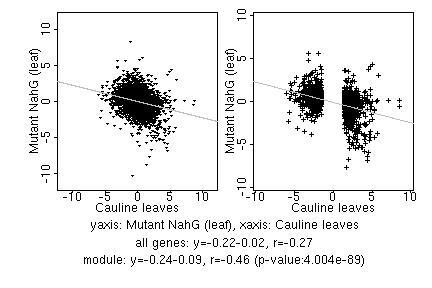
marker gene list
| Cauline leaves |
| Mutant NahG(leaf) |
| probeID | AGICode | Annotation | gene expression |
|
|---|---|---|---|---|
| 1 | 261569_at | AT1G01060 | LHY (LATE ELONGATED HYPOCOTYL); DNA binding / transcription factor | 2.31 |
| 2 | 261578_at | AT1G01100 | 60S acidic ribosomal protein P1 (RPP1A) | -1.61 |
| 3 | 261581_at | AT1G01140 | CIPK9 (CBL-INTERACTING PROTEIN KINASE 9); kinase | 1.75 |
| 4 | 261574_at | AT1G01190 | CYP78A8 (cytochrome P450, family 78, subfamily A, polypeptide 8); oxygen binding | -2.86 |
| 5 | 261042_at | AT1G01200 | AtRABA3 (Arabidopsis Rab GTPase homolog A3); GTP binding | -2.52 |
| 6 | 261056_at | AT1G01360 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G01026.1); similar to hypothetical protein [Arabidopsis suecica] (GB:BAF63139.1); contains domain SSF55961 (SSF55961) | 1.75 |
| 7 | 261046_at | AT1G01390 | UDP-glucoronosyl/UDP-glucosyl transferase family protein | 2.81 |
| 8 | 259429_at | AT1G01600 | CYP86A4 (cytochrome P450, family 86, subfamily A, polypeptide 4); oxygen binding | -1.77 |
| 9 | 261586_at | AT1G01640 | speckle-type POZ protein-related | 1.84 |
| 10 | 261538_at | AT1G01830 | armadillo/beta-catenin repeat family protein | 1.65 |
| 11 | 260933_at | AT1G02470 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G02475.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO40177.1); contains InterPro domain Streptomyces cyclase/dehydrase (InterPro:IPR005031) | 3.19 |
| 12 | 260910_at | AT1G02690 | importin alpha-2 subunit, putative | -1.76 |
| 13 | 262109_at | AT1G02730 | ATCSLD5 (CELLULOSE SYNTHASE-LIKE D5); 1,4-beta-D-xylan synthase/ cellulose synthase | -2.08 |
| 14 | 262121_at | AT1G02800 | ATCEL2 (Arabidopsis thaliana Cellulase 2); hydrolase, hydrolyzing O-glycosyl compounds | -2.56 |
| 15 | 262118_at | AT1G02850 | glycosyl hydrolase family 1 protein | 2.48 |
| 16 | 264843_at | AT1G03400 | 2-oxoglutarate-dependent dioxygenase, putative | 1.83 |
| 17 | 264822_at | AT1G03457 | RNA-binding protein, putative | -2.05 |
| 18 | 265085_at | AT1G03780 | targeting protein-related | -3.92 |
| 19 | 265066_at | AT1G03870 | FLA9 | -1.47 |
| 20 | 265097_at | AT1G04020 | ATBARD1/BARD1 (BREAST CANCER ASSOCIATED RING 1); transcription coactivator | -1.89 |
| 21 | 263668_at | AT1G04350 | 2-oxoglutarate-dependent dioxygenase, putative | 1.78 |
| 22 | 263677_at | AT1G04520 | 33 kDa secretory protein-related | -3.08 |
| 23 | 264606_at | AT1G04660 | glycine-rich protein | -1.82 |
| 24 | 261129_at | AT1G04820 | TUA4 (tubulin alpha-4 chain) | -2.26 |
| 25 | 265194_at | AT1G05010 | EFE (ETHYLENE FORMING ENZYME) | 1.67 |
| 26 | 264581_at | AT1G05210 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G32380.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO24140.1); contains InterPro domain Transmembrane protein 97, predicted (InterPro:IPR016964) | -4.06 |
| 27 | 263184_at | AT1G05560 | UGT1 (UDP-glucosyl transferase 75B1); UDP-glycosyltransferase/ transferase, transferring glycosyl groups | 1.70 |
| 28 | 257451_at | AT1G05690 | BT3 (BTB and TAZ domain protein 3); protein binding / transcription regulator | 1.76 |
| 29 | 260955_at | AT1G06000 | UDP-glucoronosyl/UDP-glucosyl transferase family protein | -2.70 |
| 30 | 260957_at | AT1G06080 | ADS1 (DELTA 9 DESATURASE 1); oxidoreductase | -1.43 |
| 31 | 260783_at | AT1G06160 | ethylene-responsive factor, putative | 2.85 |
| 32 | 259394_at | AT1G06420 | unknown protein | -2.96 |
| 33 | 262619_at | AT1G06550 | enoyl-CoA hydratase/isomerase family protein | -1.79 |
| 34 | 262635_at | AT1G06570 | PDS1 (PHYTOENE DESATURATION 1) | 1.70 |
| 35 | 256066_at | AT1G06980 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G30230.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO21365.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN59819.1) | -2.07 |
| 36 | 256050_at | AT1G07000 | ATEXO70B2 (exocyst subunit EXO70 family protein B2); protein binding | 2.02 |
| 37 | 256065_at | AT1G07070 | 60S ribosomal protein L35a (RPL35aA) | -2.13 |
| 38 | 261080_at | AT1G07370 | PCNA1 (PROLIFERATING CELLULAR NUCLEAR ANTIGEN); DNA binding / DNA polymerase processivity factor | -2.44 |
| 39 | 261077_at | AT1G07430 | protein phosphatase 2C, putative / PP2C, putative | 2.66 |
| 40 | 261410_at | AT1G07610 | MT1C (metallothionein 1C) | -1.50 |
| 41 | 261692_at | AT1G08450 | CRT3 (CALRETICULIN 3); calcium ion binding | 2.39 |
| 42 | 264802_at | AT1G08560 | SYP111 (syntaxin 111); SNAP receptor | -3.22 |
| 43 | 264648_at | AT1G09080 | BIP3; ATP binding | 3.08 |
| 44 | 264263_at | AT1G09155 | ATPP2-B15 (Phloem protein 2-B15); carbohydrate binding | -2.86 |
| 45 | 264266_at | AT1G09160 | protein phosphatase 2C-related / PP2C-related | -1.61 |
| 46 | 264262_at | AT1G09200 | histone H3 | -2.88 |
| 47 | 264261_at | AT1G09240 | nicotianamine synthase, putative | 4.45 |
| 48 | 264672_at | AT1G09750 | chloroplast nucleoid DNA-binding protein-related | -1.82 |
| 49 | 264523_at | AT1G10030 | ERG28 (ARABIDOPSIS HOMOLOG OF YEAST ERGOSTEROL28) | -1.42 |
| 50 | 264522_at | AT1G10050 | glycosyl hydrolase family 10 protein / carbohydrate-binding domain-containing protein | 1.58 |
| 51 | 264524_at | AT1G10070 | ATBCAT-2; branched-chain-amino-acid transaminase/ catalytic | 2.90 |
| 52 | 264453_at | AT1G10300 | GTP-binding protein-related | -2.30 |
| 53 | 264434_at | AT1G10340 | ankyrin repeat family protein | 2.18 |
| 54 | 261834_at | AT1G10640 | polygalacturonase, putative / pectinase, putative | -3.35 |
| 55 | 262788_at | AT1G10690 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G60783.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN59885.1) | 2.44 |
| 56 | 262758_at | AT1G10780 | F-box family protein | -1.62 |
| 57 | 262783_at | AT1G10850 | ATP binding / protein serine/threonine kinase | -1.86 |
| 58 | 260483_at | AT1G11000 | MLO4 (MILDEW RESISTANCE LOCUS O 4); calmodulin binding | -3.10 |
| 59 | 262452_at | AT1G11210 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G11220.1); similar to cotton fiber expressed protein 1 [Gossypium hirsutum] (GB:AAC33276.1); contains InterPro domain Protein of unknown function DUF761, plant (InterPro:IPR008480) | 2.07 |
| 60 | 262508_at | AT1G11300 | carbohydrate binding / kinase | -2.58 |
| 61 | 261873_at | AT1G11350 | S-locus lectin protein kinase family protein | 1.98 |
| 62 | 261825_at | AT1G11545 | xyloglucan:xyloglucosyl transferase, putative / xyloglucan endotransglycosylase, putative / endo-xyloglucan transferase, putative | -1.74 |
| 63 | 262811_at | AT1G11700 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G61930.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO17934.1); contains InterPro domain Protein of unknown function DUF584 (InterPro:IPR007608) | 3.08 |
| 64 | 264372_at | AT1G11840 | ATGLX1 (GLYOXALASE I HOMOLOG); lactoylglutathione lyase | -1.47 |
| 65 | 264343_at | AT1G11850 | unknown protein | -1.70 |
| 66 | 264386_at | AT1G12000 | pyrophosphate--fructose-6-phosphate 1-phosphotransferase beta subunit, putative / pyrophosphate-dependent 6-phosphofructose-1-kinase, putative | -1.63 |
| 67 | 264347_at | AT1G12040 | LRX1 (LEUCINE-RICH REPEAT/EXTENSIN 1); protein binding / structural constituent of cell wall | -2.18 |
| 68 | 264342_at | AT1G12080 | contains domain PTHR22683 (PTHR22683) | -3.07 |
| 69 | 264348_at | AT1G12110 | NRT1.1 (NITRATE TRANSPORTER 1.1); transporter | 1.68 |
| 70 | 259535_at | AT1G12280 | disease resistance protein (CC-NBS-LRR class), putative | 1.62 |
| 71 | 259520_at | AT1G12320 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G62840.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO24498.1); contains InterPro domain Protein of unknown function DUF1442 (InterPro:IPR009902) | 1.66 |
| 72 | 259514_at | AT1G12480 | CDI3/OZS1/RCD3/SLAC1 (SLOW ANION CHANNEL-ASSOCIATED 1); transporter | 1.59 |
| 73 | 255937_at | AT1G12610 | DDF1 (DWARF AND DELAYED FLOWERING 1); DNA binding / transcription factor | 1.89 |
| 74 | 261203_at | AT1G12845 | similar to hypothetical protein MtrDRAFT_AC149131g9v2 [Medicago truncatula] (GB:ABD32556.1) | -2.19 |
| 75 | 261198_at | AT1G12940 | ATNRT2.5 (NITRATE TRANSPORTER2.5); nitrate transmembrane transporter | 2.83 |
| 76 | 262772_at | AT1G13210 | ACA.L (AUTOINHIBITED CA2+/ATPASE II); ATPase, coupled to transmembrane movement of ions, phosphorylative mechanism / calmodulin binding | 1.72 |
| 77 | 259358_at | AT1G13250 | GATL3 (Galacturonosyltransferase-like 3); polygalacturonate 4-alpha-galacturonosyltransferase/ transferase, transferring glycosyl groups / transferase, transferring hexosyl groups | -1.56 |
| 78 | 259385_at | AT1G13470 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G13520.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO42040.1); contains InterPro domain Protein of unknown function DUF1262 (InterPro:IPR010683) | 4.58 |
| 79 | 262607_at | AT1G13990 | similar to unnamed protein product [Vitis vinifera] (GB:CAO68469.1) | 2.18 |
| 80 | 261526_at | AT1G14370 | APK2A (PROTEIN KINASE 2A); kinase | 1.86 |
| 81 | 262849_at | AT1G14710 | hydroxyproline-rich glycoprotein family protein | -1.69 |
| 82 | 262840_at | AT1G14900 | HMGA (HIGH MOBILITY GROUP A); DNA binding | -2.39 |
| 83 | 260714_at | AT1G14980 | CPN10 (CHAPERONIN 10) | -1.48 |
| 84 | 262594_at | AT1G15250 | 60S ribosomal protein L37 (RPL37A) | -1.56 |
| 85 | 262603_at | AT1G15380 | lactoylglutathione lyase family protein / glyoxalase I family protein | -1.53 |
| 86 | 262582_at | AT1G15410 | aspartate-glutamate racemase family | 1.93 |
| 87 | 259500_at | AT1G15740 | leucine-rich repeat family protein | 1.62 |
| 88 | 259489_at | AT1G15790 | similar to protein binding / transcription cofactor [Arabidopsis thaliana] (TAIR:AT1G15780.1); similar to Os08g0564800 [Oryza sativa (japonica cultivar-group)] (GB:NP_001062533.1); similar to hypothetical protein OsJ_027171 [Oryza sativa (japonica cultivar-group)] (GB:EAZ43688.1); similar to CTV.22 [Poncirus trifoliata] (GB:AAN62354.1) | 1.61 |
| 89 | 261789_at | AT1G15930 | 40S ribosomal protein S12 (RPS12A) | -1.55 |
| 90 | 261790_at | AT1G16000 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G80890.1); similar to Os12g0556400 [Oryza sativa (japonica cultivar-group)] (GB:NP_001067003.1); similar to hypothetical protein OsI_037443 [Oryza sativa (indica cultivar-group)] (GB:EAY83484.1); similar to hypothetical protein LOC_Os12g36930 [Oryza sativa (japonica cultivar-group)] (GB:ABA99557.1) | -1.72 |
| 91 | 262754_at | AT1G16350 | inosine-5'-monophosphate dehydrogenase, putative | -1.46 |
| 92 | 262730_at | AT1G16390 | ATOCT3 (ARABIDOPSIS THALIANA ORGANIC CATION/CARNITINE TRANSPORTER2); carbohydrate transmembrane transporter/ sugar:hydrogen ion symporter | -2.27 |
| 93 | 255768_at | AT1G16705 | p300/CBP acetyltransferase-related protein-related | -2.73 |
| 94 | 255763_at | AT1G16730 | similar to unknown [Picea sitchensis] (GB:ABK21208.1) | 3.27 |
| 95 | 256112_at | AT1G16920 | RAB11 (ARABIDOPSIS RAB GTPASE HOMOLOG A1B); GTP binding | -1.93 |
| 96 | 256109_at | AT1G16950 | unknown protein | -1.51 |
| 97 | 262518_at | AT1G17170 | ATGSTU24 (ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE (CLASS TAU) 24); glutathione transferase | 2.04 |
| 98 | 262516_at | AT1G17190 | ATGSTU26 (Arabidopsis thaliana Glutathione S-transferase (class tau) 26); glutathione transferase | -1.86 |
| 99 | 261087_at | AT1G17350 | auxin-induced-related / indole-3-acetic acid induced-related | -1.51 |
| 100 | 260735_at | AT1G17610 | disease resistance protein-related | 1.67 |
| 101 | 256072_at | AT1G18080 | ATARCA (Arabidopsis thaliana Homolog of the Tobacco ArcA); nucleotide binding | -1.48 |
| 102 | 256125_at | AT1G18250 | ATLP-1 (Arabidopsis thaumatin-like protein 1) | -3.16 |
| 103 | 261660_at | AT1G18370 | HIK (HINKEL); microtubule motor | -2.37 |
| 104 | 255776_at | AT1G18540 | 60S ribosomal protein L6 (RPL6A) | -1.54 |
| 105 | 255753_at | AT1G18570 | MYB51 (MYB DOMAIN PROTEIN 51); DNA binding / transcription factor | 1.85 |
| 106 | 261421_at | AT1G18840 | IQD30; calmodulin binding | -1.79 |
| 107 | 259479_at | AT1G19020 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G48180.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO40966.1) | 1.72 |
| 108 | 259466_at | AT1G19050 | ARR7 (RESPONSE REGULATOR 7); transcription regulator/ two-component response regulator | -1.87 |
| 109 | 256039_at | AT1G19190 | hydrolase | -1.59 |
| 110 | 256012_at | AT1G19250 | FMO1 (FLAVIN-DEPENDENT MONOOXYGENASE 1); monooxygenase | 3.34 |
| 111 | 260667_at | AT1G19440 | very-long-chain fatty acid condensing enzyme, putative | -1.65 |
| 112 | 260664_at | AT1G19510 | myb family transcription factor | 2.88 |
| 113 | 261134_at | AT1G19630 | CYP722A1 (cytochrome P450, family 722, subfamily A, polypeptide 1); oxygen binding | 2.61 |
| 114 | 255782_at | AT1G19850 | MP (MONOPTEROS); transcription factor | -1.57 |
| 115 | 261221_at | AT1G19960 | similar to transmembrane receptor [Arabidopsis thaliana] (TAIR:AT2G32140.1) | 2.18 |
| 116 | 261248_at | AT1G20030 | pathogenesis-related thaumatin family protein | -3.28 |
| 117 | 261247_at | AT1G20070 | unknown protein | 3.10 |
| 118 | 261224_at | AT1G20160 | ATSBT5.2; subtilase | 1.71 |
| 119 | 255941_at | AT1G20350 | ATTIM17-1 (Arabidopsis thaliana translocase inner membrane subunit 17-1); P-P-bond-hydrolysis-driven protein transmembrane transporter | 2.41 |
| 120 | 262854_at | AT1G20870 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G54850.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO66281.1); contains InterPro domain HSP20-like chaperone (InterPro:IPR008978) | -1.54 |
| 121 | 262803_at | AT1G21000 | zinc-binding family protein | 3.18 |
| 122 | 261455_at | AT1G21070 | transporter-related | -1.64 |
| 123 | 261459_at | AT1G21100 | O-methyltransferase, putative | 4.94 |
| 124 | 261453_at | AT1G21130 | O-methyltransferase, putative | 2.43 |
| 125 | 259559_at | AT1G21240 | WAK3 (WALL ASSOCIATED KINASE 3); kinase/ protein serine/threonine kinase | 4.13 |
| 126 | 259561_at | AT1G21250 | WAK1 (CELL WALL-ASSOCIATED KINASE); kinase | 3.45 |
| 127 | 259560_at | AT1G21270 | WAK2 (wall-associated kinase 2); protein serine/threonine kinase | 2.45 |
| 128 | 262486_at | AT1G21740 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G77500.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN81539.1); contains InterPro domain Protein of unknown function DUF632 (InterPro:IPR006867); contains InterPro domain Protein of unknown function DUF630 (InterPro:IPR006868) | -3.11 |
| 129 | 260857_at | AT1G21880 | LYM1 (LYSM DOMAIN GPI-ANCHORED PROTEIN 1 PRECURSOR) | -1.77 |
| 130 | 255957_at | AT1G22160 | senescence-associated protein-related | 3.32 |
| 131 | 255924_at | AT1G22170 | phosphoglycerate/bisphosphoglycerate mutase family protein | -1.78 |
| 132 | 255962_at | AT1G22330 | RNA binding | -2.41 |
| 133 | 255969_at | AT1G22330 | RNA binding | -2.06 |
| 134 | 255943_at | AT1G22370 | ATUGT85A5 (UDP-GLUCOSYL TRANSFERASE 85A5); transferase, transferring glycosyl groups | 1.88 |
| 135 | 261937_at | AT1G22570 | proton-dependent oligopeptide transport (POT) family protein | 1.85 |
| 136 | 264203_at | AT1G22780 | PFL (POINTED FIRST LEAVES); structural constituent of ribosome | -1.99 |
| 137 | 264774_at | AT1G22890 | unknown protein | 2.41 |
| 138 | 264772_at | AT1G22930 | T-complex protein 11 | 2.16 |
| 139 | 264894_at | AT1G23040 | hydroxyproline-rich glycoprotein family protein | 1.70 |
| 140 | 264901_at | AT1G23090 | AST91 (SULFATE TRANSPORTER 91); sulfate transmembrane transporter | 1.58 |
| 141 | 264899_at | AT1G23130 | Bet v I allergen family protein | 4.78 |
| 142 | 264898_at | AT1G23205 | invertase/pectin methylesterase inhibitor family protein | -2.23 |
| 143 | 263043_at | AT1G23350 | invertase/pectin methylesterase inhibitor family protein | -1.87 |
| 144 | 265189_at | AT1G23840 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G23830.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO14801.1) | 2.31 |
| 145 | 263034_at | AT1G24020 | MLP423 (MLP-LIKE PROTEIN 423) | -4.83 |
| 146 | 264866_at | AT1G24140 | matrixin family protein | 2.41 |
| 147 | 255732_at | AT1G25450 | very-long-chain fatty acid condensing enzyme, putative | -1.44 |
| 148 | 255727_at | AT1G25510 | aspartyl protease family protein | -4.43 |
| 149 | 255735_at | AT1G25520 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G68650.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO42107.1); contains InterPro domain Protein of unknown function UPF0016; (InterPro:IPR001727) | 1.61 |
| 150 | 261007_at | AT1G26400 | FAD-binding domain-containing protein | -1.88 |
| 151 | 261005_at | AT1G26420 | FAD-binding domain-containing protein | 2.62 |
| 152 | 261004_at | AT1G26450 | beta-1,3-glucanase-related | -1.66 |
| 153 | 261017_at | AT1G26570 | ATUGD1/UGD1 (UDP-glucose dehydrogenase 1); UDP-glucose 6-dehydrogenase | -1.60 |
| 154 | 261263_at | AT1G26790 | Dof-type zinc finger domain-containing protein | 1.71 |
| 155 | 263681_at | AT1G26840 | ATORC6/ORC6 (Origin recognition complex protein 6); DNA binding | -3.73 |
| 156 | 263691_at | AT1G26880 | 60S ribosomal protein L34 (RPL34A) | -1.53 |
| 157 | 263686_at | AT1G26910 | 60S ribosomal protein L10 (RPL10B) | -1.62 |
| 158 | 264987_at | AT1G27030 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G27020.1); similar to unknown protein [Oryza sativa (japonica cultivar-group)] (GB:AAN05517.1); similar to Os10g0463800 [Oryza sativa (japonica cultivar-group)] (GB:NP_001064789.1) | -1.94 |
| 159 | 264978_at | AT1G27120 | galactosyltransferase family protein | -2.28 |
| 160 | 264444_at | AT1G27360 | squamosa promoter-binding protein-like 11 (SPL11) | 1.69 |
| 161 | 264438_at | AT1G27400 | 60S ribosomal protein L17 (RPL17A) | -1.73 |
| 162 | 264449_at | AT1G27460 | NPGR1 (NO POLLEN GERMINATION RELATED 1); calmodulin binding | -1.68 |
| 163 | 261641_at | AT1G27670 | unknown protein | -1.96 |
| 164 | 259592_at | AT1G27950 | lipid transfer protein-related | -1.52 |
| 165 | 259596_at | AT1G28130 | GH3.17; indole-3-acetic acid amido synthetase | -3.82 |
| 166 | 245671_at | AT1G28230 | PUP1 (PURINE PERMEASE 1); purine transmembrane transporter | 2.25 |
| 167 | 245688_at | AT1G28290 | AGP31 (ARABINOGALACTAN-PROTEIN 31); structural constituent of cell wall | -5.42 |
| 168 | 261439_at | AT1G28395 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G33847.2); similar to unnamed protein product [Vitis vinifera] (GB:CAO39903.1) | -1.51 |
| 169 | 261467_at | AT1G28520 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G42400.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO66378.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO69825.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN71043.1) | 1.60 |
| 170 | 262736_at | AT1G28570 | GDSL-motif lipase, putative | 1.92 |
| 171 | 260840_at | AT1G29050 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G34070.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO48076.1); contains InterPro domain Protein of unknown function DUF231, plant (InterPro:IPR004253) | 2.67 |
| 172 | 260884_at | AT1G29240 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G34170.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO48048.1); contains InterPro domain Protein of unknown function DUF688 (InterPro:IPR007789) | 1.74 |
| 173 | 260895_at | AT1G29250 | nucleic acid binding | -1.47 |
| 174 | 259769_at | AT1G29400 | AML5 (ARABIDOPSIS MEI2-LIKE PROTEIN 5); RNA binding | 1.71 |
| 175 | 259784_at | AT1G29450 | auxin-responsive protein, putative | 2.75 |
| 176 | 259787_at | AT1G29460 | auxin-responsive protein, putative | 1.80 |
| 177 | 246633_at | AT1G29720 | protein kinase family protein | 3.55 |
| 178 | 260028_at | AT1G29980 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G34510.1); similar to unknown [Populus trichocarpa] (GB:ABK95079.1); contains InterPro domain Protein of unknown function DUF642 (InterPro:IPR006946); contains InterPro domain Galactose-binding like (InterPro:IPR008979) | -1.99 |
| 179 | 245776_at | AT1G30260 | AGL79 (AGAMOUS-LIKE 79) | 3.11 |
| 180 | 261803_at | AT1G30500 | CCAAT-binding transcription factor (CBF-B/NF-YA) family protein | 1.58 |
| 181 | 263219_at | AT1G30600 | subtilase family protein | -1.56 |
| 182 | 265128_at | AT1G30860 | protein binding / zinc ion binding | 1.66 |
| 183 | 265161_at | AT1G30900 | vacuolar sorting receptor, putative | 1.56 |
| 184 | 265158_at | AT1G31040 | zinc ion binding | -2.40 |
| 185 | 262558_at | AT1G31335 | unknown protein | -1.54 |
| 186 | 246601_at | AT1G31710 | copper amine oxidase, putative | -3.03 |
| 187 | 246602_at | AT1G31710 | copper amine oxidase, putative | -1.79 |
| 188 | 246582_at | AT1G31750 | proline-rich family protein | 1.74 |
| 189 | 246581_at | AT1G31760 | SWIB complex BAF60b domain-containing protein | -2.30 |
| 190 | 246260_at | AT1G31820 | amino acid permease family protein | 3.70 |
| 191 | 246265_at | AT1G31860 | AT-IE (Arabidopsis thaliana bifunctional HisI-HisE protein) | -2.43 |
| 192 | 245789_at | AT1G32090 | early-responsive to dehydration protein-related / ERD protein-related | -1.84 |
| 193 | 260706_at | AT1G32350 | AOX1D (ALTERNATIVE OXIDASE 1D); alternative oxidase | 2.35 |
| 194 | 260696_at | AT1G32520 | similar to unnamed protein product [Vitis vinifera] (GB:CAO63428.1) | 1.64 |
| 195 | 261242_at | AT1G32960 | ATSBT3.3; subtilase | 2.71 |
| 196 | 261188_at | AT1G33000 | transposable element gene | -1.53 |
| 197 | 261216_at | AT1G33030 | O-methyltransferase family 2 protein | 1.88 |
| 198 | 261615_at | AT1G33050 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G10470.1) | 1.59 |
| 199 | 261567_at | AT1G33055 | unknown protein | -3.88 |
| 200 | 261618_at | AT1G33110 | MATE efflux family protein | 1.81 |
| 201 | 261984_at | AT1G33760 | AP2 domain-containing transcription factor, putative | 4.15 |
| 202 | 260116_at | AT1G33960 | AIG1 (AVRRPT2-INDUCED GENE 1); GTP binding | 2.25 |
| 203 | 259930_at | AT1G34355 | forkhead-associated domain-containing protein / FHA domain-containing protein | -3.56 |
| 204 | 262408_at | AT1G34750 | protein phosphatase 2C, putative / PP2C, putative | 1.59 |
| 205 | 262412_at | AT1G34760 | GRF11 (General regulatory factor 11); amino acid binding / protein phosphorylated amino acid binding | 1.93 |
| 206 | 259550_at | AT1G35230 | AGP5 (ARABINOGALACTAN-PROTEIN 5) | 2.81 |
