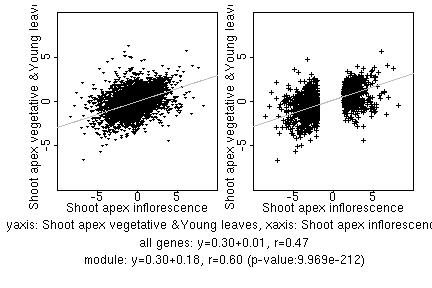
marker gene list
| Shoot apex inflorescence (after bolting) |
| Shoot apex vegetative &Young leaves |
| probeID | AGICode | Annotation | gene expression |
|
|---|---|---|---|---|
| 1 | 261576_at | AT1G01070 | nodulin MtN21 family protein | -5.33 |
| 2 | 261580_at | AT1G01110 | IQD18 (IQ-domain 18) | 2.78 |
| 3 | 261574_at | AT1G01190 | CYP78A8 (cytochrome P450, family 78, subfamily A, polypeptide 8); oxygen binding | -3.12 |
| 4 | 261055_at | AT1G01300 | aspartyl protease family protein | 1.77 |
| 5 | 261058_at | AT1G01370 | HTR12 (CENTROMERIC HISTONE H3); DNA binding | 2.46 |
| 6 | 259426_at | AT1G01470 | LEA14 (LATE EMBRYOGENESIS ABUNDANT 14) | -5.46 |
| 7 | 264180_at | AT1G02190 | CER1 protein, putative | 6.51 |
| 8 | 264148_at | AT1G02220 | ANAC003 (Arabidopsis NAC domain containing protein 3); transcription factor | -2.33 |
| 9 | 259445_at | AT1G02400 | ATGA2OX6/DTA1 (GIBBERELLIN 2-OXIDASE 6); gibberellin 2-beta-dioxygenase | -2.09 |
| 10 | 260916_at | AT1G02475 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G01883.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO40177.1); contains InterPro domain Streptomyces cyclase/dehydrase (InterPro:IPR005031) | -1.95 |
| 11 | 260915_at | AT1G02660 | lipase class 3 family protein | -2.02 |
| 12 | 260909_at | AT1G02670 | DNA repair protein, putative | 2.29 |
| 13 | 260910_at | AT1G02690 | importin alpha-2 subunit, putative | 2.44 |
| 14 | 262109_at | AT1G02730 | ATCSLD5 (CELLULOSE SYNTHASE-LIKE D5); 1,4-beta-D-xylan synthase/ cellulose synthase | 2.50 |
| 15 | 262121_at | AT1G02800 | ATCEL2 (Arabidopsis thaliana Cellulase 2); hydrolase, hydrolyzing O-glycosyl compounds | 5.74 |
| 16 | 262113_at | AT1G02820 | late embryogenesis abundant 3 family protein / LEA3 family protein | -5.71 |
| 17 | 262131_at | AT1G02900 | RALFL1 (RALF-LIKE 1) | -1.96 |
| 18 | 262106_at | AT1G02970 | WEE1 (ARABIDOPSIS WEE1 KINASE HOMOLOG); kinase/ protein kinase | 1.46 |
| 19 | 263168_at | AT1G03020 | glutaredoxin family protein | -2.46 |
| 20 | 263114_at | AT1G03130 | PSAD-2 (photosystem I subunit D-2) | -2.92 |
| 21 | 264363_at | AT1G03170 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G02810.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO22484.1) | 3.04 |
| 22 | 264822_at | AT1G03457 | RNA-binding protein, putative | 1.45 |
| 23 | 264815_at | AT1G03620 | phagocytosis and cell motility protein ELMO1-related | -2.06 |
| 24 | 264842_at | AT1G03700 | integral membrane family protein | 2.69 |
| 25 | 264830_at | AT1G03710 | cysteine protease inhibitor | 4.57 |
| 26 | 264825_at | AT1G03720 | cathepsin-related | 3.66 |
| 27 | 265085_at | AT1G03780 | targeting protein-related | 3.08 |
| 28 | 265082_at | AT1G03830 | guanylate-binding family protein | 1.48 |
| 29 | 265067_at | AT1G03850 | glutaredoxin family protein | -2.41 |
| 30 | 265066_at | AT1G03870 | FLA9 | -3.36 |
| 31 | 265044_at | AT1G03950 | VPS2.3 | -2.78 |
| 32 | 265097_at | AT1G04020 | ATBARD1/BARD1 (BREAST CANCER ASSOCIATED RING 1); transcription coactivator | 1.93 |
| 33 | 265042_at | AT1G04040 | acid phosphatase class B family protein | -5.80 |
| 34 | 257404_at | AT1G04050 | SUVR1 (Arabidopsis homolog of SU(VAR)3-9 1); histone-lysine N-methyltransferase/ zinc ion binding | 1.67 |
| 35 | 265089_at | AT1G04050 | SUVR1 (Arabidopsis homolog of SU(VAR)3-9 1); histone-lysine N-methyltransferase/ zinc ion binding | 2.81 |
| 36 | 263656_at | AT1G04240 | SHY2 (SHORT HYPOCOTYL 2); transcription factor | -3.13 |
| 37 | 263668_at | AT1G04350 | 2-oxoglutarate-dependent dioxygenase, putative | -3.35 |
| 38 | 263677_at | AT1G04520 | 33 kDa secretory protein-related | 1.98 |
| 39 | 264610_at | AT1G04645 | self-incompatibility protein-related | 1.60 |
| 40 | 264606_at | AT1G04660 | glycine-rich protein | 1.77 |
| 41 | 264603_at | AT1G04670 | similar to pollen-preferential protein [Brassica rapa subsp. chinensis] (GB:ABH09777.1) | -2.21 |
| 42 | 265194_at | AT1G05010 | EFE (ETHYLENE FORMING ENZYME) | -3.19 |
| 43 | 264573_at | AT1G05310 | pectinesterase family protein | -3.16 |
| 44 | 264580_at | AT1G05340 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G32210.1) | -2.60 |
| 45 | 261384_at | AT1G05440 | similar to unnamed protein product [Vitis vinifera] (GB:CAO71189.1) | 2.67 |
| 46 | 261381_at | AT1G05460 | SDE3 (SILENCING DEFECTIVE) | 1.46 |
| 47 | 263203_at | AT1G05490 | CHR31 (chromatin remodeling 31); ATP binding / DNA binding / helicase/ nucleic acid binding | 1.70 |
| 48 | 263204_at | AT1G05490 | CHR31 (chromatin remodeling 31); ATP binding / DNA binding / helicase/ nucleic acid binding | 3.73 |
| 49 | 263182_at | AT1G05575 | unknown protein | -2.48 |
| 50 | 263179_at | AT1G05710 | ethylene-responsive protein, putative | 1.64 |
| 51 | 260945_at | AT1G05950 | similar to unnamed protein product [Vitis vinifera] (GB:CAO64784.1); contains domain dsRNA-binding domain-like (SSF54768) | 2.67 |
| 52 | 260955_at | AT1G06000 | UDP-glucoronosyl/UDP-glucosyl transferase family protein | 1.75 |
| 53 | 260957_at | AT1G06080 | ADS1 (DELTA 9 DESATURASE 1); oxidoreductase | 1.78 |
| 54 | 257491_at | AT1G06170 | basic helix-loop-helix (bHLH) family protein | 2.24 |
| 55 | 260784_at | AT1G06180 | ATMYB13 (myb domain protein 13); DNA binding / transcription factor | 1.54 |
| 56 | 259394_at | AT1G06420 | unknown protein | 3.93 |
| 57 | 262626_at | AT1G06430 | FTSH8 (FtsH protease 8); ATP-dependent peptidase/ ATPase/ metallopeptidase/ zinc ion binding | -2.10 |
| 58 | 262629_at | AT1G06460 | ACD32.1 (ALPHA-CRYSTALLIN DOMAIN 31.2) | -2.67 |
| 59 | 262635_at | AT1G06570 | PDS1 (PHYTOENE DESATURATION 1) | -1.92 |
| 60 | 262638_at | AT1G06650 | 2-oxoglutarate-dependent dioxygenase, putative | -2.49 |
| 61 | 260830_at | AT1G06760 | histone H1, putative | 1.47 |
| 62 | 260831_at | AT1G06830 | glutaredoxin family protein | -3.43 |
| 63 | 256062_at | AT1G07090 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G58500.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN60458.1); contains InterPro domain Protein of unknown function DUF640 (InterPro:IPR006936) | -2.32 |
| 64 | 256057_at | AT1G07180 | ATNDI1/NDA1 (ALTERNATIVE NAD(P)H DEHYDROGENASE 1); NADH dehydrogenase | -3.68 |
| 65 | 256053_at | AT1G07260 | UDP-glucoronosyl/UDP-glucosyl transferase family protein | 1.84 |
| 66 | 261080_at | AT1G07370 | PCNA1 (PROLIFERATING CELLULAR NUCLEAR ANTIGEN); DNA binding / DNA polymerase processivity factor | 2.49 |
| 67 | 261065_at | AT1G07500 | unknown protein | -2.41 |
| 68 | 261410_at | AT1G07610 | MT1C (metallothionein 1C) | -3.76 |
| 69 | 261420_at | AT1G07720 | beta-ketoacyl-CoA synthase family protein | -1.91 |
| 70 | 261411_at | AT1G07790 | HTB1; DNA binding | 2.69 |
| 71 | 261815_at | AT1G08320 | bZIP family transcription factor | 4.92 |
| 72 | 264802_at | AT1G08560 | SYP111 (syntaxin 111); SNAP receptor | 2.52 |
| 73 | 264782_at | AT1G08810 | MYB60 (myb domain protein 60); DNA binding / transcription factor | -2.35 |
| 74 | 264646_at | AT1G08860 | BON3 (BONZAI 3) | 3.28 |
| 75 | 264652_at | AT1G08920 | sugar transporter, putative | -2.64 |
| 76 | 264624_at | AT1G08930 | ERD6 (EARLY RESPONSE TO DEHYDRATION 6); carbohydrate transmembrane transporter/ sugar:hydrogen ion symporter | -3.36 |
| 77 | 264655_at | AT1G09070 | (AT)SRC2/SRC2 (SOYBEAN GENE REGULATED BY COLD-2); protein binding | -1.96 |
| 78 | 264251_at | AT1G09190 | pentatricopeptide (PPR) repeat-containing protein | 2.89 |
| 79 | 264262_at | AT1G09200 | histone H3 | 1.90 |
| 80 | 263676_at | AT1G09340 | CRB; binding / catalytic/ coenzyme binding | -2.45 |
| 81 | 264550_at | AT1G09450 | haspin-related | 2.18 |
| 82 | 264467_at | AT1G10140 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G58420.1); contains InterPro domain Uncharacterised conserved protein UCP031279 (InterPro:IPR016972) | -2.14 |
| 83 | 264465_at | AT1G10230 | ASK18 (ARABIDOPSIS SKP1-LIKE 18); protein binding / ubiquitin-protein ligase | 2.30 |
| 84 | 264452_at | AT1G10270 | GRP23 (GLUTAMINE-RICH PROTEIN23); binding | 1.55 |
| 85 | 264434_at | AT1G10340 | ankyrin repeat family protein | -2.46 |
| 86 | 264435_at | AT1G10360 | ATGSTU18 (GLUTATHIONE S-TRANSFERASE 29); glutathione transferase | -2.41 |
| 87 | 263258_at | AT1G10540 | xanthine/uracil permease family protein | 1.67 |
| 88 | 261834_at | AT1G10640 | polygalacturonase, putative / pectinase, putative | 3.10 |
| 89 | 262758_at | AT1G10780 | F-box family protein | 1.85 |
| 90 | 262783_at | AT1G10850 | ATP binding / protein serine/threonine kinase | 1.60 |
| 91 | 260461_at | AT1G10980 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G61670.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO41520.1); contains InterPro domain Transmembrane receptor, eukaryota; (InterPro:IPR009637) | 3.51 |
| 92 | 260470_at | AT1G11120 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G61170.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN77872.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO24110.1) | 1.83 |
| 93 | 262451_at | AT1G11130 | SUB (STRUBBELIG); protein binding | 1.93 |
| 94 | 262479_at | AT1G11130 | SUB (STRUBBELIG); protein binding | 1.54 |
| 95 | 262452_at | AT1G11210 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G11220.1); similar to cotton fiber expressed protein 1 [Gossypium hirsutum] (GB:AAC33276.1); contains InterPro domain Protein of unknown function DUF761, plant (InterPro:IPR008480) | -2.24 |
| 96 | 262477_at | AT1G11220 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G11230.1); similar to fiber expressed protein [Gossypium hirsutum] (GB:AAY85179.1); contains InterPro domain Protein of unknown function DUF761, plant (InterPro:IPR008480) | 2.40 |
| 97 | 261821_at | AT1G11530 | ATCXXS1 (C-TERMINAL CYSTEINE RESIDUE IS CHANGED TO A SERINE 1); thiol-disulfide exchange intermediate | -1.94 |
| 98 | 261826_at | AT1G11580 | ATPMEPCRA; pectinesterase | -1.84 |
| 99 | 262819_at | AT1G11600 | CYP77B1 (cytochrome P450, family 77, subfamily B, polypeptide 1); oxygen binding | 3.67 |
| 100 | 262808_at | AT1G11730 | galactosyltransferase family protein | 1.60 |
| 101 | 264390_at | AT1G11950 | transcription factor | 1.69 |
| 102 | 264346_at | AT1G12010 | 1-aminocyclopropane-1-carboxylate oxidase, putative / ACC oxidase, putative | -4.56 |
| 103 | 264348_at | AT1G12110 | NRT1.1 (NITRATE TRANSPORTER 1.1); transporter | -4.18 |
| 104 | 259528_at | AT1G12330 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G12900.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO16971.1) | 2.01 |
| 105 | 259525_at | AT1G12560 | ATEXPA7 (ARABIDOPSIS THALIANA EXPANSIN A7) | -2.45 |
| 106 | 261207_at | AT1G12830 | unknown protein | 1.68 |
| 107 | 262777_at | AT1G13030 | sphere organelles protein-related | 1.52 |
| 108 | 262826_at | AT1G13080 | CYP71B2 (CYTOCHROME P450 71B2); oxygen binding | -2.84 |
| 109 | 262793_at | AT1G13110 | CYP71B7 (cytochrome P450, family 71, subfamily B, polypeptide 7); oxygen binding | -3.67 |
| 110 | 262774_at | AT1G13230 | leucine-rich repeat family protein | 1.96 |
| 111 | 259364_at | AT1G13260 | RAV1 (Related to ABI3/VP1 1); DNA binding / transcription factor | -2.66 |
| 112 | 257515_at | AT1G13290 | zinc finger (C2H2 type) family protein | 2.23 |
| 113 | 259409_at | AT1G13330 | similar to unnamed protein product [Vitis vinifera] (GB:CAO42122.1); contains InterPro domain Tat binding protein 1-interacting (InterPro:IPR010776) | 1.68 |
| 114 | 259410_at | AT1G13340 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G34220.2); similar to unknown [Carica papaya] (GB:ABS01355.1); contains InterPro domain Protein of unknown function DUF292, eukaryotic (InterPro:IPR005061) | -2.98 |
| 115 | 259386_at | AT1G13400 | JGL/NUB (NUBBIN); nucleic acid binding / zinc ion binding | 1.84 |
| 116 | 256096_at | AT1G13650 | similar to 18S pre-ribosomal assembly protein gar2-related [Arabidopsis thaliana] (TAIR:AT2G03810.4); similar to 18S pre-ribosomal assembly protein gar2-related [Arabidopsis thaliana] (TAIR:AT2G03810.2); similar to 18S pre-ribosomal assembly protein gar2-related [Arabidopsis thaliana] (TAIR:AT2G03810.3); similar to hypothetical protein Tc00.1047053503999.40 [Trypanosoma cruzi strain CL Brener] (GB:XP_813437.1) | -3.19 |
| 117 | 256102_at | AT1G13680 | phospholipase C | 5.21 |
| 118 | 256099_at | AT1G13710 | CYP78A5 (cytochrome P450, family 78, subfamily A, polypeptide 5); oxygen binding | 5.43 |
| 119 | 259448_at | AT1G13790 | XH/XS domain-containing protein / XS zinc finger domain-containing protein | 1.85 |
| 120 | 262615_at | AT1G13950 | EIF-5A (eukaryotic translation initiation factor 5A-1); translation initiation factor | -4.85 |
| 121 | 262607_at | AT1G13990 | similar to unnamed protein product [Vitis vinifera] (GB:CAO68469.1) | -2.10 |
| 122 | 262612_at | AT1G14150 | oxygen evolving enhancer 3 (PsbQ) family protein | -3.87 |
| 123 | 262654_at | AT1G14180 | unknown protein | 1.72 |
| 124 | 261480_at | AT1G14280 | PKS2 (PHYTOCHROME KINASE SUBSTRATE 2) | -3.17 |
| 125 | 261526_at | AT1G14370 | APK2A (PROTEIN KINASE 2A); kinase | -2.49 |
| 126 | 261502_at | AT1G14440 | ATHB31 (ARABIDOPSIS THALIANA HOMEOBOX PROTEIN 31); transcription factor | 2.98 |
| 127 | 261473_at | AT1G14490 | DNA-binding protein-related | 3.42 |
| 128 | 262843_at | AT1G14687 | ATHB32 (ARABIDOPSIS THALIANA HOMEOBOX PROTEIN 32); DNA binding / transcription factor | 4.41 |
| 129 | 262839_at | AT1G14770 | zinc ion binding | 1.45 |
| 130 | 262840_at | AT1G14900 | HMGA (HIGH MOBILITY GROUP A); DNA binding | 2.59 |
| 131 | 262596_at | AT1G15080 | ATPAP2 (PHOSPHATIDIC ACID PHOSPHATASE 2); phosphatidate phosphatase | 1.51 |
| 132 | 262605_at | AT1G15170 | MATE efflux family protein | -2.03 |
| 133 | 262575_at | AT1G15210 | ATPDR7/PDR7 (PLEIOTROPIC DRUG RESISTANCE 7); ATPase, coupled to transmembrane movement of substances | 1.67 |
| 134 | 262599_at | AT1G15350 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G25360.1); similar to hypothetical protein [Glycine max] (GB:AAN03467.1) | -2.05 |
| 135 | 262595_at | AT1G15360 | SHN1/WIN1 (SHINE1); DNA binding / transcription factor | 3.65 |
| 136 | 262586_at | AT1G15480 | DNA binding | 2.12 |
| 137 | 261765_at | AT1G15570 | CYCA2;3 (CYCLIN A2;3); cyclin-dependent protein kinase regulator | 1.53 |
| 138 | 261761_at | AT1G15660 | similar to CENP-C [Arabidopsis arenosa] (GB:AAU04611.1); similar to kinetochore protein [Capsella rubella] (GB:BAF63162.1); contains domain PTHR16684 (PTHR16684); contains domain PTHR16684:SF3 (PTHR16684:SF3) | 1.82 |
| 139 | 259491_at | AT1G15820 | LHCB6 (LIGHT HARVESTING COMPLEX PSII); chlorophyll binding | -1.90 |
| 140 | 261844_at | AT1G15940 | binding | 1.98 |
| 141 | 261788_at | AT1G15980 | similar to unnamed protein product [Vitis vinifera] (GB:CAO49411.1); contains domain G3DSA:3.40.50.2000 (G3DSA:3.40.50.2000); contains domain SSF53756 (SSF53756) | -2.69 |
| 142 | 261786_at | AT1G15990 | ATCNGC7 (CYCLIC NUCLEOTIDE GATED CHANNEL 7); calmodulin binding / cyclic nucleotide binding / ion channel | -2.30 |
| 143 | 261840_at | AT1G16070 | AtTLP8 (TUBBY LIKE PROTEIN 8); transcription factor | 3.09 |
| 144 | 257478_at | AT1G16130 | WAKL2 (WALL ASSOCIATED KINASE-LIKE 2); kinase | -3.12 |
| 145 | 262702_at | AT1G16220 | protein phosphatase 2C family protein / PP2C family protein | 1.48 |
| 146 | 262752_at | AT1G16330 | CYCB3;1 (CYCLIN B3;1); cyclin-dependent protein kinase | 1.81 |
| 147 | 262730_at | AT1G16390 | ATOCT3 (ARABIDOPSIS THALIANA ORGANIC CATION/CARNITINE TRANSPORTER2); carbohydrate transmembrane transporter/ sugar:hydrogen ion symporter | -2.98 |
| 148 | 246321_at | AT1G16640 | transcriptional factor B3 family protein | 1.69 |
| 149 | 255768_at | AT1G16705 | p300/CBP acetyltransferase-related protein-related | 1.50 |
| 150 | 255763_at | AT1G16730 | similar to unknown [Picea sitchensis] (GB:ABK21208.1) | -2.34 |
| 151 | 256114_at | AT1G16850 | unknown protein | -4.36 |
| 152 | 256115_at | AT1G16880 | uridylyltransferase-related | -1.90 |
| 153 | 262482_at | AT1G17020 | SRG1 (SENESCENCE-RELATED GENE 1); oxidoreductase, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors | -3.48 |
| 154 | 262538_at | AT1G17140 | tropomyosin-related | 1.73 |
| 155 | 261037_at | AT1G17420 | LOX3 (Lipoxygenase 3); iron ion binding / lipoxygenase/ metal ion binding / oxidoreductase, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen | -1.88 |
| 156 | 260683_at | AT1G17560 | HLL (HUELLENLOS); structural constituent of ribosome | 1.69 |
| 157 | 260686_at | AT1G17620 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G11890.1); similar to unknown [Populus trichocarpa] (GB:ABK96083.1); contains InterPro domain Harpin-induced 1 (InterPro:IPR010847) | -1.86 |
| 158 | 255903_at | AT1G17950 | MYB52 (myb domain protein 52); DNA binding / transcription factor | -1.96 |
| 159 | 256128_at | AT1G18140 | LAC1 (Laccase 1); copper ion binding / oxidoreductase | 3.78 |
| 160 | 256125_at | AT1G18250 | ATLP-1 (Arabidopsis thaumatin-like protein 1) | 3.43 |
| 161 | 261660_at | AT1G18370 | HIK (HINKEL); microtubule motor | 2.66 |
| 162 | 261717_at | AT1G18400 | BEE1 (BR ENHANCED EXPRESSION 1); transcription factor | -1.89 |
| 163 | 255773_at | AT1G18590 | sulfotransferase family protein | -2.49 |
| 164 | 255779_at | AT1G18650 | glycosyl hydrolase family protein 17 | 1.68 |
| 165 | 261422_at | AT1G18730 | similar to unknown [Populus trichocarpa] (GB:ABK95263.1) | -2.19 |
| 166 | 259462_at | AT1G18940 | nodulin family protein | -2.02 |
| 167 | 259479_at | AT1G19020 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G48180.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO40966.1) | -4.61 |
| 168 | 256015_at | AT1G19150 | LHCA6 (Photosystem I light harvesting complex gene 6); chlorophyll binding | -2.74 |
| 169 | 256017_at | AT1G19180 | JAZ1/TIFY10A (JASMONATE-ZIM-DOMAIN PROTEIN 1); protein binding | -3.19 |
| 170 | 260656_at | AT1G19380 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G65650.1); similar to unknown [Vitis pseudoreticulata] (GB:ABC69762.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO61173.1); contains InterPro domain Protein of unknown function DUF1195 (InterPro:IPR010608) | -1.87 |
| 171 | 261137_at | AT1G19830 | auxin-responsive protein, putative | 2.98 |
| 172 | 255782_at | AT1G19850 | MP (MONOPTEROS); transcription factor | 3.38 |
| 173 | 255814_at | AT1G19900 | glyoxal oxidase-related | -2.46 |
| 174 | 261248_at | AT1G20030 | pathogenesis-related thaumatin family protein | 2.37 |
| 175 | 261226_at | AT1G20190 | ATEXPA11 (ARABIDOPSIS THALIANA EXPANSIN A11) | -5.13 |
| 176 | 255941_at | AT1G20350 | ATTIM17-1 (Arabidopsis thaliana translocase inner membrane subunit 17-1); P-P-bond-hydrolysis-driven protein transmembrane transporter | -3.67 |
| 177 | 259570_at | AT1G20440 | COR47 (cold regulated 47) | -4.18 |
| 178 | 259565_at | AT1G20530 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G17110.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN74079.1); contains InterPro domain Protein of unknown function DUF632 (InterPro:IPR006867); contains InterPro domain Protein of unknown function DUF630 (InterPro:IPR006868) | 4.00 |
| 179 | 256082_at | AT1G20720 | helicase-related | 1.62 |
| 180 | 262796_at | AT1G20850 | XCP2 (XYLEM CYSTEINE PEPTIDASE 2); cysteine-type peptidase/ peptidase | -2.34 |
| 181 | 262802_at | AT1G20930 | CDKB2;2 (CYCLIN-DEPENDENT KINASE B2;2); kinase | 1.83 |
| 182 | 262803_at | AT1G21000 | zinc-binding family protein | -3.70 |
| 183 | 262801_at | AT1G21010 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G76600.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO66314.1) | -1.89 |
| 184 | 261454_at | AT1G21090 | hydroxyproline-rich glycoprotein family protein | 2.23 |
| 185 | 261459_at | AT1G21100 | O-methyltransferase, putative | -2.74 |
| 186 | 261453_at | AT1G21130 | O-methyltransferase, putative | -4.40 |
| 187 | 261448_at | AT1G21140 | nodulin, putative | -1.85 |
| 188 | 259553_x_at | AT1G21310 | ATEXT3 (EXTENSIN 3); structural constituent of cell wall | 1.83 |
| 189 | 260921_at | AT1G21540 | AMP-binding protein, putative | 1.73 |
| 190 | 260922_at | AT1G21560 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G01170.1); similar to hypothetical protein OsI_014365 [Oryza sativa (indica cultivar-group)] (GB:EAY93132.1); similar to Os04g0221000 [Oryza sativa (japonica cultivar-group)] (GB:NP_001052267.1) | 1.55 |
| 191 | 262503_at | AT1G21670 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G21680.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN73514.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO61906.1); contains InterPro domain WD40-like Beta Propeller (InterPro:IPR011659); contains InterPro domain Six-bladed beta-propeller, TolB-like (InterPro:IPR011042) | -2.32 |
| 192 | 262505_at | AT1G21680 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G21670.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN73514.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO61906.1); similar to hypothetical protein OsJ_012725 [Oryza sativa (japonica cultivar-group)] (GB:EAZ29242.1); contains InterPro domain WD40-like Beta Propeller (InterPro:IPR011659); contains InterPro domain Six-bladed beta-propeller, TolB-like (InterPro:IPR011042) | -2.00 |
| 193 | 262501_at | AT1G21690 | EMB1968 (EMBRYO DEFECTIVE 1968); ATPase | 1.66 |
| 194 | 262486_at | AT1G21740 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G77500.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN81539.1); contains InterPro domain Protein of unknown function DUF632 (InterPro:IPR006867); contains InterPro domain Protein of unknown function DUF630 (InterPro:IPR006868) | 3.08 |
| 195 | 262494_at | AT1G21810 | similar to myosin heavy chain-related [Arabidopsis thaliana] (TAIR:AT1G77580.2); similar to coiled-coil protein [Lycopersicon esculentum] (GB:AAN03605.1); contains InterPro domain Prefoldin (InterPro:IPR009053); contains InterPro domain Protein of unknown function DUF869, plant (InterPro:IPR008587) | 2.10 |
| 196 | 260857_at | AT1G21880 | LYM1 (LYSM DOMAIN GPI-ANCHORED PROTEIN 1 PRECURSOR) | 1.64 |
| 197 | 255926_at | AT1G22190 | AP2 domain-containing transcription factor, putative | -3.12 |
| 198 | 255964_at | AT1G22275 | ZYP1b | 1.54 |
| 199 | 261934_at | AT1G22400 | ATUGT85A1/UGT85A1 (UDP-GLUCOSYL TRANSFERASE 85A1); UDP-glycosyltransferase/ glucuronosyltransferase/ transferase, transferring glycosyl groups / transferase, transferring hexosyl groups | -2.01 |
| 200 | 261928_at | AT1G22480 | plastocyanin-like domain-containing protein | 2.36 |
| 201 | 261941_at | AT1G22490 | basic helix-loop-helix (bHLH) family protein | 2.35 |
| 202 | 261937_at | AT1G22570 | proton-dependent oligopeptide transport (POT) family protein | -2.11 |
| 203 | 264201_at | AT1G22630 | heat shock protein binding / unfolded protein binding | -2.45 |
| 204 | 264195_at | AT1G22690 | gibberellin-responsive protein, putative | -2.45 |
| 205 | 264204_at | AT1G22710 | SUC2 (SUCROSE-PROTON SYMPORTER 2); carbohydrate transmembrane transporter/ sucrose transmembrane transporter/ sucrose:hydrogen symporter/ sugar:hydrogen ion symporter | -3.96 |
| 206 | 264209_at | AT1G22740 | RAB7 (Ras-related protein 7); GTP binding | -2.66 |
| 207 | 264774_at | AT1G22890 | unknown protein | -2.05 |
| 208 | 264773_at | AT1G22900 | similar to disease resistance-responsive family protein [Arabidopsis thaliana] (TAIR:AT5G42500.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO23438.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN61168.1); contains InterPro domain Plant disease resistance response protein (InterPro:IPR004265) | -2.08 |
| 209 | 264721_at | AT1G23000 | heavy-metal-associated domain-containing protein | 3.75 |
| 210 | 264751_at | AT1G23020 | ATFRO3/FRO3 (FERRIC REDUCTION OXIDASE 3); ferric-chelate reductase | -3.58 |
| 211 | 264894_at | AT1G23040 | hydroxyproline-rich glycoprotein family protein | -2.65 |
| 212 | 264889_at | AT1G23050 | hydroxyproline-rich glycoprotein family protein | -3.38 |
| 213 | 264887_at | AT1G23120 | major latex protein-related / MLP-related | -3.46 |
| 214 | 264899_at | AT1G23130 | Bet v I allergen family protein | -6.25 |
| 215 | 264893_at | AT1G23140 | C2 domain-containing protein | -1.89 |
| 216 | 262988_at | AT1G23310 | GGT1 (ALANINE-2-OXOGLUTARATE AMINOTRANSFERASE 1) | -2.02 |
| 217 | 263013_at | AT1G23380 | KNAT6 (Knotted-like Arabidopsis thaliana 6); DNA binding / transcription factor | 2.14 |
| 218 | 262986_at | AT1G23390 | kelch repeat-containing F-box family protein | -2.13 |
| 219 | 263016_at | AT1G23410 | ubiquitin extension protein, putative / 40S ribosomal protein S27A (RPS27aA) | 2.26 |
| 220 | 263015_at | AT1G23440 | pyrrolidone-carboxylate peptidase family protein | -1.89 |
| 221 | 265179_at | AT1G23650 | unknown protein | 1.75 |
| 222 | 265171_at | AT1G23790 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G70340.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO40888.1); contains InterPro domain Protein of unknown function DUF936, plant (InterPro:IPR010341) | 2.10 |
| 223 | 263034_at | AT1G24020 | MLP423 (MLP-LIKE PROTEIN 423) | 1.70 |
| 224 | 263031_at | AT1G24070 | ATCSLA10 (Cellulose synthase-like A10); transferase, transferring glycosyl groups | 2.25 |
| 225 | 264868_at | AT1G24090 | RNase H domain-containing protein | 1.71 |
| 226 | 264866_at | AT1G24140 | matrixin family protein | -3.86 |
| 227 | 264872_at | AT1G24260 | SEP3 (SEPALLATA3); transcription factor | 3.18 |
| 228 | 265002_at | AT1G24400 | LHT2 (LYSINE HISTIDINE TRANSPORTER 2); amino acid transmembrane transporter | -1.92 |
| 229 | 265028_at | AT1G24530 | transducin family protein / WD-40 repeat family protein | -2.10 |
| 230 | 265025_at | AT1G24575 | unknown protein | -2.47 |
| 231 | 265024_at | AT1G24600 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G67920.1) | -3.29 |
| 232 | 255733_at | AT1G25400 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G68440.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO42150.1) | -2.42 |
| 233 | 255727_at | AT1G25510 | aspartyl protease family protein | 2.58 |
| 234 | 255734_at | AT1G25550 | myb family transcription factor | -2.44 |
| 235 | 245878_at | AT1G26190 | phosphoribulokinase/uridine kinase family protein | 2.14 |
| 236 | 245873_at | AT1G26260 | basic helix-loop-helix (bHLH) family protein | 1.67 |
| 237 | 245819_at | AT1G26310 | CAL (CAULIFLOWER); DNA binding / transcription factor | 2.10 |
| 238 | 245869_at | AT1G26330 | similar to unnamed protein product [Vitis vinifera] (GB:CAO41839.1); contains InterPro domain WD40 repeat-like (InterPro:IPR011046); contains InterPro domain WD40/YVTN repeat-like (InterPro:IPR015943) | 1.45 |
| 239 | 261004_at | AT1G26450 | beta-1,3-glucanase-related | 1.82 |
| 240 | 261000_at | AT1G26540 | agenet domain-containing protein | 2.55 |
| 241 | 261260_at | AT1G26680 | transcriptional factor B3 family protein | 2.94 |
| 242 | 261268_at | AT1G26740 | structural constituent of ribosome | 1.64 |
| 243 | 261262_at | AT1G26760 | binding | 1.63 |
| 244 | 261274_at | AT1G26780 | MYB117 (myb domain protein 117); transcription factor | 2.11 |
| 245 | 263681_at | AT1G26840 | ATORC6/ORC6 (Origin recognition complex protein 6); DNA binding | 2.17 |
| 246 | 263688_at | AT1G26920 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G69760.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO48418.1) | -2.67 |
| 247 | 264445_at | AT1G27290 | similar to unknown [Populus trichocarpa] (GB:ABK95086.1) | -2.29 |
| 248 | 264444_at | AT1G27360 | squamosa promoter-binding protein-like 11 (SPL11) | 1.64 |
| 249 | 264489_at | AT1G27370 | squamosa promoter-binding protein-like 10 (SPL10) | 2.35 |
| 250 | 261643_at | AT1G27720 | TAF4b (TBP-ASSOCIATED FACTOR 4b); transcription initiation factor | 2.02 |
| 251 | 261648_at | AT1G27730 | STZ (SALT TOLERANCE ZINC FINGER); nucleic acid binding / transcription factor/ zinc ion binding | -3.84 |
| 252 | 245762_at | AT1G27880 | ATP-dependent DNA helicase, putative | 1.91 |
| 253 | 245688_at | AT1G28290 | AGP31 (ARABINOGALACTAN-PROTEIN 31); structural constituent of cell wall | 2.16 |
| 254 | 261496_at | AT1G28360 | ATERF12/ERF12 (ERF domain protein 12); DNA binding / transcription factor/ transcription repressor | 2.60 |
| 255 | 262745_at | AT1G28600 | lipase, putative | -2.94 |
| 256 | 260840_at | AT1G29050 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G34070.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO48076.1); contains InterPro domain Protein of unknown function DUF231, plant (InterPro:IPR004253) | -1.84 |
| 257 | 260883_at | AT1G29270 | similar to transcription regulator [Arabidopsis thaliana] (TAIR:AT2G40435.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN79056.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO48025.1) | 2.25 |
| 258 | 259789_at | AT1G29395 | COR414-TM1 (cold regulated 414 thylakoid membrane 1) | -2.69 |
| 259 | 259784_at | AT1G29450 | auxin-responsive protein, putative | -5.46 |
| 260 | 259783_at | AT1G29510 | SAUR68 (SMALL AUXIN UPREGULATED 68) | -1.95 |
| 261 | 259780_at | AT1G29630 | Identical to Exonuclease 1 (EXO1) [Arabidopsis Thaliana] (GB:Q8L6Z7;GB:Q9C7N8); similar to exonuclease, putative [Arabidopsis thaliana] (TAIR:AT1G18090.2); similar to exonuclease, putative [Arabidopsis thaliana] (TAIR:AT1G18090.1); similar to Exonuclease 1 (OsEXO-1) (GB:Q60GC1); contains InterPro domain XPG N-terminal; (InterPro:IPR006085); contains InterPro domain XPG I; (InterPro:IPR006086); contains InterPro domain Helix-hairpin-helix motif, class 2; (InterPro:IPR008918); contains InterPro domain DNA repair protein (XPGC)/yeast Rad; (InterPro:IPR006084) | 1.45 |
| 262 | 259786_at | AT1G29660 | GDSL-motif lipase/hydrolase family protein | -5.44 |
| 263 | 259788_at | AT1G29670 | GDSL-motif lipase/hydrolase family protein | -1.98 |
| 264 | 246633_at | AT1G29720 | protein kinase family protein | -5.61 |
| 265 | 260028_at | AT1G29980 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G34510.1); similar to unknown [Populus trichocarpa] (GB:ABK95079.1); contains InterPro domain Protein of unknown function DUF642 (InterPro:IPR006946); contains InterPro domain Galactose-binding like (InterPro:IPR008979) | 1.63 |
| 266 | 260024_at | AT1G30080 | glycosyl hydrolase family 17 protein | 3.31 |
| 267 | 245776_at | AT1G30260 | AGL79 (AGAMOUS-LIKE 79) | -2.43 |
| 268 | 256311_at | AT1G30330 | ARF6 (AUXIN RESPONSE FACTOR 6); transcription factor | 1.95 |
| 269 | 261800_at | AT1G30490 | PHV (PHAVOLUTA); DNA binding / transcription factor | 1.57 |
| 270 | 261804_at | AT1G30530 | UDP-glucoronosyl/UDP-glucosyl transferase family protein | 1.55 |
| 271 | 263219_at | AT1G30600 | subtilase family protein | 1.57 |
