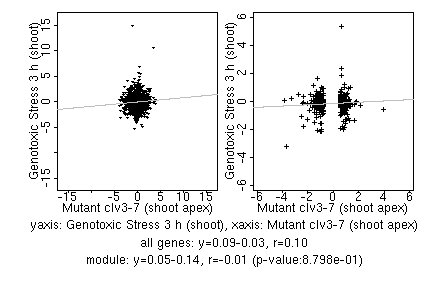
marker gene list
| Mutant clv3-7(shoot apex) |
| Genotoxic Stress 3 h (shoot) |
| probeID | AGICode | Annotation | gene expression |
|
|---|---|---|---|---|
| 1 | 259429_at | AT1G01600 | CYP86A4 (cytochrome P450, family 86, subfamily A, polypeptide 4); oxygen binding | -1.05 |
| 2 | 264171_at | AT1G02100 | leucine carboxyl methyltransferase family protein | -0.83 |
| 3 | 264363_at | AT1G03170 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G02810.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO22484.1) | 1.17 |
| 4 | 265083_at | AT1G03820 | similar to E6-4 [Gossypium hirsutum] (GB:AAY43793.1); similar to E6-3 [Gossypium hirsutum] (GB:AAY43792.1); similar to E6 (GB:AAB03081.1) | 0.69 |
| 5 | 265093_at | AT1G03905 | ABC transporter family protein | 0.67 |
| 6 | 265096_at | AT1G04030 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G44040.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO40580.1) | 0.78 |
| 7 | 263656_at | AT1G04240 | SHY2 (SHORT HYPOCOTYL 2); transcription factor | 1.10 |
| 8 | 264579_at | AT1G05205 | similar to hypothetical protein OsJ_021517 [Oryza sativa (japonica cultivar-group)] (GB:EAZ38034.1); similar to unknown protein [Oryza sativa (japonica cultivar-group)] (GB:BAD45491.1); similar to hypothetical protein OsI_023333 [Oryza sativa (indica cultivar-group)] (GB:EAZ02101.1) | -0.72 |
| 9 | 261256_at | AT1G05760 | RTM1 (RESTRICTED TEV MOVEMENT 1) | -0.71 |
| 10 | 262637_at | AT1G06640 | 2-oxoglutarate-dependent dioxygenase, putative | 0.62 |
| 11 | 256041_at | AT1G07230 | phosphoesterase family protein | 0.67 |
| 12 | 264510_at | AT1G09530 | PAP3/PIF3/POC1 (PHYTOCHROME INTERACTING FACTOR 3); DNA binding / protein binding / transcription factor/ transcription regulator | 0.67 |
| 13 | 263236_at | AT1G10470 | ARR4 (RESPONSE REGULATOR 4); transcription regulator/ two-component response regulator | 0.61 |
| 14 | 260461_at | AT1G10980 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G61670.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO41520.1); contains InterPro domain Transmembrane receptor, eukaryota; (InterPro:IPR009637) | -0.99 |
| 15 | 262479_at | AT1G11130 | SUB (STRUBBELIG); protein binding | 0.65 |
| 16 | 264343_at | AT1G11850 | unknown protein | -0.96 |
| 17 | 259529_at | AT1G12400 | DNA binding | -1.33 |
| 18 | 262575_at | AT1G15210 | ATPDR7/PDR7 (PLEIOTROPIC DRUG RESISTANCE 7); ATPase, coupled to transmembrane movement of substances | -0.79 |
| 19 | 262598_at | AT1G15260 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G16070.1) | -1.08 |
| 20 | 262751_at | AT1G16310 | cation efflux family protein | 0.93 |
| 21 | 255891_at | AT1G17870 | ATEGY3 | 0.71 |
| 22 | 261221_at | AT1G19960 | similar to transmembrane receptor [Arabidopsis thaliana] (TAIR:AT2G32140.1) | -0.90 |
| 23 | 262796_at | AT1G20850 | XCP2 (XYLEM CYSTEINE PEPTIDASE 2); cysteine-type peptidase/ peptidase | -0.81 |
| 24 | 255960_at | AT1G22140 | similar to unnamed protein product [Vitis vinifera] (GB:CAO61756.1); contains domain PTHR12681 (PTHR12681) | 0.78 |
| 25 | 261929_at | AT1G22460 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G64600.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO61677.1); contains InterPro domain Protein of unknown function DUF246, plant (InterPro:IPR004348) | -0.97 |
| 26 | 261928_at | AT1G22480 | plastocyanin-like domain-containing protein | 0.73 |
| 27 | 261942_at | AT1G22590 | AGL87; transcription factor | -0.95 |
| 28 | 264211_at | AT1G22770 | GI (GIGANTEA); binding | 0.91 |
| 29 | 263013_at | AT1G23380 | KNAT6 (Knotted-like Arabidopsis thaliana 6); DNA binding / transcription factor | 0.84 |
| 30 | 265025_at | AT1G24575 | unknown protein | 1.05 |
| 31 | 255732_at | AT1G25450 | very-long-chain fatty acid condensing enzyme, putative | -0.78 |
| 32 | 255726_at | AT1G25530 | lysine and histidine specific transporter, putative | 0.93 |
| 33 | 261260_at | AT1G26680 | transcriptional factor B3 family protein | 2.23 |
| 34 | 263681_at | AT1G26840 | ATORC6/ORC6 (Origin recognition complex protein 6); DNA binding | -0.70 |
| 35 | 264447_at | AT1G27300 | similar to unnamed protein product [Vitis vinifera] (GB:CAO66149.1) | -0.94 |
| 36 | 259583_at | AT1G28070 | protein binding | 0.65 |
| 37 | 260696_at | AT1G32520 | similar to unnamed protein product [Vitis vinifera] (GB:CAO63428.1) | 0.63 |
| 38 | 256481_at | AT1G33430 | galactosyltransferase family protein | 0.91 |