| 207 | 259546_at | AT1G35350 | similar to SHB1 (SHORT HYPOCOTYL UNDER BLUE1) [Arabidopsis thaliana] (TAIR:AT4G25350.1); similar to EXS family protein / ERD1/XPR1/SYG1 family protein [Arabidopsis thaliana] (TAIR:AT1G26730.1); similar to ATP binding / ATPase, coupled to transmembrane movement of substances [Arabidopsis thaliana] (TAIR:AT1G14040.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO68453.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO68461.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN73334.1); contains InterPro domain SPX, N-terminal (InterPro:IPR004331); contains InterPro domain EXS, C-terminal; (InterPro:IPR004342) | 1.74 |
| 208 | 262028_at | AT1G35560 | TCP family transcription factor, putative | 1.90 |
| 209 | 261339_at | AT1G35710 | leucine-rich repeat transmembrane protein kinase, putative | 4.90 |
| 210 | 256319_at | AT1G35910 | trehalose-6-phosphate phosphatase, putative | -3.58 |
| 211 | 261363_at | AT1G41830 | SKS6 (SKU5 Similar 6); pectinesterase | -1.88 |
| 212 | 264415_at | AT1G43160 | RAP2.6 (related to AP2 6); DNA binding / transcription factor | -2.38 |
| 213 | 259507_at | AT1G43910 | AAA-type ATPase family protein | 1.71 |
| 214 | 259453_at | AT1G44090 | ATGA20OX5 (GIBBERELLIN 20-OXIDASE 5); iron ion binding / oxidoreductase | -1.64 |
| 215 | 245739_at | AT1G44110 | CYCA1;1 (CYCLIN A1;1); cyclin-dependent protein kinase regulator | -1.97 |
| 216 | 260944_at | AT1G45130 | BGAL5 (beta-galactosidase 5); beta-galactosidase | -2.71 |
| 217 | 245807_at | AT1G46768 | RAP2.1 (related to AP2 1); DNA binding / transcription factor | 1.70 |
| 218 | 260506_at | AT1G47210 | CYCA3;2; cyclin-dependent protein kinase | -2.06 |
| 219 | 261688_at | AT1G47380 | protein phosphatase 2C-related / PP2C-related | -1.57 |
| 220 | 261308_at | AT1G48480 | RKL1 (Receptor-like kinase 1); ATP binding / kinase/ protein serine/threonine kinase | -2.03 |
| 221 | 256144_at | AT1G48630 | guanine nucleotide-binding family protein / activated protein kinase C receptor, putative / RACK, putative | -2.27 |
| 222 | 260771_at | AT1G49160 | WNK7 (Arabidopsis WNK kinase 7); kinase | -1.42 |
| 223 | 260770_at | AT1G49200 | zinc finger (C3HC4-type RING finger) family protein | 1.97 |
| 224 | 260753_at | AT1G49230 | zinc finger (C3HC4-type RING finger) family protein | 1.64 |
| 225 | 262414_at | AT1G49430 | LACS2 (LONG-CHAIN ACYL-COA SYNTHETASE 2) | -2.02 |
| 226 | 262396_at | AT1G49470 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G55230.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN63047.1); contains InterPro domain Protein of unknown function DUF716 (InterPro:IPR006904) | 2.16 |
| 227 | 262399_at | AT1G49500 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G19030.1) | -2.00 |
| 228 | 261605_at | AT1G49580 | calcium-dependent protein kinase, putative / CDPK, putative | -1.49 |
| 229 | 261608_at | AT1G49650 | cell death associated protein-related | -1.42 |
| 230 | 261614_at | AT1G49760 | PAB8 (POLY(A) BINDING PROTEIN 8); RNA binding / translation initiation factor | -1.68 |
| 231 | 259815_at | AT1G49870 | similar to MBD10 (methyl-CpG-binding domain 10), DNA binding [Arabidopsis thaliana] (TAIR:AT1G15340.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO63952.1) | -4.55 |
| 232 | 261639_at | AT1G50010 | TUA2 (tubulin alpha-2 chain) | -2.68 |
| 233 | 261658_at | AT1G50040 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G19680.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO61535.1); contains InterPro domain Protein of unknown function DUF1005 (InterPro:IPR010410) | -1.50 |
| 234 | 261636_at | AT1G50110 | branched-chain amino acid aminotransferase 6 / branched-chain amino acid transaminase 6 (BCAT6) | -2.37 |
| 235 | 262467_at | AT1G50240 | armadillo/beta-catenin repeat family protein | -1.53 |
| 236 | 261859_at | AT1G50490 | UBC20 (UBIQUITIN-CONJUGATING ENZYME 20); ubiquitin-protein ligase | -1.95 |
| 237 | 245749_at | AT1G51090 | heavy-metal-associated domain-containing protein | 1.96 |
| 238 | 265136_at | AT1G51270 | structural molecule | 3.39 |
| 239 | 260486_at | AT1G51550 | F-box family protein | 1.78 |
| 240 | 256183_at | AT1G51660 | ATMKK4 (MITOGEN-ACTIVATED PROTEIN KINASE KINASE 4); MAP kinase kinase/ kinase | 1.56 |
| 241 | 256168_at | AT1G51805 | leucine-rich repeat protein kinase, putative | 2.52 |
| 242 | 246373_at | AT1G51860 | leucine-rich repeat protein kinase, putative | 4.25 |
| 243 | 246368_at | AT1G51890 | leucine-rich repeat protein kinase, putative | 1.85 |
| 244 | 259841_at | AT1G52200 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G18470.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO45876.1); contains InterPro domain Protein of unknown function Cys-rich (InterPro:IPR006461) | 2.23 |
| 245 | 257504_at | AT1G52250 | dynein light chain type 1 family protein | -4.71 |
| 246 | 259612_at | AT1G52300 | 60S ribosomal protein L37 (RPL37B) | -1.72 |
| 247 | 259640_at | AT1G52400 | BGL1 (BETA-GLUCOSIDASE HOMOLOG 1); hydrolase, hydrolyzing O-glycosyl compounds | -2.60 |
| 248 | 260148_at | AT1G52800 | oxidoreductase, 2OG-Fe(II) oxygenase family protein | 2.53 |
| 249 | 260155_at | AT1G52870 | peroxisomal membrane protein-related | 2.95 |
| 250 | 260156_at | AT1G52880 | NAM (Arabidopsis NAC domain containing protein 18); transcription factor | 2.31 |
| 251 | 260203_at | AT1G52890 | ANAC019 (Arabidopsis NAC domain containing protein 19); transcription factor | 2.13 |
| 252 | 261375_at | AT1G53160 | SPL4 (SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 4); DNA binding / transcription factor | 2.97 |
| 253 | 261315_at | AT1G53170 | ATERF-8/ATERF8 (ETHYLENE RESPONSE ELEMENT BINDING FACTOR 4); DNA binding / transcription factor/ transcription repressor | 1.64 |
| 254 | 260617_at | AT1G53345 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G09580.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO63241.1); contains domain DHH phosphoesterases (SSF64182) | -2.85 |
| 255 | 260982_at | AT1G53520 | chalcone-flavanone isomerase-related | -3.38 |
| 256 | 263003_at | AT1G54450 | calcium-binding EF-hand family protein | -1.42 |
| 257 | 259660_at | AT1G55260 | lipid binding | -2.61 |
| 258 | 259661_at | AT1G55265 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G19860.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN64354.1); contains InterPro domain Protein of unknown function DUF538 (InterPro:IPR007493) | 2.05 |
| 259 | 259664_at | AT1G55330 | AGP21 (ARABINOGALACTAN PROTEIN 21) | -1.63 |
| 260 | 265075_at | AT1G55450 | embryo-abundant protein-related | 1.60 |
| 261 | 264562_at | AT1G55760 | BTB/POZ domain-containing protein | 1.86 |
| 262 | 260592_at | AT1G55850 | ATCSLE1 (Cellulose synthase-like E1); cellulose synthase/ transferase, transferring glycosyl groups | 2.51 |
| 263 | 262096_at | AT1G56010 | NAC1 (Arabidopsis NAC domain containing protein 21, Arabidopsis NAC domain containing protein 22); transcription factor | 1.78 |
| 264 | 262092_at | AT1G56150 | auxin-responsive family protein | 2.49 |
| 265 | 259629_at | AT1G56510 | disease resistance protein (TIR-NBS-LRR class), putative | 2.18 |
| 266 | 245627_at | AT1G56600 | ATGOLS2 (ARABIDOPSIS THALIANA GALACTINOL SYNTHASE 2); transferase, transferring glycosyl groups / transferase, transferring hexosyl groups | 8.62 |
| 267 | 245628_at | AT1G56650 | PAP1 (PRODUCTION OF ANTHOCYANIN PIGMENT 1); DNA binding / transcription factor | 1.93 |
| 268 | 245657_at | AT1G56720 | protein kinase family protein | -1.50 |
| 269 | 246405_at | AT1G57630 | disease resistance protein (TIR class), putative | 3.48 |
| 270 | 245828_at | AT1G57820 | ORTH2/VIM1 (VARIANT IN METHYLATION 1); DNA binding / chromatin binding / double-stranded methylated DNA binding / histone binding / methyl-CpG binding / methyl-CpNpG binding / methyl-CpNpN binding | -4.41 |
| 271 | 246396_at | AT1G58180 | carbonic anhydrase family protein / carbonate dehydratase family protein | 2.17 |
| 272 | 245840_at | AT1G58420 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G10140.1); contains InterPro domain Uncharacterised conserved protein UCP031279 (InterPro:IPR016972) | 2.06 |
| 273 | 245842_at | AT1G58430 | RXF26; carboxylesterase | -2.10 |
| 274 | 262081_at | AT1G59540 | ZCF125; microtubule motor | -1.52 |
| 275 | 262126_at | AT1G59620 | CW9; ATP binding | 1.58 |
| 276 | 262916_at | AT1G59700 | ATGSTU16 (Arabidopsis thaliana Glutathione S-transferase (class tau) 16); glutathione transferase | 1.75 |
| 277 | 262899_at | AT1G59870 | PDR8/PEN3 (PLEIOTROPIC DRUG RESISTANCE8); ATPase, coupled to transmembrane movement of substances / cadmium ion transmembrane transporter | 1.64 |
| 278 | 264217_at | AT1G60190 | armadillo/beta-catenin repeat family protein / U-box domain-containing protein | 1.78 |
| 279 | 264277_at | AT1G60390 | BURP domain-containing protein / polygalacturonase, putative | -1.92 |
| 280 | 264929_at | AT1G60730 | aldo/keto reductase family protein | 1.79 |
| 281 | 264758_at | AT1G61340 | F-box family protein | -1.88 |
| 282 | 265006_at | AT1G61570 | TIM13; P-P-bond-hydrolysis-driven protein transmembrane transporter | -1.44 |
| 283 | 264400_at | AT1G61800 | GPT2 (glucose-6-phosphate/phosphate translocator 2); antiporter/ glucose-6-phosphate transmembrane transporter | 1.72 |
| 284 | 264746_at | AT1G62300 | WRKY6 (WRKY DNA-binding protein 6); transcription factor | 1.84 |
| 285 | 260635_at | AT1G62422 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G12020.1); similar to unknown [Brassica rapa] (GB:ABL97977.1) | 1.70 |
| 286 | 265117_at | AT1G62500 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | -2.83 |
| 287 | 265121_at | AT1G62560 | flavin-containing monooxygenase family protein / FMO family protein | 2.19 |
| 288 | 261101_at | AT1G63030 | DDF2 (DWARF AND DELAYED FLOWERING 2); DNA binding / transcription factor | 2.90 |
| 289 | 259686_at | AT1G63100 | scarecrow transcription factor family protein | -2.03 |
| 290 | 260242_at | AT1G63650 | EGL3 (ENHANCER OF GLABRA3); DNA binding / transcription factor | -4.87 |
| 291 | 260296_at | AT1G63750 | ATP binding / nucleoside-triphosphatase/ nucleotide binding / protein binding | 2.50 |
| 292 | 260312_at | AT1G63880 | disease resistance protein (TIR-NBS-LRR class), putative | 1.87 |
| 293 | 260325_at | AT1G63940 | monodehydroascorbate reductase, putative | -1.82 |
| 294 | 259766_at | AT1G64360 | unknown protein | 7.22 |
| 295 | 259765_at | AT1G64370 | unknown protein | 2.43 |
| 296 | 262002_at | AT1G64450 | proline-rich family protein | -3.36 |
| 297 | 261958_at | AT1G64500 | glutaredoxin family protein | 2.81 |
| 298 | 261975_at | AT1G64640 | plastocyanin-like domain-containing protein | -1.76 |
| 299 | 261949_at | AT1G64670 | BDG1 (BODYGUARD1); hydrolase | -1.78 |
| 300 | 262870_at | AT1G64710 | alcohol dehydrogenase, putative | 1.88 |
| 301 | 262883_at | AT1G64780 | ATAMT1;2 (AMMONIUM TRANSPORTER 1;2); ammonium transmembrane transporter | 2.54 |
| 302 | 262860_at | AT1G64810 | APO1 (ACCUMULATION OF PHOTOSYSTEM ONE 1) | 1.61 |
| 303 | 262882_at | AT1G64900 | CYP89A2 (CYTOCHROME P450 89A2); oxygen binding | 1.88 |
| 304 | 262875_at | AT1G64970 | G-TMT (GAMMA-TOCOPHEROL METHYLTRANSFERASE) | 1.64 |
| 305 | 261907_at | AT1G65060 | 4CL3 (4-coumarate:CoA ligase 3); 4-coumarate-CoA ligase | -1.89 |
| 306 | 264166_at | AT1G65370 | meprin and TRAF homology domain-containing protein / MATH domain-containing protein | -1.53 |
| 307 | 264153_at | AT1G65390 | ATPP2-A5; carbohydrate binding | 2.09 |
| 308 | 264638_at | AT1G65480 | FT (FLOWERING LOCUS T) | 4.26 |
| 309 | 264636_at | AT1G65490 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G65486.1) | 2.53 |
| 310 | 264635_at | AT1G65500 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G65486.1) | 2.29 |
| 311 | 262930_at | AT1G65690 | harpin-induced protein-related / HIN1-related / harpin-responsive protein-related | 1.95 |
| 312 | 261921_at | AT1G65900 | similar to unnamed protein product [Vitis vinifera] (GB:CAO22185.1); contains domain Helical backbone metal receptor (SSF53807) | -1.90 |
| 313 | 256526_at | AT1G66090 | disease resistance protein (TIR-NBS class), putative | 1.64 |
| 314 | 256527_at | AT1G66100 | thionin, putative | 4.61 |
| 315 | 256519_at | AT1G66110 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G37410.1); contains InterPro domain Protein of unknown function DUF577 (InterPro:IPR007598) | -2.42 |
| 316 | 260140_at | AT1G66390 | PAP2 (PRODUCTION OF ANTHOCYANIN PIGMENT 2); DNA binding / transcription factor | 5.51 |
| 317 | 260107_at | AT1G66430 | pfkB-type carbohydrate kinase family protein | -2.79 |
| 318 | 256324_at | AT1G66760 | MATE efflux family protein | -2.85 |
| 319 | 256378_at | AT1G66830 | leucine-rich repeat transmembrane protein kinase, putative | 1.85 |
| 320 | 256366_at | AT1G66880 | serine/threonine protein kinase family protein | 2.61 |
| 321 | 255852_at | AT1G66970 | glycerophosphoryl diester phosphodiesterase family protein | 1.79 |
| 322 | 264968_at | AT1G67360 | rubber elongation factor (REF) family protein | 1.75 |
| 323 | 245189_at | AT1G67670 | unknown protein | -1.46 |
| 324 | 245186_at | AT1G67710 | ARR11 (RESPONSE REGULATOR 11); transcription factor/ two-component response regulator | 2.08 |
| 325 | 245196_at | AT1G67750 | pectate lyase family protein | -2.47 |
| 326 | 245192_at | AT1G67780 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G67270.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO68021.1); contains InterPro domain DDT (InterPro:IPR004022) | -2.68 |
| 327 | 245197_at | AT1G67800 | copine-related | 1.56 |
| 328 | 245193_at | AT1G67810 | Fe-S metabolism associated domain-containing protein | 2.00 |
| 329 | 245215_at | AT1G67830 | ATFXG1 (ALPHA-FUCOSIDASE 1); alpha-L-fucosidase/ carboxylesterase | -1.59 |
| 330 | 260004_at | AT1G67860 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G67865.1) | 2.98 |
| 331 | 260012_at | AT1G67865 | unknown protein | 3.50 |
| 332 | 260007_at | AT1G67870 | glycine-rich protein | 2.41 |
| 333 | 260005_at | AT1G67920 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G24600.1) | 1.68 |
| 334 | 260002_at | AT1G67940 | ATNAP3 (Arabidopsis thaliana non-intrinsic ABC protein 3) | 2.15 |
| 335 | 259992_at | AT1G67970 | AT-HSFA8 (Arabidopsis thaliana heat shock transcription factor A8); DNA binding / transcription factor | 1.84 |
| 336 | 260266_at | AT1G68520 | zinc finger (B-box type) family protein | 2.11 |
| 337 | 262281_at | AT1G68570 | proton-dependent oligopeptide transport (POT) family protein | 2.38 |
| 338 | 262286_at | AT1G68585 | metal ion binding | 1.64 |
| 339 | 262232_at | AT1G68600 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G25480.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G17470.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO42118.1); contains InterPro domain Protein of unknown function UPF0005 (InterPro:IPR006214) | 2.75 |
| 340 | 262229_at | AT1G68620 | hydrolase | 2.32 |
| 341 | 262278_at | AT1G68640 | PAN (PERIANTHIA); DNA binding / transcription factor | -2.60 |
| 342 | 259642_at | AT1G69030 | similar to BSD domain-containing protein [Arabidopsis thaliana] (TAIR:AT1G26300.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN73547.1); contains InterPro domain BSD (InterPro:IPR005607) | -1.65 |
| 343 | 260343_at | AT1G69200 | kinase | -1.65 |
| 344 | 256294_at | AT1G69450 | similar to HYP1 (HYPOTHETICAL PROTEIN 1) [Arabidopsis thaliana] (TAIR:AT3G01100.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO64743.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO15025.1); similar to Protein of unknown function DUF221 [Medicago truncatula] (GB:ABN08272.1); contains InterPro domain Protein of unknown function DUF221; (InterPro:IPR003864) | 2.15 |
| 345 | 256300_at | AT1G69490 | NAP (NAC-LIKE, ACTIVATED BY AP3/PI); transcription factor | 5.05 |
| 346 | 259834_at | AT1G69570 | Dof-type zinc finger domain-containing protein | 1.79 |
| 347 | 260420_at | AT1G69610 | structural constituent of ribosome | 2.12 |
| 348 | 260368_at | AT1G69700 | ATHVA22C (Arabidopsis thaliana HVA22 homologue C) | -1.51 |
| 349 | 260396_at | AT1G69720 | HO3 (HEME OXYGENASE 3); heme oxygenase (decyclizing) | 2.71 |
| 350 | 260417_at | AT1G69770 | CMT3 (CHROMOMETHYLASE 3) | -1.50 |
| 351 | 260410_at | AT1G69870 | proton-dependent oligopeptide transport (POT) family protein | 1.60 |
| 352 | 264694_at | AT1G70250 | receptor serine/threonine kinase, putative | 1.82 |
| 353 | 260181_at | AT1G70710 | AtGH9B1 (ARABIDOPSIS THALIANA GLYCOSYL HYDROLASE 9B1); hydrolase, hydrolyzing O-glycosyl compounds | -2.10 |
| 354 | 262312_at | AT1G70830 | MLP28 (MLP-LIKE PROTEIN 28) | -2.94 |
| 355 | 259751_at | AT1G71030 | ATMYBL2 (Arabidopsis myb-like 2); DNA binding / transcription factor | 1.62 |
| 356 | 259752_at | AT1G71040 | LPR2 (LOW PHOSPHATE ROOT2); copper ion binding | 1.57 |
| 357 | 259897_at | AT1G71380 | ATCEL3/ATGH9B3 (ARABIDOPSIS THALIANA GLYCOSYL HYDROLASE 9B3); hydrolase, hydrolyzing O-glycosyl compounds | -1.41 |
| 358 | 259943_at | AT1G71480 | nuclear transport factor 2 (NTF2) family protein | 1.58 |
| 359 | 261521_at | AT1G71830 | SERK1 (SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE 1); kinase | -2.23 |
| 360 | 256337_at | AT1G72060 | serine-type endopeptidase inhibitor | 1.63 |
| 361 | 259801_at | AT1G72230 | plastocyanin-like domain-containing protein | -1.71 |
| 362 | 260426_at | AT1G72370 | P40 (40S ribosomal protein SA); structural constituent of ribosome | -1.57 |
| 363 | 259912_at | AT1G72670 | IQD8 (IQ-domain 8); calmodulin binding | -3.39 |
| 364 | 259891_at | AT1G72730 | eukaryotic translation initiation factor 4A, putative / eIF-4A, putative | -1.75 |
| 365 | 262366_at | AT1G72890 | disease resistance protein (TIR-NBS class), putative | 1.71 |
| 366 | 262376_at | AT1G72970 | HTH (HOTHEAD); aldehyde-lyase | -2.51 |
| 367 | 262357_at | AT1G73040 | jacalin lectin family protein | 3.79 |
| 368 | 245725_at | AT1G73370 | SUS6; UDP-glycosyltransferase/ sucrose synthase | -1.67 |
| 369 | 260077_at | AT1G73620 | thaumatin-like protein, putative / pathogenesis-related protein, putative | -2.21 |
| 370 | 260061_at | AT1G73690 | AT;CDKD;1/CAK3AT/CDKD1;1 (CYCLIN-DEPENDENT KINASE D1;1); kinase | -3.70 |
| 371 | 260075_at | AT1G73700 | MATE efflux family protein | 1.64 |
| 372 | 260046_at | AT1G73805 | calmodulin binding | 1.86 |
| 373 | 260068_at | AT1G73805 | calmodulin binding | 2.03 |
| 374 | 260380_at | AT1G73870 | zinc finger (B-box type) family protein | 2.89 |
| 375 | 260390_at | AT1G73940 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G49410.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G49410.2); similar to unnamed protein product [Vitis vinifera] (GB:CAO62865.1) | -1.46 |
| 376 | 260392_at | AT1G74030 | enolase, putative | -2.69 |
| 377 | 260239_at | AT1G74360 | leucine-rich repeat transmembrane protein kinase, putative | 2.10 |
| 378 | 260225_at | AT1G74590 | ATGSTU10 (Arabidopsis thaliana Glutathione S-transferase (class tau) 10); glutathione transferase | 4.00 |
| 379 | 259925_at | AT1G75040 | PR5 (PATHOGENESIS-RELATED GENE 5) | 4.66 |
| 380 | 259927_at | AT1G75100 | JAC1 (J-DOMAIN PROTEIN REQUIRED FOR CHLOROPLAST ACCUMULATION RESPONSE 1); heat shock protein binding | 2.28 |
| 381 | 256507_at | AT1G75150 | similar to unnamed protein product [Vitis vinifera] (GB:CAO61179.1) | -2.09 |
| 382 | 256452_at | AT1G75240 | ATHB33 (ARABIDOPSIS THALIANA HOMEOBOX PROTEIN 33); DNA binding / transcription factor | -1.60 |
| 383 | 256503_at | AT1G75250 | myb family transcription factor | 4.96 |
| 384 | 261120_at | AT1G75410 | BLH3; DNA binding / transcription factor | 1.81 |
| 385 | 261118_at | AT1G75460 | ATP-dependent protease La (LON) domain-containing protein | 1.73 |
| 386 | 262980_at | AT1G75680 | ATGH9B7 (ARABIDOPSIS THALIANA GLYCOSYL HYDROLASE 9B7); hydrolase, hydrolyzing O-glycosyl compounds | -1.62 |
| 387 | 262978_at | AT1G75780 | TUB1 (tubulin beta-1 chain); structural molecule | -3.01 |
| 388 | 261754_at | AT1G76130 | AMY2/ATAMY2 (ALPHA-AMYLASE-LIKE 2); alpha-amylase | 1.86 |
| 389 | 261728_at | AT1G76160 | SKS5 (SKU5 Similar 5); copper ion binding / oxidoreductase | -1.78 |
| 390 | 261780_at | AT1G76310 | CYCB2;4 (CYCLIN B2;4); cyclin-dependent protein kinase regulator | -2.17 |
| 391 | 259980_at | AT1G76520 | auxin efflux carrier family protein | 2.05 |
| 392 | 259968_at | AT1G76530 | auxin efflux carrier family protein | 2.64 |
| 393 | 259978_at | AT1G76540 | CDKB2;1 (CYCLIN-DEPENDENT KINASE B2;1); kinase | -2.01 |
| 394 | 259969_at | AT1G76550 | pyrophosphate--fructose-6-phosphate 1-phosphotransferase alpha subunit, putative / pyrophosphate-dependent 6-phosphofructose-1-kinase, putative | -1.45 |
| 395 | 259977_at | AT1G76590 | zinc-binding family protein | 2.22 |
| 396 | 259879_at | AT1G76650 | CML38; calcium ion binding | 1.68 |
| 397 | 259878_at | AT1G76790 | O-methyltransferase family 2 protein | -2.36 |
| 398 | 259871_at | AT1G76800 | nodulin, putative | 2.22 |
| 399 | 264960_at | AT1G76930 | ATEXT4 (EXTENSIN 4) | -3.06 |
| 400 | 264958_at | AT1G76960 | unknown protein | 8.70 |
| 401 | 264477_at | AT1G77240 | AMP-binding protein, putative | -2.06 |
| 402 | 259705_at | AT1G77450 | ANAC032 (Arabidopsis NAC domain containing protein 32); transcription factor | 1.80 |
| 403 | 259763_at | AT1G77630 | peptidoglycan-binding LysM domain-containing protein | -1.62 |
| 404 | 259679_at | AT1G77720 | protein kinase family protein | -1.45 |
| 405 | 259678_at | AT1G77750 | 30S ribosomal protein S13, chloroplast, putative | -1.53 |
| 406 | 262137_at | AT1G77920 | bZIP family transcription factor | 2.60 |
| 407 | 262194_at | AT1G77930 | DNAJ heat shock N-terminal domain-containing protein | 1.81 |
| 408 | 262163_at | AT1G77940 | 60S ribosomal protein L30 (RPL30B) | -1.55 |
| 409 | 262180_at | AT1G78050 | phosphoglycerate/bisphosphoglycerate mutase family protein | -3.29 |
| 410 | 262181_at | AT1G78060 | glycosyl hydrolase family 3 protein | -1.57 |
| 411 | 260059_at | AT1G78090 | ATTPPB (TREHALOSE-6-PHOSPHATE PHOSPHATASE) | -2.85 |
| 412 | 260806_at | AT1G78260 | RNA recognition motif (RRM)-containing protein | -1.50 |
| 413 | 260799_at | AT1G78270 | ATUGT85A4 (UDP-GLUCOSYL TRANSFERASE 85A4); UDP-glycosyltransferase/ glucuronosyltransferase/ transferase, transferring hexosyl groups | 2.49 |
| 414 | 260774_at | AT1G78290 | serine/threonine protein kinase, putative | 2.97 |
| 415 | 260804_at | AT1G78410 | VQ motif-containing protein | 2.60 |