| 272 | 263225_at | AT1G30650 | WRKY14 (WRKY DNA-binding protein 14); transcription factor | 1.50 |
| 273 | 264529_at | AT1G30820 | CTP synthase, putative / UTP--ammonia ligase, putative | 1.80 |
| 274 | 265161_at | AT1G30900 | vacuolar sorting receptor, putative | -2.30 |
| 275 | 265153_at | AT1G30950 | UFO (UNUSUAL FLORAL ORGANS); ubiquitin-protein ligase | 4.07 |
| 276 | 263700_at | AT1G31150 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G36970.1); similar to hypothetical protein 23.t00036 [Brassica oleracea] (GB:ABD65624.1); contains InterPro domain Region of unknown function DUF1985 (InterPro:IPR015410) | 1.47 |
| 277 | 262550_at | AT1G31310 | hydroxyproline-rich glycoprotein family protein | 3.31 |
| 278 | 257467_at | AT1G31320 | LBD4 (LOB DOMAIN-CONTAINING PROTEIN 4) | 2.35 |
| 279 | 256487_at | AT1G31540 | disease resistance protein (TIR-NBS-LRR class), putative | -2.89 |
| 280 | 246601_at | AT1G31710 | copper amine oxidase, putative | -3.98 |
| 281 | 246581_at | AT1G31760 | SWIB complex BAF60b domain-containing protein | 3.11 |
| 282 | 255720_at | AT1G32060 | PRK (PHOSPHORIBULOKINASE); ATP binding / phosphoribulokinase/ protein binding | -2.31 |
| 283 | 255719_at | AT1G32080 | membrane protein, putative | -2.55 |
| 284 | 245792_at | AT1G32100 | pinoresinol-lariciresinol reductase, putative | -2.22 |
| 285 | 245794_at | AT1G32170 | XTR4 (XYLOGLUCAN ENDOTRANSGLYCOSYLASE 4); hydrolase, acting on glycosyl bonds | -2.92 |
| 286 | 245784_at | AT1G32190 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G24320.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO63335.1); contains InterPro domain Alpha/beta hydrolase fold-1 (InterPro:IPR000073) | 3.35 |
| 287 | 245758_at | AT1G32240 | KAN2 (KANADI 2); DNA binding / transcription factor | 1.74 |
| 288 | 260693_at | AT1G32450 | proton-dependent oligopeptide transport (POT) family protein | -3.87 |
| 289 | 261711_at | AT1G32700 | zinc-binding family protein | -2.12 |
| 290 | 261239_at | AT1G32930 | galactosyltransferase family protein | 1.86 |
| 291 | 261594_at | AT1G33240 | AT-GTL1 (Arabidopsis thaliana GT2-like 1); transcription factor | -1.95 |
| 292 | 256426_at | AT1G33420 | PHD finger family protein | 1.71 |
| 293 | 256421_at | AT1G33500 | similar to kinase interacting family protein [Arabidopsis thaliana] (TAIR:AT3G22790.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO15598.1); similar to Os08g0320300 [Oryza sativa (japonica cultivar-group)] (GB:NP_001061527.1) | 1.72 |
| 294 | 245765_at | AT1G33600 | leucine-rich repeat family protein | -2.63 |
| 295 | 261991_at | AT1G33700 | catalytic | -1.94 |
| 296 | 262001_at | AT1G33790 | jacalin lectin family protein | 2.71 |
| 297 | 261981_at | AT1G33811 | GDSL-motif lipase/hydrolase family protein | -1.85 |
| 298 | 255982_at | AT1G34000 | OHP2 (ONE-HELIX PROTEIN 2) | -2.00 |
| 299 | 262543_at | AT1G34245 | similar to EPF1 (EPIDERMAL PATTERNING FACTOR 1) [Arabidopsis thaliana] (TAIR:AT2G20875.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO64636.1); contains domain PROKAR_LIPOPROTEIN (PS51257) | 1.95 |
| 300 | 259930_at | AT1G34355 | forkhead-associated domain-containing protein / FHA domain-containing protein | 2.25 |
| 301 | 262410_at | AT1G34770 | MAGE-8 antigen-related | 1.54 |
| 302 | 245757_at | AT1G35140 | PHI-1 (PHOSPHATE-INDUCED 1) | -2.46 |
| 303 | 245756_at | AT1G35190 | oxidoreductase, 2OG-Fe(II) oxygenase family protein | -2.34 |
| 304 | 259550_at | AT1G35230 | AGP5 (ARABINOGALACTAN-PROTEIN 5) | -2.92 |
| 305 | 259549_at | AT1G35290 | thioesterase family protein | 1.67 |
| 306 | 259546_at | AT1G35350 | similar to SHB1 (SHORT HYPOCOTYL UNDER BLUE1) [Arabidopsis thaliana] (TAIR:AT4G25350.1); similar to EXS family protein / ERD1/XPR1/SYG1 family protein [Arabidopsis thaliana] (TAIR:AT1G26730.1); similar to ATP binding / ATPase, coupled to transmembrane movement of substances [Arabidopsis thaliana] (TAIR:AT1G14040.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO68453.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO68461.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN73334.1); contains InterPro domain SPX, N-terminal (InterPro:IPR004331); contains InterPro domain EXS, C-terminal; (InterPro:IPR004342) | -6.57 |
| 307 | 261285_at | AT1G35720 | ANNAT1 (ANNEXIN ARABIDOPSIS 1); calcium ion binding / calcium-dependent phospholipid binding | -3.13 |
| 308 | 256459_at | AT1G36180 | ATP binding / biotin binding / catalytic/ ligase | 1.64 |
| 309 | 261979_at | AT1G37130 | NIA2 (NITRATE REDUCTASE 2) | -3.85 |
| 310 | 262033_at | AT1G37140 | MCT1 (mei2 C-Terminal RRM only like 1); RNA binding | 3.45 |
| 311 | 261363_at | AT1G41830 | SKS6 (SKU5 Similar 6); pectinesterase | -1.88 |
| 312 | 259625_at | AT1G42970 | GAPB (GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE B SUBUNIT); glyceraldehyde-3-phosphate dehydrogenase | -2.33 |
| 313 | 260869_at | AT1G43800 | acyl-(acyl-carrier-protein) desaturase, putative / stearoyl-ACP desaturase, putative | 4.89 |
| 314 | 245740_at | AT1G44100 | AAP5 (amino acid permease 5); amino acid transmembrane transporter | -2.52 |
| 315 | 245739_at | AT1G44110 | CYCA1;1 (CYCLIN A1;1); cyclin-dependent protein kinase regulator | 2.63 |
| 316 | 261323_at | AT1G44760 | universal stress protein (USP) family protein | 2.91 |
| 317 | 261325_at | AT1G44780 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G08310.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO61850.1) | 1.73 |
| 318 | 261335_at | AT1G44800 | nodulin MtN21 family protein | -4.57 |
| 319 | 261330_at | AT1G44900 | ATP binding / DNA binding / DNA-dependent ATPase | 2.59 |
| 320 | 260941_at | AT1G44970 | peroxidase, putative | 2.10 |
| 321 | 260943_at | AT1G45145 | ATTRX5 (thioredoxin H-type 5); thiol-disulfide exchange intermediate | -2.67 |
| 322 | 245807_at | AT1G46768 | RAP2.1 (related to AP2 1); DNA binding / transcription factor | -3.62 |
| 323 | 261681_at | AT1G47340 | F-box family protein | 1.55 |
| 324 | 261684_at | AT1G47400 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G47395.1) | -2.08 |
| 325 | 262443_at | AT1G47655 | Dof-type zinc finger domain-containing protein | 1.59 |
| 326 | 259616_at | AT1G47960 | C/VIF1 (CELL WALL / VACUOLAR INHIBITOR OF FRUCTOSIDASE 1); pectinesterase inhibitor | -3.14 |
| 327 | 259617_at | AT1G47970 | unknown protein | 1.68 |
| 328 | 260727_at | AT1G48100 | glycoside hydrolase family 28 protein / polygalacturonase (pectinase) family protein | 2.28 |
| 329 | 262236_at | AT1G48330 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G17580.1); similar to unknown [Populus trichocarpa] (GB:ABK96144.1) | 1.70 |
| 330 | 261300_at | AT1G48560 | unknown protein | 1.48 |
| 331 | 261306_at | AT1G48610 | AT hook motif-containing protein | 2.12 |
| 332 | 260748_at | AT1G49210 | zinc finger (C3HC4-type RING finger) family protein | -2.90 |
| 333 | 260753_at | AT1G49230 | zinc finger (C3HC4-type RING finger) family protein | -1.96 |
| 334 | 262396_at | AT1G49470 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G55230.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN63047.1); contains InterPro domain Protein of unknown function DUF716 (InterPro:IPR006904) | -2.93 |
| 335 | 262399_at | AT1G49500 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G19030.1) | -4.45 |
| 336 | 259815_at | AT1G49870 | similar to MBD10 (methyl-CpG-binding domain 10), DNA binding [Arabidopsis thaliana] (TAIR:AT1G15340.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO63952.1) | 2.79 |
| 337 | 261631_at | AT1G49940 | similar to male sterility MS5 family protein [Arabidopsis thaliana] (TAIR:AT1G04770.1) | 1.87 |
| 338 | 261635_at | AT1G50020 | similar to unnamed protein product [Vitis vinifera] (GB:CAO49863.1) | -1.87 |
| 339 | 261636_at | AT1G50110 | branched-chain amino acid aminotransferase 6 / branched-chain amino acid transaminase 6 (BCAT6) | 1.96 |
| 340 | 262467_at | AT1G50240 | armadillo/beta-catenin repeat family protein | 2.47 |
| 341 | 261859_at | AT1G50490 | UBC20 (UBIQUITIN-CONJUGATING ENZYME 20); ubiquitin-protein ligase | 1.80 |
| 342 | 245750_at | AT1G51060 | HTA10; DNA binding | 1.57 |
| 343 | 245749_at | AT1G51090 | heavy-metal-associated domain-containing protein | -3.84 |
| 344 | 265147_at | AT1G51380 | eukaryotic translation initiation factor 4A, putative / eIF-4A, putative | 2.03 |
| 345 | 260515_at | AT1G51460 | ABC transporter family protein | 2.74 |
| 346 | 256175_at | AT1G51670 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G48180.1); contains domain Cysteine proteinases (SSF54001) | 2.60 |
| 347 | 256186_at | AT1G51680 | 4CL1 (4-COUMARATE:COA LIGASE 1); 4-coumarate-CoA ligase | -2.19 |
| 348 | 256168_at | AT1G51805 | leucine-rich repeat protein kinase, putative | -3.31 |
| 349 | 259841_at | AT1G52200 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G18470.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO45876.1); contains InterPro domain Protein of unknown function Cys-rich (InterPro:IPR006461) | -1.86 |
| 350 | 262129_at | AT1G52500 | ATFPG-1/ATFPG-2/ATMMH-1/ATMMH-2/FPG-1/FPG-2 (FORMAMIDOPYRIMIDINE-DNA GLYCOSYLASE 1, FORMAMIDOPYRIMIDINE-DNA GLYCOSYLASE 2); DNA N-glycosylase | 1.57 |
| 351 | 262151_at | AT1G52510 | hydrolase, alpha/beta fold family protein | -1.92 |
| 352 | 262160_at | AT1G52590 | Identical to DCC family protein At1g52590, chloroplast precursor [Arabidopsis Thaliana] (GB:Q9SSR1;GB:Q8L9T1); similar to RNase H domain-containing protein [Arabidopsis thaliana] (TAIR:AT1G24090.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO62758.1); contains InterPro domain Putative thiol-disulphide oxidoreductase DCC (InterPro:IPR007263) | -2.40 |
| 353 | 262156_at | AT1G52680 | late embryogenesis abundant protein-related / LEA protein-related | -3.87 |
| 354 | 260155_at | AT1G52870 | peroxisomal membrane protein-related | -2.62 |
| 355 | 260156_at | AT1G52880 | NAM (Arabidopsis NAC domain containing protein 18); transcription factor | -2.71 |
| 356 | 260157_at | AT1G52930 | brix domain-containing protein | 1.48 |
| 357 | 261368_at | AT1G53070 | legume lectin family protein | 2.38 |
| 358 | 261364_at | AT1G53140 | dynamin family protein | 2.84 |
| 359 | 261375_at | AT1G53160 | SPL4 (SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 4); DNA binding / transcription factor | 4.09 |
| 360 | 260617_at | AT1G53345 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G09580.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO63241.1); contains domain DHH phosphoesterases (SSF64182) | 1.48 |
| 361 | 260983_at | AT1G53560 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G17080.1); similar to unknown [Populus trichocarpa] (GB:ABK95219.1); contains domain PTHR10052 (PTHR10052); contains domain PTHR10052:SF2 (PTHR10052:SF2) | -3.77 |
| 362 | 259965_at | AT1G53670 | MSRB1 (METHIONINE SULFOXIDE REDUCTASE B 1); peptide-methionine-(S)-S-oxide reductase | -2.48 |
| 363 | 262257_at | AT1G53860 | remorin family protein | 2.67 |
| 364 | 263161_at | AT1G54020 | myrosinase-associated protein, putative | 3.78 |
| 365 | 262998_at | AT1G54280 | haloacid dehalogenase-like hydrolase family protein | -2.04 |
| 366 | 264188_at | AT1G54690 | G-H2AX/GAMMA-H2AX/H2AXB/HTA3; DNA binding | 1.50 |
| 367 | 264238_at | AT1G54740 | similar to structural constituent of ribosome [Arabidopsis thaliana] (TAIR:AT1G22110.1); similar to unknown [Populus trichocarpa x Populus deltoides] (GB:ABK96485.1) | -2.29 |
| 368 | 264241_at | AT1G54840 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G54850.1); similar to heat shock protein-like protein [Cucumis melo] (GB:AAO45755.1); contains InterPro domain HSP20-like chaperone (InterPro:IPR008978); contains InterPro domain Heat shock protein Hsp20 (InterPro:IPR002068) | 1.71 |
| 369 | 259656_at | AT1G55200 | protein kinase family protein | 3.25 |
| 370 | 265075_at | AT1G55450 | embryo-abundant protein-related | -3.15 |
| 371 | 260602_at | AT1G55920 | AtSerat2;1 (SERINE ACETYLTRANSFERASE 1) | -2.67 |
| 372 | 262096_at | AT1G56010 | NAC1 (Arabidopsis NAC domain containing protein 21, Arabidopsis NAC domain containing protein 22); transcription factor | -3.90 |
| 373 | 262088_at | AT1G56020 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G12970.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN69700.1) | 2.85 |
| 374 | 262094_at | AT1G56110 | NOP56 (ARABIDOPSIS HOMOLOG OF NUCLEOLAR PROTEIN NOP56) | 1.89 |
| 375 | 262098_at | AT1G56170 | HAP5B (Heme activator protein (yeast) homolog 5B); DNA binding / transcription factor | 1.58 |
| 376 | 256222_at | AT1G56210 | copper chaperone (CCH)-related | 2.16 |
| 377 | 259632_at | AT1G56430 | nicotianamine synthase, putative | -1.89 |
| 378 | 246405_at | AT1G57630 | disease resistance protein (TIR class), putative | -3.71 |
| 379 | 246380_at | AT1G57750 | CYP96A15/MAH1 (MID-CHAIN ALKANE HYDROXYLASE 1); oxygen binding | 3.76 |
| 380 | 245828_at | AT1G57820 | ORTH2/VIM1 (VARIANT IN METHYLATION 1); DNA binding / chromatin binding / double-stranded methylated DNA binding / histone binding / methyl-CpG binding / methyl-CpNpG binding / methyl-CpNpN binding | 3.11 |
| 381 | 245842_at | AT1G58430 | RXF26; carboxylesterase | 5.47 |
| 382 | 262081_at | AT1G59540 | ZCF125; microtubule motor | 2.47 |
| 383 | 262072_at | AT1G59590 | ZCF37 | -5.28 |
| 384 | 262073_at | AT1G59640 | ZCW32 (BIGPETAL, BIGPETALUB); DNA binding / transcription factor | 1.49 |
| 385 | 262124_at | AT1G59660 | nucleoporin family protein | 1.62 |
| 386 | 262912_at | AT1G59740 | proton-dependent oligopeptide transport (POT) family protein | -2.00 |
| 387 | 262899_at | AT1G59870 | PDR8/PEN3 (PLEIOTROPIC DRUG RESISTANCE8); ATPase, coupled to transmembrane movement of substances / cadmium ion transmembrane transporter | -3.03 |
| 388 | 263738_at | AT1G60060 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G53900.2); similar to unnamed protein product [Vitis vinifera] (GB:CAO40921.1); contains domain PTHR13902:SF3 (PTHR13902:SF3); contains domain PTHR13902 (PTHR13902) | 1.79 |
| 389 | 264247_at | AT1G60160 | potassium transporter family protein | 1.81 |
| 390 | 264919_at | AT1G60540 | pseudogene of the dynamin family | 2.45 |
| 391 | 264939_at | AT1G60630 | leucine-rich repeat family protein | 2.08 |
| 392 | 264929_at | AT1G60730 | aldo/keto reductase family protein | -4.87 |
| 393 | 259907_at | AT1G60860 | ARF GTPase-activating domain-containing protein | 2.03 |
| 394 | 259721_at | AT1G60890 | phosphatidylinositol-4-phosphate 5-kinase family protein | 1.65 |
| 395 | 264937_at | AT1G61130 | SCPL32; serine carboxypeptidase | 1.87 |
| 396 | 264763_at | AT1G61450 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G61415.1); similar to Os05g0275600 [Oryza sativa (japonica cultivar-group)] (GB:NP_001055074.1) | 2.42 |
| 397 | 264433_at | AT1G61810 | BGLU45; hydrolase, hydrolyzing O-glycosyl compounds | 1.91 |
| 398 | 264286_at | AT1G61870 | PPR336 (PENTATRICOPEPTIDE REPEAT 336) | 1.66 |
| 399 | 264287_at | AT1G61930 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G11700.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO17934.1); contains InterPro domain Protein of unknown function DUF584 (InterPro:IPR007608) | 1.52 |
| 400 | 264741_at | AT1G62290 | aspartyl protease family protein | -3.15 |
| 401 | 264746_at | AT1G62300 | WRKY6 (WRKY DNA-binding protein 6); transcription factor | -5.78 |
| 402 | 260632_at | AT1G62360 | STM (SHOOT MERISTEMLESS); transcription factor | 5.33 |
| 403 | 265117_at | AT1G62500 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | 5.00 |
| 404 | 265111_at | AT1G62510 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | -4.57 |
| 405 | 265118_at | AT1G62660 | beta-fructosidase (BFRUCT3) / beta-fructofuranosidase / invertase, vacuolar | -2.86 |
| 406 | 262687_at | AT1G62670 | pentatricopeptide (PPR) repeat-containing protein | 1.86 |
| 407 | 262642_at | AT1G62690 | unknown protein | 1.68 |
| 408 | 262644_at | AT1G62710 | BETA-VPE (vacuolar processing enzyme beta); cysteine-type endopeptidase | -1.99 |
| 409 | 262646_at | AT1G62800 | ASP4 (ASPARTATE AMINOTRANSFERASE 4); catalytic/ pyridoxal phosphate binding / transaminase/ transferase, transferring nitrogenous groups | -2.50 |
| 410 | 257466_at | AT1G62840 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G12320.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO24498.1); contains InterPro domain Protein of unknown function DUF1442 (InterPro:IPR009902) | -3.16 |
| 411 | 261123_at | AT1G62860 | pseudogene of pentatricopeptide (PPR) repeat-containing protein | 2.15 |
| 412 | 261100_at | AT1G63020 | NRPD1a (NUCLEAR RNA POLYMERASE D 1A); DNA binding / DNA-directed RNA polymerase | 2.12 |
| 413 | 259686_at | AT1G63100 | scarecrow transcription factor family protein | 2.22 |
| 414 | 259690_at | AT1G63160 | replication factor C 40 kDa, putative | 1.54 |
| 415 | 261549_at | AT1G63470 | DNA-binding family protein | 2.36 |
| 416 | 261548_at | AT1G63480 | DNA-binding family protein | 2.63 |
| 417 | 260242_at | AT1G63650 | EGL3 (ENHANCER OF GLABRA3); DNA binding / transcription factor | 2.87 |
| 418 | 260241_at | AT1G63710 | CYP86A7 (cytochrome P450, family 86, subfamily A, polypeptide 7); oxygen binding | 2.40 |
| 419 | 260296_at | AT1G63750 | ATP binding / nucleoside-triphosphatase/ nucleotide binding / protein binding | -3.67 |
| 420 | 262347_at | AT1G64110 | AAA-type ATPase family protein | -1.87 |
| 421 | 262337_at | AT1G64260 | zinc finger protein-related | 1.57 |
| 422 | 259765_at | AT1G64370 | unknown protein | -6.96 |
| 423 | 259737_at | AT1G64400 | long-chain-fatty-acid--CoA ligase, putative / long-chain acyl-CoA synthetase, putative | -2.27 |
| 424 | 262002_at | AT1G64450 | proline-rich family protein | 2.05 |
| 425 | 261958_at | AT1G64500 | glutaredoxin family protein | -1.91 |
| 426 | 261957_at | AT1G64660 | ATMGL; catalytic/ methionine gamma-lyase | -1.88 |
| 427 | 264157_at | AT1G65310 | ATXTH17 (XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 17); hydrolase, acting on glycosyl bonds | -3.31 |
| 428 | 264166_at | AT1G65370 | meprin and TRAF homology domain-containing protein / MATH domain-containing protein | 1.86 |
| 429 | 264153_at | AT1G65390 | ATPP2-A5; carbohydrate binding | -2.57 |
| 430 | 264163_at | AT1G65445 | transferase-related | -2.13 |
| 431 | 264636_at | AT1G65490 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G65486.1) | -3.98 |
| 432 | 264635_at | AT1G65500 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G65486.1) | -1.93 |
| 433 | 262930_at | AT1G65690 | harpin-induced protein-related / HIN1-related / harpin-responsive protein-related | -3.52 |
| 434 | 261913_at | AT1G65860 | flavin-containing monooxygenase family protein / FMO family protein | -3.49 |
| 435 | 261921_at | AT1G65900 | similar to unnamed protein product [Vitis vinifera] (GB:CAO22185.1); contains domain Helical backbone metal receptor (SSF53807) | 1.67 |
| 436 | 256522_at | AT1G66160 | U-box domain-containing protein | -2.94 |
| 437 | 256524_at | AT1G66200 | ATGSR2 (Arabidopsis thaliana glutamine synthase clone R2); glutamate-ammonia ligase | -2.29 |
| 438 | 259822_at | AT1G66230 | MYB20 (myb domain protein 20); DNA binding / transcription factor | -3.77 |
| 439 | 259823_at | AT1G66250 | glycosyl hydrolase family 17 protein | 2.35 |
| 440 | 260135_at | AT1G66400 | calmodulin-related protein, putative | -2.77 |
| 441 | 256359_at | AT1G66460 | protein kinase family protein | -2.98 |
| 442 | 256378_at | AT1G66830 | leucine-rich repeat transmembrane protein kinase, putative | -2.97 |
| 443 | 255852_at | AT1G66970 | glycerophosphoryl diester phosphodiesterase family protein | -2.25 |
| 444 | 255851_at | AT1G67040 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G26910.2); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G26910.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO49345.1) | 3.56 |
| 445 | 255854_at | AT1G67050 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G38320.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G38320.2); similar to At1g67050-like protein [Arabidopsis lyrata subsp. petraea] (GB:ABY59212.1) | -2.00 |
| 446 | 264969_at | AT1G67320 | DNA primase, large subunit family | 2.08 |
| 447 | 264998_at | AT1G67330 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G27930.1); similar to unknown [Populus trichocarpa] (GB:ABK93495.1); contains InterPro domain Protein of unknown function DUF579, plant (InterPro:IPR006514) | -3.00 |
| 448 | 260192_at | AT1G67630 | POLA2; alpha DNA polymerase | 1.67 |
| 449 | 245195_at | AT1G67740 | PSBY (photosystem II BY) | -1.89 |
| 450 | 245196_at | AT1G67750 | pectate lyase family protein | 1.47 |
| 451 | 245191_at | AT1G67770 | TEL2 (TERMINAL EAR1-LIKE 2); RNA binding | 2.06 |
| 452 | 245192_at | AT1G67780 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G67270.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO68021.1); contains InterPro domain DDT (InterPro:IPR004022) | 2.06 |
| 453 | 245193_at | AT1G67810 | Fe-S metabolism associated domain-containing protein | -3.62 |
| 454 | 245215_at | AT1G67830 | ATFXG1 (ALPHA-FUCOSIDASE 1); alpha-L-fucosidase/ carboxylesterase | 1.86 |
| 455 | 260004_at | AT1G67860 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G67865.1) | -2.80 |
| 456 | 260012_at | AT1G67865 | unknown protein | -1.90 |
| 457 | 260007_at | AT1G67870 | glycine-rich protein | -3.33 |
| 458 | 260014_at | AT1G68010 | HPR (HYDROXYPYRUVATE REDUCTASE); glycerate dehydrogenase/ poly(U) binding | -2.22 |
| 459 | 259998_at | AT1G68120 | ATBPC3/BBR/BPC3/BPC3 (BASIC PENTACYSTEINE 3); DNA binding / transcription factor | 2.16 |
| 460 | 260433_at | AT1G68170 | nodulin MtN21 family protein | 3.58 |
| 461 | 260431_at | AT1G68190 | zinc finger (B-box type) family protein | -2.25 |
| 462 | 260484_at | AT1G68360 | zinc finger protein-related | 2.92 |
| 463 | 259858_at | AT1G68400 | leucine-rich repeat transmembrane protein kinase, putative | 2.10 |
| 464 | 260263_at | AT1G68480 | JAG (JAGGED); nucleic acid binding / zinc ion binding | 2.23 |
| 465 | 260264_at | AT1G68500 | similar to hypothetical protein [Vitis vinifera] (GB:CAN66643.1) | -2.05 |
| 466 | 260266_at | AT1G68520 | zinc finger (B-box type) family protein | -3.21 |
| 467 | 262229_at | AT1G68620 | hydrolase | -3.60 |
| 468 | 262278_at | AT1G68640 | PAN (PERIANTHIA); DNA binding / transcription factor | 4.94 |
| 469 | 260041_at | AT1G68780 | leucine-rich repeat family protein | 1.51 |
| 470 | 260037_at | AT1G68840 | RAV2 (REGULATOR OF THE ATPASE OF THE VACUOLAR MEMBRANE); DNA binding / transcription factor | -4.50 |
| 471 | 257516_at | AT1G69040 | ACR4 (ACT REPEAT 4); amino acid binding | 1.52 |
| 472 | 259371_at | AT1G69080 | universal stress protein (USP) family protein | -2.97 |
| 473 | 259372_at | AT1G69120 | AP1 (APETALA1); DNA binding / transcription factor | 6.06 |
| 474 | 260355_at | AT1G69180 | CRC (CRABS CLAW); transcription factor | 8.26 |
| 475 | 260349_at | AT1G69400 | transducin family protein / WD-40 repeat family protein | 1.85 |
| 476 | 260347_at | AT1G69420 | zinc finger (DHHC type) family protein | 1.47 |
| 477 | 256293_at | AT1G69440 | AGO7 (ARGONAUTE7); nucleic acid binding | 1.69 |
| 478 | 259834_at | AT1G69570 | Dof-type zinc finger domain-containing protein | -2.52 |
| 479 | 259831_at | AT1G69600 | ATHB29/ZFHD1 (ZINC FINGER HOMEODOMAIN 1); DNA binding / transcription activator/ transcription factor | 3.05 |
| 480 | 260417_at | AT1G69770 | CMT3 (CHROMOMETHYLASE 3) | 2.06 |
| 481 | 260414_at | AT1G69850 | ATNRT1:2 (NITRATE TRANSPORTER 1:2); calcium ion binding / transporter | -2.72 |
| 482 | 260407_at | AT1G69910 | protein kinase family protein | 1.68 |
| 483 | 264696_at | AT1G70230 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G01430.1); similar to Os09g0375300 [Oryza sativa (japonica cultivar-group)] (GB:NP_001063036.1); similar to Os06g0235200 [Oryza sativa (japonica cultivar-group)] (GB:NP_001057240.1); similar to leaf senescence protein-like [Oryza sativa (japonica cultivar-group)] (GB:BAD26032.1); contains InterPro domain Protein of unknown function DUF231, plant (InterPro:IPR004253) | -5.15 |
| 484 | 264313_at | AT1G70410 | carbonic anhydrase, putative / carbonate dehydratase, putative | -5.21 |
| 485 | 264314_at | AT1G70420 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G23710.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO66069.1); contains InterPro domain Protein of unknown function DUF1645 (InterPro:IPR012442) | -1.91 |
| 486 | 260332_at | AT1G70470 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G23530.1) | 1.73 |
| 487 | 260334_at | AT1G70510 | KNAT2 (KNOTTED-LIKE FROM ARABIDOPSIS THALIANA 2); transcription factor | 3.32 |
| 488 | 260364_at | AT1G70560 | alliinase C-terminal domain-containing protein | 2.17 |
| 489 | 260181_at | AT1G70710 | AtGH9B1 (ARABIDOPSIS THALIANA GLYCOSYL HYDROLASE 9B1); hydrolase, hydrolyzing O-glycosyl compounds | 2.93 |
| 490 | 262288_at | AT1G70760 | inorganic carbon transport protein-related | -2.58 |
| 491 | 262314_at | AT1G70810 | C2 domain-containing protein | -3.29 |
| 492 | 262309_at | AT1G70820 | phosphoglucomutase, putative / glucose phosphomutase, putative | -2.10 |
| 493 | 262304_at | AT1G70890 | MLP43 (MLP-LIKE PROTEIN 43) | -5.70 |
| 494 | 262263_at | AT1G70940 | PIN3 (PIN-FORMED 3); auxin:hydrogen symporter/ transporter | -2.88 |
| 495 | 262290_at | AT1G70985 | hydroxyproline-rich glycoprotein family protein | -2.32 |
| 496 | 259751_at | AT1G71030 | ATMYBL2 (Arabidopsis myb-like 2); DNA binding / transcription factor | -2.24 |
| 497 | 259943_at | AT1G71480 | nuclear transport factor 2 (NTF2) family protein | -1.95 |
| 498 | 261506_at | AT1G71697 | ATCK1 (CHOLINE KINASE) | -2.38 |
| 499 | 261510_at | AT1G71760 | similar to unnamed protein product [Vitis vinifera] (GB:CAO67526.1) | 1.88 |
| 500 | 261521_at | AT1G71830 | SERK1 (SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE 1); kinase | 1.45 |
| 501 | 257487_at | AT1G71850 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G24320.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO64799.1); contains InterPro domain Protein of unknown function DUF860, plant (InterPro:IPR008578) | 1.74 |
| 502 | 260144_at | AT1G71960 | ABC transporter family protein | -3.63 |
| 503 | 256337_at | AT1G72060 | serine-type endopeptidase inhibitor | -1.89 |
| 504 | 259803_at | AT1G72150 | PATL1 (PATELLIN 1); transporter | -2.46 |
| 505 | 259848_at | AT1G72180 | leucine-rich repeat transmembrane protein kinase, putative | -2.21 |
| 506 | 259851_at | AT1G72250 | kinesin motor protein-related | 2.56 |
| 507 | 259802_at | AT1G72260 | THI2.1 (THIONIN 2.1); toxin receptor binding | 4.88 |
| 508 | 260427_at | AT1G72430 | auxin-responsive protein-related | -3.00 |
| 509 | 260399_at | AT1G72520 | lipoxygenase, putative | -3.75 |
| 510 | 259912_at | AT1G72670 | IQD8 (IQ-domain 8); calmodulin binding | 3.85 |
| 511 | 259864_at | AT1G72800 | nuM1-related | -2.25 |
| 512 | 262381_at | AT1G72900 | disease resistance protein (TIR-NBS class), putative | -3.65 |
| 513 | 262382_at | AT1G72920 | disease resistance protein (TIR-NBS class), putative | -1.85 |
| 514 | 262376_at | AT1G72970 | HTH (HOTHEAD); aldehyde-lyase | 2.17 |
| 515 | 262356_at | AT1G73000 | similar to Bet v I allergen family protein [Arabidopsis thaliana] (TAIR:AT2G26040.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO43790.1); contains InterPro domain Streptomyces cyclase/dehydrase (InterPro:IPR005031) | -2.43 |
| 516 | 262357_at | AT1G73040 | jacalin lectin family protein | -2.01 |
| 517 | 260101_at | AT1G73260 | trypsin and protease inhibitor family protein / Kunitz family protein | -3.52 |
| 518 | 245734_at | AT1G73480 | hydrolase, alpha/beta fold family protein | -1.93 |
| 519 | 245731_at | AT1G73500 | ATMKK9 (Arabidopsis thaliana MAP kinase kinase 9); kinase | -2.52 |
| 520 | 245777_at | AT1G73540 | ATNUDT21 (Arabidopsis thaliana Nudix hydrolase homolog 21); hydrolase | -2.05 |
| 521 | 259845_at | AT1G73590 | PIN1 (PIN-FORMED 1); transporter | 2.89 |
| 522 | 260077_at | AT1G73620 | thaumatin-like protein, putative / pathogenesis-related protein, putative | 2.73 |
| 523 | 260076_at | AT1G73630 | calcium-binding protein, putative | -2.45 |
| 524 | 260061_at | AT1G73690 | AT;CDKD;1/CAK3AT/CDKD1;1 (CYCLIN-DEPENDENT KINASE D1;1); kinase | 1.64 |
| 525 | 260048_at | AT1G73750 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G15060.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO42199.1); contains InterPro domain Uncharacterised conserved protein UCP031088, alpha/beta hydrolase, At1g15070 (InterPro:IPR016969) | -1.94 |
| 526 | 260380_at | AT1G73870 | zinc finger (B-box type) family protein | -3.07 |
| 527 | 260335_at | AT1G74000 | SS3 (STRICTOSIDINE SYNTHASE 3) | -1.89 |
| 528 | 260386_at | AT1G74010 | strictosidine synthase family protein | -2.95 |
| 529 | 260391_at | AT1G74020 | SS2 (STRICTOSIDINE SYNTHASE 2); strictosidine synthase | -2.12 |
| 530 | 260389_at | AT1G74055 | unknown protein | 2.04 |
| 531 | 260385_at | AT1G74090 | sulfotransferase family protein | -2.18 |
| 532 | 260237_at | AT1G74430 | MYB95 (myb domain protein 95); DNA binding / transcription factor | 1.64 |
| 533 | 260226_at | AT1G74660 | MIF1 (MINI ZINC FINGER 1); DNA binding / transcription factor | -2.31 |
| 534 | 260221_at | AT1G74670 | gibberellin-responsive protein, putative | -3.29 |
| 535 | 262175_at | AT1G74880 | NDH-O (NAD(P)H:PLASTOQUINONE DEHYDROGENASE COMPLEX SUBUNIT O) | -2.43 |
| 536 | 259925_at | AT1G75040 | PR5 (PATHOGENESIS-RELATED GENE 5) | -4.51 |
| 537 | 259926_at | AT1G75090 | methyladenine glycosylase family protein | 2.95 |
| 538 | 256507_at | AT1G75150 | similar to unnamed protein product [Vitis vinifera] (GB:CAO61179.1) | 1.94 |
| 539 | 256452_at | AT1G75240 | ATHB33 (ARABIDOPSIS THALIANA HOMEOBOX PROTEIN 33); DNA binding / transcription factor | 2.73 |
| 540 | 256503_at | AT1G75250 | myb family transcription factor | -2.96 |
| 541 | 261118_at | AT1G75460 | ATP-dependent protease La (LON) domain-containing protein | -3.74 |
| 542 | 262976_at | AT1G75520 | SRS5 (SHI-RELATED SEQUENCE 5) | 1.80 |
| 543 | 257460_at | AT1G75580 | auxin-responsive protein, putative | 2.98 |
| 544 | 262971_at | AT1G75640 | leucine-rich repeat family protein / protein kinase family protein | 2.67 |
| 545 | 262970_at | AT1G75690 | chaperone protein dnaJ-related | -2.28 |
| 546 | 262947_at | AT1G75750 | GASA1 (GAST1 PROTEIN HOMOLOG 1) | -5.54 |
| 547 | 261751_at | AT1G76080 | ATCDSP32/CDSP32 (CHLOROPLASTIC DROUGHT-INDUCED STRESS PROTEIN OF 32 KD); thiol-disulfide exchange intermediate | -3.05 |
| 548 | 261749_at | AT1G76180 | ERD14 (EARLY RESPONSE TO DEHYDRATION 14) | -2.35 |
| 549 | 261780_at | AT1G76310 | CYCB2;4 (CYCLIN B2;4); cyclin-dependent protein kinase regulator | 2.49 |