| 39 | 261285_at | AT1G35720 | ANNAT1 (ANNEXIN ARABIDOPSIS 1); calcium ion binding / calcium-dependent phospholipid binding | 1.39 |
| 40 | 260941_at | AT1G44970 | peroxidase, putative | 0.91 |
| 41 | 260943_at | AT1G45145 | ATTRX5 (thioredoxin H-type 5); thiol-disulfide exchange intermediate | 1.01 |
| 42 | 256145_at | AT1G48750 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | -0.75 |
| 43 | 265060_at | AT1G52150 | ATHB-15 (INCURVATA 4); DNA binding / transcription factor | 0.78 |
| 44 | 259960_at | AT1G53710 | protein serine/threonine phosphatase | 0.91 |
| 45 | 263174_at | AT1G54040 | ESP (EPITHIOSPECIFIER PROTEIN) | -0.71 |
| 46 | 263171_at | AT1G54115 | cation exchanger, putative | 0.73 |
| 47 | 245842_at | AT1G58430 | RXF26; carboxylesterase | -1.18 |
| 48 | 260632_at | AT1G62360 | STM (SHOOT MERISTEMLESS); transcription factor | 0.64 |
| 49 | 260241_at | AT1G63710 | CYP86A7 (cytochrome P450, family 86, subfamily A, polypeptide 7); oxygen binding | -1.46 |
| 50 | 260317_at | AT1G63800 | UBC5 (UBIQUITIN-CONJUGATING ENZYME 5); ubiquitin-protein ligase | 0.81 |
| 51 | 259737_at | AT1G64400 | long-chain-fatty-acid--CoA ligase, putative / long-chain acyl-CoA synthetase, putative | 0.65 |
| 52 | 261950_at | AT1G64620 | Dof-type zinc finger domain-containing protein | 1.04 |
| 53 | 245196_at | AT1G67750 | pectate lyase family protein | 0.74 |
| 54 | 260002_at | AT1G67940 | ATNAP3 (Arabidopsis thaliana non-intrinsic ABC protein 3) | -1.20 |
| 55 | 260041_at | AT1G68780 | leucine-rich repeat family protein | -0.75 |
| 56 | 260395_at | AT1G69780 | ATHB13; DNA binding / transcription factor | 0.70 |
| 57 | 259753_at | AT1G71050 | heavy-metal-associated domain-containing protein / copper chaperone (CCH)-related | -0.93 |
| 58 | 259746_at | AT1G71060 | pentatricopeptide (PPR) repeat-containing protein | -1.05 |
| 59 | 260448_at | AT1G72410 | COP1-interacting protein-related | 0.65 |
| 60 | 259892_at | AT1G72610 | GLP1 (GERMIN-LIKE PROTEIN 1); manganese ion binding / metal ion binding / nutrient reservoir | -0.82 |
| 61 | 259891_at | AT1G72730 | eukaryotic translation initiation factor 4A, putative / eIF-4A, putative | -0.69 |
| 62 | 262369_at | AT1G73010 | phosphoric monoester hydrolase | 0.79 |
| 63 | 260072_at | AT1G73650 | oxidoreductase, acting on the CH-CH group of donors | -1.28 |
| 64 | 262680_at | AT1G75880 | family II extracellular lipase 1 (EXL1) | -2.27 |
| 65 | 261749_at | AT1G76180 | ERD14 (EARLY RESPONSE TO DEHYDRATION 14) | 0.71 |
| 66 | 260806_at | AT1G78260 | RNA recognition motif (RRM)-containing protein | 0.62 |
| 67 | 262039_at | AT1G80050 | APT2 (ADENINE PHOSPHORIBOSYL TRANSFERASE 2); adenine phosphoribosyltransferase | 0.74 |
| 68 | 262041_at | AT1G80100 | AHP6 (ARABIDOPSIS HISTIDINE PHOSPHOTRANSFER PROTEIN 6); transferase, transferring phosphorus-containing groups | 1.01 |
| 69 | 261888_at | AT1G80800 | pseudogene, 40S ribosomal protein S12 (RPS12B), similar to ribosomal protein S12 GB:AAD39838 GI:5106775 from (Hordeum vulgare); blastp match of 65% identity and 4.7e-12 P-value to GP|23617253|dbj|BAC20920.1||AP005764 putative 40S ribosomal protein S12 {Oryza sativa (japonica cultivar-group)} | -1.10 |
| 70 | 265731_at | AT2G01130 | ATP binding / helicase/ nucleic acid binding | -1.08 |
| 71 | 266331_at | AT2G01570 | RGA1 (REPRESSOR OF GA1-3 1); transcription factor | 0.65 |
| 72 | 265867_at | AT2G01620 | MEE11 (maternal effect embryo arrest 11) | -0.80 |
| 73 | 263809_at | AT2G04570 | GDSL-motif lipase/hydrolase family protein | -0.89 |
| 74 | 263046_at | AT2G05380 | GRP3S (GLYCINE-RICH PROTEIN 3 SHORT ISOFORM) | -2.29 |
| 75 | 266038_at | AT2G07680 | ATMRP11 (Arabidopsis thaliana multidrug resistance-associated protein 11) | 0.72 |
| 76 | 263819_x_at | AT2G10140 | transposable element gene | -1.59 |
| 77 | 263295_at | AT2G14210 | ANR1; DNA binding / transcription factor | 0.92 |
| 78 | 263330_at | AT2G15320 | leucine-rich repeat family protein | 0.69 |
| 79 | 266529_at | AT2G16880 | pentatricopeptide (PPR) repeat-containing protein | 0.63 |
| 80 | 263073_at | AT2G17500 | auxin efflux carrier family protein | -0.73 |
| 81 | 264788_at | AT2G17880 | DNAJ heat shock protein, putative | -1.04 |
| 82 | 265821_at | AT2G17950 | WUS (WUSCHEL); DNA binding / transcription factor | 1.08 |
| 83 | 265818_at | AT2G18040 | PIN1AT (parvulin 1At) | -3.75 |