| 416 | 264300_at | AT1G78670 | ATGGH3 (GAMMA-GLUTAMYL HYDROLASE 3); gamma-glutamyl hydrolase | 1.79 |
| 417 | 264096_at | AT1G78995 | similar to unnamed protein product [Vitis vinifera] (GB:CAO41778.1) | 1.56 |
| 418 | 257427_at | AT1G79060 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G56020.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN69700.1) | -3.44 |
| 419 | 264091_at | AT1G79110 | protein binding / zinc ion binding | 2.40 |
| 420 | 264102_at | AT1G79270 | ECT8 (evolutionarily conserved C-terminal region 8) | 2.03 |
| 421 | 262945_at | AT1G79510 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G16320.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO64929.1); contains domain NTF2-like (SSF54427) | 1.93 |
| 422 | 262050_at | AT1G80130 | binding | 4.59 |
| 423 | 262049_at | AT1G80180 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G15400.2); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G15400.3); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G15400.1); similar to hypothetical protein MtrDRAFT_AC148340g12v2 [Medicago truncatula] (GB:ABD28396.1) | 1.90 |
| 424 | 260329_at | AT1G80370 | CYCA2;4 (CYCLIN A2;4); cyclin-dependent protein kinase regulator | -3.74 |
| 425 | 260287_at | AT1G80440 | kelch repeat-containing F-box family protein | 1.71 |
| 426 | 261893_at | AT1G80690 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G25170.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO45044.1); contains InterPro domain Protein of unknown function DUF862, eukaryotic (InterPro:IPR008580) | -2.14 |
| 427 | 261881_at | AT1G80760 | NIP6;1 (NOD26-like intrinsic protein 6;1); water channel | 3.07 |
| 428 | 257474_at | AT1G80850 | methyladenine glycosylase family protein | -1.54 |
| 429 | 265736_at | AT2G01250 | 60S ribosomal protein L7 (RPL7B) | -1.52 |
| 430 | 266353_at | AT2G01520 | MLP328 (MLP-LIKE PROTEIN 328) | -4.80 |
| 431 | 263595_at | AT2G01890 | PAP8 (PURPLE ACID PHOSPHATASE PRECURSOR); acid phosphatase/ protein serine/threonine phosphatase | -1.42 |
| 432 | 266119_at | AT2G02100 | LCR69/PDF2.2 (Low-molecular-weight cysteine-rich 69); protease inhibitor | -2.48 |
| 433 | 267481_at | AT2G02780 | leucine-rich repeat transmembrane protein kinase, putative | -1.57 |
| 434 | 267472_at | AT2G02850 | ARPN (PLANTACYANIN); copper ion binding | -3.49 |
| 435 | 266770_at | AT2G03090 | ATEXPA15 (ARABIDOPSIS THALIANA EXPANSIN A15) | -1.80 |
| 436 | 266707_at | AT2G03310 | unknown protein | 1.56 |
| 437 | 265713_at | AT2G03530 | UPS2 (UREIDE PERMEASE 2) | 1.73 |
| 438 | 265709_at | AT2G03540 | transposable element gene | 2.96 |
| 439 | 264041_at | AT2G03710 | SEP4 (SEPALLATA4); DNA binding / transcription factor | 1.98 |
| 440 | 263402_at | AT2G04050 | MATE efflux family protein | 1.76 |
| 441 | 263401_at | AT2G04070 | transporter | 1.85 |
| 442 | 263406_at | AT2G04160 | AIR3 (Auxin-Induced in Root cultures 3); subtilase | 1.62 |
| 443 | 263852_at | AT2G04450 | ATNUDT6 (Arabidopsis thaliana Nudix hydrolase homolog 6); ADP-ribose diphosphatase/ NAD binding / hydrolase | 1.85 |
| 444 | 263628_at | AT2G04780 | FLA7 | -2.21 |
| 445 | 263632_at | AT2G04795 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G35732.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO24128.1) | 2.66 |
| 446 | 263046_at | AT2G05380 | GRP3S (GLYCINE-RICH PROTEIN 3 SHORT ISOFORM) | 2.30 |
| 447 | 265377_at | AT2G05790 | glycosyl hydrolase family 17 protein | -2.05 |
| 448 | 266024_at | AT2G05950 | transposable element gene | 3.18 |
| 449 | 266035_at | AT2G05990 | MOD1 (MOSAIC DEATH 1); enoyl-[acyl-carrier-protein] reductase (NADH)/ oxidoreductase | -2.67 |
| 450 | 266500_at | AT2G06925 | ATSPLA2-ALPHA/PLA2-ALPHA (PHOSPHOLIPASE A2-ALPHA); phospholipase A2 | -1.65 |
| 451 | 266427_at | AT2G07170 | binding | -1.45 |
| 452 | 265400_at | AT2G10940 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | -2.03 |
| 453 | 265656_at | AT2G13820 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | -2.74 |
| 454 | 265837_at | AT2G14560 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G33840.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO48825.1); contains InterPro domain Protein of unknown function DUF567 (InterPro:IPR007612) | 4.49 |
| 455 | 266385_at | AT2G14610 | PR1 (PATHOGENESIS-RELATED GENE 1) | 4.90 |
| 456 | 266588_at | AT2G14890 | AGP9 (ARABINOGALACTAN PROTEIN 9) | -2.64 |
| 457 | 266613_at | AT2G14900 | gibberellin-regulated family protein | -2.74 |
| 458 | 265892_at | AT2G15020 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G64190.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN67723.1) | 5.15 |
| 459 | 265893_at | AT2G15040 | pseudogene, disease resistance protein-related, low similarity to disease resistance protein Cf-2.1 (Lycopersicon pimpinellifolium) GI:1184075; contains Pfam profile PF00560: Leucine Rich Repeat; blastp match of 38% identity and 4.4e-96 P-value to GP|1184075|gb|AAC15779.1||U42444 Cf-2.1 {Lycopersicon pimpinellifolium} | 3.34 |
| 460 | 265894_at | AT2G15050 | LTP; lipid binding | 1.57 |
| 461 | 265917_at | AT2G15080 | disease resistance family protein | 3.73 |
| 462 | 263297_at | AT2G15310 | ATARFB1A (ADP-ribosylation factor B1A); GTP binding | 2.32 |
| 463 | 263565_at | AT2G15390 | FUT4 (fucosyltransferase 4); fucosyltransferase/ transferase, transferring glycosyl groups | 2.52 |
| 464 | 265499_at | AT2G15480 | UGT73B5 (UDP-GLUCOSYL TRANSFERASE 73B5); UDP-glycosyltransferase/ transferase, transferring glycosyl groups | 3.14 |
| 465 | 265472_at | AT2G15580 | zinc finger (C3HC4-type RING finger) family protein | 1.69 |
| 466 | 265539_at | AT2G15830 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G33960.1) | 2.63 |
| 467 | 265481_at | AT2G15960 | unknown protein | 1.60 |
| 468 | 263612_at | AT2G16440 | DNA replication licensing factor, putative | -2.46 |
| 469 | 265405_at | AT2G16750 | protein kinase family protein | -1.59 |
| 470 | 265383_at | AT2G16780 | MSI2 (NUCLEOSOME/CHROMATIN ASSEMBLY FACTOR GROUP C 2) | -2.05 |
| 471 | 257439_at | AT2G17000 | mechanosensitive ion channel domain-containing protein / MS ion channel domain-containing protein | -1.94 |
| 472 | 263584_at | AT2G17040 | ANAC036 (Arabidopsis NAC domain containing protein 36); transcription factor | 4.80 |
| 473 | 264851_at | AT2G17290 | CPK6 (CALCIUM-DEPENDENT PROTEIN KINASE 6); calmodulin-dependent protein kinase/ kinase | 1.65 |
| 474 | 264849_at | AT2G17360 | 40S ribosomal protein S4 (RPS4A) | -1.49 |
| 475 | 263047_at | AT2G17630 | phosphoserine aminotransferase, putative | -2.07 |
| 476 | 264590_at | AT2G17710 | similar to unnamed protein product [Vitis vinifera] (GB:CAO42932.1) | 3.07 |
| 477 | 264788_at | AT2G17880 | DNAJ heat shock protein, putative | -1.82 |
| 478 | 265817_at | AT2G18050 | HIS1-3 (HISTONE H1-3); DNA binding | 2.35 |
| 479 | 265339_at | AT2G18230 | ATPPA2 (ARABIDOPSIS THALIANA PYROPHOSPHORYLASE 2); inorganic diphosphatase/ pyrophosphatase | 1.65 |
| 480 | 265379_at | AT2G18340 | late embryogenesis abundant domain-containing protein / LEA domain-containing protein | -1.97 |
| 481 | 265338_at | AT2G18400 | ribosomal protein L6 family protein | -1.59 |
| 482 | 266070_at | AT2G18660 | EXLB3 (EXPANSIN-LIKE B3 PRECURSOR) | 4.26 |
| 483 | 266017_at | AT2G18690 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G18680.1); similar to hypothetical protein OsI_029427 [Oryza sativa (indica cultivar-group)] (GB:EAZ08195.1) | 2.33 |
| 484 | 267490_at | AT2G19130 | S-locus lectin protein kinase family protein | 2.16 |
| 485 | 267465_at | AT2G19170 | SLP3 (Subtilisin-like serine protease 3); subtilase | -1.54 |
| 486 | 265939_at | AT2G19650 | DC1 domain-containing protein | 1.87 |
| 487 | 266687_at | AT2G19670 | protein arginine N-methyltransferase, putative | -3.36 |
| 488 | 266684_at | AT2G19720 | RPS15AB (ribosomal protein S15A B); structural constituent of ribosome | -1.51 |
| 489 | 266699_at | AT2G19730 | 60S ribosomal protein L28 (RPL28A) | -1.57 |
| 490 | 266693_at | AT2G19800 | MIOX2 (MYO-INOSITOL OXYGENASE 2) | -2.11 |
| 491 | 265597_at | AT2G20142 | transmembrane receptor | 1.99 |
| 492 | 263372_at | AT2G20450 | 60S ribosomal protein L14 (RPL14A) | -2.19 |
| 493 | 263369_at | AT2G20480 | similar to unknown [Populus trichocarpa] (GB:ABK93010.1); similar to hypothetical protein OsI_030526 [Oryza sativa (indica cultivar-group)] (GB:EAZ09294.1); similar to Os09g0446000 [Oryza sativa (japonica cultivar-group)] (GB:NP_001063306.1) | -3.33 |
| 494 | 263376_at | AT2G20520 | FLA6 | -1.73 |
| 495 | 263715_at | AT2G20570 | GPRI1 (GOLDEN2-LIKE 1); transcription factor | 2.52 |
| 496 | 265443_at | AT2G20750 | ATEXPB1 (ARABIDOPSIS THALIANA EXPANSIN B1) | -2.12 |
| 497 | 264025_at | AT2G21050 | amino acid permease, putative | -1.66 |
| 498 | 264007_at | AT2G21140 | ATPRP2 (PROLINE-RICH PROTEIN 2) | -2.72 |
| 499 | 263765_at | AT2G21540 | SEC14 cytosolic factor, putative / phosphoglyceride transfer protein, putative | -1.41 |
| 500 | 263519_at | AT2G21580 | 40S ribosomal protein S25 (RPS25B) | -1.78 |
| 501 | 263549_at | AT2G21650 | MEE3 (maternal effect embryo arrest 3); DNA binding / transcription factor | 1.90 |
| 502 | 263882_at | AT2G21790 | R1/RNR1 (RIBONUCLEOTIDE REDUCTASE 1); ribonucleoside-diphosphate reductase | -1.91 |
| 503 | 263875_at | AT2G21970 | SEP2 (STRESS ENHANCED PROTEIN 2) | 1.71 |
| 504 | 263431_at | AT2G22170 | lipid-associated family protein | -4.57 |
| 505 | 263486_at | AT2G22200 | AP2 domain-containing transcription factor | -1.90 |
| 506 | 263432_at | AT2G22230 | beta-hydroxyacyl-ACP dehydratase, putative | -1.62 |
| 507 | 263458_at | AT2G22290 | AtRABH1d (Arabidopsis Rab GTPase homolog H1d); GTP binding | 1.56 |
| 508 | 265350_at | AT2G22620 | lyase | -3.85 |
| 509 | 266827_at | AT2G22920 | SCPL12; serine carboxypeptidase | -2.62 |
| 510 | 245074_at | AT2G23200 | protein kinase family protein | 1.58 |
| 511 | 245071_at | AT2G23230 | terpene synthase/cyclase family protein | -1.62 |
| 512 | 245085_at | AT2G23350 | PAB4 (POLY(A) BINDING PROTEIN 4); RNA binding / translation initiation factor | -1.44 |
| 513 | 267135_at | AT2G23430 | ICK1 (KIP-RELATED PROTEIN 1) | 1.71 |
| 514 | 267120_at | AT2G23530 | similar to protein binding / zinc ion binding [Arabidopsis thaliana] (TAIR:AT4G37110.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO44617.1) | -2.93 |
| 515 | 267288_at | AT2G23680 | stress-responsive protein, putative | 2.15 |
| 516 | 267289_at | AT2G23770 | protein kinase family protein / peptidoglycan-binding LysM domain-containing protein | 1.66 |
| 517 | 266572_at | AT2G23840 | HNH endonuclease domain-containing protein | 1.96 |
| 518 | 265993_at | AT2G24160 | pseudogene, leucine rich repeat protein family, contains leucine rich-repeat domains Pfam:PF00560, INTERPRO:IPR001611; contains some similarity to Cf-4 (Lycopersicon hirsutum) gi|2808683|emb|CAA05268; blastp match of 37% identity and 8.4e-98 P-value to GP|2808683|emb|CAA05268.1||AJ002235 Cf-4 {Lycopersicon hirsutum} | 2.93 |
| 519 | 265994_at | AT2G24170 | endomembrane protein 70, putative | -1.72 |
| 520 | 265695_at | AT2G24490 | ATRPA2/ROR1/RPA2 (REPLICON PROTEIN A); protein binding | -1.41 |
| 521 | 263800_at | AT2G24600 | ankyrin repeat family protein | 2.29 |
| 522 | 263539_at | AT2G24850 | TAT3 (TYROSINE AMINOTRANSFERASE 3); transaminase | 2.69 |
| 523 | 263535_at | AT2G24970 | similar to unnamed protein product [Vitis vinifera] (GB:CAO44373.1) | -1.73 |
| 524 | 263536_at | AT2G25000 | WRKY60 (WRKY DNA-BINDING PROTEIN 60); transcription factor | 1.64 |
| 525 | 264377_at | AT2G25060 | plastocyanin-like domain-containing protein | -2.34 |
| 526 | 264379_at | AT2G25200 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G11000.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO44287.1); contains InterPro domain Protein of unknown function DUF868, plant (InterPro:IPR008586) | 1.57 |
| 527 | 263585_at | AT2G25210 | 60S ribosomal protein L39 (RPL39A) | -2.47 |
| 528 | 263640_at | AT2G25270 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G12400.1); similar to Os02g0799300 [Oryza sativa (japonica cultivar-group)] (GB:NP_001048408.1); similar to hypothetical protein OsJ_008445 [Oryza sativa (japonica cultivar-group)] (GB:EAZ24962.1) | -1.74 |
| 529 | 265659_at | AT2G25440 | leucine-rich repeat family protein | 2.25 |
| 530 | 265913_at | AT2G25625 | similar to unnamed protein product [Vitis vinifera] (GB:CAO44157.1) | 2.98 |
| 531 | 266658_at | AT2G25735 | unknown protein | 1.76 |
| 532 | 266655_at | AT2G25880 | ATAUR2 (ATAURORA2); histone serine kinase(H3-S10 specific) / kinase | -2.50 |
| 533 | 266849_at | AT2G25940 | ALPHA-VPE (ALPHA-VACUOLAR PROCESSING ENZYME); cysteine-type endopeptidase | -1.59 |
| 534 | 257365_x_at | AT2G26020 | PDF1.2b (plant defensin 1.2b) | 4.96 |
| 535 | 245038_at | AT2G26560 | PLP2 (PHOSPHOLIPASE A 2A); nutrient reservoir | 3.90 |
| 536 | 267618_at | AT2G26760 | CYCB1;4; cyclin-dependent protein kinase regulator | -4.55 |
| 537 | 266305_at | AT2G27120 | POL2B/TIL2 (TILTED2); DNA-directed DNA polymerase | -2.63 |
| 538 | 265649_at | AT2G27510 | ATFD3 (FERREDOXIN 3); electron carrier | -2.08 |
| 539 | 266256_at | AT2G27710 | 60S acidic ribosomal protein P2 (RPP2B) | -1.80 |
| 540 | 266258_at | AT2G27720 | 60S acidic ribosomal protein P2 (RPP2A) | -1.58 |
| 541 | 266253_at | AT2G27840 | HDT4 (histone deacetylase 13) | -1.50 |
| 542 | 264061_at | AT2G27970 | CKS2 (CDK-SUBUNIT 2); cyclin-dependent protein kinase | -1.45 |
| 543 | 265573_at | AT2G28200 | nucleic acid binding / transcription factor/ zinc ion binding | 1.77 |
| 544 | 265547_at | AT2G28305 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G37210.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO21862.1); contains InterPro domain Conserved hypothetical protein CHP00730 (InterPro:IPR005269) | 1.81 |
| 545 | 263441_at | AT2G28620 | kinesin motor protein-related | -2.29 |
| 546 | 266226_at | AT2G28740 | HIS4 (Histone H4) | -2.61 |
| 547 | 266223_at | AT2G28790 | osmotin-like protein, putative | -3.48 |
| 548 | 266774_at | AT2G29050 | ATRBL1 (ARABIDOPSIS THALIANA RHOMBOID-LIKE 1) | -1.67 |
| 549 | 266782_at | AT2G29120 | ATGLR2.7 (Arabidopsis thaliana glutamate receptor 2.7) | 3.60 |
| 550 | 266783_at | AT2G29130 | LAC2 (laccase 2); copper ion binding / oxidoreductase | 4.10 |
| 551 | 266291_at | AT2G29320 | tropinone reductase, putative / tropine dehydrogenase, putative | 1.57 |
| 552 | 266292_at | AT2G29350 | SAG13 (Senescence-associated gene 13); oxidoreductase | 4.27 |
| 553 | 266296_at | AT2G29420 | ATGSTU7 (GLUTATHIONE S-TRANSFERASE 25); glutathione transferase | 1.61 |
| 554 | 266267_at | AT2G29460 | ATGSTU4 (GLUTATHIONE S-TRANSFERASE 22); glutathione transferase | 2.81 |
| 555 | 266290_at | AT2G29490 | ATGSTU1 (GLUTATHIONE S-TRANSFERASE 19); glutathione transferase | 1.72 |
| 556 | 266295_at | AT2G29550 | TUB7 (tubulin beta-7 chain) | -2.19 |
| 557 | 266297_at | AT2G29570 | PCNA2 (PROLIFERATING CELL NUCLEAR ANTIGEN 2); DNA binding / DNA polymerase processivity factor | -2.35 |
| 558 | 263485_at | AT2G29890 | VLN1 (VILLIN 1); actin binding | -2.09 |
| 559 | 266835_at | AT2G29990 | NDA2 (ALTERNATIVE NAD(P)H DEHYDROGENASE 2); NADH dehydrogenase | 1.79 |
| 560 | 267299_at | AT2G30150 | UDP-glucoronosyl/UDP-glucosyl transferase family protein | -1.68 |
| 561 | 267308_at | AT2G30200 | [acyl-carrier-protein] S-malonyltransferase/ binding / catalytic/ transferase | -1.59 |
| 562 | 267246_at | AT2G30250 | WRKY25 (WRKY DNA-binding protein 25); transcription factor | 2.13 |
| 563 | 267523_at | AT2G30600 | BTB/POZ domain-containing protein | -1.43 |
| 564 | 264114_at | AT2G31270 | ATCDT1A/CDT1/CDT1A (ARABIDOPSIS HOMOLOG OF YEAST CDT1 A); cyclin-dependent protein kinase/ protein binding | -1.61 |
| 565 | 263249_at | AT2G31360 | ADS2 (16:0DELTA9 ARABIDOPSIS DESATURASE 2); oxidoreductase | -1.69 |
| 566 | 263474_at | AT2G31725 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G05730.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN73933.1); contains InterPro domain Protein of unknown function DUF842, eukaryotic (InterPro:IPR008560) | -3.72 |
| 567 | 263478_at | AT2G31880 | leucine-rich repeat transmembrane protein kinase, putative | 2.30 |
| 568 | 265672_at | AT2G31980 | cysteine proteinase inhibitor-related | -1.47 |
| 569 | 265728_at | AT2G31990 | exostosin family protein | 1.84 |
| 570 | 265671_at | AT2G32060 | 40S ribosomal protein S12 (RPS12C) | -1.80 |
| 571 | 265723_at | AT2G32140 | transmembrane receptor | 2.21 |
| 572 | 265698_at | AT2G32160 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G32170.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO64660.1); contains InterPro domain N2227-like (InterPro:IPR012901) | 4.46 |
| 573 | 265730_at | AT2G32220 | 60S ribosomal protein L27 (RPL27A) | -1.45 |
| 574 | 266357_at | AT2G32290 | BAM6/BMY5 (BETA-AMYLASE 6); beta-amylase | 2.45 |
| 575 | 266334_at | AT2G32380 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G05210.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO24140.1); contains InterPro domain Transmembrane protein 97, predicted (InterPro:IPR016964) | -1.57 |
| 576 | 267118_at | AT2G32590 | similar to unnamed protein product [Vitis vinifera] (GB:CAO24036.1); contains InterPro domain Barren; (InterPro:IPR008418) | -1.82 |
| 577 | 267546_at | AT2G32680 | disease resistance family protein | 4.13 |
| 578 | 267550_at | AT2G32800 | AP4.3A; ATP binding / protein kinase | 2.08 |
| 579 | 267645_at | AT2G32860 | glycosyl hydrolase family 1 protein | 1.62 |
| 580 | 267595_at | AT2G32990 | ATGH9B8 (ARABIDOPSIS THALIANA GLYCOSYL HYDROLASE 9B8); hydrolase, hydrolyzing O-glycosyl compounds | -1.47 |
| 581 | 255817_at | AT2G33330 | 33 kDa secretory protein-related | -1.53 |
| 582 | 255789_at | AT2G33370 | 60S ribosomal protein L23 (RPL23B) | -1.91 |
| 583 | 255795_at | AT2G33380 | RD20 (RESPONSIVE TO DESSICATION 20); calcium ion binding | 1.94 |
| 584 | 255794_at | AT2G33480 | ANAC041 (Arabidopsis NAC domain containing protein 41); transcription factor | 2.06 |
| 585 | 255818_at | AT2G33570 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G44670.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO66312.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN76802.1); contains InterPro domain Protein of unknown function DUF23 (InterPro:IPR008166) | -2.09 |
| 586 | 267459_at | AT2G33850 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G28400.1); similar to unknown [Brassica napus] (GB:AAC06020.1) | -5.32 |
| 587 | 267007_at | AT2G34480 | 60S ribosomal protein L18A (RPL18aB) | -1.50 |
| 588 | 266996_at | AT2G34490 | CYP710A2 (cytochrome P450, family 710, subfamily A, polypeptide 2); C-22 sterol desaturase/ oxygen binding | -1.45 |
| 589 | 266905_at | AT2G34560 | katanin, putative | -1.55 |
| 590 | 267413_at | AT2G34960 | CAT5 (CATIONIC AMINO ACID TRANSPORTER 5); cationic amino acid transmembrane transporter | 2.04 |
| 591 | 266517_at | AT2G35120 | glycine cleavage system H protein, mitochondrial, putative | -1.89 |
| 592 | 266489_at | AT2G35190 | NPSN11 (NOVEL PLANT SNARE 11); protein transporter | -2.93 |
| 593 | 266548_at | AT2G35210 | AGD10/MEE28/RPA (MATERNAL EFFECT EMBRYO ARREST 28); DNA binding | -2.12 |
| 594 | 266640_at | AT2G35585 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G31940.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN79460.1) | -1.49 |
| 595 | 265823_at | AT2G35760 | integral membrane family protein | 1.76 |
| 596 | 263942_at | AT2G35860 | FLA16 (FASCICLIN-LIKE ARABINOGALACTAN PROTEIN 16 PRECURSOR) | -1.46 |
| 597 | 263956_at | AT2G35940 | BLH1 (embryo sac development arrest 29); DNA binding / transcription factor | 2.95 |
| 598 | 263289_at | AT2G36170 | ubiquitin extension protein 2 (UBQ2) / 60S ribosomal protein L40 (RPL40A) | -1.68 |
| 599 | 263960_at | AT2G36200 | kinesin motor protein-related | -2.45 |
| 600 | 265210_at | AT2G36620 | RPL24A (RIBOSOMAL PROTEIN L24); structural constituent of ribosome | -2.33 |
| 601 | 265203_at | AT2G36630 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G25737.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO16008.1); contains InterPro domain Protein of unknown function DUF81; (InterPro:IPR002781) | 1.77 |
| 602 | 263841_at | AT2G36870 | xyloglucan:xyloglucosyl transferase, putative / xyloglucan endotransglycosylase, putative / endo-xyloglucan transferase, putative | -1.92 |
| 603 | 263840_at | AT2G36885 | similar to unnamed protein product [Vitis vinifera] (GB:CAO63025.1) | -1.46 |
| 604 | 263847_at | AT2G36970 | UDP-glucoronosyl/UDP-glucosyl transferase family protein | 3.43 |
| 605 | 263845_at | AT2G37040 | PAL1 (PHE AMMONIA LYASE 1); phenylalanine ammonia-lyase | -1.60 |
| 606 | 265445_at | AT2G37190 | 60S ribosomal protein L12 (RPL12A) | -2.31 |
| 607 | 266007_at | AT2G37380 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G39370.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO41570.1) | -2.34 |
| 608 | 265960_at | AT2G37470 | histone H2B, putative | -1.77 |
| 609 | 267170_at | AT2G37585 | glycosyltransferase family 14 protein / core-2/I-branching enzyme family protein | -1.48 |
| 610 | 267174_at | AT2G37600 | 60S ribosomal protein L36 (RPL36A) | -1.66 |
| 611 | 267158_at | AT2G37640 | ATEXPA3 (ARABIDOPSIS THALIANA EXPANSIN A3) | -2.22 |
| 612 | 267160_at | AT2G37670 | WD-40 repeat family protein | -2.43 |
| 613 | 267165_at | AT2G37710 | RLK (RECEPTOR LECTIN KINASE); kinase | 1.99 |
| 614 | 267168_at | AT2G37770 | aldo/keto reductase family protein | 3.18 |
| 615 | 266087_at | AT2G37790 | aldo/keto reductase family protein | -1.98 |
| 616 | 266097_at | AT2G37970 | SOUL-1; binding | 1.70 |
| 617 | 266086_at | AT2G38060 | transporter-related | -2.75 |
| 618 | 267144_at | AT2G38110 | ATGPAT6/GPAT6 (GLYCEROL-3-PHOSPHATE ACYLTRANSFERASE 6); 1-acylglycerol-3-phosphate O-acyltransferase/ acyltransferase | -1.75 |
| 619 | 267146_at | AT2G38160 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G40070.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G40070.2); similar to hypothetical protein [Vitis vinifera] (GB:CAN77490.1) | -2.34 |
| 620 | 267026_at | AT2G38340 | AP2 domain-containing transcription factor, putative (DRE2B) | 1.78 |
| 621 | 267055_at | AT2G38360 | prenylated rab acceptor (PRA1) family protein | -1.71 |
| 622 | 267036_at | AT2G38465 | similar to unnamed protein product [Vitis vinifera] (GB:CAO61577.1) | 1.57 |
| 623 | 266415_at | AT2G38530 | LTP2 (LIPID TRANSFER PROTEIN 2); lipid binding | -3.07 |
