterase family protein | 2.29 |
| 1944 | 248408_at | AT5G51520 | invertase/pectin methylesterase inhibitor family protein | 2.55 |
| 1945 | 248416_at | AT5G51630 | disease resistance protein (TIR-NBS-LRR class), putative | 2.43 |
| 1946 | 248377_at | AT5G51720 | similar to Os07g0467200 [Oryza sativa (japonica cultivar-group)] (GB:NP_001059590.1); similar to hypothetical protein OsI_025030 [Oryza sativa (indica cultivar-group)] (GB:EAZ03798.1); contains domain PTHR13680 (PTHR13680); contains domain PTHR13680:SF1 (PTHR13680:SF1) | -8.58 |
| 1947 | 248427_at | AT5G51750 | ATSBT1.3; subtilase | -2.76 |
| 1948 | 248400_at | AT5G52020 | AP2 domain-containing protein | 3.27 |
| 1949 | 248392_at | AT5G52050 | MATE efflux protein-related | 2.52 |
| 1950 | 248402_at | AT5G52100 | CRR1 (CHLORORESPIRATION REDUCTION 1); dihydrodipicolinate reductase | -3.43 |
| 1951 | 248348_at | AT5G52190 | sugar isomerase (SIS) domain-containing protein | -2.64 |
| 1952 | 248337_at | AT5G52310 | COR78 (COLD REGULATED 78) | -2.75 |
| 1953 | 248353_at | AT5G52320 | CYP96A4 (cytochrome P450, family 96, subfamily A, polypeptide 4); oxygen binding | -2.67 |
| 1954 | 248338_at | AT5G52440 | HCF106 (High chlorophyll fluorescence 106) | -2.63 |
| 1955 | 248332_at | AT5G52640 | HSP81-1 (HEAT SHOCK PROTEIN 81-1); ATP binding / unfolded protein binding | 3.50 |
| 1956 | 248329_at | AT5G52780 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G19100.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO49500.1) | -4.41 |
| 1957 | 248287_at | AT5G52970 | thylakoid lumen 15.0 kDa protein | -6.38 |
| 1958 | 248252_at | AT5G53250 | AGP22/ATAGP22 (ARABINOGALACTAN PROTEINS 22) | 7.15 |
| 1959 | 248254_at | AT5G53320 | leucine-rich repeat transmembrane protein kinase, putative | 3.38 |
| 1960 | 248224_at | AT5G53490 | thylakoid lumenal 17.4 kDa protein, chloroplast | -3.70 |
| 1961 | 248242_at | AT5G53580 | aldo/keto reductase family protein | -3.04 |
| 1962 | 248207_at | AT5G53970 | aminotransferase, putative | -5.58 |
| 1963 | 248209_at | AT5G53990 | glycosyltransferase family protein | 4.02 |
| 1964 | 248203_at | AT5G54230 | MYB49 (myb domain protein 49); transcription factor | 2.56 |
| 1965 | 248153_at | AT5G54250 | ATCNGC4 (DEFENSE, NO DEATH 2); calmodulin binding / cation channel/ cyclic nucleotide binding | -7.39 |
| 1966 | 248151_at | AT5G54270 | LHCB3 (LIGHT-HARVESTING CHLOROPHYLL BINDING PROTEIN 3) | -11.40 |
| 1967 | 248181_at | AT5G54290 | cytochrome c biogenesis protein family | -3.20 |
| 1968 | 248178_at | AT5G54370 | late embryogenesis abundant protein-related / LEA protein-related | 5.60 |
| 1969 | 248164_at | AT5G54490 | PBP1 (PINOID-BINDING PROTEIN 1); calcium ion binding | 2.72 |
| 1970 | 248174_at | AT5G54600 | 50S ribosomal protein L24, chloroplast (CL24) | -2.86 |
| 1971 | 248177_at | AT5G54630 | zinc finger protein-related | -3.26 |
| 1972 | 248128_at | AT5G54770 | THI1 (THIAZOLE REQUIRING) | -5.80 |
| 1973 | 248094_at | AT5G55220 | trigger factor type chaperone family protein | -4.07 |
| 1974 | 248062_at | AT5G55450 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | -6.71 |
| 1975 | 248028_at | AT5G55620 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G09950.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO39940.1) | -3.55 |