| 84 | 265809_at | AT2G18100 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G36210.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G36210.3); similar to unnamed protein product [Vitis vinifera] (GB:CAO48261.1); contains InterPro domain Protein of unknown function DUF726 (InterPro:IPR007941) | 1.38 |
| 85 | 266690_at | AT2G19900 | ATNADP-ME1 (NADP-MALIC ENZYME 1); malate dehydrogenase (oxaloacetate-decarboxylating) (NADP+)/ malic enzyme/ oxidoreductase, acting on NADH or NADPH, NAD or NADP as acceptor | 1.35 |
| 86 | 265427_at | AT2G20740 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G20230.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO23953.1) | 0.66 |
| 87 | 265441_at | AT2G20870 | cell wall protein precursor, putative | -0.87 |
| 88 | 263757_at | AT2G21430 | cysteine proteinase A494, putative / thiol protease, putative | 1.59 |
| 89 | 263545_at | AT2G21560 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G39190.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN77202.1) | 0.71 |
| 90 | 264054_at | AT2G22540 | SVP (SHORT VEGETATIVE PHASE); transcription factor | 0.92 |
| 91 | 265345_at | AT2G22680 | zinc finger (C3HC4-type RING finger) family protein | -0.78 |
| 92 | 267298_at | AT2G23760 | BLH4 (SAWTOOTH 2); DNA binding / transcription factor | -0.82 |
| 93 | 264403_at | AT2G25150 | transferase family protein | -1.46 |
| 94 | 245035_at | AT2G26400 | ARD/ATARD3 (ACIREDUCTONE DIOXYGENASE); acireductone dioxygenase [iron(II)-requiring]/ heteroglycan binding / metal ion binding | -1.73 |
| 95 | 245056_at | AT2G26480 | UDP-glucoronosyl/UDP-glucosyl transferase family protein | 0.85 |
| 96 | 266309_at | AT2G27140 | heat shock family protein | -1.27 |
| 97 | 264066_at | AT2G27880 | argonaute protein, putative / AGO, putative | 1.31 |
| 98 | 264068_at | AT2G27990 | BLH8 (BEL1-LIKE HOMEODOMAIN 8); DNA binding / transcription factor | 0.64 |
| 99 | 265255_at | AT2G28420 | lactoylglutathione lyase family protein / glyoxalase I family protein | -0.80 |
| 100 | 263442_at | AT2G28605 | Identical to PsbP-related thylakoid lumenal protein 3, chloroplast precursor [Arabidopsis Thaliana] (GB:Q8VY52;GB:P83050;GB:Q8S8B1); similar to unnamed protein product [Vitis vinifera] (GB:CAO71682.1); contains InterPro domain Mog1/PsbP/DUF1795, alpha/beta/alpha sandwich (InterPro:IPR016124); contains InterPro domain Mog1/PsbP, alpha/beta/alpha sandwich (InterPro:IPR016123) | -0.73 |
| 101 | 266229_at | AT2G28840 | ankyrin repeat family protein | 0.69 |
| 102 | 267192_at | AT2G30890 | membrane protein, putative | 1.03 |
| 103 | 266481_at | AT2G31070 | TCP10 (TCP DOMAIN PROTEIN 10); transcription factor | -0.76 |
| 104 | 263465_at | AT2G31940 | oxidoreductase/ transition metal ion binding | -0.71 |
| 105 | 265698_at | AT2G32160 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G32170.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO64660.1); contains InterPro domain N2227-like (InterPro:IPR012901) | 0.63 |
| 106 | 267040_at | AT2G34300 | dehydration-responsive protein-related | -1.12 |
| 107 | 266549_at | AT2G35150 | phosphate-responsive 1 family protein | 0.95 |
| 108 | 263931_at | AT2G36220 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G52710.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO15901.1) | 1.44 |
| 109 | 267144_at | AT2G38110 | ATGPAT6/GPAT6 (GLYCEROL-3-PHOSPHATE ACYLTRANSFERASE 6); 1-acylglycerol-3-phosphate O-acyltransferase/ acyltransferase | -0.72 |
| 110 | 267096_at | AT2G38180 | GDSL-motif lipase/hydrolase family protein | 0.67 |
| 111 | 263296_at | AT2G38800 | calmodulin-binding protein-related | 0.98 |
| 112 | 266168_at | AT2G38870 | protease inhibitor, putative | 0.63 |
| 113 | 266979_at | AT2G39470 | PPL2 (PSBP-LIKE PROTEIN 2); calcium ion binding | -1.10 |
| 114 | 267349_at | AT2G40010 | 60S acidic ribosomal protein P0 (RPP0A) | -0.73 |
| 115 | 266054_at | AT2G40640 | similar to ubiquitin-protein ligase [Arabidopsis thaliana] (TAIR:AT5G05230.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO41513.1); contains domain SSF57850 (SSF57850) | 0.64 |
| 116 | 257382_at | AT2G40750 | WRKY54 (WRKY DNA-binding protein 54); transcription factor | -0.88 |
| 117 | 267077_at | AT2G40970 | myb family transcription factor | 1.30 |
| 118 | 263497_at | AT2G42540 | COR15A (COLD-REGULATED 15A) | 1.44 |
| 119 | 263498_at | AT2G42610 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G07090.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN66524.1); contains InterPro domain Protein of unknown function DUF640 (InterPro:IPR006936) | 0.93 |