| 624 | 257375_at | AT2G38640 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G41590.1); similar to unknown [Populus trichocarpa] (GB:ABK94888.1); contains InterPro domain Protein of unknown function DUF567 (InterPro:IPR007612) | 1.73 |
| 625 | 266418_at | AT2G38750 | ANNAT4 (ANNEXIN ARABIDOPSIS 4); calcium ion binding / calcium-dependent phospholipid binding | -1.42 |
| 626 | 266396_at | AT2G38790 | unknown protein | 2.03 |
| 627 | 263296_at | AT2G38800 | calmodulin-binding protein-related | 1.73 |
| 628 | 263264_at | AT2G38810 | HTA8; DNA binding | -5.23 |
| 629 | 266168_at | AT2G38870 | protease inhibitor, putative | -1.43 |
| 630 | 266188_at | AT2G39000 | GCN5-related N-acetyltransferase (GNAT) family protein | 1.79 |
| 631 | 266191_at | AT2G39040 | peroxidase, putative | -1.42 |
| 632 | 266980_at | AT2G39390 | 60S ribosomal protein L35 (RPL35B) | -1.67 |
| 633 | 266962_at | AT2G39435 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G53540.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO41581.1); contains domain PTHR21726:SF5 (PTHR21726:SF5); contains domain PTHR21726 (PTHR21726) | 1.58 |
| 634 | 266981_at | AT2G39460 | ATRPL23A (RIBOSOMAL PROTEIN L23A); RNA binding / structural constituent of ribosome | -1.45 |
| 635 | 266965_at | AT2G39510 | nodulin MtN21 family protein | -3.81 |
| 636 | 267592_at | AT2G39710 | aspartyl protease family protein | 1.69 |
| 637 | 245088_at | AT2G39850 | subtilase | 2.29 |
| 638 | 267355_at | AT2G39900 | LIM domain-containing protein | -2.60 |
| 639 | 267349_at | AT2G40010 | 60S acidic ribosomal protein P0 (RPP0A) | -1.83 |
| 640 | 263384_at | AT2G40130 | heat shock protein-related | 4.10 |
| 641 | 263386_at | AT2G40150 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G55990.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO41384.1); contains InterPro domain Protein of unknown function DUF231, plant (InterPro:IPR004253) | -1.41 |
| 642 | 263805_at | AT2G40400 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G56140.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN64033.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO41449.1); similar to hypothetical protein OsI_004191 [Oryza sativa (indica cultivar-group)] (GB:EAY76344.1); contains InterPro domain Protein of unknown function DUF399 (InterPro:IPR007314) | -2.22 |
| 643 | 263829_at | AT2G40435 | transcription regulator | 2.56 |
| 644 | 255874_at | AT2G40550 | similar to unnamed protein product [Vitis vinifera] (GB:CAO41481.1); contains domain PTHR13489 (PTHR13489) | -1.43 |
| 645 | 255822_at | AT2G40610 | ATEXPA8 (ARABIDOPSIS THALIANA EXPANSIN A8) | 1.86 |
| 646 | 266078_at | AT2G40670 | ARR16 (response regulator 16); transcription regulator/ two-component response regulator | 1.97 |
| 647 | 267076_at | AT2G41090 | calmodulin-like calcium-binding protein, 22 kDa (CaBP-22) | 2.86 |
| 648 | 267063_at | AT2G41120 | similar to unnamed protein product [Vitis vinifera] (GB:CAO18305.1); contains InterPro domain Protein of unknown function DUF309 (InterPro:IPR005500) | 1.75 |
| 649 | 267084_at | AT2G41180 | sigA-binding protein-related | 1.82 |
| 650 | 266363_at | AT2G41250 | haloacid dehalogenase-like hydrolase family protein | 1.84 |
| 651 | 266368_at | AT2G41380 | embryo-abundant protein-related | 3.59 |
| 652 | 266371_at | AT2G41410 | calmodulin, putative | 1.63 |
| 653 | 245112_at | AT2G41540 | GPDHC1; NAD binding / glycerol-3-phosphate dehydrogenase (NAD+) | -2.46 |
| 654 | 245106_at | AT2G41650 | unknown protein | -1.88 |
| 655 | 267538_at | AT2G41870 | remorin family protein | 2.69 |
| 656 | 267535_at | AT2G41940 | ZFP8 (ZINC FINGER PROTEIN 8); nucleic acid binding / transcription factor/ zinc ion binding | 2.21 |
| 657 | 267580_at | AT2G41990 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G35170.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN73167.1) | -1.67 |
| 658 | 267636_at | AT2G42110 | similar to unknown [Populus trichocarpa] (GB:ABK94541.1); similar to unknown [Populus trichocarpa] (GB:ABK92893.1); contains domain PTHR10270:SF18 (PTHR10270:SF18); contains domain PTHR10270 (PTHR10270) | -4.57 |
| 659 | 265852_at | AT2G42350 | zinc finger (C3HC4-type RING finger) family protein | 2.70 |
| 660 | 265853_at | AT2G42360 | zinc finger (C3HC4-type RING finger) family protein | 1.64 |
| 661 | 265877_at | AT2G42380 | bZIP transcription factor family protein | -2.66 |
| 662 | 263987_at | AT2G42690 | lipase, putative | 1.80 |
| 663 | 263973_at | AT2G42740 | RPL16A (ribosomal protein large subunit 16A); structural constituent of ribosome | -2.61 |
| 664 | 263985_at | AT2G42750 | DNAJ heat shock N-terminal domain-containing protein | 1.74 |
| 665 | 263986_at | AT2G42790 | CSY3 (CITRATE SYNTHASE 3); citrate (SI)-synthase | 1.66 |
| 666 | 263979_at | AT2G42840 | PDF1 (PROTODERMAL FACTOR 1) | -4.08 |
| 667 | 265266_at | AT2G42890 | AML2; RNA binding | 1.57 |
| 668 | 265243_at | AT2G43040 | NPG1 (NO POLLEN GERMINATION 1); calmodulin binding | -1.47 |
| 669 | 260548_at | AT2G43360 | BIO2 (BIOTIN AUXOTROPH 2); biotin synthase | -1.63 |
| 670 | 260538_at | AT2G43460 | 60S ribosomal protein L38 (RPL38A) | -1.50 |
| 671 | 260546_at | AT2G43520 | ATTI2 (ARABIDOPSIS THALIANA TRYPSIN INHIBITOR PROTEIN 2); trypsin inhibitor | -1.90 |
| 672 | 260549_at | AT2G43535 | trypsin inhibitor, putative | -2.28 |
| 673 | 260547_at | AT2G43550 | trypsin inhibitor, putative | -3.12 |
| 674 | 260568_at | AT2G43570 | chitinase, putative | 3.33 |
| 675 | 260562_at | AT2G43850 | ankyrin protein kinase, putative (APK1) | 1.95 |
| 676 | 267224_at | AT2G43990 | similar to unnamed protein product [Vitis vinifera] (GB:CAO46373.1) | -2.79 |
| 677 | 267213_at | AT2G44120 | 60S ribosomal protein L7 (RPL7C) | -1.76 |
| 678 | 267367_at | AT2G44210 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G55360.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO67164.1); contains InterPro domain Protein of unknown function DUF239, plant (InterPro:IPR004314) | 1.57 |
| 679 | 267388_at | AT2G44450 | glycosyl hydrolase family 1 protein | -1.78 |
| 680 | 266889_at | AT2G44640 | similar to PDE320 (PIGMENT DEFECTIVE 320) [Arabidopsis thaliana] (TAIR:AT3G06960.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO70456.1) | -1.81 |
| 681 | 245136_at | AT2G45210 | auxin-responsive protein-related | 3.79 |
| 682 | 245141_at | AT2G45400 | BEN1; oxidoreductase, acting on CH-OH group of donors | -2.27 |
| 683 | 251395_at | AT2G45470 | FLA8 (Arabinogalactan protein 8) | -2.80 |
| 684 | 267559_at | AT2G45570 | CYP76C2 (cytochrome P450, family 76, subfamily C, polypeptide 2); oxygen binding | 1.83 |
| 685 | 267503_at | AT2G45600 | hydrolase | 2.08 |
| 686 | 267509_at | AT2G45660 | AGL20 (AGAMOUS-LIKE 20); transcription factor | 2.34 |
| 687 | 263777_at | AT2G46450 | ATCNGC12 (cyclic nucleotide gated channel 12); cyclic nucleotide binding / ion channel | 1.83 |
| 688 | 265460_at | AT2G46600 | calcium-binding protein, putative | 1.62 |
| 689 | 266327_at | AT2G46680 | ATHB-7 (ARABIDOPSIS THALIANA HOMEOBOX 7); transcription factor | 1.84 |
| 690 | 266719_at | AT2G46830 | CCA1 (CIRCADIAN CLOCK ASSOCIATED 1); transcription factor | 2.39 |
| 691 | 266752_at | AT2G47000 | MDR4/PGP4 (P-GLYCOPROTEIN4); ATPase, coupled to transmembrane movement of substances / xenobiotic-transporting ATPase | 2.01 |
| 692 | 260581_at | AT2G47190 | MYB2 (myb domain protein 2); DNA binding / transcription factor | 1.93 |
| 693 | 260527_at | AT2G47270 | transcription factor/ transcription regulator | 1.61 |
| 694 | 245174_at | AT2G47500 | microtubule motor | -1.48 |
| 695 | 245171_at | AT2G47560 | zinc finger (C3HC4-type RING finger) family protein | -1.42 |
| 696 | 245121_at | AT2G47610 | 60S ribosomal protein L7A (RPL7aA) | -1.72 |
| 697 | 266485_at | AT2G47630 | esterase/lipase/thioesterase family protein | -1.86 |
| 698 | 266464_at | AT2G47800 | ATMRP4 (Arabidopsis thaliana multidrug resistance-associated protein 4) | 2.02 |
| 699 | 266516_at | AT2G47880 | glutaredoxin family protein | 4.98 |
| 700 | 259272_at | AT3G01290 | band 7 family protein | 1.67 |
| 701 | 257524_at | AT3G01330 | DEL3 (DP-E2F-like 3); transcription factor | -1.68 |
| 702 | 258952_at | AT3G01410 | RNase H domain-containing protein | -3.46 |
| 703 | 258947_at | AT3G01830 | calmodulin-related protein, putative | 1.82 |
| 704 | 259001_at | AT3G01960 | unknown protein | 2.50 |
| 705 | 258975_at | AT3G01970 | WRKY45 (WRKY DNA-binding protein 45); transcription factor | 4.50 |
| 706 | 258856_at | AT3G02040 | SRG3 (SENESCENCE-RELATED GENE 3); glycerophosphodiester phosphodiesterase | -4.03 |
| 707 | 258858_at | AT3G02080 | 40S ribosomal protein S19 (RPS19A) | -1.51 |
| 708 | 258859_at | AT3G02120 | hydroxyproline-rich glycoprotein family protein | -2.56 |
| 709 | 258497_at | AT3G02380 | COL2 (CONSTANS-LIKE 2); transcription factor/ zinc ion binding | 2.35 |
| 710 | 258503_at | AT3G02500 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G16030.1); similar to hypothetical protein [Cleome spinosa] (GB:ABD96956.1) | -2.78 |
| 711 | 258487_at | AT3G02550 | LBD41 (LOB DOMAIN-CONTAINING PROTEIN 41) | -1.44 |
| 712 | 258486_at | AT3G02560 | 40S ribosomal protein S7 (RPS7B) | -1.66 |
| 713 | 258480_at | AT3G02640 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G16250.1); similar to unknown [Populus trichocarpa] (GB:ABK93711.1) | -2.91 |
| 714 | 258617_at | AT3G03000 | calmodulin, putative | 1.67 |
| 715 | 258871_at | AT3G03060 | ATPase | -1.48 |
| 716 | 259058_at | AT3G03470 | CYP89A9 (cytochrome P450, family 87, subfamily A, polypeptide 9); oxygen binding | 3.33 |
| 717 | 259054_at | AT3G03480 | CHAT (ACETYL COA:(Z)-3-HEXEN-1-OL ACETYLTRANSFERASE); acetyl CoA:(Z)-3-hexen-1-ol acetyltransferase | 2.34 |
| 718 | 259340_at | AT3G03870 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G18130.2) | 3.30 |
| 719 | 258815_at | AT3G04000 | short-chain dehydrogenase/reductase (SDR) family protein | 1.66 |
| 720 | 258537_at | AT3G04210 | disease resistance protein (TIR-NBS class), putative | 4.02 |
| 721 | 258589_at | AT3G04290 | ATLTL1/LTL1 (LI-TOLERANT LIPASE 1); carboxylesterase | -5.96 |
| 722 | 258840_at | AT3G04620 | nucleic acid binding | -1.62 |
| 723 | 258796_at | AT3G04630 | WDL1 (WVD2-LIKE 1) | -1.61 |
| 724 | 259096_at | AT3G04840 | 40S ribosomal protein S3A (RPS3aA) | -1.74 |
| 725 | 259090_at | AT3G04920 | 40S ribosomal protein S24 (RPS24A) | -1.70 |
| 726 | 259095_at | AT3G05020 | ACP1 (ACYL CARRIER PROTEIN 1) | -2.54 |
| 727 | 259133_at | AT3G05400 | sugar transporter, putative | -2.16 |
| 728 | 259136_at | AT3G05470 | formin homology 2 domain-containing protein / FH2 domain-containing protein | -1.46 |
| 729 | 259112_at | AT3G05560 | 60S ribosomal protein L22-2 (RPL22B) | -1.49 |
| 730 | 258887_at | AT3G05630 | PLDP2 (PHOSPHOLIPASE D ZETA 2); phospholipase D | 1.95 |
| 731 | 258894_at | AT3G05650 | disease resistance family protein | 1.97 |
| 732 | 258890_at | AT3G05690 | ATHAP2B/HAP2B/UNE8 (HEME ACTIVATOR PROTEIN (YEAST) HOMOLOG 2B); transcription factor | 1.60 |
| 733 | 258747_at | AT3G05810 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G26800.1); similar to unknown [Populus trichocarpa] (GB:ABK93627.1) | -1.75 |
| 734 | 258751_at | AT3G05890 | RCI2B (RARE-COLD-INDUCIBLE 2B) | -2.56 |
| 735 | 258472_at | AT3G06080 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G19160.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO71110.1); contains InterPro domain Protein of unknown function DUF231, plant (InterPro:IPR004253) | 1.81 |
| 736 | 258914_at | AT3G06360 | AGP27 (ARABINOGALACTAN PROTEIN 27) | -1.91 |
| 737 | 258521_at | AT3G06680 | 60S ribosomal protein L29 (RPL29B) | -2.51 |
| 738 | 258532_at | AT3G06700 | 60S ribosomal protein L29 (RPL29A) | -1.69 |
| 739 | 258530_at | AT3G06840 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G49170.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO44815.1) | -3.83 |
| 740 | 259014_at | AT3G07320 | glycosyl hydrolase family 17 protein | -2.97 |
| 741 | 259015_at | AT3G07350 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G25240.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO38687.1); contains InterPro domain Protein of unknown function DUF506, plant (InterPro:IPR006502) | -2.33 |
| 742 | 259021_at | AT3G07540 | formin homology 2 domain-containing protein / FH2 domain-containing protein | -1.42 |
| 743 | 259251_at | AT3G07600 | heavy-metal-associated domain-containing protein | 1.98 |
| 744 | 259226_at | AT3G07700 | ABC1 family protein | 1.93 |
| 745 | 259228_at | AT3G07720 | kelch repeat-containing protein | 1.63 |
| 746 | 258647_at | AT3G07870 | F-box family protein | 1.62 |
| 747 | 258641_at | AT3G08030 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G41800.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO40802.1); contains InterPro domain Protein of unknown function DUF642 (InterPro:IPR006946) | -1.94 |
| 748 | 258691_at | AT3G08630 | similar to alphavirus core protein family [Arabidopsis thaliana] (TAIR:AT3G08640.1); similar to unknown [Populus trichocarpa] (GB:ABK93697.1) | -1.53 |
| 749 | 258675_at | AT3G08770 | LTP6 (Lipid transfer protein 6); lipid binding | -3.90 |
| 750 | 258982_at | AT3G08870 | lectin protein kinase, putative | 3.25 |
| 751 | 259211_at | AT3G09020 | alpha 1,4-glycosyltransferase family protein / glycosyltransferase sugar-binding DXD motif-containing protein | 2.70 |
| 752 | 258709_at | AT3G09500 | 60S ribosomal protein L35 (RPL35A) | -1.69 |
| 753 | 258723_at | AT3G09600 | myb family transcription factor | 1.70 |
| 754 | 258658_at | AT3G09820 | ADK1 (ADENOSINE KINASE 1) | -1.83 |
| 755 | 258650_at | AT3G09830 | protein kinase, putative | 1.78 |
| 756 | 258651_at | AT3G09920 | PIP5K9 (PHOSPHATIDYL INOSITOL MONOPHOSPHATE 5 KINASE); 1-phosphatidylinositol-4-phosphate 5-kinase | 1.58 |
| 757 | 259145_at | AT3G10180 | kinesin motor protein-related | -2.66 |
| 758 | 259150_at | AT3G10320 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G41640.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO23616.1); contains InterPro domain Protein of unknown function DUF563 (InterPro:IPR007657) | 3.51 |
| 759 | 259149_at | AT3G10340 | phenylalanine ammonia-lyase, putative | -3.11 |
| 760 | 258925_at | AT3G10420 | sporulation protein-related | 2.22 |
| 761 | 258919_at | AT3G10525 | similar to SIM (SIAMESE) [Arabidopsis thaliana] (TAIR:AT5G04470.1) | 1.67 |
| 762 | 258922_at | AT3G10610 | 40S ribosomal protein S17 (RPS17C) | -2.06 |
| 763 | 258945_at | AT3G10660 | CPK2 (CALMODULIN-DOMAIN PROTEIN KINASE CDPK ISOFORM 2); calmodulin-dependent protein kinase/ kinase | -2.09 |
| 764 | 256428_at | AT3G11080 | disease resistance family protein | 1.78 |
| 765 | 259231_at | AT3G11410 | AHG3/ATPP2CA (ARABIDOPSIS THALIANA PROTEIN PHOSPHATASE 2CA); protein binding / protein serine/threonine phosphatase | 1.70 |
| 766 | 259286_at | AT3G11480 | BSMT1; S-adenosylmethionine-dependent methyltransferase | 1.98 |
| 767 | 259239_at | AT3G11510 | 40S ribosomal protein S14 (RPS14B) | -1.41 |
| 768 | 259290_at | AT3G11520 | CYCB1;3 (CYCLIN B1;3); cyclin-dependent protein kinase regulator | -1.69 |
| 769 | 259235_at | AT3G11600 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G06270.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO15841.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN79170.1) | -1.67 |
| 770 | 259102_at | AT3G11660 | NHL1 (NDR1/HIN1-like 1) | 2.44 |
| 771 | 259070_at | AT3G11670 | DGD1 (DIGALACTOSYL DIACYLGLYCEROL DEFICIENT 1); galactolipid galactosyltransferase/ transferase, transferring glycosyl groups | 1.88 |
| 772 | 256275_at | AT3G12110 | ACT11 (ACTIN-11); structural constituent of cytoskeleton | -2.27 |
| 773 | 256290_at | AT3G12203 | SCPL17 (serine carboxypeptidase-like 17); serine carboxypeptidase | -2.00 |
| 774 | 256266_at | AT3G12320 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G06980.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN75675.1) | 1.77 |
| 775 | 257697_at | AT3G12700 | aspartyl protease family protein | -2.07 |
| 776 | 257701_at | AT3G12710 | methyladenine glycosylase family protein | -1.82 |
| 777 | 257853_at | AT3G12960 | similar to seed maturation protein [Glycine tomentella] (GB:ABB72392.1) | -2.30 |
| 778 | 257860_at | AT3G13062 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G55960.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO41766.1); contains InterPro domain Lipid-binding START (InterPro:IPR002913) | 3.06 |
| 779 | 257850_at | AT3G13065 | SRF4 (STRUBBELIG-RECEPTOR FAMILY 4); ATP binding / kinase/ protein serine/threonine kinase | 3.91 |
| 780 | 256957_at | AT3G13420 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G55535.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G55535.2); similar to unnamed protein product [Vitis vinifera] (GB:CAO22125.1) | -1.59 |
| 781 | 256983_at | AT3G13470 | chaperonin, putative | -2.96 |
| 782 | 256964_at | AT3G13520 | AGP12 (ARABINOGALACTAN PROTEIN 12) | -1.88 |
| 783 | 256647_at | AT3G13610 | oxidoreductase, 2OG-Fe(II) oxygenase family protein | 2.03 |
| 784 | 256789_at | AT3G13672 | seven in absentia (SINA) family protein | 1.92 |
| 785 | 256784_at | AT3G13674 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G55205.1) | -3.58 |
| 786 | 257610_at | AT3G13810 | ATIDD11 (ARABIDOPSIS THALIANA INDETERMINATE(ID)-DOMAIN 11); nucleic acid binding / transcription factor/ zinc ion binding | 2.25 |
| 787 | 258203_at | AT3G13950 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G13266.1); similar to Ankyrin [Medicago truncatula] (GB:ABN08906.1) | 2.83 |
| 788 | 258197_at | AT3G14000 | ATBRXL2/BRX-LIKE2 | -1.57 |
| 789 | 256997_at | AT3G14067 | subtilase family protein | 1.95 |
| 790 | 257008_at | AT3G14210 | ESM1 (EPITHIOSPECIFIER MODIFIER 1); carboxylesterase | 1.84 |
| 791 | 257072_at | AT3G14220 | GDSL-motif lipase/hydrolase family protein | -1.85 |
| 792 | 258368_at | AT3G14240 | subtilase family protein | -1.83 |
| 793 | 258362_at | AT3G14280 | similar to hypothetical protein [Vitis vinifera] (GB:CAN62135.1) | 3.30 |
| 794 | 257280_at | AT3G14440 | NCED3 (NINE-CIS-EPOXYCAROTENOID DIOXYGENASE3) | 1.63 |
| 795 | 258063_at | AT3G14620 | CYP72A8 (cytochrome P450, family 72, subfamily A, polypeptide 8); oxygen binding | 2.46 |
| 796 | 258117_at | AT3G14700 | similar to SART-1 family protein [Arabidopsis thaliana] (TAIR:AT5G16780.1); similar to hypothetical protein OsJ_006675 [Oryza sativa (japonica cultivar-group)] (GB:EAZ23192.1); similar to Os02g0511500 [Oryza sativa (japonica cultivar-group)] (GB:NP_001046936.1); similar to hypothetical protein OsI_007241 [Oryza sativa (indica cultivar-group)] (GB:EAY86008.1); contains InterPro domain SART-1 protein (InterPro:IPR005011) | 1.66 |
| 797 | 257237_at | AT3G14890 | phosphoesterase | -1.71 |
| 798 | 256858_at | AT3G15140 | exonuclease family protein | -2.96 |
| 799 | 257051_at | AT3G15270 | SPL5 (SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 5); DNA binding / transcription factor | 1.87 |
| 800 | 257057_at | AT3G15310 | transposable element gene | 3.54 |
| 801 | 257054_at | AT3G15353 | MT3 (METALLOTHIONEIN 3) | 1.79 |
| 802 | 258397_at | AT3G15357 | unknown protein | -1.88 |
| 803 | 258387_at | AT3G15550 | similar to unnamed protein product [Vitis vinifera] (GB:CAO46019.1) | -2.06 |
| 804 | 257290_at | AT3G15560 | similar to unnamed protein product [Vitis vinifera] (GB:CAO46019.1) | -1.44 |
| 805 | 258222_at | AT3G15680 | zinc finger (Ran-binding) family protein | -1.73 |
| 806 | 258263_at | AT3G15780 | unknown protein | 2.36 |
| 807 | 258223_at | AT3G15840 | PIFI (POST-ILLUMINATION CHLOROPHYLL FLUORESCENCE INCREASE) | 1.64 |
| 808 | 257798_at | AT3G15950 | (TSA1-LIKE); protein binding | -2.31 |
| 809 | 258284_at | AT3G16080 | 60S ribosomal protein L37 (RPL37C) | -1.47 |
| 810 | 258049_at | AT3G16220 | similar to RNA binding / catalytic [Arabidopsis thaliana] (TAIR:AT3G16230.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO45862.1); contains InterPro domain Predicted eukaryotic LigT; (InterPro:IPR009210) | 1.88 |
| 811 | 258054_at | AT3G16240 | DELTA-TIP (delta tonoplast integral protein); water channel | -1.81 |
| 812 | 259376_at | AT3G16320 | binding | -3.71 |
| 813 | 257517_at | AT3G16330 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G52140.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN64915.1) | -2.30 |
| 814 | 259329_at | AT3G16360 | AHP4 (HPT PHOSPHOTRANSMITTER 4); histidine phosphotransfer kinase/ transferase, transferring phosphorus-containing groups | 5.78 |
| 815 | 259383_at | AT3G16470 | JR1 (Jacalin lectin family protein) | -3.62 |
| 816 | 258410_at | AT3G16780 | 60S ribosomal protein L19 (RPL19B) | -1.94 |
| 817 | 257883_at | AT3G16940 | calmodulin-binding protein | 1.62 |
| 818 | 257895_at | AT3G16950 | LPD1 (LIPOAMIDE DEHYDROGENASE 1) | -1.46 |
| 819 | 257875_at | AT3G17120 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G01960.1); similar to hypothetical protein MtrDRAFT_AC173289g6v1 [Medicago truncatula] (GB:ABN09095.1) | -1.43 |
| 820 | 258464_at | AT3G17360 | POK1 (PHRAGMOPLAST ORIENTING KINESIN 1); microtubule motor | -4.12 |
| 821 | 258347_at | AT3G17520 | late embryogenesis abundant domain-containing protein / LEA domain-containing protein | 2.76 |
| 822 | 258351_at | AT3G17700 | CNBT1 (CYCLIC NUCLEOTIDE-BINDING TRANSPORTER 1); calmodulin binding / cyclic nucleotide binding / ion channel | 1.70 |
| 823 | 258159_at | AT3G17840 | RLK902 (receptor-like kinase 902); ATP binding / kinase/ protein serine/threonine kinase | -1.90 |
| 824 | 258155_at | AT3G18130 | guanine nucleotide-binding family protein / activated protein kinase C receptor (RACK1) | -2.12 |
| 825 | 257734_at | AT3G18370 | ATSYTF/NTMC2T3/NTMC2TYPE3/SYTF | 1.56 |
| 826 | 257755_at | AT3G18760 | ribosomal protein S6 family protein | -1.61 |
| 827 | 256652_at | AT3G18850 | LPAT5; acyltransferase | -1.98 |
| 828 | 256891_at | AT3G19030 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G49500.1) | -1.57 |
| 829 | 256569_at | AT3G19550 | similar to hypothetical protein [Vitis vinifera] (GB:CAN60715.1) | 3.50 |
| 830 | 257020_at | AT3G19590 | WD-40 repeat family protein / mitotic checkpoint protein, putative | -1.46 |
| 831 | 257074_at | AT3G19660 | similar to unnamed protein product [Vitis vinifera] (GB:CAO49873.1) | 2.16 |
| 832 | 257021_at | AT3G19710 | BCAT4 (BRANCHED-CHAIN AMINOTRANSFERASE4); catalytic/ methionine-oxo-acid transaminase | 1.80 |