| 550 | 259972_at | AT1G76420 | CUC3 (CUP SHAPED COTYLEDON3); transcription factor | 3.06 |
| 551 | 259980_at | AT1G76520 | auxin efflux carrier family protein | -2.09 |
| 552 | 259978_at | AT1G76540 | CDKB2;1 (CYCLIN-DEPENDENT KINASE B2;1); kinase | 1.76 |
| 553 | 259977_at | AT1G76590 | zinc-binding family protein | -2.41 |
| 554 | 259867_at | AT1G76740 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G76840.1); similar to cell wall-anchored protein [Staphylococcus saprophyticus subsp. saprophyticus ATCC 15305] (GB:YP_300225.1) | 1.67 |
| 555 | 259878_at | AT1G76790 | O-methyltransferase family 2 protein | 1.98 |
| 556 | 259871_at | AT1G76800 | nodulin, putative | -2.92 |
| 557 | 256332_at | AT1G76890 | GT2; transcription factor | -4.67 |
| 558 | 264962_at | AT1G77110 | PIN6 (PIN-FORMED 6); auxin:hydrogen symporter/ transporter | 2.19 |
| 559 | 264482_at | AT1G77210 | sugar transporter, putative | -3.27 |
| 560 | 264478_at | AT1G77270 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G07730.1); similar to predicted protein [Physcomitrella patens subsp. patens] (GB:EDQ75856.1); similar to Os04g0539100 [Oryza sativa (japonica cultivar-group)] (GB:NP_001053436.1); similar to Hypothetical protein CBG09238 [Caenorhabditis briggsae AF16] (GB:XP_001674299.1) | 1.81 |
| 561 | 259760_at | AT1G77580 | myosin heavy chain-related | 1.53 |
| 562 | 259763_at | AT1G77630 | peptidoglycan-binding LysM domain-containing protein | 1.78 |
| 563 | 259679_at | AT1G77720 | protein kinase family protein | 1.46 |
| 564 | 259681_at | AT1G77760 | NIA1 (NITRATE REDUCTASE 1) | -2.85 |
| 565 | 262164_at | AT1G78070 | WD-40 repeat family protein | -1.90 |
| 566 | 257502_at | AT1G78110 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G22230.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO61735.1) | -2.90 |
| 567 | 260056_at | AT1G78140 | methyltransferase-related | -2.22 |
| 568 | 260051_at | AT1G78210 | hydrolase, alpha/beta fold family protein | 1.57 |
| 569 | 260774_at | AT1G78290 | serine/threonine protein kinase, putative | -2.78 |
| 570 | 260801_at | AT1G78430 | Identical to Uncharacterized protein At1g78430 [Arabidopsis Thaliana] (GB:Q9M9F9); similar to tropomyosin-related [Arabidopsis thaliana] (TAIR:AT1G17140.1); similar to tropomyosin-related [Arabidopsis thaliana] (TAIR:AT1G17140.2); similar to hypothetical protein [Vitis vinifera] (GB:CAN73256.1); contains InterPro domain Prefoldin (InterPro:IPR009053) | 2.95 |
| 571 | 263133_at | AT1G78450 | SOUL heme-binding family protein | -3.35 |
| 572 | 263126_at | AT1G78460 | SOUL heme-binding family protein | -2.46 |
| 573 | 263130_at | AT1G78650 | POLD3 | 1.80 |
| 574 | 264293_at | AT1G78770 | cell division cycle family protein | 2.15 |
| 575 | 257427_at | AT1G79060 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G56020.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN69700.1) | 1.57 |
| 576 | 264118_at | AT1G79150 | binding | 1.48 |
| 577 | 264102_at | AT1G79270 | ECT8 (evolutionarily conserved C-terminal region 8) | -6.13 |
| 578 | 262921_at | AT1G79430 | APL (ALTERED PHLOEM DEVELOPMENT); transcription factor | -1.84 |
| 579 | 262945_at | AT1G79510 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G16320.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO64929.1); contains domain NTF2-like (SSF54427) | -2.50 |
| 580 | 261353_at | AT1G79600 | ABC1 family protein | -2.02 |
| 581 | 262040_at | AT1G80080 | TMM (TOO MANY MOUTHS); protein binding | 1.47 |
| 582 | 262041_at | AT1G80100 | AHP6 (ARABIDOPSIS HISTIDINE PHOSPHOTRANSFER PROTEIN 6); transferase, transferring phosphorus-containing groups | 4.34 |
| 583 | 262049_at | AT1G80180 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G15400.2); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G15400.3); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G15400.1); similar to hypothetical protein MtrDRAFT_AC148340g12v2 [Medicago truncatula] (GB:ABD28396.1) | -5.44 |
| 584 | 260331_at | AT1G80270 | DNA-binding protein, putative | 1.83 |
| 585 | 260297_at | AT1G80280 | hydrolase, alpha/beta fold family protein | 2.60 |
| 586 | 260329_at | AT1G80370 | CYCA2;4 (CYCLIN A2;4); cyclin-dependent protein kinase regulator | 2.79 |
| 587 | 260284_at | AT1G80380 | phosphoribulokinase/uridine kinase-related | -2.55 |
| 588 | 261881_at | AT1G80760 | NIP6;1 (NOD26-like intrinsic protein 6;1); water channel | -4.01 |
| 589 | 261899_at | AT1G80820 | CCR2 (CINNAMOYL COA REDUCTASE) | -2.84 |
| 590 | 261892_at | AT1G80840 | WRKY40 (WRKY DNA-binding protein 40); transcription factor | -2.13 |
| 591 | 265789_at | AT2G01210 | leucine-rich repeat transmembrane protein kinase, putative | 2.04 |
| 592 | 265732_at | AT2G01300 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G15010.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO42242.1) | -1.84 |
| 593 | 266300_at | AT2G01420 | PIN4 (PIN-FORMED 4); auxin:hydrogen symporter/ transporter | -2.92 |
| 594 | 266353_at | AT2G01520 | MLP328 (MLP-LIKE PROTEIN 328) | 3.90 |
| 595 | 265873_at | AT2G01630 | glycosyl hydrolase family 17 protein / beta-1,3-glucanase, putative | 1.65 |
| 596 | 265872_at | AT2G01670 | ATNUDT17 (Arabidopsis thaliana Nudix hydrolase homolog 17); hydrolase | -2.69 |
| 597 | 265864_at | AT2G01750 | ATMAP70-3 (microtubule-associated proteins 70-3); microtubule binding | 1.86 |
| 598 | 265253_at | AT2G02020 | proton-dependent oligopeptide transport (POT) family protein | -2.89 |
| 599 | 266141_at | AT2G02120 | LCR70/PDF2.1 (Low-molecular-weight cysteine-rich 70); protease inhibitor | 1.99 |
| 600 | 266175_at | AT2G02450 | ANAC034/ANAC035 (Arabidopsis NAC domain containing protein 34, Arabidopsis NAC domain containing protein 35); transcription factor | -1.84 |
| 601 | 267271_at | AT2G02540 | ATHB21/ZFHD4 (ZINC FINGER HOMEODOMAIN 4); DNA binding / transcription factor | 2.18 |
| 602 | 266745_at | AT2G02950 | PKS1 (PHYTOCHROME KINASE SUBSTRATE 1) | -3.15 |
| 603 | 266770_at | AT2G03090 | ATEXPA15 (ARABIDOPSIS THALIANA EXPANSIN A15) | 1.93 |
| 604 | 266730_at | AT2G03110 | nucleic acid binding | 2.45 |
| 605 | 266732_at | AT2G03240 | EXS family protein / ERD1/XPR1/SYG1 family protein | 1.72 |
| 606 | 266707_at | AT2G03310 | unknown protein | -4.44 |
| 607 | 265713_at | AT2G03530 | UPS2 (UREIDE PERMEASE 2) | -2.49 |
| 608 | 263410_at | AT2G04039 | similar to unnamed protein product [Vitis vinifera] (GB:CAO41070.1) | -2.23 |
| 609 | 263406_at | AT2G04160 | AIR3 (Auxin-Induced in Root cultures 3); subtilase | -2.71 |
| 610 | 263809_at | AT2G04570 | GDSL-motif lipase/hydrolase family protein | 2.46 |
| 611 | 263674_at | AT2G04790 | similar to unnamed protein product [Vitis vinifera] (GB:CAO23994.1) | -1.88 |
| 612 | 263632_at | AT2G04795 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G35732.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO24128.1) | -2.46 |
| 613 | 263106_at | AT2G05160 | zinc finger (CCCH-type) family protein / RNA recognition motif (RRM)-containing protein | -2.48 |
| 614 | 265531_at | AT2G06200 | AtGRF6 (GROWTH-REGULATING FACTOR 6) | 4.16 |
| 615 | 266427_at | AT2G07170 | binding | 2.81 |
| 616 | 266959_at | AT2G07690 | minichromosome maintenance family protein / MCM family protein | 2.36 |
| 617 | 263350_at | AT2G13360 | AGT (ALANINE:GLYOXYLATE AMINOTRANSFERASE) | -3.77 |
| 618 | 263295_at | AT2G14210 | ANR1; DNA binding / transcription factor | 1.96 |
| 619 | 265837_at | AT2G14560 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G33840.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO48825.1); contains InterPro domain Protein of unknown function DUF567 (InterPro:IPR007612) | -2.56 |
| 620 | 265892_at | AT2G15020 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G64190.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN67723.1) | -3.58 |
| 621 | 263565_at | AT2G15390 | FUT4 (fucosyltransferase 4); fucosyltransferase/ transferase, transferring glycosyl groups | -2.49 |
| 622 | 265499_at | AT2G15480 | UGT73B5 (UDP-GLUCOSYL TRANSFERASE 73B5); UDP-glycosyltransferase/ transferase, transferring glycosyl groups | -3.00 |
| 623 | 265472_at | AT2G15580 | zinc finger (C3HC4-type RING finger) family protein | -3.24 |
| 624 | 265475_at | AT2G15620 | NIR1 (NITRITE REDUCTASE); ferredoxin-nitrate reductase | -2.85 |
| 625 | 265539_at | AT2G15830 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G33960.1) | -1.95 |
| 626 | 263092_at | AT2G16210 | transcriptional factor B3 family protein | 4.73 |
| 627 | 263607_at | AT2G16270 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G16630.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN75064.1) | 3.20 |
| 628 | 263600_at | AT2G16390 | DRD1 (DEFECTIVE IN RNA-DIRECTED DNA METHYLATION 1); ATP binding / DNA binding / helicase/ nucleic acid binding | 1.58 |
| 629 | 263612_at | AT2G16440 | DNA replication licensing factor, putative | 2.43 |
| 630 | 263239_at | AT2G16570 | ATASE (GLN PHOSPHORIBOSYL PYROPHOSPHATE AMIDOTRANSFERASE 1); amidophosphoribosyltransferase | 2.23 |
| 631 | 263238_at | AT2G16580 | auxin-responsive protein, putative | 2.10 |
| 632 | 265357_at | AT2G16740 | UBC29 (UBIQUITIN-CONJUGATING ENZYME 29); ubiquitin-protein ligase | -2.29 |
| 633 | 265405_at | AT2G16750 | protein kinase family protein | -3.84 |
| 634 | 265383_at | AT2G16780 | MSI2 (NUCLEOSOME/CHROMATIN ASSEMBLY FACTOR GROUP C 2) | 1.93 |
| 635 | 263584_at | AT2G17040 | ANAC036 (Arabidopsis NAC domain containing protein 36); transcription factor | -3.32 |
| 636 | 264907_at | AT2G17280 | phosphoglycerate/bisphosphoglycerate mutase family protein | 1.97 |
| 637 | 263071_at | AT2G17490 | transposable element gene | 3.10 |
| 638 | 263074_at | AT2G17560 | HMGB4 (HIGH MOBILITY GROUP B 4); transcription factor | 1.74 |
| 639 | 263017_at | AT2G17620 | CYCB2;1 (CYCLIN B2;1); cyclin-dependent protein kinase regulator | 1.93 |
| 640 | 264590_at | AT2G17710 | similar to unnamed protein product [Vitis vinifera] (GB:CAO42932.1) | -2.02 |
| 641 | 264620_at | AT2G17770 | ATBZIP27/FDP (FD PARALOG); transcription factor | 1.54 |
| 642 | 265817_at | AT2G18050 | HIS1-3 (HISTONE H1-3); DNA binding | -1.89 |
| 643 | 265327_at | AT2G18210 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G36500.1); similar to hypothetical protein [Thellungiella halophila] (GB:ABB45855.1) | -2.78 |
| 644 | 265326_at | AT2G18220 | Identical to Nucleolar complex protein 2 homolog [Arabidopsis Thaliana] (GB:Q9ZPV5); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G55510.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN81729.1); contains InterPro domain Protein of unknown function UPF0120 (InterPro:IPR005343) | 1.48 |
| 645 | 265321_at | AT2G18280 | AtTLP2 (TUBBY LIKE PROTEIN 2); phosphoric diester hydrolase/ transcription factor | -1.88 |
| 646 | 265342_at | AT2G18300 | basic helix-loop-helix (bHLH) family protein | -6.93 |
| 647 | 265340_at | AT2G18330 | AAA-type ATPase family protein | 1.46 |
| 648 | 265923_at | AT2G18470 | protein kinase family protein | 1.73 |
| 649 | 265951_at | AT2G18500 | ATOFP7/OFP7 (Arabidopsis thaliana ovate family protein 7) | 1.81 |
| 650 | 265931_at | AT2G18520 | pentatricopeptide (PPR) repeat-containing protein | 1.76 |
| 651 | 266017_at | AT2G18690 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G18680.1); similar to hypothetical protein OsI_029427 [Oryza sativa (indica cultivar-group)] (GB:EAZ08195.1) | -3.17 |
| 652 | 266934_at | AT2G18900 | transducin family protein / WD-40 repeat family protein | 1.87 |
| 653 | 267465_at | AT2G19170 | SLP3 (Subtilisin-like serine protease 3); subtilase | 1.72 |
| 654 | 267336_at | AT2G19310 | similar to HSP18.2 (HEAT SHOCK PROTEIN 18.2) [Arabidopsis thaliana] (TAIR:AT5G59720.1); similar to 18.5 kDa class I heat shock protein (HSP 18.5) (GB:P05478); contains InterPro domain HSP20-like chaperone (InterPro:IPR008978) | -3.96 |
| 655 | 266688_at | AT2G19660 | DC1 domain-containing protein | -3.23 |
| 656 | 266687_at | AT2G19670 | protein arginine N-methyltransferase, putative | 1.98 |
| 657 | 263369_at | AT2G20480 | similar to unknown [Populus trichocarpa] (GB:ABK93010.1); similar to hypothetical protein OsI_030526 [Oryza sativa (indica cultivar-group)] (GB:EAZ09294.1); similar to Os09g0446000 [Oryza sativa (japonica cultivar-group)] (GB:NP_001063306.1) | 1.46 |
| 658 | 263370_at | AT2G20500 | unknown protein | -2.13 |
| 659 | 263373_at | AT2G20515 | similar to unnamed protein product [Vitis vinifera] (GB:CAO40634.1) | 3.50 |
| 660 | 265429_at | AT2G20710 | pentatricopeptide (PPR) repeat-containing protein | 1.46 |
| 661 | 265441_at | AT2G20870 | cell wall protein precursor, putative | 5.68 |
| 662 | 265417_at | AT2G20920 | similar to unnamed protein product [Vitis vinifera] (GB:CAO47410.1) | -2.01 |
| 663 | 264025_at | AT2G21050 | amino acid permease, putative | 2.06 |
| 664 | 264026_at | AT2G21060 | ATGRP2B (GLYCINE-RICH PROTEIN 2B); nucleic acid binding | 2.46 |
| 665 | 264019_at | AT2G21130 | peptidyl-prolyl cis-trans isomerase / cyclophilin (CYP2) / rotamase | 2.61 |
| 666 | 264007_at | AT2G21140 | ATPRP2 (PROLINE-RICH PROTEIN 2) | -4.26 |
| 667 | 264022_at | AT2G21185 | unknown protein | -2.89 |
| 668 | 264014_at | AT2G21210 | auxin-responsive protein, putative | -2.50 |
| 669 | 263751_at | AT2G21300 | kinesin motor family protein | 1.87 |
| 670 | 263761_at | AT2G21330 | fructose-bisphosphate aldolase, putative | -2.14 |
| 671 | 263544_at | AT2G21590 | APL4 (large subunit of AGP 4); glucose-1-phosphate adenylyltransferase | 2.11 |
| 672 | 263882_at | AT2G21790 | R1/RNR1 (RIBONUCLEOTIDE REDUCTASE 1); ribonucleoside-diphosphate reductase | 1.79 |
| 673 | 263926_at | AT2G21800 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G22140.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO71735.1) | 1.91 |
| 674 | 263875_at | AT2G21970 | SEP2 (STRESS ENHANCED PROTEIN 2) | -2.21 |
| 675 | 264052_at | AT2G22330 | CYP79B3 (cytochrome P450, family 79, subfamily B, polypeptide 3); oxygen binding | -1.99 |
| 676 | 264001_at | AT2G22420 | peroxidase 17 (PER17) (P17) | 2.66 |
| 677 | 264005_at | AT2G22470 | AGP2 (ARABINOGALACTAN-PROTEIN 2) | -4.77 |
| 678 | 264000_at | AT2G22500 | mitochondrial substrate carrier family protein | -1.89 |
| 679 | 265349_at | AT2G22610 | kinesin motor protein-related | 2.66 |
| 680 | 266797_at | AT2G22840 | AtGRF1 (GROWTH-REGULATING FACTOR 1) | 2.54 |
| 681 | 266799_at | AT2G22860 | ATPSK2 (PHYTOSULFOKINE 2 PRECURSOR); growth factor | -2.90 |
| 682 | 266800_at | AT2G22880 | VQ motif-containing protein | -3.84 |
| 683 | 267262_at | AT2G22990 | SNG1 (SINAPOYLGLUCOSE 1); serine carboxypeptidase | -5.47 |
| 684 | 257353_at | AT2G23050 | phototropic-responsive NPH3 family protein | 3.57 |
| 685 | 267250_at | AT2G23060 | GCN5-related N-acetyltransferase (GNAT) family protein | 3.61 |
| 686 | 245076_at | AT2G23170 | GH3.3; indole-3-acetic acid amido synthetase | 2.31 |
| 687 | 267135_at | AT2G23430 | ICK1 (KIP-RELATED PROTEIN 1) | -2.60 |
| 688 | 267120_at | AT2G23530 | similar to protein binding / zinc ion binding [Arabidopsis thaliana] (TAIR:AT4G37110.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO44617.1) | 2.68 |
| 689 | 267121_at | AT2G23540 | GDSL-motif lipase/hydrolase family protein | -3.78 |
| 690 | 267127_at | AT2G23610 | esterase, putative | -2.06 |
| 691 | 267288_at | AT2G23680 | stress-responsive protein, putative | -2.47 |
| 692 | 267284_at | AT2G23700 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G66600.2); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G66600.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G66600.3); similar to unnamed protein product [Vitis vinifera] (GB:CAO45439.1); contains InterPro domain Protein of unknown function DUF547 (InterPro:IPR006869) | 1.98 |
| 693 | 267293_at | AT2G23810 | TET8 (TETRASPANIN8) | -2.65 |
| 694 | 266572_at | AT2G23840 | HNH endonuclease domain-containing protein | -1.87 |
| 695 | 265694_at | AT2G24440 | selenium binding | 2.00 |
| 696 | 265695_at | AT2G24490 | ATRPA2/ROR1/RPA2 (REPLICON PROTEIN A); protein binding | 1.74 |
| 697 | 263796_at | AT2G24540 | AFR (ATTENUATED FAR-RED RESPONSE) | -2.29 |
| 698 | 263799_at | AT2G24550 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G31510.1); similar to unknown [Populus trichocarpa x Populus deltoides] (GB:ABK96563.1) | -3.96 |
| 699 | 263318_at | AT2G24762 | ATGDU4 (ARABIDOPSIS THALIANA GLUTAMINE DUMPER 4) | -4.83 |
| 700 | 263533_at | AT2G24820 | TIC55 (TRANSLOCON AT THE INNER ENVELOPE MEMBRANE OF CHLOROPLASTS 55); oxidoreductase | -1.90 |
| 701 | 263541_at | AT2G24860 | chaperone protein dnaJ-related | -2.01 |
| 702 | 264377_at | AT2G25060 | plastocyanin-like domain-containing protein | 2.03 |
| 703 | 263640_at | AT2G25270 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G12400.1); similar to Os02g0799300 [Oryza sativa (japonica cultivar-group)] (GB:NP_001048408.1); similar to hypothetical protein OsJ_008445 [Oryza sativa (japonica cultivar-group)] (GB:EAZ24962.1) | 2.37 |
| 704 | 265609_at | AT2G25420 | similar to TPR4/WSIP2 (TOPLESS-RELATED 4) [Arabidopsis thaliana] (TAIR:AT3G15880.2); similar to TPR4/WSIP2 (TOPLESS-RELATED 4), protein binding [Arabidopsis thaliana] (TAIR:AT3G15880.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN64663.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO62716.1); contains InterPro domain WD40 repeat-like (InterPro:IPR011046); contains InterPro domain WD40/YVTN repeat-like (InterPro:IPR015943); contains InterPro domain WD40 repeat (InterPro:IPR001680); contains InterPro domain LisH dimerisation motif (InterPro:IPR006594); contains InterPro domain CTLH, C-terminal to LisH motif (InterPro:IPR006595) | 1.48 |
| 705 | 265615_at | AT2G25450 | 2-oxoglutarate-dependent dioxygenase, putative | -7.89 |
| 706 | 265913_at | AT2G25625 | similar to unnamed protein product [Vitis vinifera] (GB:CAO44157.1) | -4.16 |
| 707 | 266658_at | AT2G25735 | unknown protein | -4.03 |
| 708 | 266655_at | AT2G25880 | ATAUR2 (ATAURORA2); histone serine kinase(H3-S10 specific) / kinase | 2.07 |
| 709 | 267402_at | AT2G26180 | IQD6 (IQ-domain 6); calmodulin binding | 1.98 |
| 710 | 267376_at | AT2G26330 | ER (ERECTA) | 3.14 |
| 711 | 245035_at | AT2G26400 | ARD/ATARD3 (ACIREDUCTONE DIOXYGENASE); acireductone dioxygenase [iron(II)-requiring]/ heteroglycan binding / metal ion binding | 4.04 |
| 712 | 245041_at | AT2G26530 | AR781 | -3.05 |
| 713 | 245038_at | AT2G26560 | PLP2 (PHOSPHOLIPASE A 2A); nutrient reservoir | -4.94 |
| 714 | 267610_at | AT2G26650 | AKT1 (ARABIDOPSIS K TRANSPORTER 1); cyclic nucleotide binding / inward rectifier potassium channel | -2.80 |
| 715 | 267617_at | AT2G26670 | HY1 (HEME OXYGENASE 1) | -2.04 |
| 716 | 267612_at | AT2G26690 | nitrate transporter (NTP2) | -2.86 |
| 717 | 267614_at | AT2G26710 | BAS1/CYP734A1 (PHYB ACTIVATION TAGGED SUPPRESSOR 1); oxygen binding / steroid hydroxylase | 2.04 |
| 718 | 267618_at | AT2G26760 | CYCB1;4; cyclin-dependent protein kinase regulator | 2.16 |
| 719 | 266853_at | AT2G26790 | pentatricopeptide (PPR) repeat-containing protein | 2.30 |
| 720 | 266314_at | AT2G27040 | AGO4 (ARGONAUTE 4); nucleic acid binding | 1.98 |
| 721 | 266316_at | AT2G27080 | harpin-induced protein-related / HIN1-related / harpin-responsive protein-related | -2.88 |
| 722 | 265620_at | AT2G27310 | F-box family protein | -2.24 |
| 723 | 265648_at | AT2G27500 | glycosyl hydrolase family 17 protein | -3.52 |
| 724 | 266246_at | AT2G27690 | CYP94C1 (cytochrome P450, family 94, subfamily C, polypeptide 1); oxygen binding | -3.50 |
| 725 | 266259_at | AT2G27830 | similar to pentatricopeptide (PPR) repeat-containing protein [Arabidopsis thaliana] (TAIR:AT4G22760.1); similar to hypothetical protein [Catharanthus roseus] (GB:CAC09928.1) | -2.56 |
| 726 | 264066_at | AT2G27880 | argonaute protein, putative / AGO, putative | 4.96 |
| 727 | 264061_at | AT2G27970 | CKS2 (CDK-SUBUNIT 2); cyclin-dependent protein kinase | 1.82 |
| 728 | 266156_at | AT2G28110 | FRA8 (FRAGILE FIBER8); transferase | -1.89 |
| 729 | 265573_at | AT2G28200 | nucleic acid binding / transcription factor/ zinc ion binding | -2.73 |
| 730 | 265544_at | AT2G28260 | ATCNGC15 (cyclic nucleotide gated channel 15); calmodulin binding / cation channel/ cyclic nucleotide binding | -2.15 |
| 731 | 265547_at | AT2G28305 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G37210.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO21862.1); contains InterPro domain Conserved hypothetical protein CHP00730 (InterPro:IPR005269) | -1.89 |
| 732 | 264057_at | AT2G28550 | RAP2.7/TOE1 (TARGET OF EAT1 1); DNA binding / transcription factor | -3.67 |
| 733 | 263434_at | AT2G28610 | PRS (PRESSED FLOWER); transcription factor | 2.60 |
| 734 | 263441_at | AT2G28620 | kinesin motor protein-related | 2.15 |
| 735 | 266226_at | AT2G28740 | HIS4 (Histone H4) | 1.82 |
| 736 | 266223_at | AT2G28790 | osmotin-like protein, putative | 2.83 |
| 737 | 266782_at | AT2G29120 | ATGLR2.7 (Arabidopsis thaliana glutamate receptor 2.7) | -3.69 |
| 738 | 266279_at | AT2G29290 | tropinone reductase, putative / tropine dehydrogenase, putative | -3.22 |
| 739 | 266265_at | AT2G29340 | short-chain dehydrogenase/reductase (SDR) family protein | -4.04 |
| 740 | 266292_at | AT2G29350 | SAG13 (Senescence-associated gene 13); oxidoreductase | -3.00 |
| 741 | 266293_at | AT2G29360 | tropinone reductase, putative / tropine dehydrogenase, putative | -2.84 |
| 742 | 266299_at | AT2G29450 | ATGSTU5 (Arabidopsis thaliana Glutathione S-transferase (class tau) 5); glutathione transferase | -2.68 |
| 743 | 266237_at | AT2G29540 | ATRPC14 (Arabidopsis thaliana RNA polymerase I(A) and III(C) 14 kDa subunit); DNA binding / DNA-directed RNA polymerase | 1.48 |
| 744 | 266297_at | AT2G29570 | PCNA2 (PROLIFERATING CELL NUCLEAR ANTIGEN 2); DNA binding / DNA polymerase processivity factor | 2.25 |
| 745 | 266617_at | AT2G29670 | binding | -2.71 |
| 746 | 263485_at | AT2G29890 | VLN1 (VILLIN 1); actin binding | 2.33 |
| 747 | 266866_at | AT2G29940 | ATPDR3/PDR3 (PLEIOTROPIC DRUG RESISTANCE 3); ATPase, coupled to transmembrane movement of substances | -3.10 |
| 748 | 266832_at | AT2G30040 | MAPKKK14 (Mitogen-activated protein kinase kinase kinase 14); kinase | -2.38 |
| 749 | 267300_at | AT2G30140 | UDP-glucoronosyl/UDP-glucosyl transferase family protein | -3.16 |
| 750 | 267246_at | AT2G30250 | WRKY25 (WRKY DNA-binding protein 25); transcription factor | -2.18 |
| 751 | 267516_at | AT2G30520 | RPT2 (ROOT PHOTOTROPISM 2); protein binding | -2.67 |
| 752 | 267517_at | AT2G30520 | RPT2 (ROOT PHOTOTROPISM 2); protein binding | -2.73 |
| 753 | 267496_at | AT2G30550 | lipase class 3 family protein | -2.92 |
| 754 | 267565_at | AT2G30750 | CYP71A12 (CYTOCHROME P450, FAMILY 71, SUBFAMILY A, POLYPEPTIDE 12); oxygen binding | -2.20 |
| 755 | 267205_at | AT2G30820 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G06660.1); similar to hypothetical protein [Oryza sativa (japonica cultivar-group)] (GB:AAM19018.1) | 1.64 |
| 756 | 267209_at | AT2G30930 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G06540.1) | -3.40 |
| 757 | 267196_at | AT2G30950 | VAR2 (VARIEGATED 2); ATP-dependent peptidase/ ATPase/ metallopeptidase/ zinc ion binding | -1.96 |
| 758 | 267203_at | AT2G31035 | similar to oxysterol-binding family protein [Arabidopsis thaliana] (TAIR:AT2G31030.1) | 5.01 |
| 759 | 266479_at | AT2G31160 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G23290.2); similar to unknown [Populus trichocarpa] (GB:ABK94898.1); contains InterPro domain Protein of unknown function DUF640 (InterPro:IPR006936) | 2.45 |
| 760 | 264114_at | AT2G31270 | ATCDT1A/CDT1/CDT1A (ARABIDOPSIS HOMOLOG OF YEAST CDT1 A); cyclin-dependent protein kinase/ protein binding | 2.07 |
| 761 | 263474_at | AT2G31725 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G05730.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN73933.1); contains InterPro domain Protein of unknown function DUF842, eukaryotic (InterPro:IPR008560) | 1.74 |
| 762 | 263467_at | AT2G31730 | ethylene-responsive protein, putative | 1.91 |
| 763 | 263478_at | AT2G31880 | leucine-rich repeat transmembrane protein kinase, putative | -2.39 |
| 764 | 265725_at | AT2G32030 | GCN5-related N-acetyltransferase (GNAT) family protein | -4.16 |
| 765 | 265723_at | AT2G32140 | transmembrane receptor | -2.24 |
| 766 | 265680_at | AT2G32150 | haloacid dehalogenase-like hydrolase family protein | -3.39 |
| 767 | 266358_at | AT2G32280 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G21310.1); similar to unknown [Populus trichocarpa] (GB:ABK92874.1); contains InterPro domain Protein of unknown function DUF1218 (InterPro:IPR009606) | 2.02 |
| 768 | 266357_at | AT2G32290 | BAM6/BMY5 (BETA-AMYLASE 6); beta-amylase | -3.34 |
| 769 | 257376_at | AT2G32350 | similar to ubiquitin family protein [Arabidopsis thaliana] (TAIR:AT4G05230.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO24150.1); contains InterPro domain Ubiquitin; (InterPro:IPR000626) | 1.79 |
| 770 | 267117_at | AT2G32560 | F-box family protein | -4.01 |
| 771 | 267118_at | AT2G32590 | similar to unnamed protein product [Vitis vinifera] (GB:CAO24036.1); contains InterPro domain Barren; (InterPro:IPR008418) | 2.52 |
| 772 | 267555_at | AT2G32765 | SUM5 (SUMO 5) | 2.88 |
| 773 | 267550_at | AT2G32800 | AP4.3A; ATP binding / protein kinase | -2.92 |
| 774 | 267645_at | AT2G32860 | glycosyl hydrolase family 1 protein | -4.51 |
| 775 | 255790_at | AT2G33560 | spindle checkpoint protein-related | 1.98 |
| 776 | 255792_at | AT2G33620 | DNA-binding family protein / AT-hook protein 1 (AHP1) | 1.76 |
| 777 | 255848_at | AT2G33640 | zinc finger (DHHC type) family protein | 1.96 |
| 778 | 267460_at | AT2G33810 | SPL3 (SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 3); transcription factor | 2.90 |
| 779 | 267452_at | AT2G33860 | ETT (ETTIN); transcription factor | 2.81 |
| 780 | 267006_at | AT2G34190 | xanthine/uracil permease family protein | 2.60 |
| 781 | 266901_at | AT2G34600 | JAZ7/TIFY5B (JASMONATE-ZIM-DOMAIN PROTEIN 7) | -2.70 |
| 782 | 266908_at | AT2G34650 | PID (PINOID); kinase | 3.64 |
| 783 | 267414_at | AT2G34790 | EDA28/MEE23 (MATERNAL EFFECT EMBRYO ARREST 23); electron carrier | -2.86 |
| 784 | 267425_at | AT2G34810 | FAD-binding domain-containing protein | 1.75 |
| 785 | 267427_at | AT2G34830 | WRKY35 (WRKY DNA-BINDING PROTEIN 35); transcription factor | 1.73 |
| 786 | 266544_at | AT2G35300 | late embryogenesis abundant group 1 domain-containing protein / LEA group 1 domain-containing protein | 2.65 |
| 787 | 266540_at | AT2G35310 | transcriptional factor B3 family protein | 3.21 |
| 788 | 266636_at | AT2G35370 | GDCH (Glycine decarboxylase complex H) | -1.97 |
| 789 | 266641_at | AT2G35605 | SWIB complex BAF60b domain-containing protein | 1.86 |
| 790 | 265846_at | AT2G35770 | SCPL28 (serine carboxypeptidase-like 28); serine carboxypeptidase | -1.88 |
| 791 | 263956_at | AT2G35940 | BLH1 (embryo sac development arrest 29); DNA binding / transcription factor | -2.98 |
| 792 | 263963_at | AT2G36080 | DNA-binding protein, putative | -3.73 |
| 793 | 263285_at | AT2G36120 | pseudogene, glycine-rich protein | -4.49 |
| 794 | 263287_at | AT2G36145 | similar to unnamed protein product [Vitis vinifera] (GB:CAO15876.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN74693.1) | -1.89 |
| 795 | 263960_at | AT2G36200 | kinesin motor protein-related | 2.17 |
| 796 | 263931_at | AT2G36220 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G52710.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO15901.1) | -3.25 |
| 797 | 263907_at | AT2G36270 | ABI5 (ABA INSENSITIVE 5); DNA binding / transcription activator/ transcription factor | 2.00 |
| 798 | 263962_at | AT2G36350 | protein kinase, putative | 1.76 |
| 799 | 263914_at | AT2G36400 | AtGRF3 (GROWTH-REGULATING FACTOR 3) | 2.50 |
| 800 | 263915_at | AT2G36430 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G22540.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO15956.1); contains InterPro domain Protein of unknown function DUF247, plant (InterPro:IPR004158) | -2.84 |
| 801 | 265205_at | AT2G36660 | PAB7 (POLY(A) BINDING PROTEIN 7); RNA binding / translation initiation factor | 2.32 |
| 802 | 263847_at | AT2G36970 | UDP-glucoronosyl/UDP-glucosyl transferase family protein | -2.04 |
| 803 | 265465_at | AT2G37070 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G53320.1) | 1.55 |
| 804 | 265464_at | AT2G37080 | myosin heavy chain-related | 2.22 |
| 805 | 265471_at | AT2G37130 | peroxidase 21 (PER21) (P21) (PRXR5) | -2.81 |
| 806 | 265959_at | AT2G37240 | antioxidant/ oxidoreductase | -1.91 |
| 807 | 265957_at | AT2G37300 | unknown protein | 1.89 |
| 808 | 266009_at | AT2G37420 | kinesin motor protein-related | 2.59 |
| 809 | 266010_at | AT2G37430 | zinc finger (C2H2 type) family protein (ZAT11) | -2.84 |
| 810 | 265962_at | AT2G37460 | nodulin MtN21 family protein | -2.54 |
| 811 | 265960_at | AT2G37470 | histone H2B, putative | 1.75 |
| 812 | 265964_at | AT2G37510 | RNA-binding protein, putative | 1.64 |
| 813 | 267169_at | AT2G37540 | short-chain dehydrogenase/reductase (SDR) family protein | -3.08 |
| 814 | 267157_at | AT2G37630 | AS1/ATMYB91/ATPHAN/MYB91 (ASYMMETRIC LEAVES 1, MYB DOMAIN PROTEIN 91); DNA binding / protein homodimerization/ transcription factor | 2.54 |
| 815 | 267181_at | AT2G37760 | aldo/keto reductase family protein | -2.26 |
| 816 | 267094_at | AT2G38080 | IRX12/LAC4 (laccase 4); copper ion binding / oxidoreductase | -2.16 |
| 817 | 267146_at | AT2G38160 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G40070.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G40070.2); similar to hypothetical protein [Vitis vinifera] (GB:CAN77490.1) | 3.08 |
| 818 | 267093_at | AT2G38170 | CAX1 (CATION EXCHANGER 1); calcium ion transmembrane transporter/ calcium:hydrogen antiporter | -1.96 |
| 819 | 267089_at | AT2G38300 | DNA binding / transcription factor | -4.21 |
| 820 | 267028_at | AT2G38470 | WRKY33 (WRKY DNA-binding protein 33); transcription factor | -2.86 |
| 821 | 257375_at | AT2G38640 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G41590.1); similar to unknown [Populus trichocarpa] (GB:ABK94888.1); contains InterPro domain Protein of unknown function DUF567 (InterPro:IPR007612) | -2.90 |
| 822 | 263264_at | AT2G38810 | HTA8; DNA binding | 2.87 |
| 823 | 266172_at | AT2G39010 | PIP2;6/PIP2E (plasma membrane intrinsic protein 2;6); water channel | -2.15 |
| 824 | 266993_at | AT2G39210 | nodulin family protein | -2.43 |
| 825 | 267010_at | AT2G39250 | SNZ (SCHNARCHZAPFEN); DNA binding / transcription factor | -1.95 |
| 826 | 266989_at | AT2G39330 | jacalin lectin family protein | 2.51 |