| 120 | 263971_at | AT2G42800 | leucine-rich repeat family protein | 0.62 |
| 121 | 265261_at | AT2G42990 | GDSL-motif lipase/hydrolase family protein | -0.88 |
| 122 | 260551_at | AT2G43510 | ATTI1 (ARABIDOPSIS THALIANA TRYPSIN INHIBITOR PROTEIN 1) | -1.13 |
| 123 | 260610_at | AT2G43680 | IQD14; calmodulin binding | 0.73 |
| 124 | 267389_at | AT2G44460 | glycosyl hydrolase family 1 protein | -1.07 |
| 125 | 267509_at | AT2G45660 | AGL20 (AGAMOUS-LIKE 20); transcription factor | 0.97 |
| 126 | 266760_at | AT2G46870 | NGA1 (NGATHA1); transcription factor | -0.82 |
| 127 | 259124_at | AT3G02310 | SEP2 (SEPALLATA2); DNA binding / transcription factor | -0.77 |
| 128 | 259133_at | AT3G05400 | sugar transporter, putative | -0.71 |
| 129 | 258894_at | AT3G05650 | disease resistance family protein | 1.07 |
| 130 | 258560_at | AT3G06020 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G19260.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN75990.1) | 1.14 |
| 131 | 258639_at | AT3G07820 | polygalacturonase 3 (PGA3) / pectinase | -2.30 |
| 132 | 259199_at | AT3G08980 | signal peptidase I family protein | -0.89 |
| 133 | 259009_at | AT3G09260 | PYK10 (phosphate starvation-response 3.1); hydrolase, hydrolyzing O-glycosyl compounds | -1.14 |
| 134 | 258699_at | AT3G09470 | Identical to UNC93-like protein [Arabidopsis Thaliana] (GB:Q94AA1;GB:Q67XL1;GB:Q67Y57;GB:Q9SF56); similar to hypothetical protein [Vitis vinifera] (GB:CAN76819.1); contains InterPro domain Protein of unknown function DUF895, eukaryotic (InterPro:IPR010291); contains InterPro domain MFS general substrate transporter (InterPro:IPR016196) | -0.84 |
| 135 | 258707_at | AT3G09480 | histone H2B, putative | -0.96 |
| 136 | 258719_at | AT3G09540 | pectate lyase family protein | 1.06 |
| 137 | 258942_at | AT3G09960 | calcineurin-like phosphoesterase family protein | -1.43 |
| 138 | 258962_at | AT3G10570 | CYP77A6 (cytochrome P450, family 77, subfamily A, polypeptide 6); oxygen binding | -1.08 |
| 139 | 256266_at | AT3G12320 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G06980.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN75675.1) | 1.03 |
| 140 | 257702_at | AT3G12670 | EMB2742 (EMBRYO DEFECTIVE 2742); CTP synthase | -0.80 |
| 141 | 257848_at | AT3G13030 | hAT dimerisation domain-containing protein | -0.73 |
| 142 | 257008_at | AT3G14210 | ESM1 (EPITHIOSPECIFIER MODIFIER 1); carboxylesterase | -1.37 |
| 143 | 258371_at | AT3G14410 | transporter-related | 0.65 |
| 144 | 258114_at | AT3G14660 | CYP72A13 (cytochrome P450, family 72, subfamily A, polypeptide 13); oxygen binding | 1.04 |
| 145 | 257798_at | AT3G15950 | (TSA1-LIKE); protein binding | -0.95 |
| 146 | 258434_at | AT3G16770 | ATEBP/ERF72/RAP2.3 (RELATED TO AP2 3); DNA binding / protein binding / transcription activator/ transcription factor | 1.00 |
| 147 | 257894_at | AT3G17100 | transcription factor | 0.74 |
| 148 | 257035_at | AT3G19270 | CYP707A4 (cytochrome P450, family 707, subfamily A, polypeptide 4); oxygen binding | 0.90 |
| 149 | 257093_at | AT3G20570 | plastocyanin-like domain-containing protein | -0.89 |
| 150 | 256796_at | AT3G22210 | unknown protein | -0.82 |
| 151 | 258102_at | AT3G23600 | dienelactone hydrolase family protein | -0.71 |
| 152 | 257619_at | AT3G24810 | ICK3 (kip-related protein 5); cyclin-dependent protein kinase inhibitor | 0.62 |
| 153 | 256759_at | AT3G25640 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G23100.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO42071.1); contains InterPro domain Protein of unknown function DUF617, plant (InterPro:IPR006460) | 0.98 |
| 154 | 256753_at | AT3G27160 | GHS1 (GLUCOSE HYPERSENSITIVE 1); structural constituent of ribosome | -0.75 |
| 155 | 257228_at | AT3G27890 | NQR (NADPH:QUINONE OXIDOREDUCTASE); FMN reductase | -0.87 |
| 156 | 256987_at | AT3G28560 | similar to AAA-type ATPase family protein [Arabidopsis thaliana] (TAIR:AT3G28510.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN79616.1); contains domain PTHR23070 (PTHR23070); contains domain PTHR23070:SF1 (PTHR23070:SF1) | 1.05 |
| 157 | 258245_at | AT3G29075 | glycine-rich protein | 0.86 |
| 158 | 257562_at | AT3G30480 | transposable element gene | -0.88 |
| 159 | 256940_at | AT3G30720 | unknown protein | -1.96 |
| 160 | 252709_at | AT3G43840 | oxidoreductase, acting on the CH-CH group of donors | -1.22 |