| 1976 | 248075_at | AT5G55740 | CRR21 (CHLORORESPIRATORY REDUCTION 21) | -3.43 |
| 1977 | 247991_at | AT5G56320 | ATEXPA14 (ARABIDOPSIS THALIANA EXPANSIN A14) | 6.07 |
| 1978 | 247965_at | AT5G56540 | AGP14 (ARABINOGALACTAN PROTEIN 14) | 5.10 |
| 1979 | 247977_at | AT5G56850 | similar to unknown [Populus trichocarpa x Populus deltoides] (GB:ABK96358.1) | -5.16 |
| 1980 | 247980_at | AT5G56860 | GNC (GATA, NITRATE-INDUCIBLE, CARBON METABOLISM-INVOLVED); transcription factor | -5.84 |
| 1981 | 247954_at | AT5G56870 | BGAL4 (beta-galactosidase 4); beta-galactosidase | 2.40 |
| 1982 | 247936_at | AT5G57030 | LUT2 (LUTEIN DEFICIENT 2); lycopene epsilon cyclase | -3.25 |
| 1983 | 247931_at | AT5G57040 | lactoylglutathione lyase family protein / glyoxalase I family protein | -3.08 |
| 1984 | 247947_at | AT5G57090 | EIR1 (ETHYLENE INSENSITIVE ROOT 1); auxin:hydrogen symporter/ transporter | 3.86 |
| 1985 | 247946_at | AT5G57180 | CIA2 (CHLOROPLAST IMPORT APPARATUS 2) | -3.94 |
| 1986 | 247899_at | AT5G57345 | similar to unknown [Populus trichocarpa x Populus deltoides] (GB:ABK96592.1) | -4.63 |
| 1987 | 247912_at | AT5G57480 | AAA-type ATPase family protein | 2.31 |
| 1988 | 247913_at | AT5G57510 | unknown protein | 3.15 |
| 1989 | 247871_at | AT5G57530 | xyloglucan:xyloglucosyl transferase, putative / xyloglucan endotransglycosylase, putative / endo-xyloglucan transferase, putative | 3.51 |
| 1990 | 247868_at | AT5G57620 | MYB36 (myb domain protein 36); DNA binding / transcription factor | 4.25 |
| 1991 | 247920_at | AT5G57670 | protein kinase family protein | 2.32 |
| 1992 | 247874_at | AT5G57710 | heat shock protein-related | -2.84 |
| 1993 | 247879_at | AT5G57770 | similar to unnamed protein product [Vitis vinifera] (GB:CAO66578.1); contains InterPro domain Pleckstrin-like, plant (InterPro:IPR013666); contains InterPro domain Protein of unknown function DUF828, plant (InterPro:IPR008546) | 2.38 |
| 1994 | 247882_at | AT5G57785 | unknown protein | -3.97 |
| 1995 | 247884_at | AT5G57800 | CER3/FLP1/WAX2/YRE (ECERIFERUM 3); catalytic | -3.33 |
| 1996 | 247893_at | AT5G57980 | eukaryotic rpb5 RNA polymerase subunit family protein | 2.51 |
| 1997 | 247853_at | AT5G58140 | PHOT2 (NON PHOTOTROPIC HYPOCOTYL 1-LIKE); kinase | -5.20 |
| 1998 | 247862_at | AT5G58250 | similar to hypothetical protein [Vitis vinifera] (GB:CAN75840.1); contains domain PD020337 (PD020337) | -3.00 |
| 1999 | 247816_at | AT5G58260 | Encodes subunit NDH-N of NAD(P)H:plastoquinone dehydrogenase complex (Ndh complex) present in the thylakoid membrane of chloroplasts. This subunit is thought to be required for Ndh complex assembly. | -4.32 |
| 2000 | 247814_at | AT5G58310 | hydrolase, alpha/beta fold family protein | -4.04 |
| 2001 | 247813_at | AT5G58330 | malate dehydrogenase (NADP), chloroplast, putative | -7.75 |
| 2002 | 247789_at | AT5G58680 | armadillo/beta-catenin repeat family protein | 4.35 |
| 2003 | 247778_at | AT5G58750 | wound-responsive protein-related | 3.30 |
| 2004 | 247780_at | AT5G58770 | dehydrodolichyl diphosphate synthase, putative / DEDOL-PP synthase, putative | -2.68 |
| 2005 | 247765_at | AT5G58860 | CYP86A1 (cytochrome P450, family 86, subfamily A, polypeptide 1); oxygen binding | 3.95 |