| 827 | 266968_at | AT2G39360 | protein kinase family protein | -1.90 |
| 828 | 266983_at | AT2G39400 | hydrolase, alpha/beta fold family protein | -3.09 |
| 829 | 266977_at | AT2G39420 | esterase/lipase/thioesterase family protein | -2.95 |
| 830 | 266979_at | AT2G39470 | PPL2 (PSBP-LIKE PROTEIN 2); calcium ion binding | -2.38 |
| 831 | 267592_at | AT2G39710 | aspartyl protease family protein | -2.37 |
| 832 | 245063_at | AT2G39795 | mitochondrial glycoprotein family protein / MAM33 family protein | 1.58 |
| 833 | 245087_at | AT2G39830 | zinc ion binding | 1.80 |
| 834 | 245088_at | AT2G39850 | subtilase | -2.26 |
| 835 | 267339_at | AT2G39870 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G55690.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO69095.1) | 1.71 |
| 836 | 267337_at | AT2G39980 | transferase family protein | -2.37 |
| 837 | 267357_at | AT2G40000 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G55840.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO41329.1); contains InterPro domain Hs1pro-1, C-terminal (InterPro:IPR009743); contains InterPro domain Hs1pro-1, N-terminal (InterPro:IPR009869) | -4.07 |
| 838 | 267350_at | AT2G40030 | NRPD1b (nuclear RNA polymerase D 1b); DNA binding / DNA-directed RNA polymerase | 1.80 |
| 839 | 263384_at | AT2G40130 | heat shock protein-related | 2.03 |
| 840 | 263804_at | AT2G40270 | protein kinase family protein | -2.01 |
| 841 | 263836_at | AT2G40330 | Bet v I allergen family protein | -2.33 |
| 842 | 255877_at | AT2G40460 | proton-dependent oligopeptide transport (POT) family protein | -2.29 |
| 843 | 255824_at | AT2G40530 | unknown protein | -3.11 |
| 844 | 255874_at | AT2G40550 | similar to unnamed protein product [Vitis vinifera] (GB:CAO41481.1); contains domain PTHR13489 (PTHR13489) | 1.71 |
| 845 | 255822_at | AT2G40610 | ATEXPA8 (ARABIDOPSIS THALIANA EXPANSIN A8) | -2.00 |
| 846 | 267069_at | AT2G41010 | ATCAMBP25 (ARABIDOPSIS THALIANA CALMODULIN (CAM)-BINDING PROTEIN OF 25 KDA); calmodulin binding | -2.28 |
| 847 | 257357_at | AT2G41050 | PQ-loop repeat family protein / transmembrane family protein | 1.78 |
| 848 | 267083_at | AT2G41100 | TCH3 (TOUCH 3) | -3.76 |
| 849 | 267063_at | AT2G41120 | similar to unnamed protein product [Vitis vinifera] (GB:CAO18305.1); contains InterPro domain Protein of unknown function DUF309 (InterPro:IPR005500) | -2.66 |
| 850 | 266369_at | AT2G41370 | BOP2 (BLADE ON PETIOLE2); protein binding | 1.74 |
| 851 | 266368_at | AT2G41380 | embryo-abundant protein-related | -4.14 |
| 852 | 245110_at | AT2G41550 | transcription termination factor | 2.53 |
| 853 | 245119_at | AT2G41640 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G57380.1); similar to glycosyltransferase [Medicago truncatula] (GB:CAI30145.1); contains InterPro domain Protein of unknown function DUF563 (InterPro:IPR007657) | -4.54 |
| 854 | 245106_at | AT2G41650 | unknown protein | 2.29 |
| 855 | 260522_x_at | AT2G41730 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G24640.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO14635.1) | -3.42 |
| 856 | 267538_at | AT2G41870 | remorin family protein | -3.36 |
| 857 | 267535_at | AT2G41940 | ZFP8 (ZINC FINGER PROTEIN 8); nucleic acid binding / transcription factor/ zinc ion binding | -1.97 |
| 858 | 267580_at | AT2G41990 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G35170.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN73167.1) | 1.50 |
| 859 | 267636_at | AT2G42110 | similar to unknown [Populus trichocarpa] (GB:ABK94541.1); similar to unknown [Populus trichocarpa] (GB:ABK92893.1); contains domain PTHR10270:SF18 (PTHR10270:SF18); contains domain PTHR10270 (PTHR10270) | 2.36 |
| 860 | 267639_at | AT2G42200 | squamosa promoter-binding protein-like 9 (SPL9) | 3.19 |
| 861 | 267628_at | AT2G42280 | basic helix-loop-helix (bHLH) family protein | -2.05 |
| 862 | 265877_at | AT2G42380 | bZIP transcription factor family protein | -4.28 |
| 863 | 263497_at | AT2G42540 | COR15A (COLD-REGULATED 15A) | -3.42 |
| 864 | 263496_at | AT2G42570 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G31110.2); similar to unknown [Pisum sativum] (GB:ABA29158.1); contains InterPro domain Protein of unknown function DUF231, plant (InterPro:IPR004253) | 2.68 |
| 865 | 263973_at | AT2G42740 | RPL16A (ribosomal protein large subunit 16A); structural constituent of ribosome | 1.45 |
| 866 | 263971_at | AT2G42800 | leucine-rich repeat family protein | 3.17 |
| 867 | 263979_at | AT2G42840 | PDF1 (PROTODERMAL FACTOR 1) | 2.53 |
| 868 | 265261_at | AT2G42990 | GDSL-motif lipase/hydrolase family protein | 3.00 |
| 869 | 265248_at | AT2G43010 | PIF4 (PHYTOCHROME INTERACTING FACTOR 4); DNA binding / transcription factor | -2.19 |
| 870 | 265246_at | AT2G43050 | ATPMEPCRD; pectinesterase | -3.95 |
| 871 | 265245_at | AT2G43060 | transcription factor | -5.87 |
| 872 | 266395_at | AT2G43100 | aconitase C-terminal domain-containing protein | -3.30 |
| 873 | 260546_at | AT2G43520 | ATTI2 (ARABIDOPSIS THALIANA TRYPSIN INHIBITOR PROTEIN 2); trypsin inhibitor | 2.71 |
| 874 | 260568_at | AT2G43570 | chitinase, putative | -3.23 |
| 875 | 260560_at | AT2G43590 | chitinase, putative | -4.17 |
| 876 | 260585_at | AT2G43650 | EMB2777 (EMBRYO DEFECTIVE 2777) | 1.78 |
| 877 | 260565_at | AT2G43800 | formin homology 2 domain-containing protein / FH2 domain-containing protein | 2.59 |
| 878 | 260567_at | AT2G43820 | GT/UGT74F2 (UDP-GLUCOSYLTRANSFERASE 74F2); UDP-glucosyltransferase/ UDP-glycosyltransferase/ transferase, transferring glycosyl groups / transferase, transferring hexosyl groups | -5.75 |
| 879 | 267230_at | AT2G44080 | ARL (ARGOS-LIKE) | -1.86 |
| 880 | 267238_at | AT2G44130 | kelch repeat-containing F-box family protein | -2.54 |
| 881 | 267232_at | AT2G44190 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G60000.2); similar to unnamed protein product [Vitis vinifera] (GB:CAO46272.1); contains InterPro domain Protein of unknown function DUF566 (InterPro:IPR007573) | 2.17 |
| 882 | 267367_at | AT2G44210 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G55360.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO67164.1); contains InterPro domain Protein of unknown function DUF239, plant (InterPro:IPR004314) | -1.94 |
| 883 | 267344_at | AT2G44230 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G44260.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G44260.2); similar to Protein of unknown function DUF946, plant [Medicago truncatula] (GB:ABN08108.1); contains InterPro domain Protein of unknown function DUF946, plant (InterPro:IPR009291) | -2.58 |
| 884 | 267391_at | AT2G44480 | glycosyl hydrolase family 1 protein | -2.08 |
| 885 | 267392_at | AT2G44490 | PEN2 (PENETRATION 2); hydrolase, hydrolyzing O-glycosyl compounds | -3.06 |
| 886 | 267393_at | AT2G44500 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G07900.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO15104.1); contains InterPro domain Protein of unknown function DUF246, plant (InterPro:IPR004348) | -2.92 |
| 887 | 266812_at | AT2G44830 | protein kinase, putative | 1.74 |
| 888 | 266814_at | AT2G44910 | homeobox-leucine zipper protein 4 (HB-4) / HD-ZIP protein 4 | 2.94 |
| 889 | 266126_at | AT2G45040 | matrix metalloproteinase | -3.50 |
| 890 | 245138_at | AT2G45190 | AFO (ABNORMAL FLORAL ORGANS); transcription factor | 3.02 |
| 891 | 245136_at | AT2G45210 | auxin-responsive protein-related | -2.02 |
| 892 | 245148_at | AT2G45220 | pectinesterase family protein | -3.45 |
| 893 | 245130_at | AT2G45340 | leucine-rich repeat transmembrane protein kinase, putative | 1.48 |
| 894 | 267529_at | AT2G45490 | ATAUR3 (ATAURORA3); ATP binding / histone serine kinase(H3-S10 specific) / protein kinase | 1.72 |
| 895 | 267505_at | AT2G45560 | CYP76C1 (cytochrome P450, family 76, subfamily C, polypeptide 1); heme binding / iron ion binding / monooxygenase | -3.81 |
| 896 | 267559_at | AT2G45570 | CYP76C2 (cytochrome P450, family 76, subfamily C, polypeptide 2); oxygen binding | -2.34 |
| 897 | 267528_at | AT2G45650 | AGL6 (AGAMOUS LIKE-6); DNA binding / transcription factor | 4.30 |
| 898 | 266925_at | AT2G45740 | PEX11D | -2.15 |
| 899 | 266603_at | AT2G46040 | ARID/BRIGHT DNA-binding domain-containing protein / ELM2 domain-containing protein | 2.78 |
| 900 | 266583_at | AT2G46220 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G79510.2); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G16320.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G79510.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO39696.1); contains domain NTF2-like (SSF54427) | -2.86 |
| 901 | 265460_at | AT2G46600 | calcium-binding protein, putative | -2.05 |
| 902 | 266320_at | AT2G46640 | unknown protein | -2.33 |
| 903 | 266326_at | AT2G46650 | B5 #1 (cytochrome b5 family protein #1); heme binding / transition metal ion binding | -2.31 |
| 904 | 266327_at | AT2G46680 | ATHB-7 (ARABIDOPSIS THALIANA HOMEOBOX 7); transcription factor | -2.12 |
| 905 | 266322_at | AT2G46690 | auxin-responsive family protein | -3.37 |
| 906 | 266717_at | AT2G46735 | unknown protein | -4.89 |
| 907 | 266713_at | AT2G46760 | FAD-binding domain-containing protein | 1.97 |
| 908 | 266767_at | AT2G46910 | plastid-lipid associated protein PAP / fibrillin family protein | -1.96 |
| 909 | 266753_at | AT2G46990 | IAA20 (indoleacetic acid-induced protein 20); transcription factor | 2.19 |
| 910 | 266738_at | AT2G47010 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G17030.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN67131.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO40236.1) | 1.86 |
| 911 | 260524_at | AT2G47230 | agenet domain-containing protein | 1.96 |
| 912 | 260527_at | AT2G47270 | transcription factor/ transcription regulator | -3.87 |
| 913 | 245126_at | AT2G47460 | ATMYB12/MYB12 (MYB DOMAIN PROTEIN 12); DNA binding / transcription activator/ transcription factor | 2.04 |
| 914 | 266511_at | AT2G47680 | zinc finger (CCCH type) helicase family protein | 1.58 |
| 915 | 266502_at | AT2G47720 | similar to AtATG18a (Arabidopsis thaliana homolog of yeast autophagy 18 (ATG18) a) [Arabidopsis thaliana] (TAIR:AT3G62770.2); contains domain PTHR11227 (PTHR11227); contains domain PTHR11227:SF1 (PTHR11227:SF1) | 1.86 |
| 916 | 266461_at | AT2G47730 | ATGSTF8 (GLUTATHIONE S-TRANSFERASE 8); glutathione transferase | -1.95 |
| 917 | 266462_at | AT2G47770 | benzodiazepine receptor-related | -2.18 |
| 918 | 266514_at | AT2G47890 | zinc finger (B-box type) family protein | -3.78 |
| 919 | 266483_at | AT2G47910 | CRR6 (CHLORORESPIRATORY REDUCTION 6) | -2.02 |
| 920 | 266510_at | AT2G47990 | SWA1 (SLOW WALKER1); nucleotide binding | 1.48 |
| 921 | 259275_at | AT3G01060 | similar to unnamed protein product [Vitis vinifera] (GB:CAO15045.1); similar to unknown [Brassica rapa] (GB:ABC41272.1) | -1.97 |
| 922 | 259272_at | AT3G01290 | band 7 family protein | -8.41 |
| 923 | 257524_at | AT3G01330 | DEL3 (DP-E2F-like 3); transcription factor | 1.72 |
| 924 | 259116_at | AT3G01350 | proton-dependent oligopeptide transport (POT) family protein | -3.29 |
| 925 | 258952_at | AT3G01410 | RNase H domain-containing protein | 2.09 |
| 926 | 259161_at | AT3G01500 | CA1 (CARBONIC ANHYDRASE 1); carbonate dehydratase/ zinc ion binding | -4.93 |
| 927 | 259180_at | AT3G01680 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G01670.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO14653.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO14657.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO14661.1); contains domain PTHR13871:SF2 (PTHR13871:SF2); contains domain PTHR13871 (PTHR13871) | -2.04 |
| 928 | 259189_at | AT3G01700 | AGP11 (ARABINOGALACTAN PROTEIN 11) | 2.60 |
| 929 | 258947_at | AT3G01830 | calmodulin-related protein, putative | -2.63 |
| 930 | 259000_at | AT3G01860 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G27210.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO14583.1) | -1.89 |
| 931 | 259002_at | AT3G02000 | ROXY1; thiol-disulfide exchange intermediate | 3.64 |
| 932 | 258857_at | AT3G02110 | SCPL25 (serine carboxypeptidase-like 25); serine carboxypeptidase | 2.77 |
| 933 | 258859_at | AT3G02120 | hydroxyproline-rich glycoprotein family protein | 1.68 |
| 934 | 259122_at | AT3G02210 | COBL1 (COBRA-LIKE PROTEIN 1 PRECURSOR) | 3.25 |
| 935 | 259124_at | AT3G02310 | SEP2 (SEPALLATA2); DNA binding / transcription factor | 5.11 |
| 936 | 258487_at | AT3G02550 | LBD41 (LOB DOMAIN-CONTAINING PROTEIN 41) | 2.37 |
| 937 | 258480_at | AT3G02640 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G16250.1); similar to unknown [Populus trichocarpa] (GB:ABK93711.1) | 2.57 |
| 938 | 258623_at | AT3G02790 | zinc finger (C2H2 type) family protein | 1.47 |
| 939 | 258630_at | AT3G02820 | zinc knuckle (CCHC-type) family protein | 1.64 |
| 940 | 258609_at | AT3G02910 | Identical to UPF0131 protein At3g02910 [Arabidopsis Thaliana] (GB:Q9M8T3); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G46720.1); similar to Os03g0854000 [Oryza sativa (japonica cultivar-group)] (GB:NP_001051934.1); similar to unknown [Picea sitchensis] (GB:ABK22133.1); contains InterPro domain Butirosin biosynthesis, BtrG-like (InterPro:IPR013024); contains InterPro domain Protein of unknown function UPF0131 (InterPro:IPR005347) | -2.89 |
| 941 | 258617_at | AT3G03000 | calmodulin, putative | -4.63 |
| 942 | 258871_at | AT3G03060 | ATPase | 1.86 |
| 943 | 258867_at | AT3G03130 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G17160.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO62214.1) | 2.42 |
| 944 | 258851_at | AT3G03190 | ATGSTF11 (GLUTATHIONE S-TRANSFERASE F11); glutathione transferase | -2.08 |
| 945 | 259058_at | AT3G03470 | CYP89A9 (cytochrome P450, family 87, subfamily A, polypeptide 9); oxygen binding | -2.81 |
| 946 | 259169_at | AT3G03520 | phosphoesterase family protein | -4.83 |
| 947 | 259338_at | AT3G03800 | SYP131 (syntaxin 131); SNAP receptor | -2.21 |
| 948 | 259331_at | AT3G03840 | auxin-responsive protein, putative | -4.03 |
| 949 | 259342_at | AT3G03890 | FMN binding | -2.27 |
| 950 | 258805_at | AT3G04010 | glycosyl hydrolase family 17 protein | -5.01 |
| 951 | 258807_at | AT3G04030 | myb family transcription factor | -2.27 |
| 952 | 258589_at | AT3G04290 | ATLTL1/LTL1 (LI-TOLERANT LIPASE 1); carboxylesterase | 1.73 |
| 953 | 258571_at | AT3G04420 | ANAC048 (Arabidopsis NAC domain containing protein 48); transcription factor | -1.85 |
| 954 | 258791_at | AT3G04720 | PR4 (PATHOGENESIS-RELATED 4) | -3.90 |
| 955 | 259083_at | AT3G04810 | ATNEK2; kinase | 2.37 |
| 956 | 259089_at | AT3G04960 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G27980.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO71037.1); contains InterPro domain Molecular chaperone, heat shock protein, Hsp40, DnaJ (InterPro:IPR015609) | 3.31 |
| 957 | 259087_at | AT3G04980 | DNAJ heat shock N-terminal domain-containing protein | 2.10 |
| 958 | 259303_at | AT3G05130 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G27330.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO68600.1); contains InterPro domain Prefoldin (InterPro:IPR009053) | 1.50 |
| 959 | 259353_at | AT3G05190 | aminotransferase class IV family protein | 1.69 |
| 960 | 259312_at | AT3G05200 | ATL6 (Arabidopsis T?xicos en Levadura 6); protein binding / zinc ion binding | -3.02 |
| 961 | 259325_at | AT3G05320 | similar to unnamed protein product [Vitis vinifera] (GB:CAO44440.1) | -2.56 |
| 962 | 259136_at | AT3G05470 | formin homology 2 domain-containing protein / FH2 domain-containing protein | 2.02 |
| 963 | 258895_at | AT3G05600 | epoxide hydrolase, putative | 3.14 |
| 964 | 258887_at | AT3G05630 | PLDP2 (PHOSPHOLIPASE D ZETA 2); phospholipase D | -2.22 |
| 965 | 258898_at | AT3G05740 | RECQI1 (Arabidopsis RecQ helicase l1); ATP-dependent helicase | 2.02 |
| 966 | 258741_at | AT3G05790 | Lon protease, putative | 4.12 |
| 967 | 258744_at | AT3G05830 | Encodes alpha-helical IF (intermediate filament)-like protein. | 1.75 |
| 968 | 258751_at | AT3G05890 | RCI2B (RARE-COLD-INDUCIBLE 2B) | -2.13 |
| 969 | 258471_at | AT3G06030 | ANP3 (Arabidopsis NPK1-related protein kinase 3); kinase | 2.44 |
| 970 | 258468_at | AT3G06070 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G19190.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO71112.1) | -2.89 |
| 971 | 256418_at | AT3G06160 | transcriptional factor B3 family protein | 3.17 |
| 972 | 256389_at | AT3G06220 | DNA binding / transcription factor | 7.60 |
| 973 | 258880_at | AT3G06420 | ATG8H (AUTOPHAGY 8H); microtubule binding | -2.95 |
| 974 | 258535_at | AT3G06750 | hydroxyproline-rich glycoprotein family protein | -2.60 |
| 975 | 258545_at | AT3G07050 | GTP-binding family protein | 1.52 |
| 976 | 259014_at | AT3G07320 | glycosyl hydrolase family 17 protein | 1.67 |
| 977 | 259010_at | AT3G07340 | basic helix-loop-helix (bHLH) family protein | -2.94 |
| 978 | 259021_at | AT3G07540 | formin homology 2 domain-containing protein / FH2 domain-containing protein | 1.74 |
| 979 | 259225_at | AT3G07590 | small nuclear ribonucleoprotein D1, putative / snRNP core protein D1, putative / Sm protein D1, putative | 1.54 |
| 980 | 259251_at | AT3G07600 | heavy-metal-associated domain-containing protein | -2.82 |
| 981 | 259226_at | AT3G07700 | ABC1 family protein | -2.03 |
| 982 | 258639_at | AT3G07820 | polygalacturonase 3 (PGA3) / pectinase | 2.46 |
| 983 | 258647_at | AT3G07870 | F-box family protein | -2.36 |
| 984 | 258672_at | AT3G08570 | phototropic-responsive protein, putative | 1.55 |
| 985 | 258675_at | AT3G08770 | LTP6 (Lipid transfer protein 6); lipid binding | 3.05 |
| 986 | 258982_at | AT3G08870 | lectin protein kinase, putative | -2.99 |
| 987 | 258993_at | AT3G08940 | LHCB4.2 (LIGHT HARVESTING COMPLEX PSII); chlorophyll binding | -2.54 |
| 988 | 259200_at | AT3G09070 | glycine-rich protein | 2.02 |
| 989 | 258707_at | AT3G09480 | histone H2B, putative | 1.78 |
| 990 | 258702_at | AT3G09730 | similar to unnamed protein product [Vitis vinifera] (GB:CAO18124.1) | 1.51 |
| 991 | 258941_at | AT3G09940 | ATMDAR3/MDHAR (MONODEHYDROASCORBATE REDUCTASE); monodehydroascorbate reductase (NADH) | -2.60 |
| 992 | 258931_at | AT3G10010 | DML2 (DEMETER-LIKE 2); 4 iron, 4 sulfur cluster binding / catalytic/ endonuclease | 1.64 |
| 993 | 258930_at | AT3G10040 | transcription factor | 2.69 |
| 994 | 259145_at | AT3G10180 | kinesin motor protein-related | 2.41 |
| 995 | 259142_at | AT3G10200 | dehydration-responsive protein-related | 1.95 |
| 996 | 259151_at | AT3G10310 | kinesin motor protein-related | 2.08 |
| 997 | 258919_at | AT3G10525 | similar to SIM (SIAMESE) [Arabidopsis thaliana] (TAIR:AT5G04470.1) | -2.38 |
| 998 | 258962_at | AT3G10570 | CYP77A6 (cytochrome P450, family 77, subfamily A, polypeptide 6); oxygen binding | 2.85 |
| 999 | 258768_at | AT3G10880 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G05180.1); similar to hypothetical protein OsI_017925 [Oryza sativa (indica cultivar-group)] (GB:EAY96692.1); similar to Os05g0168800 [Oryza sativa (japonica cultivar-group)] (GB:NP_001054757.1) | 2.00 |
| 1000 | 256442_at | AT3G10930 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G05300.1) | -2.60 |
| 1001 | 257579_at | AT3G11000 | similar to kelch repeat-containing protein [Arabidopsis thaliana] (TAIR:AT5G01660.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO41447.1); contains InterPro domain Kelch related (InterPro:IPR013089); contains InterPro domain Development and cell death (InterPro:IPR013989) | 4.22 |
| 1002 | 256255_at | AT3G11280 | myb family transcription factor | -2.88 |
| 1003 | 256252_at | AT3G11340 | UDP-glucoronosyl/UDP-glucosyl transferase family protein | -3.60 |
| 1004 | 259286_at | AT3G11480 | BSMT1; S-adenosylmethionine-dependent methyltransferase | -2.75 |
| 1005 | 259290_at | AT3G11520 | CYCB1;3 (CYCLIN B1;3); cyclin-dependent protein kinase regulator | 1.75 |
| 1006 | 259102_at | AT3G11660 | NHL1 (NDR1/HIN1-like 1) | -5.36 |
| 1007 | 258788_at | AT3G11780 | MD-2-related lipid recognition domain-containing protein / ML domain-containing protein | -2.11 |
| 1008 | 258727_at | AT3G11930 | universal stress protein (USP) family protein | -5.04 |
| 1009 | 256273_at | AT3G12090 | TET6 (TETRASPANIN6) | -3.11 |
| 1010 | 256261_at | AT3G12160 | AtRABA4d (Arabidopsis Rab GTPase homolog A4d); GTP binding | 2.62 |
| 1011 | 256320_at | AT3G12170 | DNAJ heat shock N-terminal domain-containing protein | 2.53 |
| 1012 | 256288_at | AT3G12270 | methyltransferase | 1.78 |
| 1013 | 256266_at | AT3G12320 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G06980.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN75675.1) | -2.36 |
| 1014 | 256259_at | AT3G12460 | 3'-5' exonuclease/ nucleic acid binding | 1.93 |
| 1015 | 256243_at | AT3G12500 | ATHCHIB (BASIC CHITINASE); chitinase | -3.78 |
| 1016 | 256284_at | AT3G12530 | PSF2 | 1.77 |
| 1017 | 257694_at | AT3G12860 | nucleolar protein Nop56, putative | 1.52 |
| 1018 | 257134_at | AT3G12870 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G56120.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO47924.1) | 1.95 |
| 1019 | 257191_at | AT3G13175 | similar to unnamed protein product [Vitis vinifera] (GB:CAO45116.1) | 2.53 |
| 1020 | 257181_at | AT3G13190 | myosin heavy chain-related | 2.88 |
| 1021 | 256960_at | AT3G13510 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G55360.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G56530.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO22094.1); similar to unknown [Populus trichocarpa] (GB:ABK93966.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN72570.1); contains InterPro domain Protein of unknown function DUF239, plant (InterPro:IPR004314) | 2.31 |
| 1022 | 256780_at | AT3G13640 | ATRLI1 (Arabidopsis thaliana RNase L inhibitor protein 1) | 1.63 |
| 1023 | 257570_at | AT3G13662 | disease resistance-responsive protein-related / dirigent protein-related | 2.67 |
| 1024 | 256784_at | AT3G13674 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G55205.1) | 1.72 |
| 1025 | 256785_at | AT3G13720 | prenylated rab acceptor (PRA1) family protein | -1.84 |
| 1026 | 256788_at | AT3G13730 | CYP90D1 (CYTOCHROME P450, FAMILY 90, SUBFAMILY D, POLYPEPTIDE 1); oxidoreductase, acting on paired donors, with incorporation or reduction of molecular oxygen, NADH or NADPH as one donor, and incorporation of one atom of oxygen / oxygen binding | -2.58 |
| 1027 | 256787_at | AT3G13790 | ATBFRUCT1/ATCWINV1 (ARABIDOPSIS THALIANA CELL WALL INVERTASE 1); beta-fructofuranosidase/ hydrolase, hydrolyzing O-glycosyl compounds | -2.20 |
| 1028 | 258204_at | AT3G13960 | AtGRF5 (GROWTH-REGULATING FACTOR 5) | 4.54 |
| 1029 | 257000_at | AT3G14120 | similar to hypothetical protein OsJ_033267 [Oryza sativa (japonica cultivar-group)] (GB:EAZ19058.1); similar to predicted protein [Physcomitrella patens subsp. patens] (GB:EDQ65731.1); similar to hypothetical protein OsI_021263 [Oryza sativa (indica cultivar-group)] (GB:EAZ00031.1); contains InterPro domain Nuclear pore protein 84/107; (InterPro:IPR007252) | 1.51 |
| 1030 | 257005_at | AT3G14190 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G12360.1) | 2.67 |
| 1031 | 258368_at | AT3G14240 | subtilase family protein | 1.72 |
| 1032 | 257280_at | AT3G14440 | NCED3 (NINE-CIS-EPOXYCAROTENOID DIOXYGENASE3) | -3.69 |
| 1033 | 258091_at | AT3G14560 | unknown protein | -4.50 |
| 1034 | 258092_at | AT3G14595 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G17080.1); similar to unknown [Populus trichocarpa] (GB:ABK95219.1); contains domain PTHR10052 (PTHR10052); contains domain PTHR10052:SF2 (PTHR10052:SF2) | -2.65 |
| 1035 | 258063_at | AT3G14620 | CYP72A8 (cytochrome P450, family 72, subfamily A, polypeptide 8); oxygen binding | -3.86 |
| 1036 | 258114_at | AT3G14660 | CYP72A13 (cytochrome P450, family 72, subfamily A, polypeptide 13); oxygen binding | -3.26 |
| 1037 | 258094_at | AT3G14690 | CYP72A15 (cytochrome P450, family 72, subfamily A, polypeptide 15); oxygen binding | -2.46 |
| 1038 | 258089_at | AT3G14740 | PHD finger family protein | 1.78 |
| 1039 | 257237_at | AT3G14890 | phosphoesterase | 1.98 |
| 1040 | 256857_at | AT3G15170 | CUC1 (CUP-SHAPED COTYLEDON1); transcription factor | 4.77 |
| 1041 | 257051_at | AT3G15270 | SPL5 (SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 5); DNA binding / transcription factor | 3.92 |
| 1042 | 258398_at | AT3G15360 | ATHM4 (Arabidopsis thioredoxin M-type 4); thiol-disulfide exchange intermediate | -2.10 |
| 1043 | 258385_at | AT3G15510 | ATNAC2 (Arabidopsis thaliana NAC domain containing protein 2); transcription factor | -3.31 |
| 1044 | 258387_at | AT3G15550 | similar to unnamed protein product [Vitis vinifera] (GB:CAO46019.1) | 2.42 |
| 1045 | 257290_at | AT3G15560 | similar to unnamed protein product [Vitis vinifera] (GB:CAO46019.1) | 2.43 |
| 1046 | 257294_at | AT3G15570 | phototropic-responsive NPH3 family protein | -3.86 |
| 1047 | 258271_at | AT3G15605 | nucleic acid binding / nucleotide binding | 1.49 |
| 1048 | 258270_at | AT3G15650 | phospholipase/carboxylesterase family protein | 2.13 |
| 1049 | 258275_at | AT3G15760 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G52565.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO62742.1) | -2.39 |
| 1050 | 258262_at | AT3G15770 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G25360.1); similar to unknown [Populus trichocarpa] (GB:ABK94402.1) | -1.88 |
| 1051 | 258223_at | AT3G15840 | PIFI (POST-ILLUMINATION CHLOROPHYLL FLUORESCENCE INCREASE) | -2.85 |
| 1052 | 258332_at | AT3G16180 | proton-dependent oligopeptide transport (POT) family protein | 1.54 |
| 1053 | 258049_at | AT3G16220 | similar to RNA binding / catalytic [Arabidopsis thaliana] (TAIR:AT3G16230.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO45862.1); contains InterPro domain Predicted eukaryotic LigT; (InterPro:IPR009210) | -4.64 |
| 1054 | 257517_at | AT3G16330 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G52140.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN64915.1) | 1.97 |
| 1055 | 259328_at | AT3G16440 | ATMLP-300B (MYROSINASE-BINDING PROTEIN-LIKE PROTEIN-300B) | -1.99 |
| 1056 | 257229_at | AT3G16490 | IQD26 (IQ-domain 26); calmodulin binding | 2.47 |
| 1057 | 257206_at | AT3G16530 | legume lectin family protein | -4.63 |
| 1058 | 258436_at | AT3G16720 | ATL2 (Arabidopsis T?xicos en Levadura 2); protein binding / zinc ion binding | -2.43 |
| 1059 | 258423_at | AT3G16730 | similar to unnamed protein product [Vitis vinifera] (GB:CAO44817.1); contains InterPro domain Protein of unknown function DUF1032 (InterPro:IPR009378) | 1.55 |
| 1060 | 257650_at | AT3G16800 | protein phosphatase 2C, putative / PP2C, putative | -2.50 |
| 1061 | 257571_at | AT3G16870 | zinc finger (GATA type) family protein | 1.71 |
| 1062 | 257896_at | AT3G16920 | chitinase | -3.44 |
| 1063 | 257929_at | AT3G16980 | DNA-directed RNA polymerase II, putative | 2.75 |
| 1064 | 257930_at | AT3G17010 | transcriptional factor B3 family protein | 4.77 |
| 1065 | 257874_at | AT3G17110 | pseudogene, glycine-rich protein | -2.47 |
| 1066 | 257879_at | AT3G17160 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G47970.1) | 2.91 |
| 1067 | 258464_at | AT3G17360 | POK1 (PHRAGMOPLAST ORIENTING KINESIN 1); microtubule motor | 1.98 |
| 1068 | 258376_at | AT3G17680 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G48405.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN67913.1); contains InterPro domain KIP1-like (InterPro:IPR011684) | 2.69 |
| 1069 | 258159_at | AT3G17840 | RLK902 (receptor-like kinase 902); ATP binding / kinase/ protein serine/threonine kinase | 2.31 |
| 1070 | 257061_at | AT3G18250 | contains domain PROKAR_LIPOPROTEIN (PS51257) | -3.95 |
| 1071 | 257724_at | AT3G18510 | unknown protein | 1.65 |
| 1072 | 257725_at | AT3G18524 | MSH2 (MUTS HOMOLOG 2); ATP binding / damaged DNA binding | 1.75 |
| 1073 | 256799_at | AT3G18560 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G49000.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO68009.1) | -2.68 |
| 1074 | 256797_at | AT3G18600 | DEAD/DEAH box helicase, putative | 1.52 |
| 1075 | 256798_at | AT3G18630 | uracil DNA glycosylase family protein | 1.95 |
| 1076 | 256652_at | AT3G18850 | LPAT5; acyltransferase | 1.61 |
| 1077 | 256922_at | AT3G19010 | oxidoreductase, 2OG-Fe(II) oxygenase family protein | -2.19 |
| 1078 | 256891_at | AT3G19030 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G49500.1) | -5.99 |
| 1079 | 257034_at | AT3G19184 | DNA binding | 4.23 |
| 1080 | 257026_at | AT3G19200 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G34420.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN61148.1) | 1.56 |
| 1081 | 258010_at | AT3G19300 | protein kinase family protein | 1.51 |
| 1082 | 258017_at | AT3G19370 | similar to transport protein-related [Arabidopsis thaliana] (TAIR:AT2G23360.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO47839.1); contains InterPro domain Protein of unknown function DUF869, plant (InterPro:IPR008587) | -2.04 |
| 1083 | 258025_at | AT3G19480 | D-3-phosphoglycerate dehydrogenase, putative / 3-PGDH, putative | -2.16 |
| 1084 | 256567_at | AT3G19553 | amino acid permease family protein | -2.24 |
| 1085 | 257022_at | AT3G19580 | AZF2 (ARABIDOPSIS ZINC-FINGER PROTEIN 2); nucleic acid binding / transcription factor/ zinc ion binding | -2.16 |
| 1086 | 257020_at | AT3G19590 | WD-40 repeat family protein / mitotic checkpoint protein, putative | 2.25 |
| 1087 | 257017_at | AT3G19620 | glycosyl hydrolase family 3 protein | -4.43 |
| 1088 | 257074_at | AT3G19660 | similar to unnamed protein product [Vitis vinifera] (GB:CAO49873.1) | -4.25 |
| 1089 | 257021_at | AT3G19710 | BCAT4 (BRANCHED-CHAIN AMINOTRANSFERASE4); catalytic/ methionine-oxo-acid transaminase | -5.21 |
| 1090 | 257966_at | AT3G19800 | similar to unnamed protein product [Vitis vinifera] (GB:CAO15360.1) | -2.06 |
| 1091 | 257939_at | AT3G19930 | STP4 (SUGAR TRANSPORTER 4); carbohydrate transmembrane transporter/ sugar:hydrogen ion symporter | -3.11 |
| 1092 | 256625_at | AT3G20010 | SNF2 domain-containing protein / helicase domain-containing protein / RING finger domain-containing protein | 1.60 |
| 1093 | 256626_at | AT3G20015 | pepsin A | 1.99 |
| 1094 | 257128_at | AT3G20080 | CYP705A15 (cytochrome P450, family 705, subfamily A, polypeptide 15); oxygen binding | 1.50 |
| 1095 | 257112_at | AT3G20120 | CYP705A21 (cytochrome P450, family 705, subfamily A, polypeptide 21); oxygen binding | 1.49 |
| 1096 | 257115_at | AT3G20150 | kinesin motor family protein | 2.16 |
| 1097 | 257118_at | AT3G20180 | metal ion binding | 5.38 |