| 161 | 252618_at | AT3G45140 | LOX2 (LIPOXYGENASE 2) | -0.84 |
| 162 | 252612_at | AT3G45160 | contains domain PROKAR_LIPOPROTEIN (PS51257) | 4.00 |
| 163 | 252575_at | AT3G45440 | lectin protein kinase family protein | 1.32 |
| 164 | 252483_at | AT3G46600 | scarecrow transcription factor family protein | 0.68 |
| 165 | 252374_at | AT3G48100 | ARR5 (ARABIDOPSIS RESPONSE REGULATOR 5); transcription regulator/ two-component response regulator | 0.74 |
| 166 | 252181_at | AT3G50685 | similar to unnamed protein product [Vitis vinifera] (GB:CAO22868.1) | -1.03 |
| 167 | 252131_at | AT3G50930 | AAA-type ATPase family protein | 0.97 |
| 168 | 252095_at | AT3G51000 | epoxide hydrolase, putative | -1.20 |
| 169 | 251940_at | AT3G53450 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G37210.1); similar to Conserved hypothetical protein 730 [Medicago truncatula] (GB:ABD32794.1); contains InterPro domain Conserved hypothetical protein CHP00730 (InterPro:IPR005269) | 1.25 |
| 170 | 251928_at | AT3G53980 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | 0.77 |
| 171 | 251927_at | AT3G53990 | universal stress protein (USP) family protein | 0.81 |
| 172 | 251785_at | AT3G55130 | ATWBC19 (WHITE-BROWN COMPLEX HOMOLOG 19); ATPase, coupled to transmembrane movement of substances | -0.84 |
| 173 | 251590_at | AT3G57690 | AGP23/ATAGP23 (ARABINOGALACTAN-PROTEIN 23) | -1.02 |
| 174 | 251515_at | AT3G59270 | syntaxin-related family protein | 1.08 |
| 175 | 251422_at | AT3G60540 | sec61beta family protein | -0.72 |
| 176 | 251169_at | AT3G63210 | MARD1 (MEDIATOR OF ABA-REGULATED DORMANCY 1) | 0.65 |
| 177 | 251178_at | AT3G63440 | ATCKX6/ATCKX7/CKX6 (CYTOKININ OXIDASE/DEHYDROGENASE 6); cytokinin dehydrogenase | 0.76 |
| 178 | 251119_at | AT3G63510 | FAD binding / catalytic/ tRNA dihydrouridine synthase | -0.88 |
| 179 | 255694_at | AT4G00050 | UNE10 (unfertilized embryo sac 10); DNA binding / transcription factor | 0.72 |
| 180 | 255527_at | AT4G02360 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G02813.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO40037.1); contains InterPro domain Protein of unknown function DUF538 (InterPro:IPR007493) | -1.29 |
| 181 | 255172_x_at | AT4G08000 | transposable element gene | -2.43 |
| 182 | 255131_at | AT4G08280 | similar to unnamed protein product [Vitis vinifera] (GB:CAO61844.1); contains InterPro domain Glutaredoxin 2 (InterPro:IPR008554); contains InterPro domain Thioredoxin-like fold (InterPro:IPR012336); contains InterPro domain Thioredoxin fold (InterPro:IPR012335) | -0.83 |
| 183 | 255080_at | AT4G09030 | AGP10 (Arabinogalactan protein 10) | -0.92 |
| 184 | 254998_at | AT4G09760 | choline kinase, putative | 0.91 |
| 185 | 254955_at | AT4G10920 | KELP; DNA binding / transcription coactivator | 0.78 |
| 186 | 254944_at | AT4G10930 | similar to hypothetical protein [Vitis vinifera] (GB:CAN74396.1) | 0.66 |
| 187 | 254877_at | AT4G11640 | ATSR (ARABIDOPSIS THALIANA SIGNAL-RESPONSIVE); serine racemase | -1.30 |
| 188 | 254777_at | AT4G12960 | gamma interferon responsive lysosomal thiol reductase family protein / GILT family protein | -2.70 |
| 189 | 254687_at | AT4G13770 | CYP83A1 (CYTOCHROME P450 83A1); oxygen binding | 1.21 |
| 190 | 245329_at | AT4G14365 | zinc finger (C3HC4-type RING finger) family protein / ankyrin repeat family protein | 0.91 |
| 191 | 245501_at | AT4G15620 | integral membrane family protein | -0.80 |
| 192 | 245485_at | AT4G16230 | GDSL-motif lipase/hydrolase family protein | -1.15 |
| 193 | 245460_at | AT4G16990 | disease resistance protein (TIR-NBS class), putative | -0.78 |
| 194 | 254702_at | AT4G17940 | binding | 0.90 |
| 195 | 254629_at | AT4G18425 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G46090.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO64139.1); contains InterPro domain Protein of unknown function DUF679 (InterPro:IPR007770) | -0.85 |
| 196 | 254609_at | AT4G18970 | GDSL-motif lipase/hydrolase family protein | -0.93 |
| 197 | 254391_at | AT4G21590 | ENDO3 (ENDONUCLEASE 3); T/G mismatch-specific endonuclease/ endonuclease/ nucleic acid binding / single-stranded DNA specific endodeoxyribonuclease | -0.72 |
| 198 | 254387_at | AT4G21850 | methionine sulfoxide reductase domain-containing protein / SeIR domain-containing protein | -0.80 |
| 199 | 254293_at | AT4G23060 | IQD22 (IQ-domain 22); calmodulin binding | 0.64 |