| 833 | 257938_at | AT3G19820 | DWF1 (DIMINUTO 1); catalytic | -2.97 |
| 834 | 257964_at | AT3G19850 | phototropic-responsive NPH3 family protein | 2.18 |
| 835 | 256627_at | AT3G19970 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G18245.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO66476.1); contains InterPro domain Protein of unknown function DUF829, eukaryotic (InterPro:IPR008547) | 1.57 |
| 836 | 257128_at | AT3G20080 | CYP705A15 (cytochrome P450, family 705, subfamily A, polypeptide 15); oxygen binding | 1.80 |
| 837 | 257122_at | AT3G20250 | APUM5 (ARABIDOPSIS PUMILIO 5); RNA binding | 2.02 |
| 838 | 257663_at | AT3G20260 | structural constituent of ribosome | -4.18 |
| 839 | 256666_at | AT3G20670 | HTA13; DNA binding | -1.71 |
| 840 | 258040_at | AT3G21190 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G51630.1); similar to unknown [Populus trichocarpa] (GB:ABK93128.1); contains InterPro domain Protein of unknown function DUF246, plant (InterPro:IPR004348) | -1.54 |
| 841 | 258179_at | AT3G21690 | MATE efflux family protein | 1.66 |
| 842 | 257949_at | AT3G21750 | UDP-glucoronosyl/UDP-glucosyl transferase family protein | 1.65 |
| 843 | 257950_at | AT3G21780 | UGT71B6 (UDP-GLUCOSYL TRANSFERASE 71B6); UDP-glycosyltransferase/ abscisic acid glucosyltransferase/ transferase, transferring glycosyl groups | 5.19 |
| 844 | 256794_at | AT3G22230 | 60S ribosomal protein L27 (RPL27B) | -1.49 |
| 845 | 256766_at | AT3G22231 | PCC1 (PATHOGEN AND CIRCADIAN CONTROLLED 1) | 2.15 |
| 846 | 256930_at | AT3G22460 | cysteine synthase, putative / O-acetylserine (thiol)-lyase, putative / O-acetylserine sulfhydrylase, putative | 2.73 |
| 847 | 258322_at | AT3G22740 | HMT3 (Homocysteine S-methyltransferase 3); homocysteine S-methyltransferase | 2.04 |
| 848 | 258341_at | AT3G22790 | kinase interacting family protein | -2.17 |
| 849 | 258325_at | AT3G22830 | AT-HSFA6B (Arabidopsis thaliana heat shock transcription factor A6B); DNA binding / transcription factor | -1.89 |
| 850 | 256832_at | AT3G22880 | ATDMC1 (RECA-LIKE GENE); ATP binding / DNA-dependent ATPase/ damaged DNA binding | -1.94 |
| 851 | 256839_at | AT3G22930 | calmodulin, putative | 1.96 |
| 852 | 257764_at | AT3G23010 | disease resistance family protein / LRR family protein | 3.22 |
| 853 | 258296_at | AT3G23390 | 60S ribosomal protein L36a/L44 (RPL36aA) | -1.66 |
| 854 | 258100_at | AT3G23550 | MATE efflux family protein | 2.57 |
| 855 | 258098_at | AT3G23670 | KINESIN-12B/PAKRP1L; microtubule motor/ plus-end-directed microtubule motor | -3.79 |
| 856 | 257203_at | AT3G23730 | xyloglucan:xyloglucosyl transferase, putative / xyloglucan endotransglycosylase, putative / endo-xyloglucan transferase, putative | -3.08 |
| 857 | 257196_at | AT3G23790 | AMP-binding protein, putative | 2.38 |
| 858 | 257204_at | AT3G23805 | RALFL24 (RALF-LIKE 24) | -1.56 |
| 859 | 257173_at | AT3G23810 | SAHH2 (S-ADENOSYL-L-HOMOCYSTEINE (SAH) HYDROLASE 2); adenosylhomocysteinase | -2.94 |
| 860 | 256890_at | AT3G23830 | GR-RBP4/GRP4 (GLYCINE-RICH RNA-BINDING PROTEIN 4); RNA binding / double-stranded DNA binding / single-stranded DNA binding | -2.69 |
| 861 | 256861_at | AT3G23920 | BAM1/BMY7/TR-BAMY (BETA-AMYLASE 1); beta-amylase | 1.81 |
| 862 | 256862_at | AT3G23940 | dehydratase family | -1.74 |
| 863 | 257591_at | AT3G24900 | disease resistance family protein / LRR family protein | 2.08 |
| 864 | 257100_at | AT3G25010 | disease resistance family protein | 4.11 |
| 865 | 257101_at | AT3G25020 | disease resistance family protein | 1.59 |
| 866 | 257906_at | AT3G25520 | ATL5 (A. THALIANA RIBOSOMAL PROTEIN L5); structural constituent of ribosome | -1.75 |
| 867 | 256756_at | AT3G25610 | haloacid dehalogenase-like hydrolase family protein | 3.03 |
| 868 | 256754_at | AT3G25690 | CHUP1 (CHLOROPLAST UNUSUAL POSITIONING 1) | 1.67 |
| 869 | 258086_at | AT3G25860 | LTA2 (PLASTID E2 SUBUNIT OF PYRUVATE DECARBOXYLASE); dihydrolipoyllysine-residue acetyltransferase | -1.67 |
| 870 | 258075_at | AT3G25900 | ATHMT-1/HMT-1; homocysteine S-methyltransferase | -1.60 |
| 871 | 258067_at | AT3G25980 | mitotic spindle checkpoint protein, putative (MAD2) | -2.36 |
| 872 | 257623_at | AT3G26210 | CYP71B23 (cytochrome P450, family 71, subfamily B, polypeptide 23); oxygen binding | 3.20 |
| 873 | 257624_at | AT3G26220 | CYP71B3 (cytochrome P450, family 71, subfamily B, polypeptide 3); oxygen binding | 3.64 |
| 874 | 257625_at | AT3G26230 | CYP71B24 (cytochrome P450, family 71, subfamily B, polypeptide 24); oxygen binding | 3.48 |
| 875 | 257635_at | AT3G26280 | CYP71B4 (cytochrome P450, family 71, subfamily B, polypeptide 4); oxygen binding | 4.58 |
| 876 | 256874_at | AT3G26320 | CYP71B36 (cytochrome P450, family 71, subfamily B, polypeptide 36); oxygen binding | 2.02 |
| 877 | 257615_at | AT3G26510 | octicosapeptide/Phox/Bem1p (PB1) domain-containing protein | 1.77 |
| 878 | 257830_at | AT3G26690 | ATNUDT13 (ARABIDOPSIS THALIANA NUDIX HYDROLASE HOMOLOG 13); hydrolase | -2.04 |
| 879 | 257789_at | AT3G27020 | YSL6 (YELLOW STRIPE LIKE 6); oligopeptide transporter | 1.59 |
| 880 | 257809_at | AT3G27060 | TSO2 (TSO MEANING 'UGLY' IN CHINESE); ribonucleoside-diphosphate reductase | -1.96 |
| 881 | 257791_at | AT3G27110 | peptidase M48 family protein | 2.03 |
| 882 | 257151_at | AT3G27200 | plastocyanin-like domain-containing protein | -1.51 |
| 883 | 257740_at | AT3G27330 | zinc finger (C3HC4-type RING finger) family protein | -4.12 |
| 884 | 257714_at | AT3G27360 | histone H3 | -2.96 |
| 885 | 257222_at | AT3G27925 | DEGP1 (DEGP PROTEASE 1); serine-type peptidase | 1.71 |
| 886 | 257300_at | AT3G28080 | nodulin MtN21 family protein | 2.43 |
| 887 | 256577_at | AT3G28220 | meprin and TRAF homology domain-containing protein / MATH domain-containing protein | -1.51 |
| 888 | 256603_at | AT3G28270 | Identical to UPF0496 protein At3g28270 [Arabidopsis Thaliana] (GB:Q9LHD9;GB:Q8H7D0;GB:Q8H7E9;GB:Q944H3); similar to AT14A [Arabidopsis thaliana] (TAIR:AT3G28290.1); similar to AT14A [Arabidopsis thaliana] (TAIR:AT3G28300.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO22906.1); contains InterPro domain Protein of unknown function DUF677 (InterPro:IPR007749) | 3.63 |
| 889 | 256593_at | AT3G28510 | AAA-type ATPase family protein | 3.78 |
| 890 | 256596_at | AT3G28540 | AAA-type ATPase family protein | 4.23 |
| 891 | 256989_at | AT3G28580 | AAA-type ATPase family protein | 2.56 |
| 892 | 257137_at | AT3G28860 | ATMDR11/ATPGP19/MDR1/MDR11/PGP19 (P-GLYCOPROTEIN 19); ATPase, coupled to transmembrane movement of substances / auxin efflux transmembrane transporter | -1.46 |
| 893 | 257139_at | AT3G28890 | leucine-rich repeat family protein | 1.90 |
| 894 | 257141_at | AT3G28900 | 60S ribosomal protein L34 (RPL34C) | -2.53 |
| 895 | 258000_at | AT3G28940 | avirulence-responsive protein, putative / avirulence induced gene (AIG) protein, putative | 1.67 |
| 896 | 258059_at | AT3G29035 | ANAC059/ATNAC3 (Arabidopsis NAC domain containing protein 59); protein heterodimerization/ transcription factor | 3.50 |
| 897 | 256743_at | AT3G29370 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G39240.1) | -2.09 |
| 898 | 257288_at | AT3G29670 | transferase family protein | 2.13 |
| 899 | 252698_at | AT3G43670 | copper amine oxidase, putative | 1.78 |
| 900 | 252692_at | AT3G43960 | cysteine proteinase, putative | -2.79 |
| 901 | 252691_at | AT3G44050 | kinesin motor protein-related | -4.72 |
| 902 | 252681_at | AT3G44350 | ANAC061 (Arabidopsis NAC domain containing protein 61); transcription factor | 3.34 |
| 903 | 252643_at | AT3G44590 | 60S acidic ribosomal protein P2 (RPP2D) | -1.83 |
| 904 | 252648_at | AT3G44630 | disease resistance protein RPP1-WsB-like (TIR-NBS-LRR class), putative | 1.57 |
| 905 | 252625_at | AT3G44750 | HD2A (HISTONE DEACETYLASE 2A); nucleic acid binding / zinc ion binding | -1.61 |
| 906 | 246335_at | AT3G44880 | ACD1 (ACCELERATED CELL DEATH 1) | 1.98 |
| 907 | 252608_at | AT3G45090 | 2-phosphoglycerate kinase-related | -1.79 |
| 908 | 252573_at | AT3G45260 | zinc finger (C2H2 type) family protein | 1.66 |
| 909 | 252572_at | AT3G45290 | MLO3 (MILDEW RESISTANCE LOCUS O 3); calmodulin binding | 2.07 |
| 910 | 252586_at | AT3G45610 | Dof-type zinc finger domain-containing protein | -1.68 |
| 911 | 252549_at | AT3G45860 | receptor-like protein kinase, putative | 5.89 |
| 912 | 252534_at | AT3G46130 | MYB111 (myb domain protein 111); DNA binding / transcription factor | 2.39 |
| 913 | 252502_at | AT3G46900 | COPT2 (Copper transporter 2); copper ion transmembrane transporter | 1.57 |
| 914 | 252442_at | AT3G46940 | deoxyuridine 5'-triphosphate nucleotidohydrolase family | -2.39 |
| 915 | 252454_at | AT3G47130 | F-box family protein-related | -2.18 |
| 916 | 252462_at | AT3G47250 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G47200.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G47200.2); similar to unnamed protein product [Vitis vinifera] (GB:CAO15956.1); contains InterPro domain Protein of unknown function DUF247, plant (InterPro:IPR004158) | 2.11 |
| 917 | 252415_at | AT3G47340 | ASN1 (DARK INDUCIBLE 6) | -2.44 |
| 918 | 252413_at | AT3G47370 | 40S ribosomal protein S20 (RPS20B) | -1.59 |
| 919 | 252414_at | AT3G47420 | glycerol-3-phosphate transporter, putative / glycerol 3-phosphate permease, putative | 2.35 |
| 920 | 252411_at | AT3G47430 | PEX11B | 1.69 |
| 921 | 252440_at | AT3G47440 | TIP5;1 (tonoplast intrinsic protein 5;1); water channel | -2.66 |
| 922 | 252417_at | AT3G47480 | calcium-binding EF hand family protein | 3.72 |
| 923 | 252428_at | AT3G47660 | regulator of chromosome condensation (RCC1) family protein | -1.77 |
| 924 | 252403_at | AT3G48080 | lipase class 3 family protein / disease resistance protein-related | 2.28 |
| 925 | 252374_at | AT3G48100 | ARR5 (ARABIDOPSIS RESPONSE REGULATOR 5); transcription regulator/ two-component response regulator | -2.09 |
| 926 | 252367_at | AT3G48360 | BT2 (BTB AND TAZ DOMAIN PROTEIN 2); protein binding / transcription factor/ transcription regulator | -3.54 |
| 927 | 252361_at | AT3G48490 | similar to unknown [Populus trichocarpa] (GB:ABK93013.1) | -1.46 |
| 928 | 252345_at | AT3G48640 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G66670.2); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G66670.1) | 1.86 |
| 929 | 252346_at | AT3G48650 | pseudogene, At14a-related protein, similar to At14a (GI:11994571 and GI:11994573) (Arabidopsis thaliana) | 3.24 |
| 930 | 252317_at | AT3G48720 | transferase family protein | 2.67 |
| 931 | 252327_at | AT3G48740 | nodulin MtN3 family protein | 2.37 |
| 932 | 252297_at | AT3G48930 | EMB1080 (EMBRYO DEFECTIVE 1080); structural constituent of ribosome | -1.42 |
| 933 | 252300_at | AT3G49160 | pyruvate kinase family protein | 2.01 |
| 934 | 252303_at | AT3G49210 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G49200.1); similar to condensation domain-containing protein [Arabidopsis thaliana] (TAIR:AT3G49190.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO48523.1); contains InterPro domain Protein of unknown function UPF0089 (InterPro:IPR004255); contains InterPro domain Protein of unknown function DUF1298 (InterPro:IPR009721) | 1.70 |
| 935 | 252309_at | AT3G49340 | cysteine proteinase, putative | 2.67 |
| 936 | 252235_at | AT3G49910 | 60S ribosomal protein L26 (RPL26A) | -1.45 |
| 937 | 252220_at | AT3G49940 | LBD38 (LOB DOMAIN-CONTAINING PROTEIN 38) | -2.63 |
| 938 | 252240_at | AT3G50010 | DC1 domain-containing protein | 1.91 |
| 939 | 252189_at | AT3G50070 | CYCD3;3 (CYCLIN D3;3); cyclin-dependent protein kinase | -2.24 |
| 940 | 252170_at | AT3G50480 | HR4 (HOMOLOG OF RPW8 4) | 4.15 |
| 941 | 252183_at | AT3G50740 | UGT72E1 (UDP-glucosyl transferase 72E1); UDP-glycosyltransferase/ coniferyl-alcohol glucosyltransferase/ transferase, transferring glycosyl groups | 1.69 |
| 942 | 252178_at | AT3G50750 | brassinosteroid signalling positive regulator-related | 2.23 |
| 943 | 252136_at | AT3G50770 | calmodulin-related protein, putative | 3.23 |
| 944 | 252131_at | AT3G50930 | AAA-type ATPase family protein | 1.60 |
| 945 | 252126_at | AT3G50950 | disease resistance protein (CC-NBS-LRR class), putative | 1.70 |
| 946 | 252127_at | AT3G50960 | PLP3A (PHOSDUCIN-LIKE PROTEIN 3 HOMOLOG) | -1.71 |
| 947 | 252123_at | AT3G51240 | F3H (TRANSPARENT TESTA 6); naringenin 3-dioxygenase | -3.22 |
| 948 | 252148_at | AT3G51280 | male sterility MS5, putative | -2.27 |
| 949 | 252098_at | AT3G51330 | aspartyl protease family protein | 2.94 |
| 950 | 252117_at | AT3G51430 | YLS2 (yellow-leaf-specific gene 2); strictosidine synthase | 2.32 |
| 951 | 252105_at | AT3G51470 | protein phosphatase 2C, putative / PP2C, putative | -1.52 |
| 952 | 252077_at | AT3G51720 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G38370.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO48842.1); contains InterPro domain Protein of unknown function DUF827, plant (InterPro:IPR008545) | -1.79 |
| 953 | 246302_at | AT3G51860 | CAX3 (cation exchanger 3); cation:cation antiporter | 2.69 |
| 954 | 246305_at | AT3G51890 | protein binding / protein transporter/ structural molecule | 1.80 |
| 955 | 246310_at | AT3G51895 | SULTR3;1 (SULFATE TRANSPORTER 1); sulfate transmembrane transporter | 1.67 |
| 956 | 252089_at | AT3G52110 | similar to hypothetical protein OsI_009975 [Oryza sativa (indica cultivar-group)] (GB:EAY88742.1); similar to Os03g0174200 [Oryza sativa (japonica cultivar-group)] (GB:NP_001049125.1) | -1.88 |
| 957 | 256675_at | AT3G52170 | DNA binding | -1.83 |
| 958 | 256672_at | AT3G52310 | ABC transporter family protein | 1.78 |
| 959 | 252053_at | AT3G52400 | SYP122 (syntaxin 122); SNAP receptor | 1.75 |
| 960 | 252055_at | AT3G52580 | 40S ribosomal protein S14 (RPS14C) | -1.79 |
| 961 | 252061_at | AT3G52620 | unknown protein | -1.74 |
| 962 | 252000_at | AT3G52710 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G36220.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO15901.1) | 1.98 |
| 963 | 251995_at | AT3G52940 | FK (FACKEL); delta14-sterol reductase | -1.42 |
| 964 | 251993_at | AT3G52960 | peroxiredoxin type 2, putative | -1.43 |
| 965 | 251982_at | AT3G53190 | pectate lyase family protein | -2.43 |
| 966 | 251966_at | AT3G53380 | lectin protein kinase family protein | -2.18 |
| 967 | 251961_at | AT3G53620 | ATPPA4 (ARABIDOPSIS THALIANA PYROPHOSPHORYLASE 4); inorganic diphosphatase/ pyrophosphatase | -1.48 |
| 968 | 251952_at | AT3G53650 | histone H2B, putative | -1.46 |
| 969 | 251906_at | AT3G53720 | ATCHX20 (CATION/H+ EXCHANGER 20); monovalent cation:proton antiporter | 2.36 |
| 970 | 251926_at | AT3G53740 | 60S ribosomal protein L36 (RPL36B) | -1.70 |
| 971 | 251921_at | AT3G53890 | 40S ribosomal protein S21 (RPS21B) | -1.84 |
| 972 | 251920_at | AT3G53900 | uracil phosphoribosyltransferase, putative / UMP pyrophosphorylase, putative / UPRTase, putative | -1.51 |
| 973 | 251916_at | AT3G53960 | proton-dependent oligopeptide transport (POT) family protein | 1.86 |
| 974 | 251886_at | AT3G54260 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G42570.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO70127.1); contains InterPro domain Protein of unknown function DUF231, plant (InterPro:IPR004253) | -1.51 |
| 975 | 251899_at | AT3G54400 | aspartyl protease family protein | -2.89 |
| 976 | 251846_at | AT3G54560 | HTA11; DNA binding | -3.82 |
| 977 | 251844_at | AT3G54630 | similar to kinetochore protein [Capsella rubella] (GB:BAF63163.1); similar to kinetochore protein [Olimarabidopsis pumila] (GB:BAF63394.1); similar to kinetochore protein [Olimarabidopsis pumila] (GB:BAF63395.1); contains InterPro domain HEC/Ndc80p (InterPro:IPR005550) | -2.95 |
| 978 | 251852_at | AT3G54750 | similar to unnamed protein product [Vitis vinifera] (GB:CAO48944.1); contains domain PTHR12972 (PTHR12972) | -1.76 |
| 979 | 251827_at | AT3G55120 | A11/CFI/TT5 (TRANSPARENT TESTA 5); chalcone isomerase | -1.55 |
| 980 | 251783_at | AT3G55280 | 60S ribosomal protein L23A (RPL23aB) | -1.92 |
| 981 | 251791_at | AT3G55500 | ATEXPA16 (ARABIDOPSIS THALIANA EXPANSIN A16) | 3.94 |
| 982 | 251797_at | AT3G55560 | AGF2 (AT-HOOK PROTEIN OF GA FEEDBACK 2) | -1.53 |
| 983 | 251778_at | AT3G55660 | ATROPGEF6/ROPGEF6 (KINASE PARTNER PROTEIN-LIKE); Rho guanyl-nucleotide exchange factor/ | -1.91 |
| 984 | 251740_at | AT3G56070 | ROC2 (rotamase CyP 2); peptidyl-prolyl cis-trans isomerase | -1.83 |
| 985 | 251736_at | AT3G56130 | biotin/lipoyl attachment domain-containing protein | -1.98 |
| 986 | 251737_at | AT3G56340 | 40S ribosomal protein S26 (RPS26C) | -1.77 |
| 987 | 251714_at | AT3G56370 | leucine-rich repeat transmembrane protein kinase, putative | -1.66 |
| 988 | 251684_at | AT3G56410 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G05190.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G61670.1); similar to hypothetical protein Kpol_1058p4 [Vanderwaltozyma polyspora DSM 70294] (GB:XP_001645325.1) | 1.87 |
| 989 | 246293_at | AT3G56710 | SIB1 (SIGMA FACTOR BINDING PROTEIN 1); binding | 1.88 |
| 990 | 251668_at | AT3G57010 | strictosidine synthase family protein | -3.77 |
| 991 | 251673_at | AT3G57240 | BG3 (BETA-1,3-GLUCANASE 3); hydrolase, hydrolyzing O-glycosyl compounds | 2.37 |
| 992 | 251625_at | AT3G57260 | BGL2 (PATHOGENESIS-RELATED PROTEIN 2); glucan 1,3-beta-glucosidase/ hydrolase, hydrolyzing O-glycosyl compounds | 4.97 |
| 993 | 251640_at | AT3G57450 | similar to unnamed protein product [Vitis vinifera] (GB:CAO40798.1) | -1.73 |
| 994 | 251633_at | AT3G57460 | catalytic/ metal ion binding / metalloendopeptidase/ zinc ion binding | 1.80 |
| 995 | 251644_at | AT3G57540 | remorin family protein | 2.58 |
| 996 | 251591_at | AT3G57680 | peptidase S41 family protein | 1.82 |
| 997 | 251560_at | AT3G57920 | squamosa promoter-binding protein, putative | -2.59 |
| 998 | 251575_at | AT3G58120 | bZIP transcription factor family protein | -2.32 |
| 999 | 251580_at | AT3G58450 | universal stress protein (USP) family protein | 2.10 |
| 1000 | 251509_at | AT3G59010 | pectinesterase family protein | 2.14 |
| 1001 | 251497_at | AT3G59060 | PIL6 (PHYTOCHROME-INTERACTING FACTOR 5); DNA binding / transcription factor | 2.20 |
| 1002 | 251503_at | AT3G59140 | ATMRP14 (Arabidopsis thaliana multidrug resistance-associated protein 14) | 2.06 |
| 1003 | 251491_at | AT3G59480 | pfkB-type carbohydrate kinase family protein | -1.83 |
| 1004 | 251472_at | AT3G59580 | RWP-RK domain-containing protein | 2.01 |
| 1005 | 251483_at | AT3G59650 | mitochondrial ribosomal protein L51/S25/CI-B8 family protein | -2.14 |
| 1006 | 251479_at | AT3G59700 | ATHLECRK (ARABIDOPSIS THALIANA LECTIN-RECEPTOR KINASE); kinase | 2.26 |
| 1007 | 251426_at | AT3G60180 | uridylate kinase, putative / uridine monophosphate kinase, putative / UMP kinase, putative | 1.56 |
| 1008 | 251401_at | AT3G60270 | uclacyanin, putative | -1.62 |
| 1009 | 251400_at | AT3G60420 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G60450.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO70569.1); contains InterPro domain Phosphoglycerate mutase (InterPro:IPR013078); contains InterPro domain PRIB5 (InterPro:IPR012398) | 2.95 |
| 1010 | 251342_at | AT3G60690 | auxin-responsive family protein | 1.72 |
| 1011 | 251394_at | AT3G60900 | FLA10 (fasciclin-like arabinogalactan-protein 10) | -2.17 |
| 1012 | 251355_at | AT3G61100 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G61090.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO40196.1); contains InterPro domain Protein of unknown function DUF537 (InterPro:IPR007491) | -2.25 |
| 1013 | 251360_at | AT3G61210 | embryo-abundant protein-related | 3.07 |
| 1014 | 251317_at | AT3G61490 | glycoside hydrolase family 28 protein / polygalacturonase (pectinase) family protein | -1.46 |
| 1015 | 251281_at | AT3G61640 | AGP20 (ARABINOGALACTAN PROTEIN 20) | -1.65 |
| 1016 | 251272_at | AT3G61890 | ATHB-12 (ARABIDOPSIS THALIANA HOMEOBOX PROTEIN 12); transcription factor | 2.81 |
| 1017 | 251291_at | AT3G61900 | auxin-responsive family protein | 1.83 |
| 1018 | 251248_at | AT3G62150 | PGP21 (P-GLYCOPROTEIN 21); ATPase, coupled to transmembrane movement of substances | 1.58 |
| 1019 | 251218_at | AT3G62410 | CP12-2 | 1.95 |
| 1020 | 251181_at | AT3G62820 | invertase/pectin methylesterase inhibitor family protein | 1.70 |
| 1021 | 251197_at | AT3G62960 | glutaredoxin family protein | 5.01 |
| 1022 | 251200_at | AT3G63010 | ATGID1B/GID1B (GA INSENSITIVE DWARF1B); hydrolase | 1.80 |
| 1023 | 251174_at | AT3G63200 | PLA IIIB/PLP9 (Patatin-like protein 9); nutrient reservoir | -3.19 |
| 1024 | 251176_at | AT3G63380 | calcium-transporting ATPase, plasma membrane-type, putative / Ca(2+)-ATPase, putative (ACA12) | 1.73 |
| 1025 | 255694_at | AT4G00050 | UNE10 (unfertilized embryo sac 10); DNA binding / transcription factor | 2.13 |
| 1026 | 255630_at | AT4G00700 | C2 domain-containing protein | 2.56 |
| 1027 | 255626_at | AT4G00780 | meprin and TRAF homology domain-containing protein / MATH domain-containing protein | 2.52 |
| 1028 | 255642_at | AT4G00820 | IQD17 (IQ-domain 17); calmodulin binding | -2.16 |
| 1029 | 255644_at | AT4G00870 | basic helix-loop-helix (bHLH) family protein | 3.01 |
| 1030 | 255645_at | AT4G00880 | auxin-responsive family protein | 1.84 |
| 1031 | 255627_at | AT4G00955 | similar to protein kinase family protein [Arabidopsis thaliana] (TAIR:AT2G23450.2); similar to protein kinase family protein [Arabidopsis thaliana] (TAIR:AT2G23450.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO41616.1); contains InterPro domain EGF (InterPro:IPR006210) | 2.25 |
| 1032 | 255654_at | AT4G00970 | protein kinase family protein | 2.45 |
| 1033 | 255599_at | AT4G01010 | ATCNGC13 (cyclic nucleotide gated channel 13); calmodulin binding / cyclic nucleotide binding / ion channel | 2.13 |
| 1034 | 255612_at | AT4G01240 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G05390.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO39658.1); contains domain SSF53335 (SSF53335) | -1.98 |
| 1035 | 255621_at | AT4G01390 | meprin and TRAF homology domain-containing protein / MATH domain-containing protein | 5.64 |
| 1036 | 255577_at | AT4G01410 | harpin-induced family protein / HIN1 family protein / harpin-responsive family protein | -1.59 |
| 1037 | 255587_at | AT4G01480 | ATPPA5 (ARABIDOPSIS THALIANA PYROPHOSPHORYLASE 5); inorganic diphosphatase/ pyrophosphatase | 1.58 |