| 2006 | 247769_at | AT5G58910 | LAC16 (laccase 16); copper ion binding / oxidoreductase | 2.71 |
| 2007 | 247755_at | AT5G59090 | ATSBT4.12; subtilase | 6.97 |
| 2008 | 247760_at | AT5G59130 | subtilase family protein | -3.77 |
| 2009 | 247739_at | AT5G59240 | 40S ribosomal protein S8 (RPS8B) | 2.31 |
| 2010 | 247709_at | AT5G59250 | sugar transporter family protein | -6.50 |
| 2011 | 247719_at | AT5G59305 | similar to unknown [Populus trichocarpa] (GB:ABK94520.1) | 5.45 |
| 2012 | 247718_at | AT5G59310 | LTP4 (LIPID TRANSFER PROTEIN 4); lipid binding | -4.34 |
| 2013 | 247727_at | AT5G59490 | haloacid dehalogenase-like hydrolase family protein | 3.04 |
| 2014 | 247678_at | AT5G59520 | ZIP2 (ZINC TRANSPORTER 2 PRECURSOR); transferase, transferring glycosyl groups / zinc ion transmembrane transporter | 2.42 |
| 2015 | 247729_at | AT5G59530 | 2-oxoglutarate-dependent dioxygenase, putative | 5.00 |
| 2016 | 247679_at | AT5G59540 | oxidoreductase, 2OG-Fe(II) oxygenase family protein | -2.48 |
| 2017 | 247685_at | AT5G59680 | leucine-rich repeat protein kinase, putative | 3.46 |
| 2018 | 247694_at | AT5G59750 | riboflavin biosynthesis protein, putative | -2.89 |
| 2019 | 247696_at | AT5G59780 | MYB59 (myb domain protein 59); DNA binding / transcription factor | 2.88 |
| 2020 | 247655_at | AT5G59820 | RHL41 (RESPONSIVE TO HIGH LIGHT 41); nucleic acid binding / transcription factor/ zinc ion binding | 2.43 |
| 2021 | 247674_at | AT5G59930 | DC1 domain-containing protein / UV-B light-insensitive protein, putative | 2.91 |
| 2022 | 247634_at | AT5G60520 | late embryogenesis abundant protein-related / LEA protein-related | 4.92 |
| 2023 | 247645_at | AT5G60530 | late embryogenesis abundant protein-related / LEA protein-related | 4.04 |
| 2024 | 247637_at | AT5G60600 | GcpE (CHLOROPLAST BIOGENESIS 4); 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase | -2.87 |
| 2025 | 247586_at | AT5G60660 | PIP2;4/PIP2F (plasma membrane intrinsic protein 2;4); water channel | 4.68 |
| 2026 | 247582_at | AT5G60760 | 2-phosphoglycerate kinase-related | 3.87 |
| 2027 | 247604_at | AT5G60950 | COBL5 (COBRA-LIKE PROTEIN 5 PRECURSOR) | 3.32 |
| 2028 | 247523_at | AT5G61410 | RPE (EMBRYO DEFECTIVE 2728); ribulose-phosphate 3-epimerase | -3.50 |
| 2029 | 247543_at | AT5G61600 | ethylene-responsive element-binding family protein | 2.98 |
| 2030 | 247486_at | AT5G62140 | similar to unknown [Populus trichocarpa] (GB:ABK94834.1) | -4.27 |
| 2031 | 247469_at | AT5G62165 | AGL42 (AGAMOUS LIKE 42); transcription factor | 3.44 |
| 2032 | 247458_at | AT5G62180 | ATCXE20 (ARABIDOPSIS THALIANA CARBOXYESTERASE 20); carboxylesterase | 3.18 |
| 2033 | 247463_at | AT5G62210 | embryo-specific protein-related | -6.32 |
| 2034 | 247476_at | AT5G62330 | similar to invertase/pectin methylesterase inhibitor family protein [Arabidopsis thaliana] (TAIR:AT5G62340.1) | 5.92 |
| 2035 | 247477_at | AT5G62340 | invertase/pectin methylesterase inhibitor family protein | 5.95 |
| 2036 | 247435_at | AT5G62480 | ATGSTU9 (GLUTATHIONE S-TRANSFERASE TAU 9); glutathione transferase | 2.89 |
| 2037 | 247431_at | AT5G62520 | SRO5 (SIMILAR TO RCD ONE 5); NAD+ ADP-ribosyltransferase | 2.42 |