| 200 | 254232_at | AT4G23600 | CORI3 (CORONATINE INDUCED 1, JASMONIC ACID RESPONSIVE 2); transaminase | -1.26 |
| 201 | 254234_at | AT4G23680 | major latex protein-related / MLP-related | -1.28 |
| 202 | 254201_at | AT4G24130 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G56580.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO62919.1); contains InterPro domain Protein of unknown function DUF538 (InterPro:IPR007493) | -0.76 |
| 203 | 254098_at | AT4G25100 | FSD1 (FE SUPEROXIDE DISMUTASE 1); iron superoxide dismutase | 1.32 |
| 204 | 254096_at | AT4G25150 | acid phosphatase, putative | 0.70 |
| 205 | 254082_at | AT4G25720 | glutamine cyclotransferase family protein | 0.72 |
| 206 | 253915_at | AT4G27280 | calcium-binding EF hand family protein | 0.88 |
| 207 | 253765_at | AT4G28740 | similar to LPA1 (LOW PSII ACCUMULATION1), binding [Arabidopsis thaliana] (TAIR:AT1G02910.1); similar to OSJNBa0043L24.5 [Oryza sativa (japonica cultivar-group)] (GB:CAE04717.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO40509.1) | -0.77 |
| 208 | 253689_at | AT4G29770 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G29760.1) | 1.07 |
| 209 | 253672_at | AT4G29820 | ATCFIM-25/CFIM-25 (ARABIDOPSIS HOMOLOG OF CFIM-25) | 0.67 |
| 210 | 253671_at | AT4G29990 | light repressible receptor protein kinase | 1.93 |
| 211 | 253666_at | AT4G30270 | MERI5B (MERISTEM-5); hydrolase, acting on glycosyl bonds / xyloglucan:xyloglucosyl transferase | -0.90 |
| 212 | 253545_at | AT4G31310 | avirulence-responsive protein-related / avirulence induced gene (AIG) protein-related | -0.88 |
| 213 | 253437_at | AT4G32460 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G11420.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO44183.1); contains InterPro domain Protein of unknown function DUF642 (InterPro:IPR006946); contains InterPro domain Galactose-binding like (InterPro:IPR008979) | -1.05 |
| 214 | 253451_at | AT4G32730 | PC-MYB1 (myb domain protein 3R1); DNA binding / transcription coactivator/ transcription factor | 0.69 |
| 215 | 253367_at | AT4G33180 | hydrolase, alpha/beta fold family protein | -0.77 |
| 216 | 253161_at | AT4G35770 | SEN1 (DARK INDUCIBLE 1) | -1.23 |
| 217 | 253108_at | AT4G35900 | FD; DNA binding / transcription factor | 1.08 |
| 218 | 246219_at | AT4G36760 | ATAPP1 (aminopeptidase P1) | -0.76 |
| 219 | 246284_at | AT4G36780 | brassinosteroid signalling positive regulator-related | 0.64 |
| 220 | 246217_at | AT4G36920 | AP2 (APETALA 2); transcription factor | -0.75 |
| 221 | 253045_at | AT4G37445 | similar to calcium-binding EF hand family protein [Arabidopsis thaliana] (TAIR:AT1G64850.1); similar to Os02g0126600 [Oryza sativa (japonica cultivar-group)] (GB:NP_001045753.1); similar to unknown [Oryza sativa (indica cultivar-group)] (GB:ABR26063.1) | -0.72 |
| 222 | 253012_at | AT4G37900 | glycine-rich protein | 0.73 |
| 223 | 252971_at | AT4G38770 | PRP4 (PROLINE-RICH PROTEIN 4) | -1.23 |
| 224 | 252921_at | AT4G39030 | EDS5 (ENHANCED DISEASE SUSCEPTIBILITY 5); antiporter/ transporter | -0.75 |
| 225 | 252863_at | AT4G39800 | MI-1-P SYNTHASE (Myo-inositol-1-phosphate synthase); inositol-3-phosphate synthase | 0.64 |
| 226 | 251065_at | AT5G01870 | lipid transfer protein, putative | -0.80 |
| 227 | 251013_at | AT5G02540 | short-chain dehydrogenase/reductase (SDR) family protein | 0.80 |
| 228 | 250999_at | AT5G02630 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G18520.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G09570.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO18079.1); contains InterPro domain Transmembrane receptor, eukaryota; (InterPro:IPR009637) | -1.09 |
| 229 | 250942_at | AT5G03350 | legume lectin family protein | -0.88 |
| 230 | 250921_at | AT5G03460 | similar to unknown [Populus trichocarpa] (GB:ABK93498.1) | 0.61 |
| 231 | 250915_at | AT5G03790 | ATHB51/LMI1 (LATE MERISTEM IDENTITY1); DNA binding / sequence-specific DNA binding / transcription factor | -1.11 |
| 232 | 250869_at | AT5G03840 | TFL1 (TERMINAL FLOWER 1); phosphatidylethanolamine binding | 1.18 |
| 233 | 250723_at | AT5G06300 | carboxy-lyase | 0.62 |
| 234 | 250587_at | AT5G07640 | zinc finger (C3HC4-type RING finger) family protein | 1.74 |
| 235 | 250558_at | AT5G07990 | TT7 (TRANSPARENT TESTA 7); flavonoid 3'-monooxygenase/ oxygen binding | -0.81 |
| 236 | 250517_at | AT5G08260 | SCPL35 (serine carboxypeptidase-like 35); serine carboxypeptidase | -1.11 |