| 1038 | 255597_at | AT4G01730 | zinc finger (DHHC type) family protein | -1.41 |
| 1039 | 255513_at | AT4G02060 | PRL (PROLIFERA); ATP binding / DNA binding / DNA-dependent ATPase | -2.18 |
| 1040 | 255521_at | AT4G02280 | SUS3; UDP-glycosyltransferase/ sucrose synthase/ transferase, transferring glycosyl groups | 1.81 |
| 1041 | 255517_at | AT4G02290 | ATGH9B13 (ARABIDOPSIS THALIANA GLYCOSYL HYDROLASE 9B13); hydrolase, hydrolyzing O-glycosyl compounds | -4.12 |
| 1042 | 255502_at | AT4G02410 | lectin protein kinase family protein | 2.47 |
| 1043 | 255503_at | AT4G02420 | lectin protein kinase, putative | 2.54 |
| 1044 | 255460_at | AT4G02800 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G30050.1); similar to Os04g0228100 [Oryza sativa (japonica cultivar-group)] (GB:NP_001052288.1); similar to H0209A05.2 [Oryza sativa (indica cultivar-group)] (GB:CAH66085.1) | -3.57 |
| 1045 | 255450_at | AT4G02850 | phenazine biosynthesis PhzC/PhzF family protein | -1.44 |
| 1046 | 255462_at | AT4G02940 | oxidoreductase, 2OG-Fe(II) oxygenase family protein | 1.59 |
| 1047 | 255469_at | AT4G03030 | kelch repeat-containing F-box family protein | 1.61 |
| 1048 | 255410_at | AT4G03100 | rac GTPase activating protein, putative | -3.51 |
| 1049 | 255433_at | AT4G03210 | XTH9 (XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 9); hydrolase, acting on glycosyl bonds | -1.85 |
| 1050 | 255429_at | AT4G03410 | peroxisomal membrane protein-related | 2.10 |
| 1051 | 255406_at | AT4G03450 | ankyrin repeat family protein | 2.28 |
| 1052 | 255319_at | AT4G04220 | disease resistance family protein | 1.61 |
| 1053 | 255340_at | AT4G04490 | protein kinase family protein | 3.25 |
| 1054 | 255341_at | AT4G04500 | protein kinase family protein | 3.10 |
| 1055 | 255298_at | AT4G04840 | methionine sulfoxide reductase domain-containing protein / SeIR domain-containing protein | -3.42 |
| 1056 | 255310_at | AT4G04955 | ATALN (ARABIDOPSIS ALLANTOINASE); allantoinase/ hydrolase | 2.07 |
| 1057 | 255265_at | AT4G05190 | ATK5 (Arabidopsis thaliana kinesin 5); microtubule motor | -2.22 |
| 1058 | 255236_at | AT4G05520 | calcium-binding EF hand family protein | -1.72 |
| 1059 | 255243_at | AT4G05590 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G22310.1); similar to unknown [Populus trichocarpa] (GB:ABK93494.1); contains InterPro domain Protein of unknown function UPF0041 (InterPro:IPR005336) | 2.65 |
| 1060 | 255149_at | AT4G08150 | KNAT1 (BREVIPEDICELLUS 1); transcription factor | -2.35 |
| 1061 | 255129_at | AT4G08290 | nodulin MtN21 family protein | 1.60 |
| 1062 | 255127_at | AT4G08300 | nodulin MtN21 family protein | 3.38 |
| 1063 | 255148_at | AT4G08470 | MAPKKK10 (Mitogen-activated protein kinase kinase kinase 10); kinase | 1.91 |
| 1064 | 255104_at | AT4G08685 | SAH7 | -1.46 |
| 1065 | 255116_at | AT4G08850 | leucine-rich repeat family protein / protein kinase family protein | 1.59 |
| 1066 | 255068_at | AT4G08920 | CRY1 (CRYPTOCHROME 1) | 1.65 |
| 1067 | 255088_at | AT4G09350 | DNAJ heat shock N-terminal domain-containing protein | 1.60 |
| 1068 | 254998_at | AT4G09760 | choline kinase, putative | 1.86 |
| 1069 | 255016_at | AT4G10120 | ATSPS4F; sucrose-phosphate synthase/ transferase, transferring glycosyl groups | 2.39 |
| 1070 | 254971_at | AT4G10380 | NIP5;1/NLM6/NLM8 (NOD26-like intrinsic protein 5;1); boron transporter/ water channel | 2.70 |
| 1071 | 254975_at | AT4G10500 | oxidoreductase, 2OG-Fe(II) oxygenase family protein | 3.21 |
| 1072 | 254964_at | AT4G11080 | high mobility group (HMG1/2) family protein | -1.65 |
| 1073 | 254908_at | AT4G11200 | transposable element gene | -3.02 |
| 1074 | 254924_at | AT4G11330 | ATMPK5 (MAP KINASE 5); MAP kinase/ kinase | 1.75 |
| 1075 | 254919_at | AT4G11360 | RHA1B (RING-H2 finger A1B); protein binding / zinc ion binding | 1.66 |
| 1076 | 254845_at | AT4G11820 | MVA1 (HYDROXYMETHYLGLUTARYL-COA SYNTHASE); acetyl-CoA C-acetyltransferase/ hydroxymethylglutaryl-CoA synthase | -1.68 |
| 1077 | 254869_at | AT4G11890 | protein kinase family protein | 3.36 |
| 1078 | 254834_at | AT4G12300 | CYP706A4 (cytochrome P450, family 706, subfamily A, polypeptide 4); oxygen binding | 1.78 |
| 1079 | 254809_at | AT4G12410 | auxin-responsive family protein | -1.84 |
| 1080 | 254815_at | AT4G12420 | SKU5 (skewed 5); copper ion binding | -1.87 |
| 1081 | 254805_at | AT4G12480 | pEARLI 1; lipid binding | 1.57 |
| 1082 | 254832_at | AT4G12490 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | 2.36 |
| 1083 | 254831_at | AT4G12600 | ribosomal protein L7Ae/L30e/S12e/Gadd45 family protein | -1.82 |
| 1084 | 254785_at | AT4G12730 | FLA2 | -2.55 |
| 1085 | 254789_at | AT4G12880 | plastocyanin-like domain-containing protein | -2.61 |
| 1086 | 254794_at | AT4G12970 | similar to unnamed protein product [Vitis vinifera] (GB:CAO17947.1) | -2.20 |
| 1087 | 254759_at | AT4G13180 | short-chain dehydrogenase/reductase (SDR) family protein | 1.98 |
| 1088 | 254715_at | AT4G13550 | lipase class 3 family protein | 1.66 |
| 1089 | 254687_at | AT4G13770 | CYP83A1 (CYTOCHROME P450 83A1); oxygen binding | 1.63 |
| 1090 | 254735_at | AT4G13810 | disease resistance family protein / LRR family protein | 2.26 |
| 1091 | 254688_at | AT4G13830 | J20 (DNAJ-LIKE 20); heat shock protein binding | 1.85 |
| 1092 | 245385_at | AT4G14020 | rapid alkalinization factor (RALF) family protein | 4.18 |
| 1093 | 245325_at | AT4G14130 | XTR7 (XYLOGLUCAN ENDOTRANSGLYCOSYLASE 7); hydrolase, acting on glycosyl bonds | -1.55 |
| 1094 | 245311_at | AT4G14320 | 60S ribosomal protein L36a/L44 (RPL36aB) | -1.53 |
| 1095 | 245329_at | AT4G14365 | zinc finger (C3HC4-type RING finger) family protein / ankyrin repeat family protein | 3.03 |
| 1096 | 245265_at | AT4G14400 | ACD6 (ACCELERATED CELL DEATH 6); protein binding | 4.79 |
| 1097 | 245576_at | AT4G14770 | tesmin/TSO1-like CXC domain-containing protein | -2.58 |
| 1098 | 245270_at | AT4G14960 | TUA6 (tubulin alpha-6 chiain) | -1.47 |
| 1099 | 245585_at | AT4G14970 | similar to hypothetical protein [Vitis vinifera] (GB:CAN76433.1); similar to Os07g0154400 [Oryza sativa (japonica cultivar-group)] (GB:NP_001058915.1); similar to hypothetical protein OsI_024046 [Oryza sativa (indica cultivar-group)] (GB:EAZ02814.1) | -1.91 |
| 1100 | 245372_at | AT4G15000 | 60S ribosomal protein L27 (RPL27C) | -2.06 |
| 1101 | 245558_at | AT4G15430 | similar to early-responsive to dehydration protein-related / ERD protein-related [Arabidopsis thaliana] (TAIR:AT4G04340.1); similar to early-responsive to dehydration protein-related / ERD protein-related [Arabidopsis thaliana] (TAIR:AT4G04340.3); similar to early-responsive to dehydration protein-related / ERD protein-related [Arabidopsis thaliana] (TAIR:AT3G21620.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO24038.1); similar to CM0545.360.nc [Lotus japonicus] (GB:BAF98597.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO69661.1); contains InterPro domain Protein of unknown function DUF221; (InterPro:IPR003864) | 1.80 |
| 1102 | 245253_at | AT4G15440 | HPL1 (HYDROPEROXIDE LYASE 1); heme binding / iron ion binding / monooxygenase | 2.26 |
| 1103 | 245528_at | AT4G15530 | PPDK (PYRUVATE ORTHOPHOSPHATE DIKINASE); kinase/ pyruvate, phosphate dikinase | 2.67 |
| 1104 | 245277_at | AT4G15550 | IAGLU (INDOLE-3-ACETATE BETA-D-GLUCOSYLTRANSFERASE); UDP-glycosyltransferase/ transferase, transferring glycosyl groups | 1.78 |
| 1105 | 245343_at | AT4G15830 | binding | -3.50 |
| 1106 | 245523_at | AT4G15910 | ATDI21 (Arabidopsis thaliana drought-induced 21) | -2.34 |
| 1107 | 245524_at | AT4G15920 | nodulin MtN3 family protein | -1.91 |
| 1108 | 245353_at | AT4G16000 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G15990.1) | 3.54 |
| 1109 | 245393_at | AT4G16260 | glycosyl hydrolase family 17 protein | 1.64 |
| 1110 | 245323_at | AT4G16500 | cysteine protease inhibitor family protein / cystatin family protein | -1.74 |
| 1111 | 245453_at | AT4G16900 | ATP binding / protein binding / transmembrane receptor | 1.60 |
| 1112 | 245460_at | AT4G16990 | disease resistance protein (TIR-NBS class), putative | 1.84 |
| 1113 | 245463_at | AT4G17030 | ATEXLB1 (ARABIDOPSIS THALIANA EXPANSIN-LIKE B1) | 2.73 |
| 1114 | 245346_at | AT4G17090 | CT-BMY (BETA-AMYLASE 3, BETA-AMYLASE 8); beta-amylase | 2.64 |
| 1115 | 245301_at | AT4G17190 | FPS2 (FARNESYL DIPHOSPHATE SYNTHASE 2); dimethylallyltranstransferase/ geranyltranstransferase | -1.48 |
| 1116 | 245411_at | AT4G17240 | similar to hypothetical protein 31.t00013 [Brassica oleracea] (GB:ABD65111.1) | -3.58 |
| 1117 | 245272_at | AT4G17250 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G47580.1); similar to hypothetical protein 40.t00008 [Brassica oleracea] (GB:ABD65133.1) | 1.73 |
| 1118 | 245399_at | AT4G17340 | DELTA-TIP2/TIP2;2 (tonoplast intrinsic protein 2;2); water channel | 2.33 |
| 1119 | 245416_at | AT4G17350 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G47440.1); similar to hypothetical protein 31.t00035 [Brassica oleracea] (GB:ABD65121.1); contains InterPro domain Pleckstrin-like, plant (InterPro:IPR013666); contains InterPro domain Protein of unknown function DUF828, plant (InterPro:IPR008546); contains InterPro domain Pleckstrin-like (InterPro:IPR001849) | 1.82 |
| 1120 | 245355_at | AT4G17390 | 60S ribosomal protein L15 (RPL15B) | -2.17 |
| 1121 | 245422_at | AT4G17470 | palmitoyl protein thioesterase family protein | -1.83 |
| 1122 | 254691_at | AT4G17840 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G35260.1); similar to hypothetical protein 40.t00061 [Brassica oleracea] (GB:ABD65174.1) | 2.20 |
| 1123 | 254648_at | AT4G18550 | lipase class 3 family protein | -1.46 |
| 1124 | 254633_at | AT4G18640 | MRH1 (morphogenesis of root hair 1); ATP binding / protein serine/threonine kinase | -1.49 |
| 1125 | 254592_at | AT4G18880 | AT-HSFA4A (Arabidopsis thaliana heat shock transcription factor A4A); DNA binding / transcription factor | 2.06 |
| 1126 | 254609_at | AT4G18970 | GDSL-motif lipase/hydrolase family protein | -1.89 |
| 1127 | 254564_at | AT4G19170 | NCED4 (NINE-CIS-EPOXYCAROTENOID DIOXYGENASE 4) | 4.33 |
| 1128 | 254571_at | AT4G19370 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G31720.1); similar to unknown [Picea sitchensis] (GB:ABK21519.1); similar to Os02g0703300 [Oryza sativa (japonica cultivar-group)] (GB:NP_001047854.1); contains InterPro domain Protein of unknown function DUF1218 (InterPro:IPR009606) | 2.86 |
| 1129 | 254572_at | AT4G19380 | alcohol oxidase-related | -2.90 |
| 1130 | 254579_at | AT4G19400 | actin binding | -1.50 |
| 1131 | 254573_at | AT4G19420 | pectinacetylesterase family protein | 2.58 |
| 1132 | 254574_at | AT4G19430 | unknown protein | 3.21 |
| 1133 | 254553_at | AT4G19530 | disease resistance protein (TIR-NBS-LRR class), putative | 1.69 |
| 1134 | 254551_at | AT4G19840 | ATPP2-A1 (Arabidopsis thaliana phloem protein 2-A1) | 1.97 |
| 1135 | 254500_at | AT4G20110 | vacuolar sorting receptor, putative | 2.15 |
| 1136 | 254466_at | AT4G20430 | subtilase family protein | -1.61 |
| 1137 | 254448_at | AT4G20870 | fatty acid hydroxylase, putative | -1.70 |
| 1138 | 254457_at | AT4G21170 | pentatricopeptide (PPR) repeat-containing protein | -1.80 |
| 1139 | 254400_at | AT4G21270 | ATK1 (ARABIDOPSIS THALIANA KINESIN 1); microtubule motor | -2.64 |
| 1140 | 254413_at | AT4G21440 | ATM4/ATMYB102 (ARABIDOPSIS MYB-LIKE 102); DNA binding / transcription factor | 1.63 |
| 1141 | 254393_at | AT4G21580 | oxidoreductase, zinc-binding dehydrogenase family protein | 1.63 |
| 1142 | 254391_at | AT4G21590 | ENDO3 (ENDONUCLEASE 3); T/G mismatch-specific endonuclease/ endonuclease/ nucleic acid binding / single-stranded DNA specific endodeoxyribonuclease | -3.90 |
| 1143 | 254394_at | AT4G21630 | subtilase family protein | 2.02 |
| 1144 | 254379_at | AT4G21820 | calmodulin-binding family protein | -2.61 |
| 1145 | 254363_at | AT4G22010 | SKS4 (SKU5 Similar 4); copper ion binding / oxidoreductase | -2.29 |
| 1146 | 254328_at | AT4G22570 | APT3 (ADENINE PHOSPHORIBOSYL TRANSFERASE 3); adenine phosphoribosyltransferase | 1.61 |
| 1147 | 254299_at | AT4G22920 | ATNYE1/NYE1 (NON-YELLOWING 1) | 2.12 |
| 1148 | 254289_at | AT4G22980 | similar to catalytic/ pyridoxal phosphate binding [Arabidopsis thaliana] (TAIR:AT5G51920.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO63651.1); contains domain PTHR14237 (PTHR14237) | 1.68 |
| 1149 | 254266_at | AT4G23130 | CRK5 (CYSTEINE-RICH RLK5); kinase | 1.75 |
| 1150 | 254271_at | AT4G23150 | protein kinase family protein | 4.76 |
| 1151 | 254256_at | AT4G23180 | CRK10 (CYSTEINE-RICH RLK10); kinase | 1.98 |
| 1152 | 254255_at | AT4G23220 | protein kinase family protein | 3.05 |
| 1153 | 254247_at | AT4G23260 | protein kinase | 3.44 |
| 1154 | 254250_at | AT4G23290 | protein kinase family protein | 3.43 |
| 1155 | 254251_at | AT4G23300 | protein kinase family protein | 2.44 |
| 1156 | 254252_at | AT4G23310 | receptor-like protein kinase, putative | 2.72 |
| 1157 | 254253_at | AT4G23320 | protein kinase family protein | 3.06 |
| 1158 | 254232_at | AT4G23600 | CORI3 (CORONATINE INDUCED 1, JASMONIC ACID RESPONSIVE 2); transaminase | 1.60 |
| 1159 | 254229_at | AT4G23610 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G54200.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO39657.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN73280.1) | 2.50 |
| 1160 | 254234_at | AT4G23680 | major latex protein-related / MLP-related | -2.95 |
| 1161 | 254235_at | AT4G23750 | CRF2 (CYTOKININ RESPONSE FACTOR 2); DNA binding / transcription factor | -5.50 |
| 1162 | 254233_at | AT4G23800 | high mobility group (HMG1/2) family protein | -2.90 |
| 1163 | 254231_at | AT4G23810 | WRKY53 (WRKY DNA-binding protein 53); DNA binding / protein binding / transcription activator/ transcription factor | 2.48 |
| 1164 | 254178_at | AT4G23880 | unknown protein | 2.29 |
| 1165 | 254185_at | AT4G23990 | ATCSLG3 (Cellulose synthase-like G3); transferase/ transferase, transferring glycosyl groups | 1.95 |
| 1166 | 254189_at | AT4G24000 | ATCSLG2 (Cellulose synthase-like G2); transferase/ transferase, transferring glycosyl groups | 2.90 |
| 1167 | 254174_at | AT4G24120 | YSL1 (YELLOW STRIPE LIKE 1); oligopeptide transporter | 3.03 |
| 1168 | 254153_at | AT4G24450 | ATGWD2/GWD3/PWD (PHOSPHOGLUCAN, WATER DIKINASE); ATP binding / kinase | 2.79 |
| 1169 | 254155_at | AT4G24480 | serine/threonine protein kinase, putative | 1.68 |
| 1170 | 254101_at | AT4G25000 | AMY1/ATAMY1 (ALPHA-AMYLASE-LIKE); alpha-amylase | 3.27 |
| 1171 | 254109_at | AT4G25240 | SKS1 (SKU5 SIMILAR 1); copper ion binding | -1.47 |
| 1172 | 254080_at | AT4G25630 | FIB2 (FIBRILLARIN 2) | -1.65 |
| 1173 | 254044_at | AT4G25820 | XTR9 (XYLOGLUCAN ENDOTRANSGLYCOSYLASE 9); hydrolase, acting on glycosyl bonds | -2.18 |
| 1174 | 254041_at | AT4G25830 | integral membrane family protein | -1.55 |
| 1175 | 254027_at | AT4G25835 | AAA-type ATPase family protein | -2.05 |
| 1176 | 254030_at | AT4G25890 | 60S acidic ribosomal protein P3 (RPP3A) | -1.75 |
| 1177 | 254043_at | AT4G25990 | CIL | -3.15 |
| 1178 | 253992_at | AT4G26060 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G57060.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO65021.1); contains domain PTHR10052 (PTHR10052); contains domain PTHR10052:SF2 (PTHR10052:SF2) | 2.19 |
| 1179 | 254012_at | AT4G26230 | 60S ribosomal protein L31 (RPL31B) | -1.94 |
| 1180 | 253978_at | AT4G26660 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G55520.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G55520.2); similar to unnamed protein product [Vitis vinifera] (GB:CAO38858.1); contains InterPro domain Kinesin-related (InterPro:IPR010544) | -2.61 |
| 1181 | 253947_at | AT4G26760 | microtubule associated protein (MAP65/ASE1) family protein | -2.17 |
| 1182 | 253940_at | AT4G26950 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G04630.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO38947.1); similar to unknown [Picea sitchensis] (GB:ABK21247.1); contains InterPro domain Protein of unknown function DUF584 (InterPro:IPR007608) | 2.49 |
| 1183 | 253904_at | AT4G27140 | 2S seed storage protein 1 / 2S albumin storage protein / NWMU1-2S albumin 1 | -1.89 |
| 1184 | 253909_at | AT4G27270 | quinone reductase family protein | -1.43 |
| 1185 | 253911_at | AT4G27300 | S-locus protein kinase, putative | 2.79 |
| 1186 | 253875_at | AT4G27520 | plastocyanin-like domain-containing protein | -1.82 |
| 1187 | 253854_at | AT4G27900 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G53420.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO48522.1); contains InterPro domain CCT (InterPro:IPR010402) | 1.59 |
| 1188 | 253855_at | AT4G28050 | TET7 (TETRASPANIN7) | -1.87 |
| 1189 | 253827_at | AT4G28085 | unknown protein | 1.87 |
| 1190 | 253851_at | AT4G28110 | AtMYB41 (myb domain protein 41); DNA binding / transcription factor | -3.09 |
| 1191 | 253811_at | AT4G28190 | ULT1 (ULTRAPETALA1); DNA binding / binding | -1.91 |
| 1192 | 253815_at | AT4G28250 | ATEXPB3 (ARABIDOPSIS THALIANA EXPANSIN B3) | -2.03 |
| 1193 | 253806_at | AT4G28270 | zinc finger (C3HC4-type RING finger) family protein | 2.22 |
| 1194 | 253817_at | AT4G28310 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G52270.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO69747.1) | -2.07 |
| 1195 | 253819_at | AT4G28350 | lectin protein kinase family protein | 2.48 |
| 1196 | 253776_at | AT4G28390 | AAC3 (ADP/ATP CARRIER 3); ATP:ADP antiporter/ binding | 2.01 |
| 1197 | 253780_at | AT4G28400 | protein phosphatase 2C, putative / PP2C, putative | 1.72 |
| 1198 | 253779_at | AT4G28490 | HAESA (RECEPTOR-LIKE PROTEIN KINASE 5); ATP binding / kinase/ protein serine/threonine kinase | 2.64 |
| 1199 | 253769_at | AT4G28560 | RIC7 (ROP-INTERACTIVE CRIB MOTIF-CONTAINING PROTEIN 7); protein binding | -2.26 |
| 1200 | 253788_at | AT4G28680 | tyrosine decarboxylase, putative | -1.49 |
| 1201 | 253754_at | AT4G29020 | glycine-rich protein | -3.07 |
| 1202 | 253753_at | AT4G29030 | glycine-rich protein | -3.28 |
| 1203 | 253747_at | AT4G29050 | lectin protein kinase family protein | 2.77 |
| 1204 | 253732_at | AT4G29140 | MATE efflux protein-related | -3.23 |
| 1205 | 253710_at | AT4G29230 | ANAC075 (Arabidopsis NAC domain containing protein 75); transcription factor | -1.66 |
| 1206 | 253723_at | AT4G29240 | leucine-rich repeat family protein / extensin family protein | -1.53 |
| 1207 | 253729_at | AT4G29360 | glycosyl hydrolase family 17 protein | -2.24 |
| 1208 | 253728_at | AT4G29410 | 60S ribosomal protein L28 (RPL28C) | -1.51 |
| 1209 | 253697_at | AT4G29700 | type I phosphodiesterase/nucleotide pyrophosphatase family protein | -3.01 |
| 1210 | 253685_at | AT4G29710 | phosphodiesterase/nucleotide pyrophosphatase-related | -2.12 |
| 1211 | 253702_at | AT4G29900 | ACA10 (autoinhibited Ca2+ -ATPase 10); calcium-transporting ATPase/ calmodulin binding | 1.62 |
| 1212 | 253660_at | AT4G30140 | GDSL-motif lipase/hydrolase family protein | -4.21 |
| 1213 | 253607_at | AT4G30330 | small nuclear ribonucleoprotein E, putative / snRNP-E, putative / Sm protein E, putative | -2.27 |
| 1214 | 253579_at | AT4G30610 | BRS1 (BRI1 SUPPRESSOR 1) | -1.78 |
| 1215 | 253583_at | AT4G30680 | MA3 domain-containing protein | -3.75 |
| 1216 | 253587_at | AT4G30770 | similar to Os11g0195700 [Oryza sativa (japonica cultivar-group)] (GB:NP_001067426.1); similar to hypothetical protein OsJ_032844 [Oryza sativa (japonica cultivar-group)] (GB:EAZ18635.1); contains domain PROKAR_LIPOPROTEIN (PS51257) | -2.10 |
| 1217 | 253598_at | AT4G30800 | 40S ribosomal protein S11 (RPS11B) | -2.28 |
| 1218 | 253549_at | AT4G30930 | NFD1 (NUCLEAR FUSION DEFECTIVE 1); structural constituent of ribosome | -1.51 |
| 1219 | 253522_at | AT4G31290 | ChaC-like family protein | -2.89 |
| 1220 | 253487_at | AT4G31700 | RPS6 (RIBOSOMAL PROTEIN S6); structural constituent of ribosome | -1.97 |
| 1221 | 253480_at | AT4G31840 | plastocyanin-like domain-containing protein | -3.05 |
| 1222 | 253482_at | AT4G31985 | 60S ribosomal protein L39 (RPL39C) | -1.71 |
| 1223 | 253421_at | AT4G32340 | binding | 1.93 |
| 1224 | 253403_at | AT4G32830 | ATAUR1 (ATAURORA1); histone serine kinase(H3-S10 specific) / kinase/ protein serine/threonine kinase | -1.97 |
| 1225 | 253358_at | AT4G32940 | GAMMA-VPE (Vacuolar processing enzyme gamma); cysteine-type endopeptidase | 3.34 |
| 1226 | 253382_at | AT4G33040 | glutaredoxin family protein | 2.77 |
| 1227 | 253414_at | AT4G33050 | EDA39 (embryo sac development arrest 39); calmodulin binding | 2.95 |
| 1228 | 253309_at | AT4G33790 | acyl CoA reductase, putative | 1.95 |
| 1229 | 253270_at | AT4G34160 | CYCD3/CYCD3;1/D3 (CYCLIN D3;1); cyclin-dependent protein kinase regulator/ protein binding | -2.02 |
| 1230 | 253274_at | AT4G34200 | EDA9 (embryo sac development arrest 9); NAD binding / amino acid binding / binding / catalytic/ cofactor binding / oxidoreductase, acting on the CH-OH group of donors, NAD or NADP as acceptor / phosphoglycerate dehydrogenase | -1.69 |
| 1231 | 253285_at | AT4G34250 | fatty acid elongase, putative | -2.19 |
| 1232 | 253286_at | AT4G34260 | catalytic | -1.58 |
| 1233 | 253174_at | AT4G35090 | CAT2 (CATALASE 2); catalase | 1.58 |
| 1234 | 253148_at | AT4G35620 | CYCB2;2 (CYCLIN B2;2); cyclin-dependent protein kinase regulator | -1.85 |
| 1235 | 253156_at | AT4G35730 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G34220.2); similar to unnamed protein product [Vitis vinifera] (GB:CAO23719.1); contains InterPro domain Protein of unknown function DUF292, eukaryotic (InterPro:IPR005061) | -2.31 |
| 1236 | 253163_at | AT4G35750 | Rho-GTPase-activating protein-related | 1.74 |
| 1237 | 253161_at | AT4G35770 | SEN1 (DARK INDUCIBLE 1) | 2.27 |
| 1238 | 253078_at | AT4G36180 | leucine-rich repeat family protein | -1.80 |
| 1239 | 253090_at | AT4G36360 | BGAL3 (beta-galactosidase 3); beta-galactosidase | -2.00 |
| 1240 | 246200_at | AT4G37240 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G23690.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO45438.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN61825.1) | -2.12 |
| 1241 | 253100_at | AT4G37400 | CYP81F3 (cytochrome P450, family 81, subfamily F, polypeptide 3); oxygen binding | 1.70 |