| 2038 | 247400_at | AT5G62840 | phosphoglycerate/bisphosphoglycerate mutase family protein | -3.59 |
| 2039 | 247377_at | AT5G63180 | pectate lyase family protein | -6.54 |
| 2040 | 247359_at | AT5G63560 | transferase family protein | 2.71 |
| 2041 | 247354_at | AT5G63590 | FLS (Flavonol synthase); flavonol synthase | 5.78 |
| 2042 | 247333_at | AT5G63600 | flavonol synthase, putative | 9.69 |
| 2043 | 247337_at | AT5G63660 | LCR74/PDF2.5 (Low-molecular-weight cysteine-rich 74) | 5.41 |
| 2044 | 247320_at | AT5G64040 | PSAN (photosystem I reaction center subunit PSI-N); calmodulin binding | -6.33 |
| 2045 | 247297_at | AT5G64100 | peroxidase, putative | 6.67 |
| 2046 | 247252_at | AT5G64770 | similar to 80C09_10 [Brassica rapa subsp. pekinensis] (GB:AAZ41821.1) | -6.40 |
| 2047 | 247208_at | AT5G64870 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G25250.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G25260.1); similar to 80C09_16 [Brassica rapa subsp. pekinensis] (GB:AAZ41827.1); contains domain PTHR13806:SF3 (PTHR13806:SF3); contains domain PTHR13806 (PTHR13806) | 3.11 |
| 2048 | 247205_at | AT5G64890 | PROPEP2 (Elicitor peptide 2 precursor) | 3.86 |
| 2049 | 247213_at | AT5G64900 | ATPEP1/PROPEP1 (Elicitor peptide 1 precursor) | 2.42 |
| 2050 | 247215_at | AT5G64905 | PROPEP3 (Elicitor peptide 3 precursor) | 3.27 |
| 2051 | 247232_at | AT5G64940 | ATATH13 (ABC2 homolog 13) | -7.26 |
| 2052 | 247199_at | AT5G65210 | TGA1; DNA binding / calmodulin binding / transcription factor | 3.60 |
| 2053 | 247201_at | AT5G65220 | ribosomal protein L29 family protein | -2.48 |
| 2054 | 247177_at | AT5G65300 | unknown protein | 2.29 |
| 2055 | 247189_at | AT5G65390 | AGP7 (Arabinogalactan protein 7) | 2.39 |
| 2056 | 247182_at | AT5G65410 | ATHB25/ZFHD2 (ZINC FINGER HOMEODOMAIN 2); transcription factor | -4.26 |
| 2057 | 247162_at | AT5G65730 | xyloglucan:xyloglucosyl transferase, putative / xyloglucan endotransglycosylase, putative / endo-xyloglucan transferase, putative | -8.82 |
| 2058 | 247165_at | AT5G65790 | MYB68 (myb domain protein 68); DNA binding / transcription factor | 5.97 |
| 2059 | 247166_at | AT5G65840 | similar to antioxidant/ oxidoreductase [Arabidopsis thaliana] (TAIR:AT2G37240.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN81556.1); contains InterPro domain Thioredoxin-like fold (InterPro:IPR012336); contains InterPro domain Thioredoxin fold (InterPro:IPR012335) | -2.84 |
| 2060 | 247118_at | AT5G65890 | ACR1 (ACT DOMAIN REPEAT 1) | -3.83 |
| 2061 | 247116_at | AT5G65970 | MLO10 (MILDEW RESISTANCE LOCUS O 10); calmodulin binding | 3.47 |
| 2062 | 247120_at | AT5G65990 | amino acid transporter family protein | -3.02 |
| 2063 | 247136_at | AT5G66170 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G17850.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO48196.1); contains InterPro domain Rhodanese-like (InterPro:IPR001763) | 2.36 |
| 2064 | 247131_at | AT5G66190 | ATLFNR1 (LEAF FNR 1); NADPH dehydrogenase/ electron transporter, transferring electrons within the cyclic electron transport pathway of photosynthesis/ electron transporter, transferring electrons within the noncyclic electron transport pathway of photosy | -6.06 |