| 1098 | 257131_at | AT3G20240 | mitochondrial substrate carrier family protein | 1.54 |
| 1099 | 257663_at | AT3G20260 | structural constituent of ribosome | 1.72 |
| 1100 | 257672_at | AT3G20300 | extracellular ligand-gated ion channel | -2.55 |
| 1101 | 256666_at | AT3G20670 | HTA13; DNA binding | 2.19 |
| 1102 | 256977_at | AT3G21040 | transposable element gene | 5.35 |
| 1103 | 256970_at | AT3G21090 | ABC transporter family protein | 1.46 |
| 1104 | 256971_at | AT3G21100 | RNA recognition motif (RRM)-containing protein | 3.54 |
| 1105 | 258166_at | AT3G21540 | transducin family protein / WD-40 repeat family protein | 1.52 |
| 1106 | 258183_at | AT3G21550 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G21520.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO71058.1); contains InterPro domain Protein of unknown function DUF679 (InterPro:IPR007770) | -2.56 |
| 1107 | 258181_at | AT3G21670 | nitrate transporter (NTP3) | -1.91 |
| 1108 | 257950_at | AT3G21780 | UGT71B6 (UDP-GLUCOSYL TRANSFERASE 71B6); UDP-glycosyltransferase/ abscisic acid glucosyltransferase/ transferase, transferring glycosyl groups | -2.01 |
| 1109 | 257254_at | AT3G21950 | S-adenosyl-L-methionine:carboxyl methyltransferase family protein | 1.71 |
| 1110 | 256766_at | AT3G22231 | PCC1 (PATHOGEN AND CIRCADIAN CONTROLLED 1) | -3.06 |
| 1111 | 256617_at | AT3G22240 | unknown protein | -5.45 |
| 1112 | 258452_at | AT3G22370 | AOX1A (alternative oxidase 1A); alternative oxidase | -3.17 |
| 1113 | 256926_at | AT3G22540 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G14819.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO71190.1); contains InterPro domain Protein of unknown function DUF1677, plant (InterPro:IPR012876) | 1.64 |
| 1114 | 256927_at | AT3G22550 | senescence-associated protein-related | 1.83 |
| 1115 | 256933_at | AT3G22600 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | -3.55 |
| 1116 | 258316_at | AT3G22660 | rRNA processing protein-related | 1.64 |
| 1117 | 258322_at | AT3G22740 | HMT3 (Homocysteine S-methyltransferase 3); homocysteine S-methyltransferase | -2.96 |
| 1118 | 258323_at | AT3G22750 | protein kinase, putative | -3.84 |
| 1119 | 258324_at | AT3G22780 | TSO1 (CHINESE FOR 'UGLY'); transcription factor | 1.66 |
| 1120 | 258341_at | AT3G22790 | kinase interacting family protein | 2.47 |
| 1121 | 256832_at | AT3G22880 | ATDMC1 (RECA-LIKE GENE); ATP binding / DNA-dependent ATPase/ damaged DNA binding | 1.63 |
| 1122 | 256835_at | AT3G22890 | APS1 (ATP sulfurylase 3) | -2.23 |
| 1123 | 256839_at | AT3G22930 | calmodulin, putative | -3.56 |
| 1124 | 257766_at | AT3G23030 | IAA2 (indoleacetic acid-induced protein 2); transcription factor | -3.25 |
| 1125 | 257769_at | AT3G23050 | IAA7 (AUXIN RESISTANT 2); transcription factor | -4.11 |
| 1126 | 257772_at | AT3G23080 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G14500.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO71306.1); contains InterPro domain Lipid-binding START (InterPro:IPR002913) | -2.16 |
| 1127 | 257761_at | AT3G23090 | similar to WDL1 (WVD2-LIKE 1) [Arabidopsis thaliana] (TAIR:AT3G04630.2); similar to WDL1 (WVD2-LIKE 1) [Arabidopsis thaliana] (TAIR:AT3G04630.3); similar to seed specific protein Bn15D14A [Brassica napus] (GB:AAP37969.1); contains InterPro domain Targeting for Xklp2 (InterPro:IPR009675) | -4.71 |
| 1128 | 257925_at | AT3G23170 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G14450.1) | -2.13 |
| 1129 | 256942_at | AT3G23290 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G31160.1); similar to unknown [Populus trichocarpa] (GB:ABK96211.1); contains InterPro domain Protein of unknown function DUF640 (InterPro:IPR006936) | 2.33 |
| 1130 | 258295_at | AT3G23400 | plastid-lipid associated protein PAP / fibrillin family protein | -2.06 |
| 1131 | 258299_at | AT3G23410 | alcohol oxidase-related | -2.56 |
| 1132 | 258098_at | AT3G23670 | KINESIN-12B/PAKRP1L; microtubule motor/ plus-end-directed microtubule motor | 2.45 |
| 1133 | 257203_at | AT3G23730 | xyloglucan:xyloglucosyl transferase, putative / xyloglucan endotransglycosylase, putative / endo-xyloglucan transferase, putative | 1.72 |
| 1134 | 257201_at | AT3G23740 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G14120.1); similar to hypothetical protein OsI_000016 [Oryza sativa (indica cultivar-group)] (GB:EAY72169.1); similar to Os01g0101800 [Oryza sativa (japonica cultivar-group)] (GB:NP_001041745.1) | 1.76 |
| 1135 | 256890_at | AT3G23830 | GR-RBP4/GRP4 (GLYCINE-RICH RNA-BINDING PROTEIN 4); RNA binding / double-stranded DNA binding / single-stranded DNA binding | 2.22 |
| 1136 | 256864_at | AT3G23890 | TOPII (TOPOISOMERASE II); ATP binding / DNA binding / DNA topoisomerase (ATP-hydrolyzing) | 3.45 |
| 1137 | 257164_at | AT3G24320 | ATMSH1/CHM/CHM1/MSH1 (MUTL PROTEIN HOMOLOG 1); ATP binding / damaged DNA binding | 2.11 |
| 1138 | 257167_at | AT3G24340 | CHR40 (chromatin remodeling 40); ATP binding / DNA binding / helicase | 2.14 |
| 1139 | 256621_at | AT3G24450 | copper-binding family protein | 2.10 |
| 1140 | 258138_at | AT3G24495 | MSH7 (MUTS HOMOLOG 6-2); ATP binding / damaged DNA binding | 1.94 |
| 1141 | 258140_at | AT3G24503 | ALDH2C4 (REDUCED EPIDERMAL FLUORESCENCE1); 3-chloroallyl aldehyde dehydrogenase/ aldehyde dehydrogenase (NAD) | -2.18 |
| 1142 | 256899_at | AT3G24660 | TMKL1 (TRANSMEMBRANE KINASE-LIKE 1); ATP binding / kinase/ protein serine/threonine kinase | 2.00 |
| 1143 | 257619_at | AT3G24810 | ICK3 (kip-related protein 5); cyclin-dependent protein kinase inhibitor | 1.55 |
| 1144 | 257813_at | AT3G25100 | CDC45 (CELL DIVISION CYCLE 45) | 1.90 |
| 1145 | 257824_at | AT3G25290 | auxin-responsive family protein | -2.35 |
| 1146 | 257956_at | AT3G25400 | similar to unnamed protein product [Vitis vinifera] (GB:CAO45250.1); contains InterPro domain NTP pyrophosphohydrolase MazG, putative catalytic core (InterPro:IPR004518); contains InterPro domain NTP Pyrophosphohydrolase MazG-related, RS21-C6 (InterPro:IPR011394); contains InterPro domain EAR (InterPro:IPR009039) | 1.48 |
| 1147 | 256755_at | AT3G25600 | calmodulin, putative | -2.22 |
| 1148 | 256756_at | AT3G25610 | haloacid dehalogenase-like hydrolase family protein | -3.79 |
| 1149 | 256761_at | AT3G25670 | leucine-rich repeat family protein | 2.01 |
| 1150 | 256754_at | AT3G25690 | CHUP1 (CHLOROPLAST UNUSUAL POSITIONING 1) | -2.04 |
| 1151 | 257642_at | AT3G25710 | basic helix-loop-helix (bHLH) family protein | 2.48 |
| 1152 | 258082_at | AT3G25905 | CLE27 (CLAVATA3/ESR-RELATED 27); receptor binding | 2.27 |
| 1153 | 258067_at | AT3G25980 | mitotic spindle checkpoint protein, putative (MAD2) | 2.17 |
| 1154 | 257542_at | AT3G26050 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G27350.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO14633.1); contains InterPro domain Targeting for Xklp2 (InterPro:IPR009675) | 1.75 |
| 1155 | 257623_at | AT3G26210 | CYP71B23 (cytochrome P450, family 71, subfamily B, polypeptide 23); oxygen binding | -2.21 |
| 1156 | 257635_at | AT3G26280 | CYP71B4 (cytochrome P450, family 71, subfamily B, polypeptide 4); oxygen binding | -3.00 |
| 1157 | 256875_at | AT3G26330 | CYP71B37 (cytochrome P450, family 71, subfamily B, polypeptide 37); oxygen binding | 2.25 |
| 1158 | 256880_at | AT3G26450 | major latex protein-related / MLP-related | -2.88 |
| 1159 | 256877_at | AT3G26470 | similar to ADR1-L1 (ADR1-LIKE 1), ATP binding / protein binding [Arabidopsis thaliana] (TAIR:AT4G33300.2); similar to ADR1-L1 (ADR1-LIKE 1), ATP binding / protein binding [Arabidopsis thaliana] (TAIR:AT4G33300.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO61278.1); contains InterPro domain Disease resistance, plant (InterPro:IPR014011) | -2.98 |
| 1160 | 258277_at | AT3G26830 | PAD3 (PHYTOALEXIN DEFICIENT 3); oxygen binding | -1.96 |
| 1161 | 256980_at | AT3G26932 | DRB3 (DSRNA-BINDING PROTEIN 3); double-stranded RNA binding | 2.62 |
| 1162 | 257779_at | AT3G26940 | CDG1 (CONSTITUTIVE DIFFERENTIAL GROWTH 1); kinase | -2.77 |
| 1163 | 257793_at | AT3G26960 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G41050.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO49052.1) | -2.08 |
| 1164 | 257791_at | AT3G27110 | peptidase M48 family protein | -1.94 |
| 1165 | 257781_at | AT3G27120 | ATPase | 1.50 |
| 1166 | 257151_at | AT3G27200 | plastocyanin-like domain-containing protein | -1.94 |
| 1167 | 257153_at | AT3G27220 | kelch repeat-containing protein | 1.91 |
| 1168 | 257149_at | AT3G27280 | ATPHB4 (PROHIBITIN 4) | 1.70 |
| 1169 | 257740_at | AT3G27330 | zinc finger (C3HC4-type RING finger) family protein | 2.65 |
| 1170 | 257714_at | AT3G27360 | histone H3 | 2.33 |
| 1171 | 258239_at | AT3G27690 | LHCB2:4 (Photosystem II light harvesting complex gene 2.3); chlorophyll binding | -3.51 |
| 1172 | 257071_at | AT3G28180 | ATCSLC04 (CELLULOSE-SYNTHASE LIKE C 4); transferase, transferring glycosyl groups | -2.38 |
| 1173 | 256576_at | AT3G28210 | PMZ; zinc ion binding | -3.41 |
| 1174 | 256603_at | AT3G28270 | Identical to UPF0496 protein At3g28270 [Arabidopsis Thaliana] (GB:Q9LHD9;GB:Q8H7D0;GB:Q8H7E9;GB:Q944H3); similar to AT14A [Arabidopsis thaliana] (TAIR:AT3G28290.1); similar to AT14A [Arabidopsis thaliana] (TAIR:AT3G28300.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO22906.1); contains InterPro domain Protein of unknown function DUF677 (InterPro:IPR007749) | -5.72 |
| 1175 | 256597_at | AT3G28500 | 60S acidic ribosomal protein P2 (RPP2C) | 3.65 |
| 1176 | 256593_at | AT3G28510 | AAA-type ATPase family protein | -2.45 |
| 1177 | 256987_at | AT3G28560 | similar to AAA-type ATPase family protein [Arabidopsis thaliana] (TAIR:AT3G28510.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN79616.1); contains domain PTHR23070 (PTHR23070); contains domain PTHR23070:SF1 (PTHR23070:SF1) | 4.79 |
| 1178 | 258003_at | AT3G29030 | ATEXPA5 (ARABIDOPSIS THALIANA EXPANSIN A5) | -2.22 |
| 1179 | 256612_at | AT3G29280 | similar to unknown [Populus trichocarpa] (GB:ABK93016.1) | 1.53 |
| 1180 | 256736_at | AT3G29410 | terpene synthase/cyclase family protein | 2.09 |
| 1181 | 257288_at | AT3G29670 | transferase family protein | -4.06 |
| 1182 | 245226_at | AT3G29970 | germination protein-related | 2.22 |
| 1183 | 252847_at | AT3G42170 | transposase-like gene with conserved domains from the family of hAT transposases that includes hobo from Drosophila melanogaster, Activator (Ac) from maize, and Tam3 from snapdragon but lacks several amino acids known to be essential for Ac transposition5. The DAYSLEEPER gene lacks 8 bp duplications and TIRs (a common feature of transcriptionally silent hAT transposases), however, DAYSLEEPER expression was detected, and several expressed sequence tags are available. The expression seems to be under the control of factors determining the circadian rhythm. DAYSLEEPER was isolated as a factor binding to a motif (Kubox1) present in the upstream region of the Arabidopsis DNA repair gene Ku70. Mutant plants lacking DAYSLEEPER or strongly overexpressing this gene do not develop in a normal manner. | 1.84 |
| 1184 | 252785_at | AT3G42660 | nucleotide binding | 1.96 |
| 1185 | 252770_at | AT3G42860 | zinc knuckle (CCHC-type) family protein | 2.22 |
| 1186 | 252716_at | AT3G43920 | DCL3 (DICER-LIKE 3); RNA binding / ribonuclease III | 1.62 |
| 1187 | 252692_at | AT3G43960 | cysteine proteinase, putative | 1.47 |
| 1188 | 252691_at | AT3G44050 | kinesin motor protein-related | 2.78 |
| 1189 | 252679_at | AT3G44260 | CCR4-NOT transcription complex protein, putative | -2.31 |
| 1190 | 252661_at | AT3G44450 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G52740.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO70571.1) | -2.77 |
| 1191 | 252625_at | AT3G44750 | HD2A (HISTONE DEACETYLASE 2A); nucleic acid binding / zinc ion binding | 2.06 |
| 1192 | 252628_at | AT3G44960 | similar to unnamed protein product [Vitis vinifera] (GB:CAO16756.1) | 1.86 |
| 1193 | 252607_at | AT3G44990 | XTR8 (xyloglucan:xyloglucosyl transferase 8); hydrolase, acting on glycosyl bonds | -5.53 |
| 1194 | 252606_at | AT3G45010 | SCPL48 (serine carboxypeptidase-like 48); serine carboxypeptidase | -2.42 |
| 1195 | 252604_at | AT3G45060 | ATNRT2.6 (Arabidopsis thaliana high affinity nitrate transporter 2.6); nitrate transmembrane transporter | 1.68 |
| 1196 | 252612_at | AT3G45160 | contains domain PROKAR_LIPOPROTEIN (PS51257) | -7.67 |
| 1197 | 252591_at | AT3G45600 | TET3 (TETRASPANIN3) | -2.78 |
| 1198 | 252586_at | AT3G45610 | Dof-type zinc finger domain-containing protein | 3.50 |
| 1199 | 252592_at | AT3G45640 | ATMPK3 (MITOGEN-ACTIVATED PROTEIN KINASE 3); MAP kinase/ kinase/ protein kinase | -2.27 |
| 1200 | 252539_at | AT3G45730 | unknown protein | -4.59 |
| 1201 | 252543_at | AT3G45780 | PHOT1 (phototropin 1); kinase | -2.51 |
| 1202 | 252557_at | AT3G45960 | ATEXLA3 (ARABIDOPSIS THALIANA EXPANSIN-LIKE A3) | -2.83 |
| 1203 | 252563_at | AT3G45970 | ATEXLA1 (ARABIDOPSIS THALIANA EXPANSIN-LIKE A1) | -4.62 |
| 1204 | 252534_at | AT3G46130 | MYB111 (myb domain protein 111); DNA binding / transcription factor | -3.84 |
| 1205 | 252483_at | AT3G46600 | scarecrow transcription factor family protein | -1.85 |
| 1206 | 252441_at | AT3G46780 | PTAC16 (PLASTID TRANSCRIPTIONALLY ACTIVE18); binding / catalytic | -2.53 |
| 1207 | 252442_at | AT3G46940 | deoxyuridine 5'-triphosphate nucleotidohydrolase family | 1.51 |
| 1208 | 252462_at | AT3G47250 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G47200.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G47200.2); similar to unnamed protein product [Vitis vinifera] (GB:CAO15956.1); contains InterPro domain Protein of unknown function DUF247, plant (InterPro:IPR004158) | -3.08 |
| 1209 | 252411_at | AT3G47430 | PEX11B | -2.06 |
| 1210 | 252416_at | AT3G47460 | ATSMC2 (Arabidopsis thaliana structural maintenance of chromosome 2) | 1.48 |
| 1211 | 252419_at | AT3G47510 | unknown protein | -2.32 |
| 1212 | 252383_at | AT3G47780 | ATATH6 (ABC2 homolog 6); ATPase, coupled to transmembrane movement of substances | -2.60 |
| 1213 | 252367_at | AT3G48360 | BT2 (BTB AND TAZ DOMAIN PROTEIN 2); protein binding / transcription factor/ transcription regulator | -4.32 |
| 1214 | 252357_at | AT3G48410 | hydrolase, alpha/beta fold family protein | 1.53 |
| 1215 | 252366_at | AT3G48420 | haloacid dehalogenase-like hydrolase family protein | -2.17 |
| 1216 | 252358_at | AT3G48425 | endonuclease/exonuclease/phosphatase family protein | 1.56 |
| 1217 | 252360_at | AT3G48480 | cysteine-type peptidase | 1.92 |
| 1218 | 252316_at | AT3G48700 | ATCXE13; hydrolase | 1.73 |
| 1219 | 252319_at | AT3G48710 | GTP binding / RNA binding | 1.63 |
| 1220 | 252317_at | AT3G48720 | transferase family protein | -2.63 |
| 1221 | 252327_at | AT3G48740 | nodulin MtN3 family protein | -4.30 |
| 1222 | 252330_at | AT3G48770 | ATP binding / DNA binding | 2.03 |
| 1223 | 252293_at | AT3G48990 | AMP-dependent synthetase and ligase family protein | -3.57 |
| 1224 | 252288_at | AT3G49080 | ribosomal protein S9 family protein | 1.53 |
| 1225 | 252305_at | AT3G49240 | EMB1796 (EMBRYO DEFECTIVE 1796); binding | 1.47 |
| 1226 | 252245_at | AT3G49710 | pentatricopeptide (PPR) repeat-containing protein | 1.59 |
| 1227 | 252248_at | AT3G49750 | leucine-rich repeat family protein | 2.00 |
| 1228 | 252234_at | AT3G49780 | ATPSK4 (PHYTOSULFOKINE 4 PRECURSOR); growth factor | -4.69 |
| 1229 | 252250_at | AT3G49790 | ATP binding | -1.98 |
| 1230 | 252227_at | AT3G49900 | BTB/POZ domain-containing protein | 2.15 |
| 1231 | 252189_at | AT3G50070 | CYCD3;3 (CYCLIN D3;3); cyclin-dependent protein kinase | 3.04 |
| 1232 | 252243_at | AT3G50120 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G50150.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G50130.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G50170.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO71911.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN59797.1); contains InterPro domain Protein of unknown function DUF247, plant (InterPro:IPR004158) | -3.31 |
| 1233 | 252190_at | AT3G50170 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G50150.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G50130.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G50120.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO71911.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN59797.1); contains InterPro domain Protein of unknown function DUF247, plant (InterPro:IPR004158) | 3.83 |
| 1234 | 252197_at | AT3G50230 | leucine-rich repeat transmembrane protein kinase, putative | 3.08 |
| 1235 | 252214_at | AT3G50260 | ATERF#011/CEJ1 (COOPERATIVELY REGULATED BY ETHYLENE AND JASMONATE 1); DNA binding / transcription factor | -2.57 |
| 1236 | 252210_at | AT3G50410 | OBP1 (OBF BINDING PROTEIN 1); DNA binding / transcription factor | 1.92 |
| 1237 | 252170_at | AT3G50480 | HR4 (HOMOLOG OF RPW8 4) | -2.44 |
| 1238 | 252163_at | AT3G50610 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G66816.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO71825.1) | -2.66 |
| 1239 | 252183_at | AT3G50740 | UGT72E1 (UDP-glucosyl transferase 72E1); UDP-glycosyltransferase/ coniferyl-alcohol glucosyltransferase/ transferase, transferring glycosyl groups | -4.12 |
| 1240 | 252130_at | AT3G50820 | PSBO-2/PSBO2 (PHOTOSYSTEM II SUBUNIT O-2); oxygen evolving/ poly(U) binding | -2.97 |
| 1241 | 252128_at | AT3G50870 | MNP (MONOPOLE); transcription factor | 3.65 |
| 1242 | 252129_at | AT3G50890 | ATHB28 (ARABIDOPSIS THALIANA HOMEOBOX PROTEIN 28); DNA binding / transcription factor | 1.88 |
| 1243 | 252133_at | AT3G50900 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G66490.1); similar to hypothetical protein [Thellungiella halophila] (GB:ABB45855.1) | -5.76 |
| 1244 | 252131_at | AT3G50930 | AAA-type ATPase family protein | -2.48 |
| 1245 | 252102_at | AT3G50970 | LTI30 (LOW TEMPERATURE-INDUCED 30) | -3.27 |
| 1246 | 252095_at | AT3G51000 | epoxide hydrolase, putative | -1.85 |
| 1247 | 252123_at | AT3G51240 | F3H (TRANSPARENT TESTA 6); naringenin 3-dioxygenase | 1.60 |
| 1248 | 252148_at | AT3G51280 | male sterility MS5, putative | 2.70 |
| 1249 | 252149_at | AT3G51290 | proline-rich family protein | 3.05 |
| 1250 | 252092_at | AT3G51420 | strictosidine synthase family protein | -2.44 |
| 1251 | 252077_at | AT3G51720 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G38370.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO48842.1); contains InterPro domain Protein of unknown function DUF827, plant (InterPro:IPR008545) | 2.42 |
| 1252 | 252078_at | AT3G51740 | IMK2 (INFLORESCENCE MERISTEM RECEPTOR-LIKE KINASE 2); ATP binding / kinase/ protein serine/threonine kinase | 2.27 |
| 1253 | 252073_at | AT3G51750 | unknown protein | -2.74 |
| 1254 | 246302_at | AT3G51860 | CAX3 (cation exchanger 3); cation:cation antiporter | -1.87 |
| 1255 | 252037_at | AT3G51920 | CAM9 (CALMODULIN 9); calcium ion binding | -2.82 |
| 1256 | 252089_at | AT3G52110 | similar to hypothetical protein OsI_009975 [Oryza sativa (indica cultivar-group)] (GB:EAY88742.1); similar to Os03g0174200 [Oryza sativa (japonica cultivar-group)] (GB:NP_001049125.1) | 1.78 |
| 1257 | 256675_at | AT3G52170 | DNA binding | 1.58 |
| 1258 | 252053_at | AT3G52400 | SYP122 (syntaxin 122); SNAP receptor | -3.31 |
| 1259 | 252054_at | AT3G52540 | ATOFP18/OFP18 (Arabidopsis thaliana ovate family protein 18) | 1.49 |
| 1260 | 252011_at | AT3G52720 | carbonic anhydrase family protein | -6.00 |
| 1261 | 252010_at | AT3G52740 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G44450.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO70571.1) | -2.65 |
| 1262 | 252003_at | AT3G52770 | ZPR3 (LITTLE ZIPPER 3); protein binding | 1.48 |
| 1263 | 252009_at | AT3G52800 | zinc finger (AN1-like) family protein | -2.43 |
| 1264 | 251996_at | AT3G52840 | BGAL2 (beta-galactosidase 2); beta-galactosidase | -3.45 |
| 1265 | 251994_at | AT3G52890 | KIPK (KCBP-INTERACTING PROTEIN KINASE); kinase | 1.49 |
| 1266 | 252017_at | AT3G52970 | CYP76G1 (cytochrome P450, family 76, subfamily G, polypeptide 1); oxygen binding | -1.88 |
| 1267 | 251982_at | AT3G53190 | pectate lyase family protein | 2.10 |
| 1268 | 251977_at | AT3G53250 | auxin-responsive family protein | -2.13 |
| 1269 | 251987_at | AT3G53280 | CYP71B5 (CYTOCHROME P450 71B5); oxygen binding | -3.18 |
| 1270 | 251986_at | AT3G53310 | transcriptional factor B3 family protein | 3.32 |
| 1271 | 251990_at | AT3G53320 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G37070.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO18110.1) | 1.78 |
| 1272 | 251966_at | AT3G53380 | lectin protein kinase family protein | 1.92 |
| 1273 | 251962_at | AT3G53420 | PIP2A (PLASMA MEMBRANE INTRINSIC PROTEIN 2A); water channel | -2.23 |
| 1274 | 251952_at | AT3G53650 | histone H2B, putative | 2.40 |
| 1275 | 251906_at | AT3G53720 | ATCHX20 (CATION/H+ EXCHANGER 20); monovalent cation:proton antiporter | -3.04 |
| 1276 | 251924_at | AT3G53730 | histone H4 | 1.57 |
| 1277 | 251915_at | AT3G53940 | mitochondrial substrate carrier family protein | 1.52 |
| 1278 | 251927_at | AT3G53990 | universal stress protein (USP) family protein | -2.08 |
| 1279 | 251885_at | AT3G54050 | fructose-1,6-bisphosphatase, putative / D-fructose-1,6-bisphosphate 1-phosphohydrolase, putative / FBPase, putative | -3.13 |
| 1280 | 251882_at | AT3G54140 | proton-dependent oligopeptide transport (POT) family protein | -3.99 |
| 1281 | 251878_at | AT3G54310 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G38430.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO70793.1) | 1.90 |
| 1282 | 251898_at | AT3G54340 | AP3 (APETALA 3); DNA binding / transcription factor | 5.00 |
| 1283 | 251846_at | AT3G54560 | HTA11; DNA binding | 2.37 |
| 1284 | 251844_at | AT3G54630 | similar to kinetochore protein [Capsella rubella] (GB:BAF63163.1); similar to kinetochore protein [Olimarabidopsis pumila] (GB:BAF63394.1); similar to kinetochore protein [Olimarabidopsis pumila] (GB:BAF63395.1); contains InterPro domain HEC/Ndc80p (InterPro:IPR005550) | 3.43 |
| 1285 | 251856_at | AT3G54720 | AMP1 (ALTERED MERISTEM PROGRAM 1); dipeptidase | 1.85 |
| 1286 | 251858_at | AT3G54820 | PIP2;5/PIP2D (plasma membrane intrinsic protein 2;5); water channel | 2.67 |
| 1287 | 251826_at | AT3G55110 | ABC transporter family protein | 2.13 |
| 1288 | 251760_at | AT3G55605 | mitochondrial glycoprotein family protein / MAM33 family protein | 1.67 |
| 1289 | 251759_at | AT3G55630 | ATDFD (A. THALIANA DHFS-FPGS HOMOLOG D); tetrahydrofolylpolyglutamate synthase | -2.45 |
| 1290 | 251778_at | AT3G55660 | ATROPGEF6/ROPGEF6 (KINASE PARTNER PROTEIN-LIKE); Rho guanyl-nucleotide exchange factor/ | 2.39 |
| 1291 | 251762_at | AT3G55800 | SBPASE (SEDOHEPTULOSE-BISPHOSPHATASE); phosphoric ester hydrolase/ sedoheptulose-bisphosphatase | -2.00 |
| 1292 | 251770_at | AT3G55970 | oxidoreductase, 2OG-Fe(II) oxygenase family protein | -3.56 |
| 1293 | 251745_at | AT3G55980 | zinc finger (CCCH-type) family protein | -2.46 |
| 1294 | 251713_at | AT3G56080 | dehydration-responsive protein-related | -2.86 |
| 1295 | 251718_at | AT3G56100 | MRLK (MERISTEMATIC RECEPTOR-LIKE KINASE); ATP binding / protein serine/threonine kinase | 2.54 |
| 1296 | 251739_at | AT3G56170 | CAN (CA-2+ DEPENDENT NUCLEASE); nuclease | -2.67 |
| 1297 | 251722_at | AT3G56200 | amino acid transporter family protein | -2.57 |
| 1298 | 251733_at | AT3G56240 | CCH (COPPER CHAPERONE) | -4.85 |
| 1299 | 251727_at | AT3G56290 | similar to hypothetical protein [Vitis vinifera] (GB:CAN75527.1) | -1.94 |
| 1300 | 251704_at | AT3G56360 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G05250.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO41488.1) | -2.92 |
| 1301 | 251714_at | AT3G56370 | leucine-rich repeat transmembrane protein kinase, putative | 2.07 |
| 1302 | 251684_at | AT3G56410 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G05190.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G61670.1); similar to hypothetical protein Kpol_1058p4 [Vanderwaltozyma polyspora DSM 70294] (GB:XP_001645325.1) | -4.37 |
| 1303 | 251699_at | AT3G56630 | CYP94D2 (cytochrome P450, family 94, subfamily D, polypeptide 2); oxygen binding | -2.32 |
| 1304 | 246293_at | AT3G56710 | SIB1 (SIGMA FACTOR BINDING PROTEIN 1); binding | -2.03 |
| 1305 | 246315_at | AT3G56870 | similar to unnamed protein product [Vitis vinifera] (GB:CAO61177.1) | 1.84 |
| 1306 | 251678_at | AT3G56990 | EDA7 (embryo sac development arrest 7) | 1.61 |
| 1307 | 251658_at | AT3G57020 | strictosidine synthase family protein | -2.23 |
| 1308 | 251680_at | AT3G57060 | binding | 1.76 |
| 1309 | 251667_at | AT3G57150 | NAP57 (ARABIDOPSIS THALIANA HOMOLOGUE OF NAP57) | 1.61 |
| 1310 | 251625_at | AT3G57260 | BGL2 (PATHOGENESIS-RELATED PROTEIN 2); glucan 1,3-beta-glucosidase/ hydrolase, hydrolyzing O-glycosyl compounds | -3.54 |
| 1311 | 251636_at | AT3G57530 | CPK32 (CALCIUM-DEPENDENT PROTEIN KINASE 32); calmodulin-dependent protein kinase/ kinase | -1.99 |
| 1312 | 251592_at | AT3G57670 | NTT (NO TRANSMITTING TRACT); nucleic acid binding / transcription factor/ zinc ion binding | 2.43 |
| 1313 | 251605_at | AT3G57830 | leucine-rich repeat transmembrane protein kinase, putative | 2.49 |
| 1314 | 251608_at | AT3G57860 | UVI4-LIKE (UV-B-INSENSITIVE 4-LIKE) | 1.80 |
| 1315 | 251560_at | AT3G57920 | squamosa promoter-binding protein, putative | 3.24 |
| 1316 | 251574_at | AT3G58100 | glycosyl hydrolase family protein 17 | 1.50 |
| 1317 | 251535_at | AT3G58540 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G06190.1) | 1.52 |
| 1318 | 251527_at | AT3G58650 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G26910.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO17458.1) | 1.97 |
| 1319 | 251543_at | AT3G58770 | similar to hypothetical protein [Vitis vinifera] (GB:CAN63610.1) | 3.19 |
| 1320 | 251524_at | AT3G58990 | aconitase C-terminal domain-containing protein | -3.99 |
| 1321 | 251509_at | AT3G59010 | pectinesterase family protein | -2.71 |
| 1322 | 251497_at | AT3G59060 | PIL6 (PHYTOCHROME-INTERACTING FACTOR 5); DNA binding / transcription factor | -5.19 |
| 1323 | 251512_at | AT3G59190 | F-box family protein | 1.88 |
| 1324 | 251515_at | AT3G59270 | syntaxin-related family protein | 2.48 |
| 1325 | 251521_at | AT3G59420 | ACR4 (ARABIDOPSIS CRINKLY4); kinase | 2.24 |
| 1326 | 251485_at | AT3G59550 | SYN3 (SISTER CHROMATID COHESION 1 PROTEIN 3) | 1.60 |
| 1327 | 251443_at | AT3G59940 | kelch repeat-containing F-box family protein | -1.94 |
| 1328 | 251427_at | AT3G60130 | glycosyl hydrolase family 1 protein / beta-glucosidase, putative (YLS1) | -1.88 |
| 1329 | 251402_at | AT3G60290 | oxidoreductase, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors | -2.73 |
| 1330 | 251400_at | AT3G60420 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G60450.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO70569.1); contains InterPro domain Phosphoglycerate mutase (InterPro:IPR013078); contains InterPro domain PRIB5 (InterPro:IPR012398) | -5.62 |
| 1331 | 251342_at | AT3G60690 | auxin-responsive family protein | -1.96 |
| 1332 | 251339_at | AT3G60780 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G45360.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO24498.1); contains InterPro domain Protein of unknown function DUF1442 (InterPro:IPR009902) | -2.17 |
| 1333 | 251340_at | AT3G60830 | ATARP7 (ACTIN-RELATED PROTEIN 7); structural constituent of cytoskeleton | 1.46 |
| 1334 | 251394_at | AT3G60900 | FLA10 (fasciclin-like arabinogalactan-protein 10) | 1.69 |
| 1335 | 251354_at | AT3G61090 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G61100.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO40196.1); contains InterPro domain Protein of unknown function DUF537 (InterPro:IPR007491) | 1.71 |
| 1336 | 251355_at | AT3G61100 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G61090.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO40196.1); contains InterPro domain Protein of unknown function DUF537 (InterPro:IPR007491) | 1.49 |
| 1337 | 251336_at | AT3G61190 | BAP1 (BON ASSOCIATION PROTEIN 1) | -1.84 |
| 1338 | 251360_at | AT3G61210 | embryo-abundant protein-related | -2.84 |
| 1339 | 251361_at | AT3G61230 | LIM domain-containing protein | -1.95 |
| 1340 | 251363_at | AT3G61250 | AtMYB17 (myb domain protein 17); DNA binding / transcription factor | 3.59 |
| 1341 | 251365_at | AT3G61310 | DNA-binding family protein | 2.64 |
| 1342 | 251317_at | AT3G61490 | glycoside hydrolase family 28 protein / polygalacturonase (pectinase) family protein | 1.69 |
| 1343 | 251319_at | AT3G61610 | aldose 1-epimerase family protein | 2.18 |
| 1344 | 251289_at | AT3G61830 | ARF18 (AUXIN RESPONSE FACTOR 18); transcription factor | 1.89 |
| 1345 | 251246_at | AT3G62100 | IAA30 (indoleacetic acid-induced protein 30); transcription factor | 1.67 |
| 1346 | 251259_at | AT3G62260 | protein phosphatase 2C, putative / PP2C, putative | -2.08 |
| 1347 | 251256_at | AT3G62300 | agenet domain-containing protein | 1.77 |
| 1348 | 251218_at | AT3G62410 | CP12-2 | -4.69 |
| 1349 | 251221_at | AT3G62550 | universal stress protein (USP) family protein | -3.85 |
| 1350 | 251196_at | AT3G62950 | glutaredoxin family protein | -2.38 |
| 1351 | 251197_at | AT3G62960 | glutaredoxin family protein | -2.10 |
| 1352 | 251200_at | AT3G63010 | ATGID1B/GID1B (GA INSENSITIVE DWARF1B); hydrolase | 1.55 |
| 1353 | 251206_at | AT3G63090 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G31290.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G31290.2); similar to unnamed protein product [Vitis vinifera] (GB:CAO22503.1); contains InterPro domain Protein of unknown function DUF860, plant (InterPro:IPR008578) | 1.69 |
| 1354 | 251152_at | AT3G63130 | RANGAP1 (RAN GTPASE ACTIVATING PROTEIN 1); RAN GTPase activator | 2.19 |
| 1355 | 251157_at | AT3G63140 | mRNA-binding protein, putative | -2.57 |
| 1356 | 251155_at | AT3G63160 | unknown protein | -2.27 |
| 1357 | 251160_at | AT3G63240 | endonuclease/exonuclease/phosphatase family protein | 1.93 |
| 1358 | 251162_at | AT3G63300 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G22810.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN81204.1); contains InterPro domain Pleckstrin-like, plant (InterPro:IPR013666); contains InterPro domain Protein of unknown function DUF828, plant (InterPro:IPR008546) | 2.74 |
| 1359 | 251178_at | AT3G63440 | ATCKX6/ATCKX7/CKX6 (CYTOKININ OXIDASE/DEHYDROGENASE 6); cytokinin dehydrogenase | 2.24 |
| 1360 | 256248_at | AT3G66652 | fip1 motif-containing protein | 1.59 |