| 1242 | 253073_at | AT4G37410 | CYP81F4 (cytochrome P450, family 81, subfamily F, polypeptide 4); oxygen binding | -3.09 |
| 1243 | 253050_at | AT4G37450 | AGP18 (Arabinogalactan protein 18) | -2.87 |
| 1244 | 253051_at | AT4G37490 | CYC1 (CYCLIN 1); cyclin-dependent protein kinase regulator | -3.61 |
| 1245 | 253043_at | AT4G37540 | LBD39 (LOB DOMAIN-CONTAINING PROTEIN 39) | -3.01 |
| 1246 | 253048_at | AT4G37560 | formamidase, putative / formamide amidohydrolase, putative | 3.02 |
| 1247 | 253010_at | AT4G37750 | ANT (AINTEGUMENTA); DNA binding / transcription factor | -1.74 |
| 1248 | 253040_at | AT4G37800 | xyloglucan:xyloglucosyl transferase, putative / xyloglucan endotransglycosylase, putative / endo-xyloglucan transferase, putative | -1.41 |
| 1249 | 252984_at | AT4G37990 | ELI3-2 (ELICITOR-ACTIVATED GENE 3) | 1.83 |
| 1250 | 252997_at | AT4G38400 | ATEXLA2 (ARABIDOPSIS THALIANA EXPANSIN-LIKE A2) | -1.98 |
| 1251 | 252993_at | AT4G38540 | monooxygenase, putative (MO2) | 3.27 |
| 1252 | 252976_s_at | AT4G38550 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G20950.1); contains InterPro domain Phospholipase-like, arabidopsis (InterPro:IPR007942) | 2.31 |
| 1253 | 252977_at | AT4G38560 | similar to pEARLI4 [Arabidopsis thaliana] (TAIR:AT2G20960.1); contains InterPro domain Phospholipase-like, arabidopsis (InterPro:IPR007942) | 2.22 |
| 1254 | 252954_at | AT4G38660 | thaumatin, putative | -3.30 |
| 1255 | 252965_at | AT4G38860 | auxin-responsive protein, putative | 1.74 |
| 1256 | 252937_at | AT4G39180 | SEC14 (secretion 14) | 1.90 |
| 1257 | 252912_at | AT4G39200 | 40S ribosomal protein S25 (RPS25E) | -1.88 |
| 1258 | 252852_at | AT4G39900 | similar to unnamed protein product [Vitis vinifera] (GB:CAO67041.1) | -1.79 |
| 1259 | 252831_at | AT4G39980 | DHS1 (3-DEOXY-D-ARABINO-HEPTULOSONATE 7-PHOSPHATE SYNTHASE 1); 3-deoxy-7-phosphoheptulonate synthase | -1.41 |
| 1260 | 251142_at | AT5G01015 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G65295.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN82885.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO45347.1) | -1.88 |
| 1261 | 251141_at | AT5G01075 | beta-galactosidase | -1.78 |
| 1262 | 251113_at | AT5G01370 | unknown protein | -1.68 |
| 1263 | 251054_at | AT5G01540 | lectin protein kinase, putative | 2.30 |
| 1264 | 251109_at | AT5G01600 | ATFER1 (FERRETIN 1); ferric iron binding | 1.83 |
| 1265 | 251062_at | AT5G01840 | ATOFP1/OFP1 (ARABIDOPSIS THALIANA OVATE FAMILY PROTEIN 1); protein binding / transcription repressor | -1.80 |
| 1266 | 251065_at | AT5G01870 | lipid transfer protein, putative | -3.31 |
| 1267 | 251066_at | AT5G01880 | zinc finger (C3HC4-type RING finger) family protein | 1.85 |
| 1268 | 251075_at | AT5G01890 | leucine-rich repeat transmembrane protein kinase, putative | -1.56 |
| 1269 | 251039_at | AT5G02020 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G55646.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G59080.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO15731.1) | 3.06 |
| 1270 | 251021_at | AT5G02140 | thaumatin-like protein, putative | -2.97 |
| 1271 | 251035_at | AT5G02220 | similar to unknown [Picea sitchensis] (GB:ABK23883.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN70860.1) | -2.82 |
| 1272 | 250990_at | AT5G02290 | NAK; kinase | 1.84 |
| 1273 | 251018_at | AT5G02450 | 60S ribosomal protein L36 (RPL36C) | -1.92 |
| 1274 | 251017_at | AT5G02760 | protein phosphatase 2C family protein / PP2C family protein | 2.81 |
| 1275 | 250974_at | AT5G02820 | RHL2 (ROOT HAIRLESS 2); ATP binding / DNA binding / DNA topoisomerase (ATP-hydrolyzing) | -1.48 |
| 1276 | 250920_at | AT5G03390 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G68960.1); contains InterPro domain Protein of unknown function DUF295 (InterPro:IPR005174) | -3.58 |
| 1277 | 250948_at | AT5G03490 | UDP-glucoronosyl/UDP-glucosyl transferase family protein | 2.20 |
| 1278 | 250907_at | AT5G03670 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G36420.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO15955.1) | -1.59 |
| 1279 | 250892_at | AT5G03760 | ATCSLA09 (RESISTANT TO AGROBACTERIUM TRANSFORMATION 4); transferase, transferring glycosyl groups | -1.53 |
| 1280 | 250895_at | AT5G03850 | 40S ribosomal protein S28 (RPS28B) | -1.62 |
| 1281 | 250864_at | AT5G03870 | glutaredoxin family protein | -2.94 |
| 1282 | 245698_at | AT5G04160 | phosphate translocator-related | -1.70 |
| 1283 | 250891_at | AT5G04530 | beta-ketoacyl-CoA synthase family protein | -3.41 |
| 1284 | 250818_at | AT5G04930 | ALA1 (AMINOPHOSPHOLIPID ATPASE1); ATPase, coupled to transmembrane movement of ions, phosphorylative mechanism | 1.75 |
| 1285 | 250832_at | AT5G04950 | nicotianamine synthase, putative | -1.74 |
| 1286 | 250828_at | AT5G05250 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G56360.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO41488.1) | -3.22 |
| 1287 | 250794_at | AT5G05270 | chalcone-flavanone isomerase family protein | -5.57 |
| 1288 | 250734_at | AT5G06270 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G11600.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO15841.1) | -1.44 |
| 1289 | 250688_at | AT5G06510 | CCAAT-binding transcription factor (CBF-B/NF-YA) family protein | 2.13 |
| 1290 | 250690_at | AT5G06530 | ABC transporter family protein | 3.05 |
| 1291 | 250678_at | AT5G06540 | pentatricopeptide (PPR) repeat-containing protein | -2.29 |
| 1292 | 250680_at | AT5G06570 | hydrolase | 2.31 |
| 1293 | 250648_at | AT5G06760 | late embryogenesis abundant group 1 domain-containing protein / LEA group 1 domain-containing protein | 2.39 |
| 1294 | 250699_at | AT5G06820 | SRF2 (STRUBBELIG-RECEPTOR FAMILY 2); ATP binding / kinase/ protein serine/threonine kinase | 1.82 |
| 1295 | 250661_at | AT5G07030 | pepsin A | -4.34 |
| 1296 | 250667_at | AT5G07090 | 40S ribosomal protein S4 (RPS4B) | -2.01 |
| 1297 | 250666_at | AT5G07100 | WRKY26 (WRKY DNA-binding protein 26); transcription factor | 3.04 |
| 1298 | 250580_at | AT5G07440 | GDH2 (GLUTAMATE DEHYDROGENASE 2); oxidoreductase | -1.72 |
| 1299 | 250588_at | AT5G07660 | structural maintenance of chromosomes (SMC) family protein | -3.30 |
| 1300 | 250594_at | AT5G07760 | formin homology 2 domain-containing protein / FH2 domain-containing protein | 1.82 |
| 1301 | 250603_at | AT5G07820 | similar to chromosome scaffold protein-related [Arabidopsis thaliana] (TAIR:AT5G61260.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO15046.1); contains InterPro domain Calmodulin-binding, plant (InterPro:IPR012417) | 1.95 |
| 1302 | 250558_at | AT5G07990 | TT7 (TRANSPARENT TESTA 7); flavonoid 3'-monooxygenase/ oxygen binding | -1.78 |
| 1303 | 250565_at | AT5G08000 | E13L3 (GLUCAN ENDO-1,3-BETA-GLUCOSIDASE-LIKE PROTEIN 3) | -2.19 |
| 1304 | 250560_at | AT5G08020 | replication protein, putative | -1.81 |
| 1305 | 250562_at | AT5G08040 | TOM5 (MITOCHONDRIAL IMPORT RECEPTOR SUBUNIT TOM5 HOMOLOG) | -1.75 |
| 1306 | 250546_at | AT5G08180 | ribosomal protein L7Ae/L30e/S12e/Gadd45 family protein | -1.44 |
| 1307 | 250535_at | AT5G08480 | VQ motif-containing protein | -1.83 |
| 1308 | 250533_at | AT5G08640 | FLS (FLAVONOL SYNTHASE) | -3.77 |
| 1309 | 245891_at | AT5G09220 | AAP2 (AMINO ACID PERMEASE 2); amino acid transmembrane transporter | 2.08 |
| 1310 | 245885_at | AT5G09440 | phosphate-responsive protein, putative | 1.58 |
| 1311 | 250500_at | AT5G09530 | hydroxyproline-rich glycoprotein family protein | -3.58 |
| 1312 | 250498_at | AT5G09660 | PMDH2 (PEROXISOMAL NAD-MALATE DEHYDROGENASE 2); malate dehydrogenase | 1.59 |
| 1313 | 250470_at | AT5G10160 | beta-hydroxyacyl-ACP dehydratase, putative | -2.49 |
| 1314 | 250435_at | AT5G10380 | zinc finger (C3HC4-type RING finger) family protein | 3.05 |
| 1315 | 250434_at | AT5G10390 | histone H3 | -2.30 |
| 1316 | 250433_at | AT5G10400 | histone H3 | -3.84 |
| 1317 | 250443_at | AT5G10520 | RBK1 (ROP BINDING PROTEIN KINASES 1); kinase | 1.62 |
| 1318 | 250441_at | AT5G10540 | peptidase M3 family protein / thimet oligopeptidase family protein | -1.62 |
| 1319 | 246012_at | AT5G10650 | zinc finger (C3HC4-type RING finger) family protein | 1.59 |
| 1320 | 250445_at | AT5G10760 | aspartyl protease family protein | 5.51 |
| 1321 | 250446_at | AT5G10770 | chloroplast nucleoid DNA-binding protein, putative | 2.12 |
| 1322 | 250403_at | AT5G10920 | argininosuccinate lyase, putative / arginosuccinase, putative | -1.70 |
| 1323 | 250408_at | AT5G10930 | CIPK5 (CBL-INTERACTING PROTEIN KINASE 5); kinase | 2.55 |
| 1324 | 245901_at | AT5G11060 | KNAT4 (KNOTTED1-LIKE HOMEOBOX GENE 4); transcription factor | 2.37 |
| 1325 | 250413_at | AT5G11160 | APT5 (ADENINE PHOSPHORIBOSYLTRANSFERASE 5); adenine phosphoribosyltransferase | -1.56 |
| 1326 | 250386_at | AT5G11510 | MYB3R-4 (C-MYB-LIKE TRANSCRIPTION FACTOR 3R-4, myb domain protein 3R-4); DNA binding / transcription coactivator/ transcription factor | -2.63 |
| 1327 | 250381_at | AT5G11610 | exostosin family protein | 2.00 |
| 1328 | 250340_at | AT5G11840 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G67370.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO43828.1); contains InterPro domain Protein of unknown function DUF1230 (InterPro:IPR009631) | 1.70 |
| 1329 | 250302_at | AT5G11920 | ATCWINV6 (6-&1-FRUCTAN EXOHYDROLASE); hydrolase, hydrolyzing O-glycosyl compounds / inulinase/ levanase | 1.78 |
| 1330 | 250344_at | AT5G11930 | glutaredoxin family protein | 2.06 |
| 1331 | 250304_at | AT5G12110 | elongation factor 1B alpha-subunit 1 (eEF1Balpha1) | -3.92 |
| 1332 | 250264_at | AT5G12890 | UDP-glucoronosyl/UDP-glucosyl transferase family protein | 1.57 |
| 1333 | 245976_at | AT5G13080 | WRKY75 (WRKY DNA-BINDING PROTEIN 75); transcription factor | 1.91 |
| 1334 | 245980_at | AT5G13140 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G26960.1); similar to unknown [Euphorbia pulcherrima] (GB:ABO43960.1); contains InterPro domain Pollen Ole e 1 allergen and extensin (InterPro:IPR006041) | -2.87 |
| 1335 | 245982_at | AT5G13170 | nodulin MtN3 family protein | -2.83 |
| 1336 | 245987_at | AT5G13180 | ANAC083 (Arabidopsis NAC domain containing protein 83); transcription factor | 1.62 |
| 1337 | 250292_at | AT5G13220 | JAS1/JAZ10/TIFY9 (JASMONATE-ZIM-DOMAIN PROTEIN 10) | -1.64 |
| 1338 | 250286_at | AT5G13320 | PBS3 (AVRPPHB SUSCEPTIBLE 3) | 2.62 |
| 1339 | 250233_at | AT5G13460 | IQD11 (IQ-domain 11); calmodulin binding | -1.51 |
| 1340 | 245849_at | AT5G13520 | peptidase M1 family protein | -2.58 |
| 1341 | 250247_at | AT5G13720 | structural constituent of ribosome | 1.79 |
| 1342 | 250259_at | AT5G13800 | hydrolase, alpha/beta fold family protein | 2.46 |
| 1343 | 250228_at | AT5G13840 | WD-40 repeat family protein | -2.10 |
| 1344 | 250207_at | AT5G13930 | ATCHS/CHS/TT4 (CHALCONE SYNTHASE); naringenin-chalcone synthase | -2.02 |
| 1345 | 250208_at | AT5G14000 | ANAC084 (Arabidopsis NAC domain containing protein 84); transcription factor | -2.07 |
| 1346 | 246595_at | AT5G14780 | FDH (FORMATE DEHYDROGENASE); NAD binding / binding / catalytic/ cofactor binding / oxidoreductase, acting on the CH-OH group of donors, NAD or NADP as acceptor | 2.04 |
| 1347 | 246550_at | AT5G14920 | gibberellin-regulated family protein | -2.13 |
| 1348 | 246600_at | AT5G14930 | SAG101 (SENESCENCE-ASSOCIATED GENE 101); triacylglycerol lipase | 1.69 |
| 1349 | 246566_at | AT5G14940 | proton-dependent oligopeptide transport (POT) family protein | 1.83 |
| 1350 | 250152_at | AT5G15120 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G39890.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO14912.1); contains InterPro domain Protein of unknown function DUF1637 (InterPro:IPR012864) | -1.41 |
| 1351 | 250109_at | AT5G15230 | GASA4 (GAST1 PROTEIN HOMOLOG 4) | -2.74 |
| 1352 | 250110_at | AT5G15350 | plastocyanin-like domain-containing protein | -2.90 |
| 1353 | 246538_at | AT5G15520 | 40S ribosomal protein S19 (RPS19B) | -1.53 |
| 1354 | 246565_at | AT5G15530 | BCCP2 (biotin carboxyl carrier protein 2); biotin binding | -3.06 |
| 1355 | 246540_at | AT5G15600 | SP1L4 (SPIRAL1-LIKE4) | -1.83 |
| 1356 | 246516_at | AT5G15740 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G02250.1); similar to hypothetical protein [Cleome spinosa] (GB:ABD96934.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO49221.1); similar to hypothetical protein [Cleome spinosa] (GB:ABD96844.1); contains InterPro domain Protein of unknown function DUF246, plant (InterPro:IPR004348) | -2.22 |
| 1357 | 246531_at | AT5G15800 | SEP1 (SEPALLATA1); DNA binding / transcription factor | 3.42 |
| 1358 | 246523_at | AT5G15850 | COL1 (CONSTANS-LIKE 1); transcription factor/ zinc ion binding | 2.66 |
| 1359 | 246503_at | AT5G16130 | 40S ribosomal protein S7 (RPS7C) | -1.83 |
| 1360 | 246505_at | AT5G16250 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G02640.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO50168.1) | -3.02 |
| 1361 | 250100_at | AT5G16570 | GLN1;4 (Glutamine synthetase 1;4); glutamate-ammonia ligase | 5.49 |
| 1362 | 246415_at | AT5G17160 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G03130.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO62214.1) | -1.51 |
| 1363 | 250099_at | AT5G17300 | myb family transcription factor | 2.18 |
| 1364 | 246429_at | AT5G17450 | heavy-metal-associated domain-containing protein / copper chaperone (CCH)-related | 2.16 |
| 1365 | 246432_at | AT5G17490 | RGL3 (RGA-LIKE 3); transcription factor | -2.48 |
| 1366 | 250054_at | AT5G17860 | CAX7 (CALCIUM EXCHANGER 7); calcium:sodium antiporter/ cation:cation antiporter | 2.87 |
| 1367 | 250028_at | AT5G18130 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G03870.2); similar to unknown [Medicago truncatula] (GB:ABK28852.1) | 3.13 |
| 1368 | 249983_at | AT5G18470 | curculin-like (mannose-binding) lectin family protein | 1.91 |
| 1369 | 249987_at | AT5G18490 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G04350.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN77840.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN61465.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO40557.1); contains InterPro domain Protein of unknown function DUF946, plant (InterPro:IPR009291) | 1.95 |
| 1370 | 249996_at | AT5G18600 | glutaredoxin family protein | 2.00 |
| 1371 | 250006_at | AT5G18660 | DVR (PALE-GREEN AND CHLOROPHYLL B REDUCED 2); 3,8-divinyl protochlorophyllide a 8-vinyl reductase | -2.68 |
| 1372 | 249969_at | AT5G19090 | heavy-metal-associated domain-containing protein | -1.47 |
| 1373 | 249923_at | AT5G19120 | aspartic-type endopeptidase/ pepsin A | -2.60 |
| 1374 | 249922_at | AT5G19140 | auxin/aluminum-responsive protein, putative | 1.74 |
| 1375 | 246063_at | AT5G19340 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G05980.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN75979.1) | -2.67 |
| 1376 | 245965_at | AT5G19730 | pectinesterase family protein | -1.93 |
| 1377 | 246125_at | AT5G19875 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G31940.1); similar to hypothetical protein MtrDRAFT_AC155880g29v2 [Medicago truncatula] (GB:ABN08129.1) | 1.61 |
| 1378 | 246144_at | AT5G20110 | dynein light chain, putative | 1.62 |
| 1379 | 246098_at | AT5G20400 | oxidoreductase, 2OG-Fe(II) oxygenase family protein | 1.61 |
| 1380 | 246004_at | AT5G20630 | GLP3 (GERMIN-LIKE PROTEIN 3); manganese ion binding / metal ion binding / nutrient reservoir | -3.08 |
| 1381 | 246002_at | AT5G20740 | invertase/pectin methylesterase inhibitor family protein | -2.88 |
| 1382 | 249928_at | AT5G22250 | CCR4-NOT transcription complex protein, putative | 1.80 |
| 1383 | 249942_at | AT5G22300 | NIT4 (NITRILASE 4) | 3.67 |
| 1384 | 249940_at | AT5G22380 | ANAC090 (Arabidopsis NAC domain containing protein 90); transcription factor | 3.06 |
| 1385 | 249932_at | AT5G22390 | similar to unnamed protein product [Vitis vinifera] (GB:CAO48728.1) | 2.31 |
| 1386 | 249945_at | AT5G22440 | 60S ribosomal protein L10A (RPL10aC) | -1.81 |
| 1387 | 249895_at | AT5G22500 | acyl CoA reductase, putative / male-sterility protein, putative | -1.55 |
| 1388 | 249911_at | AT5G22740 | ATCSLA02 (Cellulose synthase-like A2); transferase, transferring glycosyl groups | -1.51 |
| 1389 | 249916_at | AT5G22880 | H2B/HTB2 (HISTONE H2B); DNA binding | -2.05 |
| 1390 | 249885_at | AT5G22940 | exostosin family protein | -1.86 |
| 1391 | 249848_at | AT5G23220 | NIC3 (NICOTINAMIDASE 3); catalytic/ nicotinamidase | -1.96 |
| 1392 | 249850_at | AT5G23240 | DNAJ heat shock N-terminal domain-containing protein | 2.86 |
| 1393 | 249832_at | AT5G23400 | disease resistance family protein / LRR family protein | -5.33 |
| 1394 | 249836_at | AT5G23420 | HMGB6 (High mobility group B 6); transcription factor | -2.05 |
| 1395 | 249794_at | AT5G23530 | ATCXE18 (ARABIDOPSIS THALIANA CARBOXYESTERASE 18); carboxylesterase | -2.84 |
| 1396 | 249800_at | AT5G23660 | MTN3 (ARABIDOPSIS HOMOLOG OF MEDICAGO TRUNCATULA MTN3) | 2.57 |
| 1397 | 249795_at | AT5G23740 | RPS11-BETA (RIBOSOMAL PROTEIN S11-BETA); structural constituent of ribosome | -1.84 |
| 1398 | 249817_at | AT5G23820 | MD-2-related lipid recognition domain-containing protein / ML domain-containing protein | -2.85 |
| 1399 | 249815_at | AT5G23900 | 60S ribosomal protein L13 (RPL13D) | -1.90 |
| 1400 | 249813_at | AT5G23940 | EMB3009 (EMBRYO DEFECTIVE 3009); transferase | -3.01 |
| 1401 | 249769_at | AT5G24120 | SIGE (RNA polymerase sigma subunit E); DNA binding / DNA-directed RNA polymerase/ sigma factor/ transcription factor | 1.68 |
| 1402 | 249774_at | AT5G24150 | SQP1 (Squalene monooxygenase 1) | 1.81 |
| 1403 | 249777_at | AT5G24210 | lipase class 3 family protein | 4.91 |
| 1404 | 249780_at | AT5G24240 | phosphatidylinositol 3- and 4-kinase family protein / ubiquitin family protein | 1.86 |
| 1405 | 249788_at | AT5G24330 | ATXR6 (Arabidopsis thaliana Trithorax- related protein 6); DNA binding | -1.83 |
| 1406 | 249732_at | AT5G24420 | glucosamine/galactosamine-6-phosphate isomerase-related | -1.98 |
| 1407 | 249754_at | AT5G24530 | oxidoreductase, 2OG-Fe(II) oxygenase family protein | 2.54 |
| 1408 | 249755_at | AT5G24580 | copper-binding family protein | -2.68 |
| 1409 | 245929_at | AT5G24760 | alcohol dehydrogenase, putative | -1.76 |
| 1410 | 246968_at | AT5G24870 | zinc finger (C3HC4-type RING finger) family protein | 1.68 |
| 1411 | 246920_at | AT5G25090 | plastocyanin-like domain-containing protein | -1.83 |
| 1412 | 246948_at | AT5G25130 | CYP71B12 (cytochrome P450, family 71, subfamily B, polypeptide 12); oxygen binding | 2.45 |
| 1413 | 246949_at | AT5G25140 | CYP71B13 (cytochrome P450, family 71, subfamily B, polypeptide 13); oxygen binding | 3.44 |
| 1414 | 246930_at | AT5G25220 | KNAT3 (KNOTTED1-LIKE HOMEOBOX GENE 3); transcription factor | 1.71 |
| 1415 | 246943_at | AT5G25440 | protein kinase family protein | 1.93 |
| 1416 | 246899_at | AT5G25590 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G52320.3); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G52320.4); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G52320.2); similar to unnamed protein product [Vitis vinifera] (GB:CAO44106.1); contains InterPro domain Actin-binding FH2 (InterPro:IPR015425); contains InterPro domain Protein of unknown function DUF632 (InterPro:IPR006867); contains InterPro domain Protein of unknown function DUF630 (InterPro:IPR006868) | -2.81 |
| 1417 | 246909_at | AT5G25770 | similar to unnamed protein product [Vitis vinifera] (GB:CAO44054.1); contains domain PTHR10992 (PTHR10992); contains domain SSF53474 (SSF53474); contains domain G3DSA:3.40.50.1820 (G3DSA:3.40.50.1820) | 1.60 |
| 1418 | 246912_at | AT5G25820 | exostosin family protein | 2.59 |
| 1419 | 246860_at | AT5G25840 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G79770.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO45837.1); contains InterPro domain Protein of unknown function DUF1677, plant (InterPro:IPR012876) | 1.57 |
| 1420 | 246888_at | AT5G26270 | unknown protein | -2.60 |
| 1421 | 246855_at | AT5G26280 | meprin and TRAF homology domain-containing protein / MATH domain-containing protein | -3.19 |
| 1422 | 246821_at | AT5G26920 | calmodulin binding | 1.99 |
| 1423 | 246778_at | AT5G27450 | MK/MVK; mevalonate kinase | -2.04 |
| 1424 | 246772_at | AT5G27490 | integral membrane Yip1 family protein | -1.41 |
| 1425 | 246758_at | AT5G27850 | 60S ribosomal protein L18 (RPL18C) | -1.57 |
| 1426 | 246701_at | AT5G28020 | ATCYSD2 (Arabidopsis thaliana cysteine synthase D2); cysteine synthase | 2.14 |
| 1427 | 246700_at | AT5G28030 | cysteine synthase, putative / O-acetylserine (thiol)-lyase, putative / O-acetylserine sulfhydrylase, putative | 3.10 |
| 1428 | 245859_at | AT5G28290 | ATNEK3; kinase | -1.46 |
| 1429 | 246103_at | AT5G28640 | AN3 (ANGUSITFOLIA3) | -2.20 |
| 1430 | 246682_at | AT5G33290 | XGD1 (XYLOGALACTURONAN DEFICIENT 1); catalytic | 2.36 |
| 1431 | 246683_at | AT5G33300 | chromosome-associated kinesin-related | -1.71 |
| 1432 | 246687_at | AT5G33370 | GDSL-motif lipase/hydrolase family protein | -2.12 |
| 1433 | 246613_at | AT5G35360 | CAC2 (acetyl co-enzyme A carboxylase biotin carboxylase subunit) | -1.55 |
| 1434 | 249726_at | AT5G35480 | unknown protein | 3.10 |
| 1435 | 249727_at | AT5G35490 | unknown protein | 2.47 |
| 1436 | 249718_at | AT5G35740 | glycosyl hydrolase family protein 17 | -1.44 |
| 1437 | 246620_at | AT5G36220 | CYP81D1 (CYTOCHROME P450 91A1); oxygen binding | 2.65 |
| 1438 | 249645_at | AT5G36910 | THI2.2 (THIONIN 2.2); toxin receptor binding | -2.42 |
| 1439 | 249639_at | AT5G36930 | disease resistance protein (TIR-NBS-LRR class), putative | 1.66 |
| 1440 | 249644_at | AT5G37010 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G65710.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN76723.1) | -1.47 |
| 1441 | 249614_at | AT5G37300 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G38995.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO48523.1); contains InterPro domain Protein of unknown function UPF0089 (InterPro:IPR004255); contains InterPro domain Protein of unknown function DUF1298 (InterPro:IPR009721) | 2.12 |
| 1442 | 249581_at | AT5G37600 | ATGSR1 (Arabidopsis thaliana glutamine synthase clone R1); glutamate-ammonia ligase | 1.77 |
| 1443 | 249550_at | AT5G38210 | serine/threonine protein kinase family protein | 2.00 |