| 2065 | 247094_at | AT5G66280 | GMD1 (GDP-D-MANNOSE 4,6-DEHYDRATASE 1); GDP-mannose 4,6-dehydratase | 3.79 |
| 2066 | 247091_at | AT5G66390 | peroxidase 72 (PER72) (P72) (PRXR8) | 6.35 |
| 2067 | 247098_at | AT5G66470 | GTP binding / RNA binding | -3.30 |
| 2068 | 247100_at | AT5G66520 | pentatricopeptide (PPR) repeat-containing protein | -2.62 |
| 2069 | 247073_at | AT5G66570 | OE33/OEE1/OEE33/PSBO-1/PSBO1 (OXYGEN-EVOLVING ENHANCER 33); oxygen evolving/ poly(U) binding | -7.15 |
| 2070 | 247047_at | AT5G66650 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G23790.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO22909.1); contains InterPro domain Protein of unknown function DUF607 (InterPro:IPR006769) | 3.21 |
| 2071 | 247059_at | AT5G66690 | UGT72E2; UDP-glycosyltransferase/ coniferyl-alcohol glucosyltransferase/ transferase, transferring glycosyl groups | 2.99 |
| 2072 | 247052_at | AT5G66700 | HB53 (homeobox-8); DNA binding / transcription factor | 2.32 |
| 2073 | 247055_at | AT5G66740 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G75160.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN74294.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO22871.1); contains InterPro domain Protein of unknown function DUF620 (InterPro:IPR006873) | -4.06 |
| 2074 | 247070_at | AT5G66815 | unknown protein | 3.98 |
| 2075 | 247024_at | AT5G66985 | unknown protein | 7.04 |
| 2076 | 247025_at | AT5G67030 | ABA1 (ABA DEFICIENT 1); zeaxanthin epoxidase | -2.61 |
| 2077 | 246998_at | AT5G67370 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G11840.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN75014.1); contains InterPro domain Protein of unknown function DUF1230 (InterPro:IPR009631) | -4.64 |
| 2078 | 246991_at | AT5G67400 | peroxidase 73 (PER73) (P73) (PRXR11) | 4.05 |
| 2079 | 246992_at | AT5G67430 | GCN5-related N-acetyltransferase (GNAT) family protein | 2.30 |
| 2080 | 245047_at | ATCG00020 | Encodes chlorophyll binding protein D1, a part of the photosystem II reaction center core | -4.94 |
| 2081 | 245049_at | ATCG00050 | Homologous to the bacterial ribosomal protein S16 | -2.60 |
| 2082 | 244939_at | ATCG00065 | chloroplast gene encoding ribosomal protein s12. The gene is located in three distinct loci on the chloroplast genome and is transpliced to make one transcript. | -2.55 |
| 2083 | 245050_at | ATCG00070 | PSII K protein | -3.06 |
| 2084 | 245024_at | ATCG00120 | Encodes the ATPase alpha subunit, which is a subunit of ATP synthase and part of the CF1 portion which catalyzes the conversion of ADP to ATP using the proton motive force. This complex is located in the thylakoid membrane of the chloroplast. | -3.11 |
| 2085 | 245025_at | ATCG00130 | ATPase F subunit. | -3.99 |
| 2086 | 245026_at | ATCG00140 | ATPase III subunit | -3.66 |
| 2087 | 244995_at | ATCG00150 | Encodes a subunit of ATPase complex CF0, which is a proton channel that supplies the proton motive force to drive ATP synthesis by CF1 portion of the complex. | -3.20 |
| 2088 | 245002_at | ATCG00270 | PSII D2 protein | -3.77 |
| 2089 | 245003_at | ATCG00280 | chloroplast gene encoding a CP43 subunit of the photosystem II reaction center. promoter contains a blue-light responsive element. | -4.69 |