| 1361 | 255694_at | AT4G00050 | UNE10 (unfertilized embryo sac 10); DNA binding / transcription factor | -1.97 |
| 1362 | 255700_at | AT4G00200 | DNA binding | 1.63 |
| 1363 | 255716_at | AT4G00330 | CRCK2 (calmodulin-binding receptor-like cytoplasmic kinase 2); kinase | -2.83 |
| 1364 | 255690_at | AT4G00360 | CYP86A2 (ABERRANT INDUCTION OF TYPE THREE GENES 1); oxygen binding | -6.31 |
| 1365 | 255675_at | AT4G00480 | ATMYC1 (Arabidopsis thaliana myc-related transcription factor 1); DNA binding / transcription factor | 1.63 |
| 1366 | 255637_at | AT4G00750 | dehydration-responsive family protein | -2.53 |
| 1367 | 255626_at | AT4G00780 | meprin and TRAF homology domain-containing protein / MATH domain-containing protein | -4.36 |
| 1368 | 255642_at | AT4G00820 | IQD17 (IQ-domain 17); calmodulin binding | 2.04 |
| 1369 | 255644_at | AT4G00870 | basic helix-loop-helix (bHLH) family protein | 6.72 |
| 1370 | 255645_at | AT4G00880 | auxin-responsive family protein | -4.24 |
| 1371 | 255599_at | AT4G01010 | ATCNGC13 (cyclic nucleotide gated channel 13); calmodulin binding / cyclic nucleotide binding / ion channel | -2.28 |
| 1372 | 255613_at | AT4G01270 | zinc finger (C3HC4-type RING finger) family protein | 1.57 |
| 1373 | 255578_at | AT4G01450 | nodulin MtN21 family protein | -2.07 |
| 1374 | 255587_at | AT4G01480 | ATPPA5 (ARABIDOPSIS THALIANA PYROPHOSPHORYLASE 5); inorganic diphosphatase/ pyrophosphatase | -2.92 |
| 1375 | 255595_at | AT4G01700 | chitinase, putative | -2.45 |
| 1376 | 255596_at | AT4G01720 | WRKY47 (WRKY DNA-binding protein 47); transcription factor | -2.53 |
| 1377 | 255597_at | AT4G01730 | zinc finger (DHHC type) family protein | 2.23 |
| 1378 | 255549_at | AT4G01950 | ATGPAT3/GPAT3 (GLYCEROL-3-PHOSPHATE ACYLTRANSFERASE 3); acyltransferase | -5.54 |
| 1379 | 255513_at | AT4G02060 | PRL (PROLIFERA); ATP binding / DNA binding / DNA-dependent ATPase | 2.43 |
| 1380 | 255562_at | AT4G02070 | MSH6 (MUTS HOMOLOG 6-1) | 1.97 |
| 1381 | 255517_at | AT4G02290 | ATGH9B13 (ARABIDOPSIS THALIANA GLYCOSYL HYDROLASE 9B13); hydrolase, hydrolyzing O-glycosyl compounds | 3.09 |
| 1382 | 255479_at | AT4G02380 | SAG21 (SENESCENCE-ASSOCIATED GENE 21) | -6.71 |
| 1383 | 255502_at | AT4G02410 | lectin protein kinase family protein | -3.76 |
| 1384 | 255491_at | AT4G02670 | ATIDD12 (ARABIDOPSIS THALIANA INDETERMINATE(ID)-DOMAIN 12); nucleic acid binding / transcription factor/ zinc ion binding | 2.42 |
| 1385 | 255460_at | AT4G02800 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G30050.1); similar to Os04g0228100 [Oryza sativa (japonica cultivar-group)] (GB:NP_001052288.1); similar to H0209A05.2 [Oryza sativa (indica cultivar-group)] (GB:CAH66085.1) | 2.61 |
| 1386 | 255448_at | AT4G02810 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G03170.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN75990.1) | 1.89 |
| 1387 | 255456_at | AT4G02920 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G03340.1); similar to hypothetical protein MtrDRAFT_AC161864g5v2 [Medicago truncatula] (GB:ABE86673.1) | -2.09 |
| 1388 | 255467_at | AT4G03010 | leucine-rich repeat family protein | 1.88 |
| 1389 | 255409_at | AT4G03090 | similar to unnamed protein product [Vitis vinifera] (GB:CAO22433.1); contains InterPro domain PUA-like (InterPro:IPR015947) | 1.49 |
| 1390 | 255410_at | AT4G03100 | rac GTPase activating protein, putative | 1.79 |
| 1391 | 255433_at | AT4G03210 | XTH9 (XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 9); hydrolase, acting on glycosyl bonds | 1.48 |
| 1392 | 255422_at | AT4G03270 | CYCD6;1 (CYCLIN D6;1); cyclin-dependent protein kinase | 1.50 |
| 1393 | 255357_at | AT4G03930 | pectinesterase | -1.95 |
| 1394 | 255331_at | AT4G04330 | similar to unnamed protein product [Vitis vinifera] (GB:CAO69665.1) | -1.88 |
| 1395 | 255345_at | AT4G04460 | aspartyl protease family protein | -2.62 |
| 1396 | 255340_at | AT4G04490 | protein kinase family protein | -2.15 |
| 1397 | 255298_at | AT4G04840 | methionine sulfoxide reductase domain-containing protein / SeIR domain-containing protein | -4.87 |
| 1398 | 255259_at | AT4G05020 | NDB2 (NAD(P)H DEHYDROGENASE B2); disulfide oxidoreductase | -2.00 |
| 1399 | 255248_at | AT4G05180 | PSBQ/PSBQ-2/PSII-Q (PHOTOSYSTEM II SUBUNIT Q-2); calcium ion binding | -1.98 |
| 1400 | 255265_at | AT4G05190 | ATK5 (Arabidopsis thaliana kinesin 5); microtubule motor | 2.41 |
| 1401 | 255266_at | AT4G05200 | protein kinase family protein | -2.97 |
| 1402 | 255275_at | AT4G05310 | ubiquitin family protein | -1.85 |
| 1403 | 255225_at | AT4G05410 | transducin family protein / WD-40 repeat family protein | 1.46 |
| 1404 | 255236_at | AT4G05520 | calcium-binding EF hand family protein | 1.94 |
| 1405 | 255172_x_at | AT4G08000 | transposable element gene | 1.64 |
| 1406 | 255149_at | AT4G08150 | KNAT1 (BREVIPEDICELLUS 1); transcription factor | 2.98 |
| 1407 | 255129_at | AT4G08290 | nodulin MtN21 family protein | -2.96 |
| 1408 | 255127_at | AT4G08300 | nodulin MtN21 family protein | -3.95 |
| 1409 | 255116_at | AT4G08850 | leucine-rich repeat family protein / protein kinase family protein | -2.37 |
| 1410 | 255061_at | AT4G08930 | ATAPRL6 (APR-LIKE 6) | -1.85 |
| 1411 | 255088_at | AT4G09350 | DNAJ heat shock N-terminal domain-containing protein | -2.04 |
| 1412 | 255038_at | AT4G09510 | beta-fructofuranosidase, putative / invertase, putative / saccharase, putative / beta-fructosidase, putative | 1.84 |
| 1413 | 255046_at | AT4G09650 | ATP synthase delta chain, chloroplast, putative / H(+)-transporting two-sector ATPase, delta (OSCP) subunit, putative | -2.36 |
| 1414 | 255028_at | AT4G09890 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G47480.1); similar to unknown [Populus trichocarpa] (GB:ABK92947.1) | -2.37 |
| 1415 | 255025_at | AT4G09900 | hydrolase, alpha/beta fold family protein | -1.85 |
| 1416 | 255011_at | AT4G10040 | CYTC-2 (CYTOCHROME C-2); electron carrier | -1.96 |
| 1417 | 255016_at | AT4G10120 | ATSPS4F; sucrose-phosphate synthase/ transferase, transferring glycosyl groups | -5.23 |
| 1418 | 255807_at | AT4G10270 | wound-responsive family protein | 3.88 |
| 1419 | 254964_at | AT4G11080 | high mobility group (HMG1/2) family protein | 1.57 |
| 1420 | 254933_at | AT4G11130 | RDR2 (RNA-DEPENDENT RNA POLYMERASE 2); RNA-directed RNA polymerase | 1.70 |
| 1421 | 254907_at | AT4G11190 | disease resistance-responsive family protein / dirigent family protein | 6.86 |
| 1422 | 254908_at | AT4G11200 | transposable element gene | 1.90 |
| 1423 | 254886_at | AT4G11760 | LCR17 (Low-molecular-weight cysteine-rich 17) | -2.32 |
| 1424 | 254862_at | AT4G12030 | bile acid:sodium symporter family protein | -3.48 |
| 1425 | 254853_at | AT4G12080 | DNA-binding family protein | -3.76 |
| 1426 | 254823_at | AT4G12580 | unknown protein | -2.03 |
| 1427 | 254808_at | AT4G12740 | adenine-DNA glycosylase-related / MYH-related | 1.45 |
| 1428 | 254783_at | AT4G12830 | hydrolase, alpha/beta fold family protein | -2.48 |
| 1429 | 254777_at | AT4G12960 | gamma interferon responsive lysosomal thiol reductase family protein / GILT family protein | 2.44 |
| 1430 | 254804_at | AT4G13010 | oxidoreductase, zinc-binding dehydrogenase family protein | -1.86 |
| 1431 | 254759_at | AT4G13180 | short-chain dehydrogenase/reductase (SDR) family protein | -2.44 |
| 1432 | 254754_at | AT4G13210 | pectate lyase family protein | 2.11 |
| 1433 | 254744_at | AT4G13345 | MEE55 (maternal effect embryo arrest 55) | -2.17 |
| 1434 | 254716_at | AT4G13560 | UNE15 (unfertilized embryo sac 15) | -4.25 |
| 1435 | 254728_at | AT4G13690 | similar to hypothetical protein MtrDRAFT_AC161864g11v2 [Medicago truncatula] (GB:ABE86675.1) | 2.73 |
| 1436 | 254687_at | AT4G13770 | CYP83A1 (CYTOCHROME P450 83A1); oxygen binding | -4.27 |
| 1437 | 254688_at | AT4G13830 | J20 (DNAJ-LIKE 20); heat shock protein binding | -1.94 |
| 1438 | 254737_at | AT4G13840 | transferase family protein | -2.15 |
| 1439 | 245385_at | AT4G14020 | rapid alkalinization factor (RALF) family protein | -4.55 |
| 1440 | 245259_at | AT4G14150 | PAKRP1 (PHRAGMOPLAST-ASSOCIATED KINESIN-RELATED PROTEIN 1); microtubule motor/ plus-end-directed microtubule motor | 2.29 |
| 1441 | 245598_at | AT4G14200 | similar to unnamed protein product [Vitis vinifera] (GB:CAO66985.1); contains domain PENTATRICOPEPTIDE REPEAT-CONTAINING PROTEIN-RELATED (PTHR10483:SF9); contains domain PENTATRICOPEPTIDE REPEAT-CONTAINING PROTEIN (PTHR10483) | 2.18 |
| 1442 | 245607_at | AT4G14330 | phragmoplast-associated kinesin-related protein 2 (PAKRP2) | 2.40 |
| 1443 | 245265_at | AT4G14400 | ACD6 (ACCELERATED CELL DEATH 6); protein binding | -3.56 |
| 1444 | 245617_at | AT4G14490 | forkhead-associated domain-containing protein / FHA domain-containing protein | 1.64 |
| 1445 | 245566_at | AT4G14610 | pseudogene, disease resistance protein (CC-NBS-LRR class), putative, domain signature CC-NBS-LRR exists, suggestive of a disease resistance protein.; blastp match of 45% identity and 2.2e-162 P-value to GP|24461866|gb|AAN62353.1|AF506028_20|AF506028 NBS-LRR type disease resistance protein {Poncirus trifoliata} | -3.71 |
| 1446 | 245571_at | AT4G14695 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G22310.1); similar to unknown [Populus trichocarpa] (GB:ABK93494.1); contains InterPro domain Protein of unknown function UPF0041 (InterPro:IPR005336) | 2.39 |
| 1447 | 245572_at | AT4G14720 | PPD2 (PEAPOD 2) | 1.68 |
| 1448 | 245576_at | AT4G14770 | tesmin/TSO1-like CXC domain-containing protein | 1.65 |
| 1449 | 245585_at | AT4G14970 | similar to hypothetical protein [Vitis vinifera] (GB:CAN76433.1); similar to Os07g0154400 [Oryza sativa (japonica cultivar-group)] (GB:NP_001058915.1); similar to hypothetical protein OsI_024046 [Oryza sativa (indica cultivar-group)] (GB:EAZ02814.1) | 2.14 |
| 1450 | 245586_at | AT4G14980 | DC1 domain-containing protein | 3.29 |
| 1451 | 245309_at | AT4G15140 | similar to unknown [Populus trichocarpa] (GB:ABK96081.1) | 2.23 |
| 1452 | 245275_at | AT4G15210 | ATBETA-AMY (BETA-AMYLASE); beta-amylase | 2.57 |
| 1453 | 245560_at | AT4G15480 | UGT84A1; UDP-glycosyltransferase/ sinapate 1-glucosyltransferase/ transferase, transferring glycosyl groups | 6.18 |
| 1454 | 245528_at | AT4G15530 | PPDK (PYRUVATE ORTHOPHOSPHATE DIKINASE); kinase/ pyruvate, phosphate dikinase | -6.64 |
| 1455 | 245317_at | AT4G15610 | integral membrane family protein | -2.04 |
| 1456 | 245505_at | AT4G15690 | glutaredoxin family protein | -5.68 |
| 1457 | 245506_at | AT4G15700 | glutaredoxin family protein | -3.52 |
| 1458 | 245364_at | AT4G15790 | similar to unnamed protein product [Vitis vinifera] (GB:CAO70981.1) | 1.64 |
| 1459 | 245343_at | AT4G15830 | binding | 2.68 |
| 1460 | 245522_at | AT4G15890 | binding | 1.82 |
| 1461 | 245523_at | AT4G15910 | ATDI21 (Arabidopsis thaliana drought-induced 21) | 1.80 |
| 1462 | 245353_at | AT4G16000 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G15990.1) | -3.63 |
| 1463 | 245483_at | AT4G16190 | cysteine proteinase, putative | -1.94 |
| 1464 | 245488_at | AT4G16270 | peroxidase 40 (PER40) (P40) | 3.24 |
| 1465 | 245336_at | AT4G16515 | unknown protein | -2.58 |
| 1466 | 245262_at | AT4G16563 | aspartyl protease family protein | -2.35 |
| 1467 | 245314_at | AT4G16745 | exostosin family protein | -1.85 |
| 1468 | 245276_at | AT4G16780 | ATHB-2 (ARABIDOPSIS THALIANA HOMEOBOX PROTEIN 2); DNA binding / transcription factor | -2.21 |
| 1469 | 245448_at | AT4G16860 | RPP4 (RECOGNITION OF PERONOSPORA PARASITICA 4) | -2.57 |
| 1470 | 245452_at | AT4G16910 | transposable element gene | -2.20 |
| 1471 | 245460_at | AT4G16990 | disease resistance protein (TIR-NBS class), putative | -3.04 |
| 1472 | 245461_at | AT4G17000 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G28770.1); similar to hypothetical protein [Capsella rubella] (GB:CAC82614.1) | 2.04 |
| 1473 | 245463_at | AT4G17030 | ATEXLB1 (ARABIDOPSIS THALIANA EXPANSIN-LIKE B1) | -2.75 |
| 1474 | 245407_at | AT4G17170 | AT-RAB2 (Arabidopsis Rab GTPase homolog B1c); GTP binding | -2.16 |
| 1475 | 245411_at | AT4G17240 | similar to hypothetical protein 31.t00013 [Brassica oleracea] (GB:ABD65111.1) | 1.78 |
| 1476 | 245264_at | AT4G17245 | zinc finger (C3HC4-type RING finger) family protein | -3.40 |
| 1477 | 245272_at | AT4G17250 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G47580.1); similar to hypothetical protein 40.t00008 [Brassica oleracea] (GB:ABD65133.1) | -4.17 |
| 1478 | 245412_at | AT4G17280 | auxin-responsive family protein | 1.50 |
| 1479 | 245399_at | AT4G17340 | DELTA-TIP2/TIP2;2 (tonoplast intrinsic protein 2;2); water channel | -3.86 |
| 1480 | 245418_at | AT4G17370 | oxidoreductase family protein | -2.00 |
| 1481 | 245419_at | AT4G17380 | MSH4 (MUTS-LIKE PROTEIN 4); ATP binding / damaged DNA binding | 1.68 |
| 1482 | 245389_at | AT4G17480 | palmitoyl protein thioesterase family protein | 2.08 |
| 1483 | 245250_at | AT4G17490 | ATERF6 (ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 6); DNA binding / transcription factor | -2.26 |
| 1484 | 245252_at | AT4G17500 | ATERF-1 (ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 1); DNA binding / transcription activator/ transcription factor | -3.52 |
| 1485 | 245404_at | AT4G17610 | tRNA/rRNA methyltransferase (SpoU) family protein | 1.63 |
| 1486 | 254691_at | AT4G17840 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G35260.1); similar to hypothetical protein 40.t00061 [Brassica oleracea] (GB:ABD65174.1) | -1.95 |
| 1487 | 254652_at | AT4G18170 | WRKY28 (WRKY DNA-binding protein 28); transcription factor | -2.88 |
| 1488 | 254667_at | AT4G18280 | glycine-rich cell wall protein-related | -2.32 |
| 1489 | 254665_at | AT4G18340 | glycosyl hydrolase family 17 protein | 2.05 |
| 1490 | 254638_at | AT4G18740 | transcription termination factor | -2.68 |
| 1491 | 254643_at | AT4G18820 | ATP binding / DNA binding / DNA-directed DNA polymerase/ nucleoside-triphosphatase/ nucleotide binding | 1.45 |
| 1492 | 254594_at | AT4G18930 | cyclic phosphodiesterase | -1.92 |
| 1493 | 254595_at | AT4G18960 | AG (AGAMOUS); transcription factor | 3.85 |
| 1494 | 254564_at | AT4G19170 | NCED4 (NINE-CIS-EPOXYCAROTENOID DIOXYGENASE 4) | -3.58 |
| 1495 | 254559_at | AT4G19200 | proline-rich family protein | -2.42 |
| 1496 | 254573_at | AT4G19420 | pectinacetylesterase family protein | -1.91 |
| 1497 | 254553_at | AT4G19530 | disease resistance protein (TIR-NBS-LRR class), putative | -2.67 |
| 1498 | 254550_at | AT4G19690 | IRT1 (IRON-REGULATED TRANSPORTER 1); cadmium ion transmembrane transporter/ iron ion transmembrane transporter/ manganese ion transmembrane transporter/ zinc ion transmembrane transporter | -2.12 |
| 1499 | 254551_at | AT4G19840 | ATPP2-A1 (Arabidopsis thaliana phloem protein 2-A1) | -5.28 |
| 1500 | 254548_at | AT4G19865 | kelch repeat-containing F-box family protein | 1.60 |
| 1501 | 254520_at | AT4G19960 | potassium ion transmembrane transporter | -2.59 |
| 1502 | 254496_at | AT4G20070 | ATAAH (ARABIDOPSIS THALIANA ALLANTOATE AMIDOHYDROLASE); allantoate deiminase/ metallopeptidase | -1.96 |
| 1503 | 254500_at | AT4G20110 | vacuolar sorting receptor, putative | -2.58 |
| 1504 | 254492_at | AT4G20260 | DREPP plasma membrane polypeptide family protein | -2.65 |
| 1505 | 254490_at | AT4G20320 | CTP synthase | 3.00 |
| 1506 | 254487_at | AT4G20780 | calcium-binding protein, putative | -2.05 |
| 1507 | 254430_at | AT4G20820 | FAD-binding domain-containing protein | -2.15 |
| 1508 | 254432_at | AT4G20830 | FAD-binding domain-containing protein | -2.99 |
| 1509 | 254431_at | AT4G20840 | FAD-binding domain-containing protein | -3.49 |
| 1510 | 254447_at | AT4G20860 | FAD-binding domain-containing protein | -5.19 |
| 1511 | 254449_at | AT4G20910 | HEN1 (HUA ENHANCER 1) | 1.59 |
| 1512 | 254443_at | AT4G21070 | ATBRCA1 (BREAST CANCER SUSCEPTIBILITY1); ubiquitin-protein ligase | 2.32 |
| 1513 | 254459_at | AT4G21200 | ATGA2OX8 (GIBBERELLIN 2-OXIDASE 8); gibberellin 2-beta-dioxygenase | 1.47 |
| 1514 | 254460_at | AT4G21210 | phosphoprotein phosphatase/ protein kinase | -2.30 |
| 1515 | 254400_at | AT4G21270 | ATK1 (ARABIDOPSIS THALIANA KINESIN 1); microtubule motor | 2.54 |
| 1516 | 254391_at | AT4G21590 | ENDO3 (ENDONUCLEASE 3); T/G mismatch-specific endonuclease/ endonuclease/ nucleic acid binding / single-stranded DNA specific endodeoxyribonuclease | 4.76 |
| 1517 | 254379_at | AT4G21820 | calmodulin-binding family protein | 2.93 |
| 1518 | 254349_at | AT4G22250 | zinc finger (C3HC4-type RING finger) family protein | 1.91 |
| 1519 | 254324_at | AT4G22640 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G22666.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G22666.2); contains InterPro domain Bifunctional inhibitor/plant lipid transfer protein/seed storage (InterPro:IPR016140) | -1.90 |
| 1520 | 254274_at | AT4G22770 | DNA-binding family protein | 1.47 |
| 1521 | 254291_at | AT4G23010 | ATUTR2/UTR2 (UDP-GALACTOSE TRANSPORTER 2) | -2.88 |
| 1522 | 254293_at | AT4G23060 | IQD22 (IQ-domain 22); calmodulin binding | 1.49 |
| 1523 | 254247_at | AT4G23260 | protein kinase | -2.96 |
| 1524 | 254248_at | AT4G23270 | protein kinase family protein | -3.11 |
| 1525 | 254250_at | AT4G23290 | protein kinase family protein | -4.53 |
| 1526 | 254239_at | AT4G23400 | PIP1;5/PIP1D (plasma membrane intrinsic protein 1;5); water channel | -2.06 |
| 1527 | 254234_at | AT4G23680 | major latex protein-related / MLP-related | 2.93 |
| 1528 | 254235_at | AT4G23750 | CRF2 (CYTOKININ RESPONSE FACTOR 2); DNA binding / transcription factor | 1.71 |
| 1529 | 254233_at | AT4G23800 | high mobility group (HMG1/2) family protein | 2.68 |
| 1530 | 254177_at | AT4G23860 | PHD finger protein-related | 1.59 |
| 1531 | 254187_at | AT4G23890 | similar to unnamed protein product [Vitis vinifera] (GB:CAO69542.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN73381.1) | -1.87 |
| 1532 | 254200_at | AT4G24110 | similar to unnamed protein product [Vitis vinifera] (GB:CAO62924.1) | 2.18 |
| 1533 | 254203_at | AT4G24150 | AtGRF8 (GROWTH-REGULATING FACTOR 8) | 5.27 |
| 1534 | 254204_at | AT4G24160 | hydrolase, alpha/beta fold family protein | -2.09 |
| 1535 | 254206_at | AT4G24180 | similar to thaumatin, putative [Arabidopsis thaliana] (TAIR:AT4G38660.2); similar to unnamed protein product [Vitis vinifera] (GB:CAO48203.1); contains InterPro domain Thaumatin, pathogenesis-related (InterPro:IPR001938) | 2.20 |
| 1536 | 254120_at | AT4G24570 | mitochondrial substrate carrier family protein | -2.55 |
| 1537 | 254125_at | AT4G24670 | alliinase family protein | 2.42 |
| 1538 | 254085_at | AT4G24960 | ATHVA22D (Arabidopsis thaliana HVA22 homologue D) | 1.55 |
| 1539 | 254100_at | AT4G25020 | KOW domain-containing protein / D111/G-patch domain-containing protein | 1.72 |
| 1540 | 254098_at | AT4G25100 | FSD1 (FE SUPEROXIDE DISMUTASE 1); iron superoxide dismutase | -6.12 |
| 1541 | 254096_at | AT4G25150 | acid phosphatase, putative | 1.48 |
| 1542 | 254109_at | AT4G25240 | SKS1 (SKU5 SIMILAR 1); copper ion binding | 2.74 |
| 1543 | 254075_at | AT4G25470 | CBF2 (FREEZING TOLERANCE QTL 4); DNA binding / transcription activator/ transcription factor | -2.34 |
| 1544 | 254074_at | AT4G25490 | CBF1 (C-REPEAT/DRE BINDING FACTOR 1); DNA binding / transcription activator/ transcription factor | -2.80 |
| 1545 | 245229_at | AT4G25620 | hydroxyproline-rich glycoprotein family protein | -2.15 |
| 1546 | 254080_at | AT4G25630 | FIB2 (FIBRILLARIN 2) | 1.98 |
| 1547 | 254021_at | AT4G25650 | ACD1-LIKE; electron carrier | -2.13 |
| 1548 | 254042_at | AT4G25810 | XTR6 (XYLOGLUCAN ENDOTRANSGLYCOSYLASE 6); hydrolase, acting on glycosyl bonds | -2.61 |
| 1549 | 254040_at | AT4G25900 | aldose 1-epimerase family protein | -2.02 |
| 1550 | 254032_at | AT4G25940 | epsin N-terminal homology (ENTH) domain-containing protein | -3.63 |
| 1551 | 254033_at | AT4G25950 | VATG3 (VACUOLAR ATP SYNTHASE G3) | 3.68 |
| 1552 | 254046_at | AT4G26020 | similar to ATGRIP/GRIP, protein binding [Arabidopsis thaliana] (TAIR:AT5G66030.1); similar to ATGRIP/GRIP [Arabidopsis thaliana] (TAIR:AT5G66030.2); similar to unnamed protein product [Vitis vinifera] (GB:CAO69578.1) | 1.61 |
| 1553 | 254016_at | AT4G26150 | CGA1 (CYTOKININ-RESPONSIVE GATA FACTOR 1); transcription factor | -2.25 |
| 1554 | 254017_at | AT4G26170 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G56780.1); similar to effector of transcription [Brassica napus] (GB:AAT00536.1) | 1.98 |
| 1555 | 254001_at | AT4G26260 | MIOX4 (MYO-INOSITOL OXYGENASE 4) | 4.35 |
| 1556 | 253987_at | AT4G26270 | phosphofructokinase family protein | 1.63 |
| 1557 | 253971_at | AT4G26530 | fructose-bisphosphate aldolase, putative | -6.47 |
| 1558 | 253978_at | AT4G26660 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G55520.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G55520.2); similar to unnamed protein product [Vitis vinifera] (GB:CAO38858.1); contains InterPro domain Kinesin-related (InterPro:IPR010544) | 1.83 |
| 1559 | 253947_at | AT4G26760 | microtubule associated protein (MAP65/ASE1) family protein | 2.32 |
| 1560 | 253943_at | AT4G27030 | small conjugating protein ligase | -2.36 |
| 1561 | 253920_at | AT4G27230 | HTA2 (HISTONE H2A); DNA binding | 1.61 |
| 1562 | 253916_at | AT4G27240 | zinc finger (C2H2 type) family protein | 2.17 |
| 1563 | 253915_at | AT4G27280 | calcium-binding EF hand family protein | -2.42 |
| 1564 | 253862_at | AT4G27330 | SPL (SPOROCYTELESS) | 3.83 |
| 1565 | 253872_at | AT4G27410 | RD26 (RESPONSIVE TO DESSICATION 26); transcription factor | -3.03 |
| 1566 | 253877_at | AT4G27435 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G15480.1); similar to fiber protein Fb34 [Gossypium barbadense] (GB:AAR07596.1); contains InterPro domain Protein of unknown function DUF1218 (InterPro:IPR009606) | -2.15 |
| 1567 | 253869_at | AT4G27510 | similar to hypothetical protein [Vitis vinifera] (GB:CAN77561.1) | 1.76 |
| 1568 | 253880_at | AT4G27590 | copper-binding protein-related | 3.40 |
| 1569 | 253830_at | AT4G27652 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G27657.1) | -2.92 |
| 1570 | 253859_at | AT4G27657 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G27652.1) | -4.32 |
| 1571 | 253883_at | AT4G27660 | similar to RIN13 (RPM1 INTERACTING PROTEIN 13) [Arabidopsis thaliana] (TAIR:AT2G20310.1); similar to unknown [Populus trichocarpa] (GB:ABK92668.1) | 1.62 |
| 1572 | 253860_at | AT4G27700 | rhodanese-like domain-containing protein | -1.91 |
| 1573 | 253887_at | AT4G27730 | ATOPT6 (oligopeptide transporter 6); oligopeptide transporter | 1.48 |
| 1574 | 253857_at | AT4G27990 | YGGT family protein | -1.96 |
| 1575 | 253825_at | AT4G28025 | similar to unnamed protein product [Vitis vinifera] (GB:CAO66511.1); contains domain Chlorophyll a-b binding protein (SSF103511) | -2.07 |
| 1576 | 253823_at | AT4G28030 | GCN5-related N-acetyltransferase (GNAT) family protein | -2.01 |
| 1577 | 253804_at | AT4G28230 | similar to unnamed protein product [Vitis vinifera] (GB:CAO41238.1) | 1.59 |
| 1578 | 253806_at | AT4G28270 | zinc finger (C3HC4-type RING finger) family protein | -2.35 |
| 1579 | 253817_at | AT4G28310 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G52270.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO69747.1) | 2.28 |
| 1580 | 253777_at | AT4G28450 | transducin family protein / WD-40 repeat family protein | 1.55 |
| 1581 | 253769_at | AT4G28560 | RIC7 (ROP-INTERACTIVE CRIB MOTIF-CONTAINING PROTEIN 7); protein binding | 3.24 |
| 1582 | 253788_at | AT4G28680 | tyrosine decarboxylase, putative | 2.34 |
| 1583 | 253792_at | AT4G28700 | ammonium transporter, putative | 2.79 |
| 1584 | 253757_at | AT4G28950 | ARAC7/ATRAC7/ATROP9/RAC7/ROP9 (RHO-RELATED PROTEIN FROM PLANTS 9); GTP binding | 2.50 |
| 1585 | 253753_at | AT4G29030 | glycine-rich protein | 6.16 |
| 1586 | 253747_at | AT4G29050 | lectin protein kinase family protein | -3.52 |
| 1587 | 253729_at | AT4G29360 | glycosyl hydrolase family 17 protein | 2.55 |
| 1588 | 253697_at | AT4G29700 | type I phosphodiesterase/nucleotide pyrophosphatase family protein | 3.17 |
| 1589 | 253696_at | AT4G29740 | CKX4 (CYTOKININ OXIDASE 4); cytokinin dehydrogenase | -1.98 |
| 1590 | 253689_at | AT4G29770 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G29760.1) | 1.63 |
| 1591 | 253643_at | AT4G29780 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G12010.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO43835.1); contains domain PTHR22930 (PTHR22930) | -2.37 |
| 1592 | 253656_at | AT4G30090 | EMB1353 (EMBRYO DEFECTIVE 1353) | 2.73 |
| 1593 | 253657_at | AT4G30110 | HMA2 (Heavy metal ATPase 2); cadmium-transporting ATPase | -4.62 |
| 1594 | 253661_at | AT4G30130 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G19090.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN72045.1); contains InterPro domain Protein of unknown function DUF632 (InterPro:IPR006867); contains InterPro domain Protein of unknown function DUF630 (InterPro:IPR006868) | 4.29 |
| 1595 | 253660_at | AT4G30140 | GDSL-motif lipase/hydrolase family protein | -3.11 |
| 1596 | 253609_at | AT4G30190 | AHA2 (Arabidopsis H(+)-ATPase 2); ATPase | -2.01 |
| 1597 | 253666_at | AT4G30270 | MERI5B (MERISTEM-5); hydrolase, acting on glycosyl bonds / xyloglucan:xyloglucosyl transferase | -3.53 |
| 1598 | 253628_at | AT4G30280 | ATXTH18/XTH18 (XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 18); hydrolase, acting on glycosyl bonds | -2.33 |
| 1599 | 253608_at | AT4G30290 | ATXTH19 (XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 19); hydrolase, acting on glycosyl bonds | -3.24 |
| 1600 | 253607_at | AT4G30330 | small nuclear ribonucleoprotein E, putative / snRNP-E, putative / Sm protein E, putative | 1.50 |
| 1601 | 253631_at | AT4G30440 | GAE1 (UDP-D-GLUCURONATE 4-EPIMERASE 1); UDP-glucuronate 4-epimerase/ catalytic | -2.05 |
| 1602 | 253620_at | AT4G30520 | leucine-rich repeat family protein / protein kinase family protein | 1.80 |
| 1603 | 253579_at | AT4G30610 | BRS1 (BRI1 SUPPRESSOR 1) | 1.62 |
| 1604 | 253598_at | AT4G30800 | 40S ribosomal protein S11 (RPS11B) | 1.67 |
| 1605 | 253599_at | AT4G30860 | ASHR3 (ASH1-RELATED 3); protein binding / zinc ion binding | 1.61 |
| 1606 | 253549_at | AT4G30930 | NFD1 (NUCLEAR FUSION DEFECTIVE 1); structural constituent of ribosome | 1.50 |
| 1607 | 253573_at | AT4G31020 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G24320.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO22751.1); contains domain PTHR12277:SF8 (PTHR12277:SF8); contains domain PTHR12277 (PTHR12277); contains domain SSF53474 (SSF53474); contains domain G3DSA:3.40.50.1820 (G3DSA:3.40.50.1820) | 1.88 |
| 1608 | 253516_at | AT4G31360 | selenium binding | 2.88 |
| 1609 | 253488_at | AT4G31610 | REM1 (REPRODUCTIVE MERISTEM 1); DNA binding / transcription factor | 5.93 |
| 1610 | 253540_at | AT4G31615 | DNA binding / transcription factor | 1.74 |
| 1611 | 253513_at | AT4G31760 | peroxidase, putative | 1.88 |
| 1612 | 253485_at | AT4G31800 | WRKY18 (WRKY DNA-binding protein 18); transcription factor | -1.97 |
| 1613 | 253514_at | AT4G31805 | WRKY family transcription factor | 3.17 |
| 1614 | 253480_at | AT4G31840 | plastocyanin-like domain-containing protein | 2.81 |
| 1615 | 253483_at | AT4G31910 | transferase family protein | 2.29 |
| 1616 | 253397_at | AT4G32710 | kinase | 1.95 |
| 1617 | 253403_at | AT4G32830 | ATAUR1 (ATAURORA1); histone serine kinase(H3-S10 specific) / kinase/ protein serine/threonine kinase | 1.77 |
| 1618 | 253401_at | AT4G32870 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G25770.2); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G25770.1); similar to unknown [Populus trichocarpa x Populus deltoides] (GB:ABK96434.1); contains domain SSF55961 (SSF55961) | -3.78 |
| 1619 | 253414_at | AT4G33050 | EDA39 (embryo sac development arrest 39); calmodulin binding | -3.55 |
| 1620 | 253363_at | AT4G33130 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G13000.2); similar to transcription factor [Arabidopsis thaliana] (TAIR:AT3G13000.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO65161.1) | 1.75 |
| 1621 | 253357_at | AT4G33400 | dem protein-related / defective embryo and meristems protein-related | 2.22 |
| 1622 | 253332_at | AT4G33420 | peroxidase, putative | -2.66 |
| 1623 | 253298_at | AT4G33560 | unknown protein | 3.04 |
| 1624 | 253305_at | AT4G33666 | unknown protein | -3.32 |
| 1625 | 253281_at | AT4G34138 | UGT73B1 (UDP-GLUCOSYL TRANSFERASE 73B1); UDP-glycosyltransferase/ abscisic acid glucosyltransferase | -2.69 |
| 1626 | 253284_at | AT4G34150 | C2 domain-containing protein | -2.02 |
| 1627 | 253270_at | AT4G34160 | CYCD3/CYCD3;1/D3 (CYCLIN D3;1); cyclin-dependent protein kinase regulator/ protein binding | 2.15 |
| 1628 | 253285_at | AT4G34250 | fatty acid elongase, putative | -4.77 |
| 1629 | 253258_at | AT4G34400 | DNA binding / transcription factor | 5.00 |
| 1630 | 253246_at | AT4G34600 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G16385.1) | -2.44 |
| 1631 | 253215_at | AT4G34950 | nodulin family protein | -2.55 |
| 1632 | 253175_at | AT4G35050 | MSI3 (NUCLEOSOME/CHROMATIN ASSEMBLY FACTOR GROUP C 3); protein binding | 1.82 |
| 1633 | 253172_at | AT4G35060 | heavy-metal-associated domain-containing protein / copper chaperone (CCH)-related | -3.38 |
| 1634 | 253168_at | AT4G35070 | similar to SBP1 (S-RIBONUCLEASE BINDING PROTEIN 1), protein binding / zinc ion binding [Arabidopsis thaliana] (TAIR:AT1G45976.1); similar to unknown [Populus trichocarpa] (GB:ABK93023.1); contains InterPro domain S-ribonuclease binding protein, SBP1, pollen (InterPro:IPR017066) | 2.28 |
| 1635 | 253174_at | AT4G35090 | CAT2 (CATALASE 2); catalase | -2.58 |
| 1636 | 253173_at | AT4G35110 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G16900.1); contains InterPro domain Phospholipase-like, arabidopsis (InterPro:IPR007942) | -1.95 |
| 1637 | 253182_at | AT4G35190 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G37210.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO47480.1); contains InterPro domain Conserved hypothetical protein CHP00730 (InterPro:IPR005269) | 3.56 |
| 1638 | 253197_at | AT4G35250 | vestitone reductase-related | -2.10 |
| 1639 | 253191_at | AT4G35350 | XCP1 (XYLEM CYSTEINE PEPTIDASE 1); cysteine-type peptidase | -2.48 |
| 1640 | 253148_at | AT4G35620 | CYCB2;2 (CYCLIN B2;2); cyclin-dependent protein kinase regulator | 1.75 |
| 1641 | 253108_at | AT4G35900 | FD; DNA binding / transcription factor | 1.46 |
| 1642 | 253078_at | AT4G36180 | leucine-rich repeat family protein | 2.88 |