| 1444 | 249527_at | AT5G38710 | proline oxidase, putative / osmotic stress-responsive proline dehydrogenase, putative | 2.32 |
| 1445 | 249483_at | AT5G38895 | zinc finger (C3HC4-type RING finger) family protein | 1.59 |
| 1446 | 249485_at | AT5G39020 | protein kinase family protein | 2.09 |
| 1447 | 249486_at | AT5G39030 | protein kinase family protein | 2.69 |
| 1448 | 249493_at | AT5G39080 | transferase family protein | 1.87 |
| 1449 | 249489_at | AT5G39090 | transferase family protein | 2.98 |
| 1450 | 249469_at | AT5G39320 | UDP-glucose 6-dehydrogenase, putative | -3.36 |
| 1451 | 249448_at | AT5G39420 | CDC2CAT (ARABIDOPSIS THALIANA CDC2C); kinase | -2.21 |
| 1452 | 249454_at | AT5G39520 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G39530.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO15021.1) | 4.40 |
| 1453 | 249467_at | AT5G39610 | ANAC092/ATNAC2/ATNAC6 (Arabidopsis NAC domain containing protein 92); protein heterodimerization/ protein homodimerization/ transcription factor | 3.90 |
| 1454 | 249415_at | AT5G39660 | CDF2 (CYCLING DOF FACTOR 2); DNA binding / protein binding / transcription factor | 1.63 |
| 1455 | 249417_at | AT5G39670 | calcium-binding EF hand family protein | 2.16 |
| 1456 | 249425_at | AT5G39790 | 5'-AMP-activated protein kinase beta-1 subunit-related | -1.58 |
| 1457 | 249378_at | AT5G40450 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G28770.1); similar to hypothetical protein Kpol_1058p4 [Vanderwaltozyma polyspora DSM 70294] (GB:XP_001645325.1) | 1.98 |
| 1458 | 249366_at | AT5G40610 | glycerol-3-phosphate dehydrogenase (NAD+) / GPDH | -2.13 |
| 1459 | 249377_at | AT5G40690 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G41730.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO14635.1) | 1.81 |
| 1460 | 249346_at | AT5G40780 | LHT1 (LYSINE HISTIDINE TRANSPORTER 1); amino acid transmembrane transporter | 2.60 |
| 1461 | 249290_at | AT5G41060 | zinc finger (DHHC type) family protein | -1.41 |
| 1462 | 249337_at | AT5G41080 | glycerophosphoryl diester phosphodiesterase family protein | -3.10 |
| 1463 | 249340_at | AT5G41140 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G63300.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO62788.1); contains domain PTHR23160 (PTHR23160) | -1.57 |
| 1464 | 249306_at | AT5G41400 | zinc finger (C3HC4-type RING finger) family protein | 2.15 |
| 1465 | 249309_at | AT5G41410 | BEL1 (BELL 1); DNA binding / transcription factor | 1.86 |
| 1466 | 249266_at | AT5G41670 | 6-phosphogluconate dehydrogenase family protein | -1.74 |
| 1467 | 249276_at | AT5G41880 | POLA3/POLA4; DNA primase | -1.61 |
| 1468 | 249206_at | AT5G42630 | ATS/KAN4 (ABERRANT TESTA SHAPE); DNA binding / transcription factor | -1.77 |
| 1469 | 249190_at | AT5G42750 | BKI1 (BRI1 KINASE INHIBITOR 1); protein heterodimerization | 1.77 |
| 1470 | 249215_at | AT5G42800 | DFR (DIHYDROFLAVONOL 4-REDUCTASE); dihydrokaempferol 4-reductase | -1.81 |
| 1471 | 249187_at | AT5G43060 | cysteine proteinase, putative / thiol protease, putative | -1.88 |
| 1472 | 249141_at | AT5G43200 | zinc finger (C3HC4-type RING finger) family protein | -2.45 |
| 1473 | 249128_at | AT5G43440 | 2-oxoglutarate-dependent dioxygenase, putative | 1.66 |
| 1474 | 249125_at | AT5G43450 | 2-oxoglutarate-dependent dioxygenase, putative | 2.71 |
| 1475 | 249112_at | AT5G43780 | APS4 | 1.97 |
| 1476 | 249071_at | AT5G44050 | MATE efflux family protein | -1.57 |
| 1477 | 249063_at | AT5G44110 | POP1 | -1.41 |
| 1478 | 249045_at | AT5G44380 | FAD-binding domain-containing protein | -2.09 |
| 1479 | 249052_at | AT5G44420 | PDF1.2 (Low-molecular-weight cysteine-rich 77) | 5.25 |
| 1480 | 249060_at | AT5G44560 | VPS2.2 | -4.27 |
| 1481 | 249010_at | AT5G44580 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G44582.1) | 3.08 |
| 1482 | 254521_at | AT5G44820 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G19970.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO46707.1); contains domain PTHR10483:SF6 (PTHR10483:SF6); contains domain PTHR10483 (PTHR10483) | 1.74 |
| 1483 | 249029_at | AT5G44870 | disease resistance protein (TIR-NBS-LRR class), putative | 1.83 |
| 1484 | 248975_at | AT5G45040 | cytochrome c6 (ATC6) | -1.84 |
| 1485 | 248994_at | AT5G45250 | RPS4 (RESISTANT TO P. SYRINGAE 4) | 1.89 |
| 1486 | 248969_at | AT5G45310 | similar to unnamed protein product [Vitis vinifera] (GB:CAO63757.1) | 1.60 |
| 1487 | 248967_at | AT5G45350 | proline-rich family protein | 1.71 |
| 1488 | 248970_at | AT5G45380 | sodium:solute symporter family protein | 3.57 |
| 1489 | 248912_at | AT5G45670 | GDSL-motif lipase/hydrolase family protein | -5.41 |
| 1490 | 248963_at | AT5G45700 | NLI interacting factor (NIF) family protein | -5.66 |
| 1491 | 248914_at | AT5G45750 | AtRABA1c (Arabidopsis Rab GTPase homolog A1c); GTP binding | -1.64 |
| 1492 | 248908_at | AT5G45800 | MEE62 (maternal effect embryo arrest 62); ATP binding / protein serine/threonine kinase | 1.77 |
| 1493 | 248932_at | AT5G46050 | ATPTR3/PTR3 (PEPTIDE TRANSPORTER PROTEIN 3); transporter | 1.61 |
| 1494 | 248879_at | AT5G46180 | delta-OAT (ornithine- delta-aminotransferase); ornithine-oxo-acid transaminase | 1.97 |
| 1495 | 248890_at | AT5G46270 | ATP binding / nucleoside-triphosphatase/ nucleotide binding / protein binding / transmembrane receptor | 1.79 |
| 1496 | 248891_at | AT5G46280 | DNA replication licensing factor, putative | -2.11 |
| 1497 | 248846_at | AT5G46500 | similar to disease resistance protein (TIR-NBS-LRR class), putative [Arabidopsis thaliana] (TAIR:AT5G46260.1) | 2.49 |
| 1498 | 248853_at | AT5G46570 | protein kinase family protein | -1.44 |
| 1499 | 248861_at | AT5G46700 | TET1/TRN2 (TETRASPANIN2) | -2.02 |
| 1500 | 248794_at | AT5G47220 | ATERF-2/ATERF2/ERF2 (ETHYLENE RESPONSE FACTOR 2); DNA binding / transcription activator/ transcription factor | 1.91 |
| 1501 | 248812_at | AT5G47330 | palmitoyl protein thioesterase family protein | 1.80 |
| 1502 | 248807_at | AT5G47500 | pectinesterase family protein | -3.44 |
| 1503 | 248759_at | AT5G47610 | zinc finger (C3HC4-type RING finger) family protein | 1.59 |
| 1504 | 248764_at | AT5G47640 | CCAAT-box binding transcription factor subunit B (NF-YB) (HAP3 ) (AHAP3) family (Hap3b) | 1.75 |
| 1505 | 248768_at | AT5G47700 | 60S acidic ribosomal protein P1 (RPP1C) | -1.87 |
| 1506 | 248775_at | AT5G47850 | protein kinase, putative | 4.26 |
| 1507 | 248777_at | AT5G47920 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G13880.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN74736.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO68720.1) | -2.88 |
| 1508 | 248691_at | AT5G48310 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G24610.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G65440.1); similar to predicted protein [Physcomitrella patens subsp. patens] (GB:EDQ82245.1); similar to predicted protein [Physcomitrella patens subsp. patens] (GB:EDQ70121.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO41110.1); contains InterPro domain C2 calcium/lipid-binding region, CaLB (InterPro:IPR008973) | -1.90 |
| 1509 | 248696_at | AT5G48360 | formin homology 2 domain-containing protein / FH2 domain-containing protein | -1.81 |
| 1510 | 248697_at | AT5G48370 | thioesterase family protein | 1.69 |
| 1511 | 248704_at | AT5G48450 | SKS3 (SKU5 Similar 3); copper ion binding | -1.64 |
| 1512 | 248683_at | AT5G48490 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | -2.12 |
| 1513 | 248708_at | AT5G48560 | basic helix-loop-helix (bHLH) family protein | 1.67 |
| 1514 | 248676_at | AT5G48850 | male sterility MS5 family protein | -2.07 |
| 1515 | 248625_at | AT5G48880 | KAT5/PKT1/PKT2 (PEROXISOMAL 3-KETO-ACYL-COA THIOLASE 1, PEROXISOMAL 3-KETO-ACYL-COA THIOLASE 2); acetyl-CoA C-acyltransferase | -2.32 |
| 1516 | 248632_at | AT5G49010 | SLD5 | -1.41 |
| 1517 | 248597_at | AT5G49160 | MET1 (DECREASED METHYLATION 2DNA) | -2.31 |
| 1518 | 248647_at | AT5G49190 | SUS2 (SUCROSE SYNTHASE 2); UDP-glycosyltransferase/ sucrose synthase/ transferase, transferring glycosyl groups | -1.85 |
| 1519 | 248608_at | AT5G49460 | ACLB-2 (ATP-citrate lyase B-2) | -1.86 |
| 1520 | 248619_at | AT5G49630 | AAP6 (AMINO ACID PERMEASE 6); amino acid transmembrane transporter | 3.33 |
| 1521 | 248590_at | AT5G49660 | leucine-rich repeat transmembrane protein kinase, putative | 1.56 |
| 1522 | 248540_at | AT5G50160 | ATFRO8/FRO8 (FERRIC REDUCTION OXIDASE 8); ferric-chelate reductase/ oxidoreductase | 2.07 |
| 1523 | 248505_at | AT5G50360 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G63350.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO68256.1) | 2.48 |
| 1524 | 248506_at | AT5G50370 | adenylate kinase, putative | -1.56 |
| 1525 | 248526_at | AT5G50740 | metal ion binding | -1.68 |
| 1526 | 248467_at | AT5G50800 | nodulin MtN3 family protein | 2.48 |
| 1527 | 248487_at | AT5G51070 | ERD1 (EARLY RESPONSIVE TO DEHYDRATION 1); ATP binding / ATPase | 2.36 |
| 1528 | 248420_at | AT5G51560 | leucine-rich repeat transmembrane protein kinase, putative | -1.63 |
| 1529 | 248413_at | AT5G51600 | PLE (PLEIADE) | -2.65 |
| 1530 | 248427_at | AT5G51750 | ATSBT1.3; subtilase | -1.62 |
| 1531 | 248371_at | AT5G51810 | AT2353/ATGA20OX2/GA20OX2 (GIBBERELLIN 20 OXIDASE 2); gibberellin 20-oxidase | -1.56 |
| 1532 | 248372_at | AT5G51850 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G25430.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO17701.1) | 1.78 |
| 1533 | 248374_at | AT5G51870 | AGL71; transcription factor | 1.89 |
| 1534 | 248355_at | AT5G52340 | ATEXO70A2 (exocyst subunit EXO70 family protein A2); protein binding | -1.96 |
| 1535 | 248333_at | AT5G52390 | photoassimilate-responsive protein, putative | 2.13 |
| 1536 | 248335_at | AT5G52450 | MATE efflux protein-related | 1.79 |
| 1537 | 248321_at | AT5G52740 | heavy-metal-associated domain-containing protein | 4.34 |
| 1538 | 248327_at | AT5G52750 | heavy-metal-associated domain-containing protein | 2.69 |
| 1539 | 248322_at | AT5G52760 | heavy-metal-associated domain-containing protein | 2.77 |
| 1540 | 248282_at | AT5G52900 | similar to unnamed protein product [Vitis vinifera] (GB:CAO49548.1) | 2.00 |
| 1541 | 248246_at | AT5G53200 | TRY (TRIPTYCHON); DNA binding / transcription factor | 1.67 |
| 1542 | 248263_at | AT5G53370 | ATPMEPCRF; pectinesterase | 2.26 |
| 1543 | 248271_at | AT5G53420 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G27900.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G27900.2); similar to Os05g0595300 [Oryza sativa (japonica cultivar-group)] (GB:NP_001056514.1); similar to hypothetical protein OsI_020507 [Oryza sativa (indica cultivar-group)] (GB:EAY99274.1); contains InterPro domain CCT (InterPro:IPR010402) | 2.16 |
| 1544 | 248276_at | AT5G53550 | YSL3 (YELLOW STRIPE LIKE 3); oligopeptide transporter | 2.06 |
| 1545 | 248207_at | AT5G53970 | aminotransferase, putative | 1.59 |
| 1546 | 248193_at | AT5G54080 | HGO (HOMOGENTISATE 1,2-DIOXYGENASE); homogentisate 1,2-dioxygenase | 2.02 |
| 1547 | 248190_at | AT5G54130 | calcium-binding EF hand family protein | 2.27 |
| 1548 | 248191_at | AT5G54130 | calcium-binding EF hand family protein | 2.41 |
| 1549 | 248200_at | AT5G54160 | ATOMT1 (O-METHYLTRANSFERASE 1) | -1.78 |
| 1550 | 248163_at | AT5G54510 | DFL1/GH3.6 (DWARF IN LIGHT 1); indole-3-acetic acid amido synthetase | -2.44 |
| 1551 | 248169_at | AT5G54610 | ANK (ANKYRIN); protein binding | 3.94 |
| 1552 | 248150_at | AT5G54670 | ATK3 (ARABIDOPSIS THALIANA KINESIN 3); microtubule motor | -1.56 |
| 1553 | 248123_at | AT5G54720 | ankyrin repeat family protein | 2.25 |
| 1554 | 248139_at | AT5G54970 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G26960.1) | -2.46 |
| 1555 | 248141_at | AT5G55010 | unknown protein | 1.64 |
| 1556 | 248100_at | AT5G55180 | glycosyl hydrolase family 17 protein | -3.32 |
| 1557 | 248062_at | AT5G55450 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | 1.87 |
| 1558 | 248085_at | AT5G55480 | glycerophosphoryl diester phosphodiesterase family protein | -2.60 |
| 1559 | 248057_at | AT5G55520 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G26660.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO38858.1); contains InterPro domain Kinesin-related (InterPro:IPR010544) | -2.77 |
| 1560 | 248074_at | AT5G55730 | FLA1 | -2.65 |
| 1561 | 248037_at | AT5G55930 | ATOPT1 (oligopeptide transporter 1); oligopeptide transporter | 2.04 |
| 1562 | 248003_at | AT5G56220 | nucleoside-triphosphatase/ nucleotide binding | -1.53 |
| 1563 | 248011_at | AT5G56300 | GAMT2; S-adenosylmethionine-dependent methyltransferase/ gibberellin carboxyl-O-methyltransferase | 1.80 |
| 1564 | 247991_at | AT5G56320 | ATEXPA14 (ARABIDOPSIS THALIANA EXPANSIN A14) | -1.67 |
| 1565 | 247965_at | AT5G56540 | AGP14 (ARABINOGALACTAN PROTEIN 14) | -1.72 |
| 1566 | 247962_at | AT5G56580 | ATMKK6 (ARABIDOPSIS NQK1); kinase | -2.10 |
| 1567 | 247981_at | AT5G56640 | MIOX5 (MYO-INOSITOL OXYGENASE 5) | -2.15 |
| 1568 | 247968_at | AT5G56670 | 40S ribosomal protein S30 (RPS30C) | -1.71 |
| 1569 | 247978_at | AT5G56710 | 60S ribosomal protein L31 (RPL31C) | -1.83 |
| 1570 | 247980_at | AT5G56860 | GNC (GATA, NITRATE-INDUCIBLE, CARBON METABOLISM-INVOLVED); transcription factor | 2.07 |
| 1571 | 247957_at | AT5G57050 | ABI2 (ABA INSENSITIVE 2); protein serine/threonine phosphatase | 1.99 |
| 1572 | 247900_at | AT5G57290 | 60S acidic ribosomal protein P3 (RPP3B) | -1.61 |
| 1573 | 247903_at | AT5G57340 | similar to hypothetical protein MtrDRAFT_AC155282g59v2 [Medicago truncatula] (GB:ABN08100.1) | 1.94 |
| 1574 | 247869_at | AT5G57520 | ZFP2 (ZINC FINGER PROTEIN 2); nucleic acid binding / transcription factor/ zinc ion binding | 2.04 |
| 1575 | 247866_at | AT5G57550 | XTR3 (XYLOGLUCAN ENDOTRANSGLYCOSYLASE 3); hydrolase, acting on glycosyl bonds | 2.03 |
| 1576 | 247873_at | AT5G57690 | diacylglycerol kinase | -2.46 |
| 1577 | 247879_at | AT5G57770 | similar to unnamed protein product [Vitis vinifera] (GB:CAO66578.1); contains InterPro domain Pleckstrin-like, plant (InterPro:IPR013666); contains InterPro domain Protein of unknown function DUF828, plant (InterPro:IPR008546) | -2.17 |
| 1578 | 247885_at | AT5G57830 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G30830.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO66637.1); contains InterPro domain Protein of unknown function DUF593 (InterPro:IPR007656) | -1.60 |
| 1579 | 247892_at | AT5G57970 | methyladenine glycosylase family protein | -4.17 |
| 1580 | 247848_at | AT5G58120 | disease resistance protein (TIR-NBS-LRR class), putative | 3.41 |
| 1581 | 247814_at | AT5G58310 | hydrolase, alpha/beta fold family protein | 2.02 |
| 1582 | 247815_at | AT5G58420 | 40S ribosomal protein S4 (RPS4D) | -1.50 |
| 1583 | 247800_at | AT5G58570 | unknown protein | 3.16 |
| 1584 | 247793_at | AT5G58650 | PSY1 (PLANT PEPTIDE CONTAINING SULFATED TYROSINE 1) | 1.87 |
| 1585 | 247789_at | AT5G58680 | armadillo/beta-catenin repeat family protein | 2.86 |
| 1586 | 247780_at | AT5G58770 | dehydrodolichyl diphosphate synthase, putative / DEDOL-PP synthase, putative | 3.35 |
| 1587 | 247740_at | AT5G58940 | CRCK1 (CALMODULIN-BINDING RECEPTOR-LIKE CYTOPLASMIC KINASE 1); kinase | 1.82 |
| 1588 | 247751_at | AT5G59050 | similar to unknown [Populus trichocarpa] (GB:ABK92460.1) | 1.74 |
| 1589 | 247754_at | AT5G59080 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G02020.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO15731.1) | 1.74 |
| 1590 | 247718_at | AT5G59310 | LTP4 (LIPID TRANSFER PROTEIN 4); lipid binding | 2.13 |
| 1591 | 247717_at | AT5G59320 | LTP3 (LIPID TRANSFER PROTEIN 3); lipid binding | 1.99 |
| 1592 | 247684_at | AT5G59670 | leucine-rich repeat protein kinase, putative | 4.20 |
| 1593 | 247685_at | AT5G59680 | leucine-rich repeat protein kinase, putative | 2.58 |
| 1594 | 247686_at | AT5G59700 | protein kinase, putative | 2.30 |
| 1595 | 247691_at | AT5G59720 | HSP18.2 (HEAT SHOCK PROTEIN 18.2) | -2.65 |
| 1596 | 247696_at | AT5G59780 | MYB59 (myb domain protein 59); DNA binding / transcription factor | 3.28 |
| 1597 | 247654_at | AT5G59850 | 40S ribosomal protein S15A (RPS15aF) | -1.55 |
| 1598 | 247651_at | AT5G59870 | HTA6; DNA binding | -2.93 |
| 1599 | 247646_at | AT5G59990 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G41380.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO21746.1); contains InterPro domain CCT (InterPro:IPR010402) | 1.96 |
| 1600 | 247584_at | AT5G60670 | 60S ribosomal protein L12 (RPL12C) | -1.77 |
| 1601 | 247602_at | AT5G60900 | RLK1 (RECEPTOR-LIKE PROTEIN KINASE 1); carbohydrate binding / kinase | 3.52 |
| 1602 | 247553_at | AT5G60910 | AGL8 (AGAMOUS-LIKE 8); transcription factor | 4.48 |
| 1603 | 247603_at | AT5G60930 | chromosome-associated kinesin, putative | -1.85 |
| 1604 | 247606_at | AT5G61000 | replication protein, putative | -1.78 |
| 1605 | 247554_at | AT5G61010 | ATEXO70E2 (EXOCYST SUBUNIT EXO70 FAMILY PROTEIN E2); protein binding | 2.20 |
| 1606 | 247573_at | AT5G61160 | AACT1 (ANTHOCYANIN 5-AROMATIC ACYLTRANSFERASE 1); transferase | 5.95 |
| 1607 | 247566_at | AT5G61170 | 40S ribosomal protein S19 (RPS19C) | -2.24 |
| 1608 | 247549_at | AT5G61420 | MYB28 (MYB DOMAIN PROTEIN 28); DNA binding / transcription factor | 2.04 |
| 1609 | 247529_at | AT5G61520 | hexose transporter, putative | 1.83 |
| 1610 | 247541_at | AT5G61660 | glycine-rich protein | 1.70 |
| 1611 | 247488_at | AT5G61820 | similar to MtN19-like protein [Pisum sativum] (GB:AAU14999.2); contains InterPro domain Stress up-regulated Nod 19 (InterPro:IPR011692) | 1.94 |
| 1612 | 247469_at | AT5G62165 | AGL42 (AGAMOUS LIKE 42); transcription factor | 3.92 |
| 1613 | 247463_at | AT5G62210 | embryo-specific protein-related | -4.14 |
| 1614 | 247474_at | AT5G62280 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G45360.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO49726.1); contains InterPro domain Protein of unknown function DUF1442 (InterPro:IPR009902) | 1.67 |
| 1615 | 247452_at | AT5G62430 | CDF1 (CYCLING DOF FACTOR 1); DNA binding / protein binding / transcription factor | 1.88 |
| 1616 | 247435_at | AT5G62480 | ATGSTU9 (GLUTATHIONE S-TRANSFERASE TAU 9); glutathione transferase | 1.93 |
| 1617 | 247432_at | AT5G62500 | ATEB1B (Arabidopsis thaliana Microtubule End Binding Protein EB1A); microtubule binding | -1.84 |
| 1618 | 247425_at | AT5G62550 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G28770.1); similar to unknown [Populus trichocarpa] (GB:ABK92769.1) | -5.38 |
| 1619 | 247427_at | AT5G62580 | binding | -1.49 |
| 1620 | 247440_at | AT5G62680 | proton-dependent oligopeptide transport (POT) family protein | 1.79 |
| 1621 | 247441_at | AT5G62710 | leucine-rich repeat family protein / protein kinase family protein | -1.41 |
| 1622 | 247406_at | AT5G62920 | ARR6 (RESPONSE REGULATOR 6); transcription regulator/ two-component response regulator | -3.74 |
| 1623 | 247362_at | AT5G63140 | ATPAP29/PAP29 (purple acid phosphatase 29); acid phosphatase/ protein serine/threonine phosphatase | -1.84 |
| 1624 | 247356_at | AT5G63800 | BGAL6/MUM2 (MUCILAGE-MODIFIED 2); beta-galactosidase | 2.11 |
| 1625 | 247304_at | AT5G63850 | AAP4 (amino acid permease 4); amino acid transmembrane transporter | 2.53 |
| 1626 | 247301_at | AT5G63920 | DNA topoisomerase III alpha, putative | -1.42 |
| 1627 | 247314_at | AT5G64000 | SAL2; 3'(2'),5'-bisphosphate nucleotidase/ inositol or phosphatidylinositol phosphatase | 3.16 |
| 1628 | 247268_at | AT5G64080 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | -2.87 |
| 1629 | 247267_at | AT5G64140 | RPS28 (RIBOSOMAL PROTEIN S28); structural constituent of ribosome | -1.55 |
| 1630 | 247252_at | AT5G64770 | similar to 80C09_10 [Brassica rapa subsp. pekinensis] (GB:AAZ41821.1) | 2.65 |
| 1631 | 247254_at | AT5G64800 | CLE21 (CLAVATA3/ESR-RELATED 21); receptor binding | 1.60 |
| 1632 | 247222_at | AT5G64840 | ATGCN5 (Arabidopsis thaliana general control non-repressible 5) | 1.72 |
| 1633 | 247218_at | AT5G65010 | ASN2 (ASPARAGINE SYNTHETASE 2); asparagine synthase (glutamine-hydrolyzing) | -1.95 |
| 1634 | 247210_at | AT5G65020 | ANNAT2 (ANNEXIN ARABIDOPSIS 2); calcium ion binding / calcium-dependent phospholipid binding | -1.93 |
| 1635 | 247192_at | AT5G65360 | histone H3 | -2.46 |
| 1636 | 247189_at | AT5G65390 | AGP7 (Arabinogalactan protein 7) | -2.60 |
| 1637 | 247190_at | AT5G65420 | CYCD4;1 (CYCLIN D4;1); cyclin-dependent protein kinase regulator | -1.95 |
| 1638 | 247145_at | AT5G65600 | legume lectin family protein / protein kinase family protein | 2.93 |
| 1639 | 247146_at | AT5G65610 | unknown protein | 1.59 |
| 1640 | 247151_at | AT5G65640 | BHLH093 (BETA HLH PROTEIN 93); DNA binding / transcription factor | -3.10 |
| 1641 | 247149_at | AT5G65660 | hydroxyproline-rich glycoprotein family protein | -2.12 |
| 1642 | 247111_at | AT5G65880 | unknown protein | -1.77 |
| 1643 | 247134_at | AT5G66230 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G51230.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO48217.1) | -2.74 |
| 1644 | 247085_at | AT5G66310 | kinesin motor family protein | -1.60 |
| 1645 | 247095_at | AT5G66400 | RAB18 (RESPONSIVE TO ABA 18) | 2.83 |
| 1646 | 247075_at | AT5G66410 | PLP3B (PHOSDUCIN-LIKE PROTEIN 3 HOMOLOG) | -1.84 |
| 1647 | 247072_at | AT5G66490 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G50900.1); similar to hypothetical protein [Thellungiella halophila] (GB:ABB45855.1) | 2.46 |
| 1648 | 247105_at | AT5G66630 | LIM domain-containing protein | 2.61 |
| 1649 | 247071_at | AT5G66640 | LIM domain-containing protein-related | 3.32 |
| 1650 | 247069_at | AT5G66920 | SKS17 (SKU5 Similar 17); copper ion binding / oxidoreductase | -1.86 |
| 1651 | 247025_at | AT5G67030 | ABA1 (ABA DEFICIENT 1); zeaxanthin epoxidase | 1.63 |
| 1652 | 247037_at | AT5G67070 | RALFL34 (RALF-LIKE 34) | -2.19 |
| 1653 | 247028_at | AT5G67100 | ICU2 (INCURVATA2); DNA-directed DNA polymerase | -1.41 |
| 1654 | 247039_at | AT5G67270 | ATEB1C (MICROTUBULE END BINDING PROTEIN 1); microtubule binding | -1.94 |
| 1655 | 246998_at | AT5G67370 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G11840.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN75014.1); contains InterPro domain Protein of unknown function DUF1230 (InterPro:IPR009631) | 2.04 |
| 1656 | 247005_at | AT5G67520 | adenylylsulfate kinase, putative | 2.82 |