| 237 | 250528_at | AT5G08600 | U3 ribonucleoprotein (Utp) family protein | -1.23 |
| 238 | 250370_at | AT5G11430 | transcription elongation factor-related | 0.95 |
| 239 | 250376_at | AT5G11550 | binding | 0.67 |
| 240 | 250167_at | AT5G15310 | AtMIXTA/AtMYB16 (myb domain protein 16); DNA binding / transcription factor | -0.98 |
| 241 | 246482_at | AT5G15930 | PAM1 (PLANT ADHESION MOLECULE 1); RAB GTPase activator | -0.83 |
| 242 | 246488_at | AT5G16010 | 3-oxo-5-alpha-steroid 4-dehydrogenase family protein / steroid 5-alpha-reductase family protein | 0.66 |
| 243 | 246459_at | AT5G16900 | leucine-rich repeat protein kinase, putative | 1.74 |
| 244 | 246099_at | AT5G20230 | ATBCB (ARABIDOPSIS BLUE-COPPER-BINDING PROTEIN); copper ion binding | -1.37 |
| 245 | 246114_at | AT5G20250 | DIN10 (DARK INDUCIBLE 10); hydrolase, hydrolyzing O-glycosyl compounds | -0.76 |
| 246 | 249939_at | AT5G22430 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G27385.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN68427.1) | -0.90 |
| 247 | 249860_at | AT5G22860 | serine carboxypeptidase S28 family protein | 0.67 |
| 248 | 249761_at | AT5G23970 | transferase family protein | -0.96 |
| 249 | 249741_at | AT5G24470 | APRR5 (PSEUDO-RESPONSE REGULATOR 5); transcription regulator | 0.63 |
| 250 | 246978_at | AT5G24910 | CYP714A1 (cytochrome P450, family 714, subfamily A, polypeptide 1); oxygen binding | -0.77 |
| 251 | 246905_at | AT5G25570 | similar to unnamed protein product [Vitis vinifera] (GB:CAO44135.1) | -1.96 |
| 252 | 246888_at | AT5G26270 | unknown protein | -3.62 |
| 253 | 246743_at | AT5G27750 | F-box family protein | 0.73 |
| 254 | 246103_at | AT5G28640 | AN3 (ANGUSITFOLIA3) | 0.63 |
| 255 | 246104_at | AT5G28650 | WRKY74 (WRKY DNA-binding protein 74); transcription factor | 1.11 |
| 256 | 246687_at | AT5G33370 | GDSL-motif lipase/hydrolase family protein | -1.22 |
| 257 | 246664_at | AT5G34800 | transposable element gene | 1.85 |
| 258 | 249271_at | AT5G41790 | CIP1 (COP1-INTERACTIVE PROTEIN 1) | 0.63 |
| 259 | 249230_at | AT5G42070 | similar to unnamed protein product [Vitis vinifera] (GB:CAO69519.1) | -0.94 |
| 260 | 249245_at | AT5G42280 | DC1 domain-containing protein | -0.86 |
| 261 | 249167_at | AT5G42860 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G45688.1); similar to H0814G11.12 [Oryza sativa (indica cultivar-group)] (GB:CAJ86345.1); similar to CAA30379.1 protein [Oryza sativa] (GB:CAB53482.1) | 1.17 |
| 262 | 249134_at | AT5G43150 | similar to unnamed protein product [Vitis vinifera] (GB:CAO61459.1) | 0.71 |
| 263 | 249147_at | AT5G43330 | malate dehydrogenase, cytosolic, putative | 0.86 |
| 264 | 249127_at | AT5G43500 | ATARP9 (ACTIN-RELATED PROTEIN 9); protein binding | 1.36 |
| 265 | 248975_at | AT5G45040 | cytochrome c6 (ATC6) | -1.11 |
| 266 | 248845_at | AT5G46470 | disease resistance protein (TIR-NBS-LRR class), putative | 1.25 |
| 267 | 248793_at | AT5G47240 | ATNUDT8 (Arabidopsis thaliana Nudix hydrolase homolog 8); hydrolase | 0.65 |
| 268 | 248779_at | AT5G47720 | acetyl-CoA C-acyltransferase, putative / 3-ketoacyl-CoA thiolase, putative | 0.78 |
| 269 | 248683_at | AT5G48490 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | -1.08 |
| 270 | 248668_at | AT5G48720 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G01990.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO23288.1) | 0.69 |
| 271 | 248607_at | AT5G49480 | ATCP1 (CA2+-BINDING PROTEIN 1); calcium ion binding | 0.68 |
| 272 | 248448_at | AT5G51190 | AP2 domain-containing transcription factor, putative | 0.98 |
| 273 | 248286_at | AT5G52870 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G64080.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO65893.1) | 0.88 |
| 274 | 248160_at | AT5G54470 | zinc finger (B-box type) family protein | 0.91 |
| 275 | 248145_at | AT5G54880 | DTW domain-containing protein | -1.19 |
| 276 | 248073_at | AT5G55720 | pectate lyase family protein | -1.40 |
| 277 | 248021_at | AT5G56500 | ATP binding / protein binding / unfolded protein binding | 1.31 |
| 278 | 247933_at | AT5G56980 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G26130.1); similar to unknown [Populus trichocarpa] (GB:ABK95362.1) | 0.73 |
| 279 | 247906_at | AT5G57420 | IAA33 (indoleacetic acid-induced protein 33); transcription factor | 0.84 |
| 280 | 247885_at | AT5G57830 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G30830.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO66637.1); contains InterPro domain Protein of unknown function DUF593 (InterPro:IPR007656) | 0.97 |