| 1643 | 246270_at | AT4G36500 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G18210.1); similar to hypothetical protein [Thellungiella halophila] (GB:ABB45855.1) | -3.50 |
| 1644 | 246275_at | AT4G36540 | BEE2 (BR ENHANCED EXPRESSION 2); DNA binding / transcription factor | -2.10 |
| 1645 | 246238_at | AT4G36670 | mannitol transporter, putative | -2.76 |
| 1646 | 246250_at | AT4G36880 | CP1 (CYSTEINE PROTEINASE1); cysteine-type peptidase | 1.60 |
| 1647 | 246197_at | AT4G37010 | caltractin, putative / centrin, putative | -3.35 |
| 1648 | 246220_at | AT4G37210 | tetratricopeptide repeat (TPR)-containing protein | 1.49 |
| 1649 | 253101_at | AT4G37430 | CYP91A2 (CYTOCHROME P450 MONOOXYGENASE 91A2); oxygen binding | 1.83 |
| 1650 | 253051_at | AT4G37490 | CYC1 (CYCLIN 1); cyclin-dependent protein kinase regulator | 3.32 |
| 1651 | 253048_at | AT4G37560 | formamidase, putative / formamide amidohydrolase, putative | -2.15 |
| 1652 | 253054_at | AT4G37580 | HLS1 (HOOKLESS 1); N-acetyltransferase | 1.53 |
| 1653 | 253064_at | AT4G37730 | ATBZIP7 (ARABIDOPSIS THALIANA BASIC LEUCINE-ZIPPER 7); DNA binding / transcription factor | 2.34 |
| 1654 | 253065_at | AT4G37740 | AtGRF2 (GROWTHREGULATING FACTOR 2) | 2.45 |
| 1655 | 253010_at | AT4G37750 | ANT (AINTEGUMENTA); DNA binding / transcription factor | 3.59 |
| 1656 | 253040_at | AT4G37800 | xyloglucan:xyloglucosyl transferase, putative / xyloglucan endotransglycosylase, putative / endo-xyloglucan transferase, putative | -3.88 |
| 1657 | 253012_at | AT4G37900 | glycine-rich protein | 2.73 |
| 1658 | 252983_at | AT4G37980 | ELI3-1 (ELICITOR-ACTIVATED GENE 3); binding / catalytic/ oxidoreductase/ zinc ion binding | -2.51 |
| 1659 | 253008_at | AT4G38210 | ATEXPA20 (ARABIDOPSIS THALIANA EXPANSIN A20) | 2.28 |
| 1660 | 252989_at | AT4G38420 | SKS9 (SKU5 Similar 9); copper ion binding / oxidoreductase | -4.94 |
| 1661 | 252976_s_at | AT4G38550 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G20950.1); contains InterPro domain Phospholipase-like, arabidopsis (InterPro:IPR007942) | -2.44 |
| 1662 | 252977_at | AT4G38560 | similar to pEARLI4 [Arabidopsis thaliana] (TAIR:AT2G20960.1); contains InterPro domain Phospholipase-like, arabidopsis (InterPro:IPR007942) | -3.16 |
| 1663 | 252954_at | AT4G38660 | thaumatin, putative | 2.31 |
| 1664 | 252957_at | AT4G38680 | CSDP2/GRP2 (COLD SHOCK DOMAIN PROTEIN 2, GLYCINE RICH PROTEIN 2); nucleic acid binding | 1.65 |
| 1665 | 252950_at | AT4G38690 | 1-phosphatidylinositol phosphodiesterase-related | -2.20 |
| 1666 | 252971_at | AT4G38770 | PRP4 (PROLINE-RICH PROTEIN 4) | -1.96 |
| 1667 | 252915_at | AT4G38810 | calcium-binding EF hand family protein | -2.19 |
| 1668 | 252972_at | AT4G38840 | auxin-responsive protein, putative | -2.37 |
| 1669 | 252965_at | AT4G38860 | auxin-responsive protein, putative | -3.33 |
| 1670 | 252914_at | AT4G39130 | dehydrin family protein | -1.87 |
| 1671 | 252896_at | AT4G39480 | CYP96A9 (cytochrome P450, family 96, subfamily A, polypeptide 9); oxygen binding | 2.72 |
| 1672 | 252877_at | AT4G39630 | similar to unnamed protein product [Vitis vinifera] (GB:CAO71785.1) | 2.08 |
| 1673 | 252906_at | AT4G39640 | GGT1 (GAMMA-GLUTAMYL TRANSPEPTIDASE 1); gamma-glutamyltransferase/ glutathione gamma-glutamylcysteinyltransferase | -2.58 |
| 1674 | 252908_at | AT4G39670 | glycolipid binding / glycolipid transporter | -4.80 |
| 1675 | 252821_at | AT4G39860 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G22270.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO39818.1) | 2.31 |
| 1676 | 251113_at | AT5G01370 | unknown protein | 2.78 |
| 1677 | 251084_at | AT5G01520 | zinc finger (C3HC4-type RING finger) family protein | -2.16 |
| 1678 | 251099_at | AT5G01660 | kelch repeat-containing protein | 2.67 |
| 1679 | 251065_at | AT5G01870 | lipid transfer protein, putative | 1.78 |
| 1680 | 251075_at | AT5G01890 | leucine-rich repeat transmembrane protein kinase, putative | 1.63 |
| 1681 | 251067_at | AT5G01910 | similar to unnamed protein product [Vitis vinifera] (GB:CAO48882.1) | 1.81 |
| 1682 | 251039_at | AT5G02020 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G55646.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G59080.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO15731.1) | -1.89 |
| 1683 | 251032_at | AT5G02030 | LSN (LARSON); DNA binding / transcription factor | 1.64 |
| 1684 | 251029_at | AT5G02050 | mitochondrial glycoprotein family protein / MAM33 family protein | 1.74 |
| 1685 | 251021_at | AT5G02140 | thaumatin-like protein, putative | 1.91 |
| 1686 | 251036_at | AT5G02160 | unknown protein | -1.88 |
| 1687 | 251024_at | AT5G02180 | amino acid transporter family protein | -1.88 |
| 1688 | 251035_at | AT5G02220 | similar to unknown [Picea sitchensis] (GB:ABK23883.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN70860.1) | 1.85 |
| 1689 | 251014_at | AT5G02520 | unknown protein | 2.18 |
| 1690 | 251010_at | AT5G02550 | unknown protein | 2.85 |
| 1691 | 251006_at | AT5G02600 | heavy-metal-associated domain-containing protein | -2.01 |
| 1692 | 250974_at | AT5G02820 | RHL2 (ROOT HAIRLESS 2); ATP binding / DNA binding / DNA topoisomerase (ATP-hydrolyzing) | 1.58 |
| 1693 | 250976_at | AT5G03060 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G11100.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G11100.2) | 1.48 |
| 1694 | 250936_at | AT5G03120 | similar to hypothetical protein [Vitis vinifera] (GB:CAN68657.1) | -2.30 |
| 1695 | 250982_at | AT5G03150 | JKD (JACKDAW); nucleic acid binding / protein binding / protein homodimerization/ transcription factor/ zinc ion binding | 1.45 |
| 1696 | 250942_at | AT5G03350 | legume lectin family protein | 1.57 |
| 1697 | 250920_at | AT5G03390 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G68960.1); contains InterPro domain Protein of unknown function DUF295 (InterPro:IPR005174) | 3.20 |
| 1698 | 250948_at | AT5G03490 | UDP-glucoronosyl/UDP-glucosyl transferase family protein | -2.04 |
| 1699 | 250907_at | AT5G03670 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G36420.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO15955.1) | 2.06 |
| 1700 | 250908_at | AT5G03680 | PTL (PETAL LOSS); transcription factor | 1.94 |
| 1701 | 250912_at | AT5G03740 | HD2C (HISTONE DEACETYLASE 2C); nucleic acid binding / zinc ion binding | 1.89 |
| 1702 | 250892_at | AT5G03760 | ATCSLA09 (RESISTANT TO AGROBACTERIUM TRANSFORMATION 4); transferase, transferring glycosyl groups | -2.64 |
| 1703 | 250915_at | AT5G03790 | ATHB51/LMI1 (LATE MERISTEM IDENTITY1); DNA binding / sequence-specific DNA binding / transcription factor | 2.98 |
| 1704 | 250869_at | AT5G03840 | TFL1 (TERMINAL FLOWER 1); phosphatidylethanolamine binding | 1.54 |
| 1705 | 250864_at | AT5G03870 | glutaredoxin family protein | 2.51 |
| 1706 | 250867_at | AT5G03880 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G10000.2); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G10000.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO15903.1); contains InterPro domain Thioredoxin-like fold (InterPro:IPR012336); contains InterPro domain Thioredoxin fold (InterPro:IPR012335) | -1.87 |
| 1707 | 250881_at | AT5G04080 | similar to unknown protein [Oryza sativa (japonica cultivar-group)] (GB:BAD87214.1) | -2.06 |
| 1708 | 245689_at | AT5G04120 | phosphoglycerate/bisphosphoglycerate mutase family protein | -2.32 |
| 1709 | 245701_at | AT5G04140 | GLS1/GLU1/GLUS (FERREDOXIN-DEPENDENT GLUTAMATE SYNTHASE 1); glutamate synthase (ferredoxin) | -2.63 |
| 1710 | 245702_at | AT5G04220 | ATSYTC/NTMC2T1.3/NTMC2TYPE1.3/SYTC | -2.92 |
| 1711 | 245711_at | AT5G04340 | C2H2 (ZINC FINGER OF ARABIDOPSIS THALIANA 6); nucleic acid binding / transcription factor/ zinc ion binding | -3.17 |
| 1712 | 250832_at | AT5G04950 | nicotianamine synthase, putative | -2.93 |
| 1713 | 250828_at | AT5G05250 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G56360.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO41488.1) | -2.04 |
| 1714 | 250777_at | AT5G05440 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G38310.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO48777.1); contains InterPro domain Bet v I allergen; (InterPro:IPR000916); contains InterPro domain Streptomyces cyclase/dehydrase (InterPro:IPR005031) | -2.22 |
| 1715 | 250793_at | AT5G05600 | oxidoreductase, 2OG-Fe(II) oxygenase family protein | -4.04 |
| 1716 | 250768_at | AT5G05670 | signal recognition particle binding | 1.56 |
| 1717 | 250750_at | AT5G05870 | UGT76C1 (UDP-GLUCOSYL TRANSFERASE 76C1); UDP-glycosyltransferase/ transferase, transferring glycosyl groups | -2.01 |
| 1718 | 250756_at | AT5G05940 | ATROPGEF5/ROPGEF5 (KINASE PARTNER PROTEIN-LIKE); Rho guanyl-nucleotide exchange factor/ | 1.97 |
| 1719 | 250764_at | AT5G05960 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | -2.70 |
| 1720 | 250758_at | AT5G06000 | EIF3G2 (eukaryotic translation initiation factor 3G2); RNA binding / translation initiation factor | 1.56 |
| 1721 | 250722_at | AT5G06190 | unknown protein | 2.20 |
| 1722 | 250734_at | AT5G06270 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G11600.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO15841.1) | 1.53 |
| 1723 | 250676_at | AT5G06320 | NHL3 (NDR1/HIN1-like 3) | -2.99 |
| 1724 | 250728_at | AT5G06440 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G11720.2); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G11720.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO39900.1); contains domain Bet v1-like (SSF55961) | -1.93 |
| 1725 | 250688_at | AT5G06510 | CCAAT-binding transcription factor (CBF-B/NF-YA) family protein | 2.65 |
| 1726 | 250678_at | AT5G06540 | pentatricopeptide (PPR) repeat-containing protein | 2.05 |
| 1727 | 250679_at | AT5G06550 | Identical to F-box protein At5g06550 [Arabidopsis Thaliana] (GB:Q67XX3;GB:Q9FG15); similar to transcription factor jumonji (jmjC) domain-containing protein [Arabidopsis thaliana] (TAIR:AT1G78280.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN75104.1); contains InterPro domain Transcription factor jumonji (InterPro:IPR013129); contains InterPro domain Cyclin-like F-box (InterPro:IPR001810); contains InterPro domain Transcription factor jumonji/aspartyl beta-hydroxylase (InterPro:IPR003347) | 1.67 |
| 1728 | 250684_at | AT5G06650 | GIS2 (GLABROUS INFLORESCENCE STEMS 2); nucleic acid binding / transcription factor/ zinc ion binding | 2.04 |
| 1729 | 250648_at | AT5G06760 | late embryogenesis abundant group 1 domain-containing protein / LEA group 1 domain-containing protein | -2.88 |
| 1730 | 250668_at | AT5G07020 | proline-rich family protein | -1.93 |
| 1731 | 250666_at | AT5G07100 | WRKY26 (WRKY DNA-binding protein 26); transcription factor | -2.88 |
| 1732 | 250663_at | AT5G07110 | prenylated rab acceptor (PRA1) family protein | -2.31 |
| 1733 | 250642_at | AT5G07180 | ERL2 (ERECTA-LIKE 2); kinase | 4.77 |
| 1734 | 250618_at | AT5G07220 | ATBAG3 (ARABIDOPSIS THALIANA BCL-2-ASSOCIATED ATHANOGENE 3); protein binding | -3.79 |
| 1735 | 250616_at | AT5G07280 | EMS1 (EXCESS MICROSPOROCYTES1); kinase | 3.16 |
| 1736 | 250633_at | AT5G07460 | PMSR2 (PEPTIDEMETHIONINE SULFOXIDE REDUCTASE 2); peptide-methionine-(S)-S-oxide reductase | -2.08 |
| 1737 | 250588_at | AT5G07660 | structural maintenance of chromosomes (SMC) family protein | 2.54 |
| 1738 | 250598_at | AT5G07690 | MYB29 (myb domain protein 29); DNA binding / transcription factor | -2.13 |
| 1739 | 250601_at | AT5G07810 | SNF2 domain-containing protein / helicase domain-containing protein / HNH endonuclease domain-containing protein | 1.91 |
| 1740 | 250603_at | AT5G07820 | similar to chromosome scaffold protein-related [Arabidopsis thaliana] (TAIR:AT5G61260.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO15046.1); contains InterPro domain Calmodulin-binding, plant (InterPro:IPR012417) | -1.84 |
| 1741 | 250565_at | AT5G08000 | E13L3 (GLUCAN ENDO-1,3-BETA-GLUCOSIDASE-LIKE PROTEIN 3) | 3.07 |
| 1742 | 250560_at | AT5G08020 | replication protein, putative | 1.96 |
| 1743 | 250575_at | AT5G08240 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G23160.1) | -3.40 |
| 1744 | 250533_at | AT5G08640 | FLS (FLAVONOL SYNTHASE) | 1.84 |
| 1745 | 245891_at | AT5G09220 | AAP2 (AMINO ACID PERMEASE 2); amino acid transmembrane transporter | -3.39 |
| 1746 | 245884_at | AT5G09300 | 2-oxoisovalerate dehydrogenase, putative / 3-methyl-2-oxobutanoate dehydrogenase, putative / branched-chain alpha-keto acid dehydrogenase E1 alpha subunit, putative | 1.60 |
| 1747 | 250498_at | AT5G09660 | PMDH2 (PEROXISOMAL NAD-MALATE DEHYDROGENASE 2); malate dehydrogenase | -2.81 |
| 1748 | 250491_at | AT5G09780 | transcriptional factor B3 family protein | 3.32 |
| 1749 | 250504_at | AT5G09840 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G64710.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO14463.1); contains InterPro domain Protein of unknown function DUF537 (InterPro:IPR007491) | 1.90 |
| 1750 | 250509_at | AT5G09970 | CYP78A7 (cytochrome P450, family 78, subfamily A, polypeptide 7); oxygen binding | 4.48 |
| 1751 | 250455_at | AT5G09980 | PROPEP4 (Elicitor peptide 4 precursor) | 1.92 |
| 1752 | 250463_at | AT5G10030 | TGA4 (TGACG MOTIF-BINDING FACTOR 4); DNA binding / calmodulin binding / transcription factor | -2.42 |
| 1753 | 250475_at | AT5G10180 | AST68 (Sulfate transporter 2.1) | -4.42 |
| 1754 | 250472_at | AT5G10210 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G65030.1); similar to 80A08_18 [Brassica rapa subsp. pekinensis] (GB:AAZ67603.1); contains InterPro domain C2 calcium-dependent membrane targeting (InterPro:IPR000008) | -2.21 |
| 1755 | 250481_at | AT5G10310 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G37810.1); similar to 80A08_28 [Brassica rapa subsp. pekinensis] (GB:AAZ67613.1) | 1.89 |
| 1756 | 250482_at | AT5G10320 | similar to 80A08_29 [Brassica rapa subsp. pekinensis] (GB:AAZ67614.1) | 1.64 |
| 1757 | 250435_at | AT5G10380 | zinc finger (C3HC4-type RING finger) family protein | -3.57 |
| 1758 | 250434_at | AT5G10390 | histone H3 | 1.79 |
| 1759 | 250433_at | AT5G10400 | histone H3 | 2.51 |
| 1760 | 250426_at | AT5G10510 | AIL6 (AINTEGUMENTA-LIKE 6); DNA binding / transcription factor | 1.80 |
| 1761 | 246018_at | AT5G10695 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G57123.1); similar to unknown [Picea sitchensis] (GB:ABK22689.1) | -3.47 |
| 1762 | 245906_at | AT5G11070 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G35090.1) | -2.53 |
| 1763 | 250413_at | AT5G11160 | APT5 (ADENINE PHOSPHORIBOSYLTRANSFERASE 5); adenine phosphoribosyltransferase | 2.74 |
| 1764 | 250389_at | AT5G11320 | YUC4 (YUCCA4); monooxygenase | 2.06 |
| 1765 | 250360_at | AT5G11360 | ATP binding / protein kinase | 1.69 |
| 1766 | 250366_at | AT5G11420 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G25460.1); similar to unknown [Ricinus communis] (GB:CAB02653.1); contains InterPro domain Protein of unknown function DUF642 (InterPro:IPR006946); contains InterPro domain Galactose-binding like (InterPro:IPR008979) | -3.64 |
| 1767 | 250386_at | AT5G11510 | MYB3R-4 (C-MYB-LIKE TRANSCRIPTION FACTOR 3R-4, myb domain protein 3R-4); DNA binding / transcription coactivator/ transcription factor | 2.32 |
| 1768 | 250376_at | AT5G11550 | binding | 1.55 |
| 1769 | 250332_at | AT5G11680 | similar to hypothetical protein OsI_031621 [Oryza sativa (indica cultivar-group)] (GB:EAY77662.1) | -1.95 |
| 1770 | 250328_at | AT5G11780 | similar to hypothetical protein [Vitis vinifera] (GB:CAN67286.1) | 1.47 |
| 1771 | 250340_at | AT5G11840 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G67370.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO43828.1); contains InterPro domain Protein of unknown function DUF1230 (InterPro:IPR009631) | -1.88 |
| 1772 | 250344_at | AT5G11930 | glutaredoxin family protein | 1.72 |
| 1773 | 250301_at | AT5G11970 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G19460.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO45667.1) | -2.04 |
| 1774 | 250326_at | AT5G12080 | mechanosensitive ion channel domain-containing protein / MS ion channel domain-containing protein | 2.02 |
| 1775 | 245204_at | AT5G12270 | oxidoreductase, 2OG-Fe(II) oxygenase family protein | 3.43 |
| 1776 | 245208_at | AT5G12330 | LRP1 (LATERAL ROOT PRIMORDIUM 1) | 2.10 |
| 1777 | 245181_at | AT5G12420 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G16350.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO48523.1); contains InterPro domain Protein of unknown function UPF0089 (InterPro:IPR004255); contains InterPro domain Protein of unknown function DUF1298 (InterPro:IPR009721) | -1.89 |
| 1778 | 250264_at | AT5G12890 | UDP-glucoronosyl/UDP-glucosyl transferase family protein | -2.03 |
| 1779 | 250277_at | AT5G12940 | leucine-rich repeat family protein | -2.32 |
| 1780 | 250269_at | AT5G12970 | C2 domain-containing protein | 2.06 |
| 1781 | 245976_at | AT5G13080 | WRKY75 (WRKY DNA-BINDING PROTEIN 75); transcription factor | -2.73 |
| 1782 | 245987_at | AT5G13180 | ANAC083 (Arabidopsis NAC domain containing protein 83); transcription factor | -2.57 |
| 1783 | 250279_at | AT5G13200 | GRAM domain-containing protein / ABA-responsive protein-related | -2.97 |
| 1784 | 250284_at | AT5G13290 | protein kinase family protein | 1.81 |
| 1785 | 250286_at | AT5G13320 | PBS3 (AVRPPHB SUSCEPTIBLE 3) | -4.41 |
| 1786 | 245849_at | AT5G13520 | peptidase M1 family protein | 2.29 |
| 1787 | 250247_at | AT5G13720 | structural constituent of ribosome | -1.92 |
| 1788 | 250257_at | AT5G13770 | pentatricopeptide (PPR) repeat-containing protein | -1.85 |
| 1789 | 250258_at | AT5G13790 | AGL15 (AGAMOUS-LIKE 15); DNA binding / transcription factor | 3.73 |
| 1790 | 250228_at | AT5G13840 | WD-40 repeat family protein | 2.77 |
| 1791 | 250212_at | AT5G13960 | SUVH4 (SU(VAR)3-9 HOMOLOG 4) | 1.95 |
| 1792 | 250217_at | AT5G14120 | nodulin family protein | -3.39 |
| 1793 | 250196_at | AT5G14580 | polyribonucleotide nucleotidyltransferase, putative | 1.55 |
| 1794 | 250138_at | AT5G14610 | ATP binding / ATP-dependent helicase | 1.96 |
| 1795 | 246584_at | AT5G14730 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G01513.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO64332.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN65788.1) | -2.45 |
| 1796 | 246596_at | AT5G14740 | CA2 (BETA CARBONIC ANHYDRASE 2); carbonate dehydratase/ zinc ion binding | -6.98 |
| 1797 | 246595_at | AT5G14780 | FDH (FORMATE DEHYDROGENASE); NAD binding / binding / catalytic/ cofactor binding / oxidoreductase, acting on the CH-OH group of donors, NAD or NADP as acceptor | -2.20 |
| 1798 | 246566_at | AT5G14940 | proton-dependent oligopeptide transport (POT) family protein | -2.70 |
| 1799 | 250152_at | AT5G15120 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G39890.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO14912.1); contains InterPro domain Protein of unknown function DUF1637 (InterPro:IPR012864) | 3.00 |
| 1800 | 250108_at | AT5G15150 | ATHB-3 (ARABIDOPSIS THALIANA HOMEOBOX 3); DNA binding / transcription factor | 1.53 |
| 1801 | 246557_at | AT5G15510 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G01015.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO15061.1); contains InterPro domain Targeting for Xklp2 (InterPro:IPR009675) | 2.37 |
| 1802 | 246562_at | AT5G15580 | LNG1 (LONGIFOLIA1) | 1.99 |
| 1803 | 246531_at | AT5G15800 | SEP1 (SEPALLATA1); DNA binding / transcription factor | 3.84 |
| 1804 | 246522_at | AT5G15830 | ATBZIP3 (ARABIDOPSIS THALIANA BASIC LEUCINE-ZIPPER 3); DNA binding / transcription factor | -3.04 |
| 1805 | 246523_at | AT5G15850 | COL1 (CONSTANS-LIKE 1); transcription factor/ zinc ion binding | -2.01 |
| 1806 | 246488_at | AT5G16010 | 3-oxo-5-alpha-steroid 4-dehydrogenase family protein / steroid 5-alpha-reductase family protein | -2.47 |
| 1807 | 246505_at | AT5G16250 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G02640.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO50168.1) | 2.21 |
| 1808 | 246419_at | AT5G17030 | UDP-glucoronosyl/UDP-glucosyl transferase family protein | 3.63 |
| 1809 | 246470_at | AT5G17080 | cathepsin-related | 3.33 |
| 1810 | 246415_at | AT5G17160 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G03130.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO62214.1) | 1.94 |
| 1811 | 250090_at | AT5G17330 | GAD (Glutamate decarboxylase 1); calmodulin binding | -3.80 |
| 1812 | 246429_at | AT5G17450 | heavy-metal-associated domain-containing protein / copper chaperone (CCH)-related | -4.13 |
| 1813 | 246439_at | AT5G17600 | zinc finger (C3HC4-type RING finger) family protein | -1.92 |
| 1814 | 246440_at | AT5G17650 | glycine/proline-rich protein | -5.17 |
| 1815 | 250051_at | AT5G17800 | AtMYB56 (myb domain protein 56); DNA binding / transcription factor | 2.11 |
| 1816 | 250071_at | AT5G18000 | transcriptional factor B3 family protein | 3.96 |
| 1817 | 250013_at | AT5G18040 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G18065.1) | 1.72 |
| 1818 | 250012_x_at | AT5G18060 | auxin-responsive protein, putative | -1.87 |
| 1819 | 250028_at | AT5G18130 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G03870.2); similar to unknown [Medicago truncatula] (GB:ABK28852.1) | -1.85 |
| 1820 | 250031_at | AT5G18240 | MYR1 (MYB-RELATED PROTEIN 1); transcription factor | -2.64 |
| 1821 | 250036_at | AT5G18340 | U-box domain-containing protein | -2.79 |
| 1822 | 249983_at | AT5G18470 | curculin-like (mannose-binding) lectin family protein | -3.67 |
| 1823 | 249987_at | AT5G18490 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G04350.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN77840.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN61465.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO40557.1); contains InterPro domain Protein of unknown function DUF946, plant (InterPro:IPR009291) | -2.02 |
| 1824 | 249996_at | AT5G18600 | glutaredoxin family protein | -4.91 |
| 1825 | 249997_at | AT5G18620 | CHR17 (CHROMATIN REMODELING FACTOR17); DNA-dependent ATPase | 1.54 |
| 1826 | 250004_at | AT5G18750 | DNAJ heat shock N-terminal domain-containing protein | 1.65 |
| 1827 | 249927_at | AT5G19220 | ADG2 (ADPG PYROPHOSPHORYLASE 2); glucose-1-phosphate adenylyltransferase | -1.85 |
| 1828 | 249918_at | AT5G19240 | Identical to Uncharacterized GPI-anchored protein At5g19240 precursor [Arabidopsis Thaliana] (GB:Q84VZ5;GB:Q8H7A4); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G19230.1); similar to unknown [Populus trichocarpa] (GB:ABK94712.1) | -3.06 |
| 1829 | 246094_at | AT5G19300 | similar to unnamed protein product [Vitis vinifera] (GB:CAO71126.1); contains InterPro domain Protein of unknown function DUF171 (InterPro:IPR003750); contains InterPro domain Nucleic acid-binding, OB-fold-like (InterPro:IPR016027) | 1.60 |
| 1830 | 246158_at | AT5G19855 | similar to unnamed protein product [Vitis vinifera] (GB:CAO21907.1) | -2.11 |
| 1831 | 246138_at | AT5G19870 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G55230.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO63741.1); contains InterPro domain Protein of unknown function DUF716 (InterPro:IPR006904) | -1.86 |
| 1832 | 246144_at | AT5G20110 | dynein light chain, putative | -2.75 |
| 1833 | 246099_at | AT5G20230 | ATBCB (ARABIDOPSIS BLUE-COPPER-BINDING PROTEIN); copper ion binding | -4.62 |
| 1834 | 246072_at | AT5G20240 | PI (PISTILLATA); DNA binding / transcription factor | 5.24 |
| 1835 | 246085_at | AT5G20540 | ATBRXL4/BRX-LIKE4 (BREVIS RADIX-LIKE 4) | 1.92 |
| 1836 | 245993_at | AT5G20700 | senescence-associated protein-related | -2.29 |
| 1837 | 246002_at | AT5G20740 | invertase/pectin methylesterase inhibitor family protein | 3.69 |
| 1838 | 246133_at | AT5G20960 | AAO1 (ALDEHYDE OXIDASE 1) | -1.97 |
| 1839 | 246025_at | AT5G21150 | PAZ domain-containing protein / piwi domain-containing protein | 3.16 |
| 1840 | 249941_at | AT5G22270 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G06270.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO15841.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN79170.1) | -3.21 |
| 1841 | 249939_at | AT5G22430 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G27385.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN68427.1) | 5.50 |
| 1842 | 249895_at | AT5G22500 | acyl CoA reductase, putative / male-sterility protein, putative | 1.91 |
| 1843 | 249916_at | AT5G22880 | H2B/HTB2 (HISTONE H2B); DNA binding | 2.90 |
| 1844 | 249866_at | AT5G23010 | MAM1 (2-isopropylmalate synthase 3); 2-isopropylmalate synthase | -5.55 |
| 1845 | 249867_at | AT5G23020 | IMS2/MAM-L/MAM3 (METHYLTHIOALKYMALATE SYNTHASE-LIKE); 2-isopropylmalate synthase/ methylthioalkylmalate synthase | -2.25 |
| 1846 | 249876_at | AT5G23060 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G59780.1); similar to extracellular Ca2+ sensing receptor [Glycine max] (GB:ABY57763.1); contains InterPro domain Rhodanese-like (InterPro:IPR001763) | -2.32 |
| 1847 | 249847_at | AT5G23210 | SCPL34; serine carboxypeptidase | -1.96 |
| 1848 | 249850_at | AT5G23240 | DNAJ heat shock N-terminal domain-containing protein | -3.47 |
| 1849 | 249836_at | AT5G23420 | HMGB6 (High mobility group B 6); transcription factor | 2.17 |
| 1850 | 249794_at | AT5G23530 | ATCXE18 (ARABIDOPSIS THALIANA CARBOXYESTERASE 18); carboxylesterase | 2.22 |
| 1851 | 249843_at | AT5G23570 | SGS3 (SUPPRESSOR OF GENE SILENCING 3) | 1.62 |
| 1852 | 249800_at | AT5G23660 | MTN3 (ARABIDOPSIS HOMOLOG OF MEDICAGO TRUNCATULA MTN3) | -3.20 |
| 1853 | 249809_at | AT5G23910 | microtubule motor | 2.21 |
| 1854 | 249813_at | AT5G23940 | EMB3009 (EMBRYO DEFECTIVE 3009); transferase | 2.89 |
| 1855 | 249760_at | AT5G23960 | terpene synthase/cyclase family protein | 1.94 |
| 1856 | 249761_at | AT5G23970 | transferase family protein | 1.98 |
| 1857 | 249765_at | AT5G24030 | SLAH3 (SLAC1 HOMOLOGUE 3); transporter | -2.58 |
| 1858 | 249770_at | AT5G24110 | WRKY30 (WRKY DNA-binding protein 30); transcription factor | -2.44 |
| 1859 | 249769_at | AT5G24120 | SIGE (RNA polymerase sigma subunit E); DNA binding / DNA-directed RNA polymerase/ sigma factor/ transcription factor | -1.87 |
| 1860 | 249774_at | AT5G24150 | SQP1 (Squalene monooxygenase 1) | -2.78 |
| 1861 | 249777_at | AT5G24210 | lipase class 3 family protein | -4.66 |
| 1862 | 249780_at | AT5G24240 | phosphatidylinositol 3- and 4-kinase family protein / ubiquitin family protein | -2.01 |
| 1863 | 249784_at | AT5G24280 | ATP binding | 1.90 |
| 1864 | 249788_at | AT5G24330 | ATXR6 (Arabidopsis thaliana Trithorax- related protein 6); DNA binding | 5.22 |
| 1865 | 249732_at | AT5G24420 | glucosamine/galactosamine-6-phosphate isomerase-related | 2.47 |
| 1866 | 249754_at | AT5G24530 | oxidoreductase, 2OG-Fe(II) oxygenase family protein | -1.85 |
| 1867 | 249752_at | AT5G24660 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G24655.1); similar to unknown protein [Brassica rapa subsp. pekinensis] (GB:AAQ92331.1) | -3.44 |
| 1868 | 246967_at | AT5G24860 | FPF1 (FLOWERING PROMOTING FACTOR 1) | 3.69 |
| 1869 | 246978_at | AT5G24910 | CYP714A1 (cytochrome P450, family 714, subfamily A, polypeptide 1); oxygen binding | 2.48 |
| 1870 | 246920_at | AT5G25090 | plastocyanin-like domain-containing protein | 2.72 |
| 1871 | 246948_at | AT5G25130 | CYP71B12 (cytochrome P450, family 71, subfamily B, polypeptide 12); oxygen binding | -3.54 |
| 1872 | 246943_at | AT5G25440 | protein kinase family protein | -2.05 |
| 1873 | 246906_at | AT5G25475 | DNA binding | 2.31 |
| 1874 | 246898_at | AT5G25580 | similar to aminoacyl-tRNA synthetase family [Arabidopsis thaliana] (TAIR:AT1G18950.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO44138.1) | 2.30 |
| 1875 | 246899_at | AT5G25590 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G52320.3); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G52320.4); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G52320.2); similar to unnamed protein product [Vitis vinifera] (GB:CAO44106.1); contains InterPro domain Actin-binding FH2 (InterPro:IPR015425); contains InterPro domain Protein of unknown function DUF632 (InterPro:IPR006867); contains InterPro domain Protein of unknown function DUF630 (InterPro:IPR006868) | 1.66 |
| 1876 | 246860_at | AT5G25840 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G79770.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO45837.1); contains InterPro domain Protein of unknown function DUF1677, plant (InterPro:IPR012876) | -4.45 |
| 1877 | 246858_at | AT5G25930 | leucine-rich repeat family protein / protein kinase family protein | -1.87 |
| 1878 | 246885_at | AT5G26230 | similar to hypothetical protein [Vitis vinifera] (GB:CAN75978.1) | 1.46 |
| 1879 | 246855_at | AT5G26280 | meprin and TRAF homology domain-containing protein / MATH domain-containing protein | -2.91 |
| 1880 | 246827_at | AT5G26330 | plastocyanin-like domain-containing protein / mavicyanin, putative | -1.86 |
| 1881 | 246831_at | AT5G26340 | MSS1 (SUGAR TRANSPORT PROTEIN 13); carbohydrate transmembrane transporter/ hexose:hydrogen symporter/ high-affinity hydrogen:glucose symporter/ sugar:hydrogen ion symporter | -3.38 |
| 1882 | 246849_at | AT5G26850 | binding | 2.38 |
| 1883 | 246821_at | AT5G26920 | calmodulin binding | -2.82 |
| 1884 | 246791_at | AT5G27280 | zinc finger (DNL type) family protein | -1.93 |
| 1885 | 246765_at | AT5G27330 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G05130.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO68600.1); contains InterPro domain Prefoldin (InterPro:IPR009053) | 1.81 |
| 1886 | 246781_at | AT5G27350 | SFP1; carbohydrate transmembrane transporter/ sugar:hydrogen ion symporter | -4.27 |
| 1887 | 246777_at | AT5G27420 | zinc finger (C3HC4-type RING finger) family protein | -2.48 |
| 1888 | 246738_at | AT5G27740 | EMB2775 (EMBRYO DEFECTIVE 2775); DNA binding / nucleoside-triphosphatase/ nucleotide binding | 1.51 |
| 1889 | 246103_at | AT5G28640 | AN3 (ANGUSITFOLIA3) | 4.22 |
| 1890 | 246104_at | AT5G28650 | WRKY74 (WRKY DNA-binding protein 74); transcription factor | 1.71 |
| 1891 | 246724_at | AT5G29000 | myb family transcription factor | -2.86 |
| 1892 | 246682_at | AT5G33290 | XGD1 (XYLOGALACTURONAN DEFICIENT 1); catalytic | -3.92 |
| 1893 | 246683_at | AT5G33300 | chromosome-associated kinesin-related | 1.82 |
| 1894 | 246687_at | AT5G33370 | GDSL-motif lipase/hydrolase family protein | 3.82 |
| 1895 | 249727_at | AT5G35490 | unknown protein | -2.52 |
| 1896 | 249709_at | AT5G35670 | IQD33 (IQ-domain 33); calmodulin binding | 1.64 |
| 1897 | 249719_at | AT5G35735 | auxin-responsive family protein | -3.19 |