| 2090 | 245004_at | ATCG00300 | encodes PsbZ, which is a subunit of photosystem II. In Chlamydomonas, this protein has been shown to be essential in the interaction between PS II and the light harvesting complex II. | -3.94 |
| 2091 | 245006_at | ATCG00340 | Encodes the D1 subunit of photosystem I and II reaction centers. | -4.02 |
| 2092 | 245007_at | ATCG00350 | Encodes psaA protein comprising the reaction center for photosystem I along with psaB protein; hydrophobic protein encoded by the chloroplast genome. | -4.73 |
| 2093 | 245009_at | ATCG00380 | Chloroplast encoded ribosomal protein S4 | -3.55 |
| 2094 | 245010_at | ATCG00420 | Encodes NADH dehydrogenase subunit J. Its transcription is increased upon sulfur depletion. | -4.25 |
| 2095 | 245011_at | ATCG00430 | Encodes a protein which was originally thought to be part of photosystem II but its wheat homolog was later shown to encode for subunit K of NADH dehydrogenase. | -4.01 |
| 2096 | 245015_at | ATCG00490 | large subunit of RUBISCO. | -3.01 |
| 2097 | 245018_at | ATCG00520 | Encodes a protein required for photosystem I assembly and stability. In cyanobacteria, loss of function mutation in this gene increases PSII/PSI ratio without any influence on photoautotrophic growth. | -2.55 |
| 2098 | 245019_at | ATCG00530 | hypothetical protein | -2.72 |
| 2099 | 245020_at | ATCG00540 | Encodes cytochrome f apoprotein; involved in photosynthetic electron transport chain; encoded by the chloroplast genome and is transcriptionally repressed by a nuclear gene HCF2. | -2.65 |
| 2100 | 244963_at | ATCG00570 | PSII cytochrome b559 | -2.52 |
| 2101 | 244964_at | ATCG00580 | PSII cytochrome b559. There have been many speculations about the function of Cyt b559, but the most favored at present is that it plays a protective role by acting as an electron acceptor or electron donor under conditions when electron flow through PSII is not optimized. | -2.83 |
| 2102 | 244965_at | ATCG00590 | hypothetical protein | -2.93 |
| 2103 | 244967_at | ATCG00630 | Encodes subunit J of photosystem I. | -2.78 |
| 2104 | 244972_at | ATCG00680 | encodes for CP47, subunit of the photosystem II reaction center. | -3.08 |
| 2105 | 244973_at | ATCG00690 | Encodes photosystem II 5 kD protein subunit PSII-T. This is a plastid-encoded gene (PsbTc) which also has a nuclear-encoded paralog (PsbTn). | -2.60 |
| 2106 | 244974_at | ATCG00700 | PSII low MW protein | -4.32 |
| 2107 | 244975_at | ATCG00710 | Encodes a 8 kD phosphoprotein that is a component of the photosystem II oxygen evolving core. Its exact molecular function has not been determined but it may play a role in mediating electron transfer between the secondary quinone acceptors, QA and QB, associated with the acceptor side of PSII. | -4.00 |
| 2108 | 244961_at | ATCG01040 | hypothetical protein | -2.87 |
| 2109 | 244962_at | ATCG01050 | Represents a plastid-encoded subunit of a NAD(P)H dehydrogenase complex. Its mRNA is edited at four positions. Translation data is not available for this gene. | -2.88 |
| 2110 | 244932_at | ATCG01060 | Encodes the PsaC subunit of photosystem I. | -3.27 |
| 2111 | 244933_at | ATCG01070 | NADH dehydrogenase ND4L | -2.79 |
| 2112 | 244936_at | ATCG01100 | NADH dehydrogenase ND1 | -2.82 |