| 281 | 247779_at | AT5G58760 | DDB2 (DAMAGED DNA-BINDING 2); nucleotide binding | 0.71 |
| 282 | 247768_at | AT5G58900 | myb family transcription factor | 0.68 |
| 283 | 247760_at | AT5G59130 | subtilase family protein | 0.89 |
| 284 | 247719_at | AT5G59305 | similar to unknown [Populus trichocarpa] (GB:ABK94520.1) | 1.44 |
| 285 | 247718_at | AT5G59310 | LTP4 (LIPID TRANSFER PROTEIN 4); lipid binding | 1.36 |
| 286 | 247682_at | AT5G59650 | leucine-rich repeat protein kinase, putative | -0.79 |
| 287 | 247691_at | AT5G59720 | HSP18.2 (HEAT SHOCK PROTEIN 18.2) | 0.99 |
| 288 | 247615_at | AT5G60250 | zinc finger (C3HC4-type RING finger) family protein | 0.73 |
| 289 | 247601_at | AT5G60850 | OBP4 (OBF BINDING PROTEIN 4); DNA binding / transcription factor | 0.80 |
| 290 | 247604_at | AT5G60950 | COBL5 (COBRA-LIKE PROTEIN 5 PRECURSOR) | 1.29 |
| 291 | 247418_at | AT5G63030 | glutaredoxin, putative | -0.85 |
| 292 | 247345_at | AT5G63760 | IBR domain-containing protein | 0.78 |
| 293 | 247208_at | AT5G64870 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G25250.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G25260.1); similar to 80C09_16 [Brassica rapa subsp. pekinensis] (GB:AAZ41827.1); contains domain PTHR13806:SF3 (PTHR13806:SF3); contains domain PTHR13806 (PTHR13806) | 0.86 |
| 294 | 247228_at | AT5G65140 | trehalose-6-phosphate phosphatase, putative | 0.93 |
| 295 | 247189_at | AT5G65390 | AGP7 (Arabinogalactan protein 7) | -0.93 |
| 296 | 247196_at | AT5G65510 | AIL7 (AINTEGUMENTA-LIKE 7); DNA binding / transcription factor | 0.92 |
| 297 | 247136_at | AT5G66170 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G17850.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO48196.1); contains InterPro domain Rhodanese-like (InterPro:IPR001763) | 0.67 |
| 298 | 247140_at | AT5G66250 | kinectin-related | -1.14 |
| 299 | 245047_at | ATCG00020 | Encodes chlorophyll binding protein D1, a part of the photosystem II reaction center core | -0.91 |
| 300 | 244997_at | ATCG00170 | RNA polymerase beta' subunit-2 | -0.74 |
| 301 | 245000_at | ATCG00210 | hypothetical protein | -1.52 |
| 302 | 245004_at | ATCG00300 | encodes PsbZ, which is a subunit of photosystem II. In Chlamydomonas, this protein has been shown to be essential in the interaction between PS II and the light harvesting complex II. | -0.83 |
| 303 | 245010_at | ATCG00420 | Encodes NADH dehydrogenase subunit J. Its transcription is increased upon sulfur depletion. | -1.51 |
| 304 | 245011_at | ATCG00430 | Encodes a protein which was originally thought to be part of photosystem II but its wheat homolog was later shown to encode for subunit K of NADH dehydrogenase. | -0.71 |
| 305 | 245015_at | ATCG00490 | large subunit of RUBISCO. | -1.14 |
| 306 | 245016_at | ATCG00500 | Encodes the carboxytransferase beta subunit of the Acetyl-CoA carboxylase (ACCase) complex in plastids. This complex catalyzes the carboxylation of acetyl-CoA to produce malonyl-CoA, the first committed step in fatty acid synthesis. | -0.86 |
| 307 | 244970_at | ATCG00660 | encodes a chloroplast ribosomal protein L20, a constituent of the large subunit of the ribosomal complex | -1.64 |
| 308 | 244977_at | ATCG00730 | A chloroplast gene encoding subunit IV of the cytochrome b6/f complex | -2.32 |
| 309 | 244994_at | ATCG01010 | Chloroplast encoded NADH dehydrogenase unit. | -3.18 |
| 310 | 244962_at | ATCG01050 | Represents a plastid-encoded subunit of a NAD(P)H dehydrogenase complex. Its mRNA is edited at four positions. Translation data is not available for this gene. | -0.75 |
| 311 | 244932_at | ATCG01060 | Encodes the PsaC subunit of photosystem I. | -1.35 |
| 312 | 244941_at | ATMG00010 | hypothetical protein | -1.17 |
| 313 | 244950_at | ATMG00160 | cytochrome c oxidase subunit 2 | -0.74 |
| 314 | 244929_at | ATMG00580 | NADH dehydrogenase subunit 4 | -1.31 |
| 315 | 244931_at | ATMG00630 | hypothetical protein | -1.74 |
| 316 | 244904_at | ATMG00670 | hypothetical protein | 0.81 |
| 317 | 244924_at | ATMG01040 | hypothetical protein | -1.10 |
| 318 | 257319_at | ATMG01100 | hypothetical protein | -1.17 |
| 319 | 257330_at | ATMG01290 | hypothetical protein | -1.78 |
| 320 | 257332_at | ATMG01350 | hypothetical protein | -1.17 |
| 321 | 257333_at | ATMG01360 | cytochrome c oxidase subunit 1 | -1.39 |