| 1898 | 249677_at | AT5G35970 | DNA-binding protein, putative | -2.11 |
| 1899 | 249688_at | AT5G36160 | aminotransferase-related | -2.62 |
| 1900 | 249645_at | AT5G36910 | THI2.2 (THIONIN 2.2); toxin receptor binding | 1.89 |
| 1901 | 249644_at | AT5G37010 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G65710.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN76723.1) | 2.54 |
| 1902 | 249651_at | AT5G37020 | ARF8 (AUXIN RESPONSE FACTOR 8); transcription factor | 1.58 |
| 1903 | 249614_at | AT5G37300 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G38995.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO48523.1); contains InterPro domain Protein of unknown function UPF0089 (InterPro:IPR004255); contains InterPro domain Protein of unknown function DUF1298 (InterPro:IPR009721) | 4.20 |
| 1904 | 249610_at | AT5G37360 | similar to unnamed protein product [Vitis vinifera] (GB:CAO38751.1) | -1.95 |
| 1905 | 249568_at | AT5G38040 | UDP-glucoronosyl/UDP-glucosyl transferase family protein | 1.51 |
| 1906 | 249544_at | AT5G38110 | ASF1B/SGA01/SGA1 (ANTI- SILENCING FUNCTION 1B) | 1.62 |
| 1907 | 249521_at | AT5G38690 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G67780.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO49292.1); contains InterPro domain DDT (InterPro:IPR004022) | 1.54 |
| 1908 | 249527_at | AT5G38710 | proline oxidase, putative / osmotic stress-responsive proline dehydrogenase, putative | -1.85 |
| 1909 | 249531_at | AT5G38770 | ATGDU7 (ARABIDOPSIS THALIANA GLUTAMINE DUMPER 7) | -3.29 |
| 1910 | 249493_at | AT5G39080 | transferase family protein | -4.51 |
| 1911 | 249497_at | AT5G39220 | hydrolase, alpha/beta fold family protein | 2.46 |
| 1912 | 249417_at | AT5G39670 | calcium-binding EF hand family protein | -3.05 |
| 1913 | 249384_at | AT5G39890 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G15120.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO14912.1); contains InterPro domain Protein of unknown function DUF1637 (InterPro:IPR012864) | 2.21 |
| 1914 | 249439_at | AT5G40020 | pathogenesis-related thaumatin family protein | -2.16 |
| 1915 | 249381_at | AT5G40040 | 60S acidic ribosomal protein P2 (RPP2E) | 3.63 |
| 1916 | 249393_at | AT5G40170 | disease resistance family protein | -2.35 |
| 1917 | 249353_at | AT5G40420 | OLEO2 (OLEOSIN 2) | -2.18 |
| 1918 | 249378_at | AT5G40450 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G28770.1); similar to hypothetical protein Kpol_1058p4 [Vanderwaltozyma polyspora DSM 70294] (GB:XP_001645325.1) | -3.80 |
| 1919 | 249379_at | AT5G40460 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G27630.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN59885.1) | 1.74 |
| 1920 | 249354_at | AT5G40480 | EMB3012 (EMBRYO DEFECTIVE 3012) | 1.89 |
| 1921 | 249377_at | AT5G40690 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G41730.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO14635.1) | -4.52 |
| 1922 | 249346_at | AT5G40780 | LHT1 (LYSINE HISTIDINE TRANSPORTER 1); amino acid transmembrane transporter | -3.80 |
| 1923 | 249336_at | AT5G41070 | DRB5 (DSRNA-BINDING PROTEIN 5); double-stranded RNA binding | 1.46 |
| 1924 | 249306_at | AT5G41400 | zinc finger (C3HC4-type RING finger) family protein | -4.37 |
| 1925 | 249309_at | AT5G41410 | BEL1 (BELL 1); DNA binding / transcription factor | -2.65 |
| 1926 | 249283_at | AT5G41800 | amino acid transporter family protein | -2.22 |
| 1927 | 249277_at | AT5G41890 | GDSL-motif lipase/hydrolase family protein | 1.62 |
| 1928 | 249237_at | AT5G42050 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G27090.1); similar to unknown [Populus trichocarpa] (GB:ABK95892.1); contains InterPro domain Kelch related (InterPro:IPR013089); contains InterPro domain Development and cell death (InterPro:IPR013989) | -2.30 |
| 1929 | 249242_at | AT5G42250 | alcohol dehydrogenase, putative | -4.61 |
| 1930 | 249244_at | AT5G42270 | VAR1 (VARIEGATED 1); ATP-dependent peptidase/ ATPase/ metallopeptidase | -1.86 |
| 1931 | 249197_at | AT5G42380 | CML37/CML39; calcium ion binding | -2.71 |
| 1932 | 249208_at | AT5G42650 | AOS (ALLENE OXIDE SYNTHASE); hydro-lyase/ oxygen binding | -2.34 |
| 1933 | 249211_at | AT5G42680 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G39610.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO61286.1); contains InterPro domain Protein of unknown function DUF617, plant (InterPro:IPR006460) | -2.52 |
| 1934 | 249184_at | AT5G43020 | leucine-rich repeat transmembrane protein kinase, putative | 3.08 |
| 1935 | 249138_at | AT5G43070 | WPP1 (WPP domain protein 1) | 1.55 |
| 1936 | 249129_at | AT5G43080 | CYCA3;1 (CYCLIN A3;1); cyclin-dependent protein kinase regulator | 1.67 |
| 1937 | 249134_at | AT5G43150 | similar to unnamed protein product [Vitis vinifera] (GB:CAO61459.1) | -2.78 |
| 1938 | 249148_at | AT5G43260 | chaperone protein dnaJ-related | -2.35 |
| 1939 | 249128_at | AT5G43440 | 2-oxoglutarate-dependent dioxygenase, putative | -2.60 |
| 1940 | 249125_at | AT5G43450 | 2-oxoglutarate-dependent dioxygenase, putative | -3.07 |
| 1941 | 249098_at | AT5G43530 | SNF2 domain-containing protein / helicase domain-containing protein / RING finger domain-containing protein | 1.93 |
| 1942 | 249115_at | AT5G43810 | ZLL (ZWILLE) | 2.30 |
| 1943 | 249080_at | AT5G43990 | SUVR2; histone-lysine N-methyltransferase/ zinc ion binding | 1.82 |
| 1944 | 249073_at | AT5G44020 | acid phosphatase class B family protein | -4.19 |
| 1945 | 249037_at | AT5G44130 | FLA13 (FASCICLIN-LIKE ARABINOGALACTAN PROTEIN 13 PRECURSOR) | -2.92 |
| 1946 | 249065_at | AT5G44260 | zinc finger (CCCH-type) family protein | -1.96 |
| 1947 | 249051_at | AT5G44390 | FAD-binding domain-containing protein | -3.17 |
| 1948 | 249046_at | AT5G44400 | FAD-binding domain-containing protein | -4.35 |
| 1949 | 249003_at | AT5G44500 | small nuclear ribonucleoprotein associated protein B, putative / snRNP-B, putative / Sm protein B, putative | 2.56 |
| 1950 | 249061_at | AT5G44550 | integral membrane family protein | -3.14 |
| 1951 | 249060_at | AT5G44560 | VPS2.2 | 2.02 |
| 1952 | 249010_at | AT5G44580 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G44582.1) | -3.44 |
| 1953 | 249012_at | AT5G44620 | CYP706A3 (cytochrome P450, family 706, subfamily A, polypeptide 3); oxygen binding | 7.03 |
| 1954 | 249005_at | AT5G44630 | terpene synthase/cyclase family protein | 5.40 |
| 1955 | 249008_at | AT5G44680 | methyladenine glycosylase family protein | -1.86 |
| 1956 | 249015_at | AT5G44730 | haloacid dehalogenase-like hydrolase family protein | 1.72 |
| 1957 | 254521_at | AT5G44820 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G19970.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO46707.1); contains domain PTHR10483:SF6 (PTHR10483:SF6); contains domain PTHR10483 (PTHR10483) | -3.57 |
| 1958 | 248964_at | AT5G45340 | CYP707A3 (cytochrome P450, family 707, subfamily A, polypeptide 3); oxygen binding | -3.26 |
| 1959 | 248940_at | AT5G45400 | replication protein, putative | 1.51 |
| 1960 | 248912_at | AT5G45670 | GDSL-motif lipase/hydrolase family protein | 2.38 |
| 1961 | 248963_at | AT5G45700 | NLI interacting factor (NIF) family protein | 3.00 |
| 1962 | 248938_at | AT5G45780 | leucine-rich repeat transmembrane protein kinase, putative | 1.62 |
| 1963 | 248910_at | AT5G45820 | CIPK20 (CBL-INTERACTING PROTEIN KINASE 20); kinase | -1.89 |
| 1964 | 248916_at | AT5G45840 | leucine-rich repeat transmembrane protein kinase, putative | -1.93 |
| 1965 | 248924_at | AT5G45960 | GDSL-motif lipase/hydrolase family protein | 3.62 |
| 1966 | 248932_at | AT5G46050 | ATPTR3/PTR3 (PEPTIDE TRANSPORTER PROTEIN 3); transporter | -2.00 |
| 1967 | 248887_at | AT5G46115 | unknown protein | -2.58 |
| 1968 | 248889_at | AT5G46230 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G09310.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO14438.1); contains InterPro domain Protein of unknown function DUF538 (InterPro:IPR007493) | -2.38 |
| 1969 | 248888_at | AT5G46240 | KAT1 (K+ ATPASE 1); cyclic nucleotide binding / inward rectifier potassium channel | -3.33 |
| 1970 | 248891_at | AT5G46280 | DNA replication licensing factor, putative | 2.49 |
| 1971 | 248853_at | AT5G46570 | protein kinase family protein | 1.48 |
| 1972 | 248855_at | AT5G46590 | ANAC096 (Arabidopsis NAC domain containing protein 96); transcription factor | 2.95 |
| 1973 | 248868_at | AT5G46780 | VQ motif-containing protein | -2.93 |
| 1974 | 248819_at | AT5G47050 | ATP binding / protein binding / shikimate kinase/ zinc ion binding | -1.91 |
| 1975 | 248821_at | AT5G47070 | protein kinase, putative | -2.24 |
| 1976 | 248831_at | AT5G47160 | YDG/SRA domain-containing protein | 3.88 |
| 1977 | 248794_at | AT5G47220 | ATERF-2/ATERF2/ERF2 (ETHYLENE RESPONSE FACTOR 2); DNA binding / transcription activator/ transcription factor | -2.32 |
| 1978 | 248807_at | AT5G47500 | pectinesterase family protein | 2.51 |
| 1979 | 248756_at | AT5G47560 | ATSDAT/ATTDT (TONOPLAST DICARBOXYLATE TRANSPORTER); malate transmembrane transporter/ sodium:dicarboxylate symporter | -3.86 |
| 1980 | 248752_at | AT5G47600 | heat shock protein-related | 6.53 |
| 1981 | 248741_at | AT5G48170 | SLY2 (SLEEPY2) | 1.51 |
| 1982 | 248691_at | AT5G48310 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G24610.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G65440.1); similar to predicted protein [Physcomitrella patens subsp. patens] (GB:EDQ82245.1); similar to predicted protein [Physcomitrella patens subsp. patens] (GB:EDQ70121.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO41110.1); contains InterPro domain C2 calcium/lipid-binding region, CaLB (InterPro:IPR008973) | 2.48 |
| 1983 | 248695_at | AT5G48350 | nucleic acid binding | 2.53 |
| 1984 | 248696_at | AT5G48360 | formin homology 2 domain-containing protein / FH2 domain-containing protein | 3.43 |
| 1985 | 248704_at | AT5G48450 | SKS3 (SKU5 Similar 3); copper ion binding | 2.06 |
| 1986 | 248710_at | AT5G48480 | Identical to Uncharacterized protein At5g48480 [Arabidopsis Thaliana] (GB:Q9LV66); similar to unknown [Populus trichocarpa] (GB:ABK95611.1); contains domain SSF54593 (SSF54593) | 2.74 |
| 1987 | 248684_at | AT5G48485 | DIR1 (DEFECTIVE IN INDUCED RESISTANCE 1); lipid binding | 2.25 |
| 1988 | 248685_at | AT5G48500 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G10930.1) | 1.88 |
| 1989 | 248658_at | AT5G48600 | ATSMC3 (ARABIDOPSIS THALIANA STRUCTURAL MAINTENANCE OF CHROMOSOME 3); ATP binding | 1.92 |
| 1990 | 248660_at | AT5G48650 | nuclear transport factor 2 (NTF2) family protein / RNA recognition motif (RRM)-containing protein | 3.00 |
| 1991 | 248675_at | AT5G48820 | ICK6/KRP3 (KIP-RELATED PROTEIN 3); cyclin binding / cyclin-dependent protein kinase inhibitor | 2.19 |
| 1992 | 248676_at | AT5G48850 | male sterility MS5 family protein | -2.04 |
| 1993 | 248642_at | AT5G49120 | senescence-associated protein-related | 1.63 |
| 1994 | 248645_at | AT5G49150 | ATGEX2/GEX2 (GAMETE EXPRESSED2) | 1.95 |
| 1995 | 248597_at | AT5G49160 | MET1 (DECREASED METHYLATION 2DNA) | 1.92 |
| 1996 | 248596_at | AT5G49330 | AtMYB111 (myb domain protein 111); DNA binding / transcription factor | 1.46 |
| 1997 | 248622_at | AT5G49360 | BXL1 (BETA-XYLOSIDASE 1); hydrolase, hydrolyzing O-glycosyl compounds | -4.12 |
| 1998 | 248607_at | AT5G49480 | ATCP1 (CA2+-BINDING PROTEIN 1); calcium ion binding | -2.06 |
| 1999 | 248611_at | AT5G49520 | WRKY48 (WRKY DNA-binding protein 48); transcription factor | -2.04 |
| 2000 | 248590_at | AT5G49660 | leucine-rich repeat transmembrane protein kinase, putative | -1.91 |
| 2001 | 248564_at | AT5G49700 | DNA-binding protein-related | 1.97 |
| 2002 | 248537_at | AT5G50100 | similar to PBng143 [Vigna radiata] (GB:BAB82450.1); contains InterPro domain Thioredoxin-like fold (InterPro:IPR012336); contains InterPro domain Putative thiol-disulphide oxidoreductase DCC (InterPro:IPR007263) | -2.86 |
| 2003 | 248540_at | AT5G50160 | ATFRO8/FRO8 (FERRIC REDUCTION OXIDASE 8); ferric-chelate reductase/ oxidoreductase | -2.43 |
| 2004 | 248551_at | AT5G50200 | WR3 (WOUND-RESPONSIVE 3); nitrate transmembrane transporter | -2.12 |
| 2005 | 248550_at | AT5G50210 | QS (QUINOLINATE SYNTHASE); quinolinate synthetase A | -1.92 |
| 2006 | 248509_at | AT5G50335 | unknown protein | -4.51 |
| 2007 | 248526_at | AT5G50740 | metal ion binding | 2.51 |
| 2008 | 248527_at | AT5G50740 | metal ion binding | 2.49 |
| 2009 | 248496_at | AT5G50790 | nodulin MtN3 family protein | 2.78 |
| 2010 | 248487_at | AT5G51070 | ERD1 (EARLY RESPONSIVE TO DEHYDRATION 1); ATP binding / ATPase | -2.15 |
| 2011 | 248448_at | AT5G51190 | AP2 domain-containing transcription factor, putative | -2.61 |
| 2012 | 248456_at | AT5G51380 | F-box family protein | 1.67 |
| 2013 | 248432_at | AT5G51390 | unknown protein | -2.43 |
| 2014 | 248420_at | AT5G51560 | leucine-rich repeat transmembrane protein kinase, putative | 2.17 |
| 2015 | 248412_at | AT5G51590 | DNA-binding protein-related | 2.60 |
| 2016 | 248413_at | AT5G51600 | PLE (PLEIADE) | 2.41 |
| 2017 | 248414_at | AT5G51610 | ribosomal protein L11 family protein | 2.59 |
| 2018 | 248377_at | AT5G51720 | similar to Os07g0467200 [Oryza sativa (japonica cultivar-group)] (GB:NP_001059590.1); similar to hypothetical protein OsI_025030 [Oryza sativa (indica cultivar-group)] (GB:EAZ03798.1); contains domain PTHR13680 (PTHR13680); contains domain PTHR13680:SF1 (PTHR13680:SF1) | -1.90 |
| 2019 | 248427_at | AT5G51750 | ATSBT1.3; subtilase | 2.56 |
| 2020 | 248428_at | AT5G51760 | AHG1 (ABA-HYPERSENSITIVE GERMINATION 1); protein serine/threonine phosphatase | 1.66 |
| 2021 | 248430_at | AT5G51800 | similar to gt-2-related [Arabidopsis thaliana] (TAIR:AT2G33550.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN62096.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO49779.1); contains InterPro domain Protein kinase-like (InterPro:IPR011009) | 3.12 |
| 2022 | 248374_at | AT5G51870 | AGL71; transcription factor | 2.94 |
| 2023 | 248385_at | AT5G51910 | TCP family transcription factor, putative | 1.97 |
| 2024 | 248398_at | AT5G51970 | sorbitol dehydrogenase, putative / L-iditol 2-dehydrogenase, putative | -2.07 |
| 2025 | 248392_at | AT5G52050 | MATE efflux protein-related | -3.03 |
| 2026 | 248348_at | AT5G52190 | sugar isomerase (SIS) domain-containing protein | -2.12 |
| 2027 | 248352_at | AT5G52300 | LTI65/RD29B (RESPONSIVE TO DESSICATION 29B) | -2.44 |
| 2028 | 248337_at | AT5G52310 | COR78 (COLD REGULATED 78) | -2.88 |
| 2029 | 248333_at | AT5G52390 | photoassimilate-responsive protein, putative | 2.30 |
| 2030 | 248327_at | AT5G52750 | heavy-metal-associated domain-containing protein | -2.32 |
| 2031 | 248322_at | AT5G52760 | heavy-metal-associated domain-containing protein | -4.76 |
| 2032 | 248277_at | AT5G52860 | ABC transporter family protein | 2.35 |
| 2033 | 248280_at | AT5G52950 | similar to unnamed protein product [Vitis vinifera] (GB:CAO69340.1) | 1.91 |
| 2034 | 248252_at | AT5G53250 | AGP22/ATAGP22 (ARABINOGALACTAN PROTEINS 22) | -2.89 |
| 2035 | 248254_at | AT5G53320 | leucine-rich repeat transmembrane protein kinase, putative | -3.14 |
| 2036 | 248263_at | AT5G53370 | ATPMEPCRF; pectinesterase | -2.98 |
| 2037 | 248276_at | AT5G53550 | YSL3 (YELLOW STRIPE LIKE 3); oligopeptide transporter | -1.86 |
| 2038 | 248213_at | AT5G53660 | AtGRF7 (GROWTH-REGULATING FACTOR 7) | 2.54 |
| 2039 | 248231_at | AT5G53770 | nucleotidyltransferase family protein | 1.77 |
| 2040 | 248227_at | AT5G53820 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G38760.1); similar to pollen coat protein [Brassica oleracea] (GB:CAA63531.1); contains domain PTHR23241:SF11 (PTHR23241:SF11); contains domain PTHR23241 (PTHR23241) | -2.01 |
| 2041 | 248240_at | AT5G53950 | CUC2 (CUP-SHAPED COTYLEDON 2); transcription factor | 2.56 |
| 2042 | 248150_at | AT5G54670 | ATK3 (ARABIDOPSIS THALIANA KINESIN 3); microtubule motor | 2.14 |
| 2043 | 248146_at | AT5G54940 | eukaryotic translation initiation factor SUI1, putative | -3.35 |
| 2044 | 248118_at | AT5G55050 | GDSL-motif lipase/hydrolase family protein | -2.85 |
| 2045 | 248090_at | AT5G55090 | MAPKKK15 (Mitogen-activated protein kinase kinase kinase 15); kinase | -3.23 |
| 2046 | 248100_at | AT5G55180 | glycosyl hydrolase family 17 protein | 1.75 |
| 2047 | 248104_at | AT5G55250 | IAMT1 (IAA CARBOXYLMETHYLTRANSFERASE 1); S-adenosylmethionine-dependent methyltransferase | 1.76 |
| 2048 | 248111_at | AT5G55330 | membrane bound O-acyl transferase (MBOAT) family protein / wax synthase-related | 2.16 |
| 2049 | 248061_at | AT5G55340 | long-chain-alcohol O-fatty-acyltransferase family protein / wax synthase family protein | 2.20 |
| 2050 | 248062_at | AT5G55450 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | 1.47 |
| 2051 | 248057_at | AT5G55520 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G26660.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO38858.1); contains InterPro domain Kinesin-related (InterPro:IPR010544) | 2.58 |
| 2052 | 248073_at | AT5G55720 | pectate lyase family protein | 4.39 |
| 2053 | 248054_at | AT5G55820 | similar to unnamed protein product [Vitis vinifera] (GB:CAO45135.1) | 2.59 |
| 2054 | 248055_at | AT5G55830 | lectin protein kinase, putative | 1.49 |
| 2055 | 248040_at | AT5G55970 | zinc finger (C3HC4-type RING finger) family protein | -1.92 |
| 2056 | 248050_at | AT5G56100 | glycine-rich protein / oleosin | -1.90 |
| 2057 | 247999_at | AT5G56150 | UBC30 (UBIQUITIN-CONJUGATING ENZYME 30); ubiquitin-protein ligase | -2.58 |
| 2058 | 248003_at | AT5G56220 | nucleoside-triphosphatase/ nucleotide binding | 2.55 |
| 2059 | 248011_at | AT5G56300 | GAMT2; S-adenosylmethionine-dependent methyltransferase/ gibberellin carboxyl-O-methyltransferase | 2.41 |
| 2060 | 247962_at | AT5G56580 | ATMKK6 (ARABIDOPSIS NQK1); kinase | 1.99 |
| 2061 | 247972_at | AT5G56740 | HAG2 (HISTONE ACETYLTRANSFERASE OF THE GNAT FAMILY 2); H4 histone acetyltransferase/ histone acetyltransferase | 2.00 |
| 2062 | 247954_at | AT5G56870 | BGAL4 (beta-galactosidase 4); beta-galactosidase | -3.36 |
| 2063 | 247931_at | AT5G57040 | lactoylglutathione lyase family protein / glyoxalase I family protein | -2.83 |
| 2064 | 247948_at | AT5G57130 | protein binding | 1.69 |
| 2065 | 247949_at | AT5G57220 | CYP81F2 (cytochrome P450, family 81, subfamily F, polypeptide 2); oxygen binding | -3.49 |
| 2066 | 247899_at | AT5G57345 | similar to unknown [Populus trichocarpa x Populus deltoides] (GB:ABK96592.1) | -3.02 |
| 2067 | 247902_at | AT5G57350 | AHA3 (Arabidopsis H(+)-ATPase 3); ATPase | -3.25 |
| 2068 | 247910_at | AT5G57410 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G18876.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO43234.1); contains domain PTHR21736 (PTHR21736) | 1.68 |
| 2069 | 247925_at | AT5G57560 | TCH4 (TOUCH 4); hydrolase, acting on glycosyl bonds / xyloglucan:xyloglucosyl transferase | -2.87 |
| 2070 | 247867_at | AT5G57630 | CIPK21 (CBL-INTERACTING PROTEIN KINASE 21); kinase | -2.28 |
| 2071 | 247921_at | AT5G57660 | zinc finger (B-box type) family protein | -2.12 |
| 2072 | 247874_at | AT5G57710 | heat shock protein-related | -2.09 |
| 2073 | 247875_at | AT5G57720 | transcriptional factor B3 family protein | 2.87 |
| 2074 | 247882_at | AT5G57785 | unknown protein | 1.47 |
| 2075 | 247883_at | AT5G57790 | unknown protein | 2.49 |
| 2076 | 247892_at | AT5G57970 | methyladenine glycosylase family protein | 1.52 |
| 2077 | 247848_at | AT5G58120 | disease resistance protein (TIR-NBS-LRR class), putative | -6.02 |
| 2078 | 247816_at | AT5G58260 | Encodes subunit NDH-N of NAD(P)H:plastoquinone dehydrogenase complex (Ndh complex) present in the thylakoid membrane of chloroplasts. This subunit is thought to be required for Ndh complex assembly. | -2.34 |
| 2079 | 247814_at | AT5G58310 | hydrolase, alpha/beta fold family protein | -4.81 |
| 2080 | 247827_at | AT5G58500 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G07090.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO21405.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN60458.1); contains InterPro domain Protein of unknown function DUF640 (InterPro:IPR006936) | 1.93 |
| 2081 | 247800_at | AT5G58570 | unknown protein | -2.56 |
| 2082 | 247772_at | AT5G58610 | PHD finger transcription factor, putative | 2.25 |
| 2083 | 247780_at | AT5G58770 | dehydrodolichyl diphosphate synthase, putative / DEDOL-PP synthase, putative | -6.80 |
| 2084 | 247765_at | AT5G58860 | CYP86A1 (cytochrome P450, family 86, subfamily A, polypeptide 1); oxygen binding | -2.11 |
| 2085 | 247768_at | AT5G58900 | myb family transcription factor | -1.91 |
| 2086 | 247747_at | AT5G59000 | zinc finger (C3HC4-type RING finger) family protein | 2.08 |
| 2087 | 247754_at | AT5G59080 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G02020.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO15731.1) | -4.85 |
| 2088 | 247719_at | AT5G59305 | similar to unknown [Populus trichocarpa] (GB:ABK94520.1) | 4.85 |
| 2089 | 247679_at | AT5G59540 | oxidoreductase, 2OG-Fe(II) oxygenase family protein | -4.46 |
| 2090 | 247685_at | AT5G59680 | leucine-rich repeat protein kinase, putative | -2.05 |
| 2091 | 247696_at | AT5G59780 | MYB59 (myb domain protein 59); DNA binding / transcription factor | -2.88 |
| 2092 | 247655_at | AT5G59820 | RHL41 (RESPONSIVE TO HIGH LIGHT 41); nucleic acid binding / transcription factor/ zinc ion binding | -3.82 |
| 2093 | 247648_at | AT5G60020 | LAC17 (laccase 17); copper ion binding / oxidoreductase | -2.54 |
| 2094 | 247667_at | AT5G60150 | similar to unnamed protein product [Vitis vinifera] (GB:CAO21780.1) | 2.09 |
| 2095 | 247625_at | AT5G60200 | Dof-type zinc finger domain-containing protein | 1.80 |
| 2096 | 247671_at | AT5G60210 | cytoplasmic linker protein-related | 2.01 |
| 2097 | 247615_at | AT5G60250 | zinc finger (C3HC4-type RING finger) family protein | 2.15 |
| 2098 | 247627_at | AT5G60360 | AALP (ARABIDOPSIS ALEURAIN-LIKE PROTEASE); cysteine-type peptidase | -1.90 |
| 2099 | 247589_at | AT5G60690 | REV (REVOLUTA); DNA binding / lipid binding / transcription factor | 1.69 |
| 2100 | 247582_at | AT5G60760 | 2-phosphoglycerate kinase-related | 2.73 |
| 2101 | 247601_at | AT5G60850 | OBP4 (OBF BINDING PROTEIN 4); DNA binding / transcription factor | -2.40 |
| 2102 | 247599_at | AT5G60880 | unknown protein | 2.26 |
| 2103 | 247553_at | AT5G60910 | AGL8 (AGAMOUS-LIKE 8); transcription factor | 3.09 |
| 2104 | 247603_at | AT5G60930 | chromosome-associated kinesin, putative | 2.34 |
| 2105 | 247606_at | AT5G61000 | replication protein, putative | 1.64 |
| 2106 | 247575_at | AT5G61030 | GR-RBP3 (glycine-rich RNA-binding protein 3); RNA binding | 2.19 |
| 2107 | 247562_at | AT5G61120 | zinc ion binding | 2.34 |
| 2108 | 247563_at | AT5G61130 | glycosyl hydrolase family protein 17 | 1.54 |
| 2109 | 247522_at | AT5G61340 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G26650.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN78131.1) | -2.57 |
| 2110 | 247581_at | AT5G61350 | protein kinase family protein | -2.55 |
| 2111 | 247549_at | AT5G61420 | MYB28 (MYB DOMAIN PROTEIN 28); DNA binding / transcription factor | -2.56 |
| 2112 | 247540_at | AT5G61590 | AP2 domain-containing transcription factor family protein | -2.56 |
| 2113 | 247543_at | AT5G61600 | ethylene-responsive element-binding family protein | -3.50 |
| 2114 | 247514_at | AT5G61640 | PMSR1 (PEPTIDEMETHIONINE SULFOXIDE REDUCTASE 1); peptide-methionine-(S)-S-oxide reductase | -1.97 |
| 2115 | 247541_at | AT5G61660 | glycine-rich protein | -3.25 |
| 2116 | 247488_at | AT5G61820 | similar to MtN19-like protein [Pisum sativum] (GB:AAU14999.2); contains InterPro domain Stress up-regulated Nod 19 (InterPro:IPR011692) | -2.70 |
| 2117 | 247490_at | AT5G61850 | LFY (LEAFY); transcription factor | 4.68 |
| 2118 | 247487_at | AT5G62150 | peptidoglycan-binding LysM domain-containing protein | -2.53 |
| 2119 | 247469_at | AT5G62165 | AGL42 (AGAMOUS LIKE 42); transcription factor | 2.35 |
| 2120 | 247463_at | AT5G62210 | embryo-specific protein-related | 2.33 |
| 2121 | 247471_at | AT5G62230 | ERL1 (ERECTA-LIKE 1); kinase | 1.87 |
| 2122 | 247449_at | AT5G62290 | nucleotide-sensitive chloride conductance regulator (ICln) family protein | 1.77 |
| 2123 | 247482_at | AT5G62410 | SMC2 (STRUCTURAL MAINTENANCE OF CHROMOSOMES 2) | 1.98 |
| 2124 | 247452_at | AT5G62430 | CDF1 (CYCLING DOF FACTOR 1); DNA binding / protein binding / transcription factor | -2.01 |
| 2125 | 247435_at | AT5G62480 | ATGSTU9 (GLUTATHIONE S-TRANSFERASE TAU 9); glutathione transferase | -1.84 |
| 2126 | 247425_at | AT5G62550 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G28770.1); similar to unknown [Populus trichocarpa] (GB:ABK92769.1) | 1.74 |
| 2127 | 247440_at | AT5G62680 | proton-dependent oligopeptide transport (POT) family protein | -2.76 |
| 2128 | 247441_at | AT5G62710 | leucine-rich repeat family protein / protein kinase family protein | 2.15 |
| 2129 | 247443_at | AT5G62720 | integral membrane HPP family protein | -2.62 |
| 2130 | 247418_at | AT5G63030 | glutaredoxin, putative | -1.84 |
| 2131 | 247362_at | AT5G63140 | ATPAP29/PAP29 (purple acid phosphatase 29); acid phosphatase/ protein serine/threonine phosphatase | 2.02 |
| 2132 | 247377_at | AT5G63180 | pectate lyase family protein | -2.46 |
| 2133 | 247352_at | AT5G63650 | SNRK2-5/SNRK2.5/SRK2H (SNF1-RELATED PROTEIN KINASE 2.5); kinase | 1.85 |
| 2134 | 247337_at | AT5G63660 | LCR74/PDF2.5 (Low-molecular-weight cysteine-rich 74) | 2.26 |
| 2135 | 247357_at | AT5G63710 | leucine-rich repeat transmembrane protein kinase, putative | -2.29 |
| 2136 | 247356_at | AT5G63800 | BGAL6/MUM2 (MUCILAGE-MODIFIED 2); beta-galactosidase | -3.06 |
| 2137 | 247301_at | AT5G63920 | DNA topoisomerase III alpha, putative | 1.83 |
| 2138 | 247310_at | AT5G63950 | CHR24 (chromatin remodeling 24); ATP binding / DNA binding / helicase | 2.08 |
| 2139 | 247320_at | AT5G64040 | PSAN (photosystem I reaction center subunit PSI-N); calmodulin binding | -2.05 |
| 2140 | 247317_at | AT5G64060 | ANAC103 (Arabidopsis NAC domain containing protein 103); transcription factor | 1.56 |
| 2141 | 247297_at | AT5G64100 | peroxidase, putative | -2.62 |
| 2142 | 247282_at | AT5G64240 | ATMC3 (METACASPASE 3); caspase | -3.47 |
| 2143 | 247244_at | AT5G64710 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G09840.1); similar to 80C09_3 [Brassica rapa subsp. pekinensis] (GB:AAZ41814.1); contains InterPro domain Protein of unknown function DUF537 (InterPro:IPR007491) | 2.02 |
| 2144 | 247215_at | AT5G64905 | PROPEP3 (Elicitor peptide 3 precursor) | -2.44 |
| 2145 | 247234_at | AT5G64980 | transcription regulator | 2.08 |
| 2146 | 247200_at | AT5G65120 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G10110.1); similar to 52O08_25 [Brassica rapa subsp. pekinensis] (GB:AAZ67571.1) | 1.46 |
| 2147 | 247199_at | AT5G65210 | TGA1; DNA binding / calmodulin binding / transcription factor | -2.65 |
| 2148 | 247177_at | AT5G65300 | unknown protein | -2.22 |
| 2149 | 247180_at | AT5G65350 | histone H3 | 1.56 |
| 2150 | 247192_at | AT5G65360 | histone H3 | 1.58 |
| 2151 | 247182_at | AT5G65410 | ATHB25/ZFHD2 (ZINC FINGER HOMEODOMAIN 2); transcription factor | 3.33 |
| 2152 | 247196_at | AT5G65510 | AIL7 (AINTEGUMENTA-LIKE 7); DNA binding / transcription factor | 3.69 |
| 2153 | 247146_at | AT5G65610 | unknown protein | -2.45 |
| 2154 | 247151_at | AT5G65640 | BHLH093 (BETA HLH PROTEIN 93); DNA binding / transcription factor | 1.90 |
| 2155 | 247149_at | AT5G65660 | hydroxyproline-rich glycoprotein family protein | -1.87 |
| 2156 | 247168_at | AT5G65860 | ankyrin repeat family protein | 1.57 |
| 2157 | 247109_at | AT5G65870 | ATPSK5 (PHYTOSULFOKINE 5 PRECURSOR); growth factor | -3.23 |
| 2158 | 247107_at | AT5G66040 | STR16 (SULFURTRANSFERASE PROTEIN 16) | -1.98 |
| 2159 | 247125_at | AT5G66070 | zinc finger (C3HC4-type RING finger) family protein | -2.47 |
| 2160 | 247129_at | AT5G66150 | glycosyl hydrolase family 38 protein | -2.61 |
| 2161 | 247134_at | AT5G66230 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G51230.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO48217.1) | 2.58 |
| 2162 | 247085_at | AT5G66310 | kinesin motor family protein | 1.57 |
| 2163 | 247087_at | AT5G66330 | leucine-rich repeat family protein | 1.68 |
| 2164 | 247095_at | AT5G66400 | RAB18 (RESPONSIVE TO ABA 18) | -2.33 |
| 2165 | 247072_at | AT5G66490 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G50900.1); similar to hypothetical protein [Thellungiella halophila] (GB:ABB45855.1) | -3.32 |
| 2166 | 247047_at | AT5G66650 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G23790.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO22909.1); contains InterPro domain Protein of unknown function DUF607 (InterPro:IPR006769) | -2.28 |
| 2167 | 247056_at | AT5G66750 | CHR01/DDM1 (DECREASED DNA METHYLATION 1); helicase | 2.58 |
| 2168 | 247068_at | AT5G66800 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G50640.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN61148.1) | 1.91 |
| 2169 | 247066_at | AT5G66940 | Dof-type zinc finger domain-containing protein | 2.83 |
| 2170 | 247024_at | AT5G66985 | unknown protein | 3.98 |
| 2171 | 247025_at | AT5G67030 | ABA1 (ABA DEFICIENT 1); zeaxanthin epoxidase | -2.11 |
| 2172 | 247028_at | AT5G67100 | ICU2 (INCURVATA2); DNA-directed DNA polymerase | 1.96 |
| 2173 | 247035_at | AT5G67110 | ALC (ALCATRAZ); DNA binding / transcription factor | 2.30 |
| 2174 | 246983_at | AT5G67200 | leucine-rich repeat transmembrane protein kinase, putative | 1.90 |
| 2175 | 247034_at | AT5G67260 | CYCD3;2 (CYCLIN D3;2); cyclin-dependent protein kinase | 2.31 |
| 2176 | 247039_at | AT5G67270 | ATEB1C (MICROTUBULE END BINDING PROTEIN 1); microtubule binding | 1.92 |
| 2177 | 246987_at | AT5G67300 | ATMYB44/ATMYBR1/MYBR1 (MYB DOMAIN PROTEIN 44); DNA binding / transcription factor | -2.54 |
| 2178 | 246998_at | AT5G67370 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G11840.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN75014.1); contains InterPro domain Protein of unknown function DUF1230 (InterPro:IPR009631) | -2.93 |
| 2179 | 246996_at | AT5G67420 | LBD37 (LOB DOMAIN-CONTAINING PROTEIN 37) | -2.21 |
| 2180 | 246994_at | AT5G67460 | glycosyl hydrolase family protein 17 | 1.83 |
| 2181 | 245047_at | ATCG00020 | Encodes chlorophyll binding protein D1, a part of the photosystem II reaction center core | -1.98 |
| 2182 | 244931_at | ATMG00630 | hypothetical protein | 1.72 |
| 2183 | 244903_at | ATMG00660 | hypothetical protein | 1.73 |