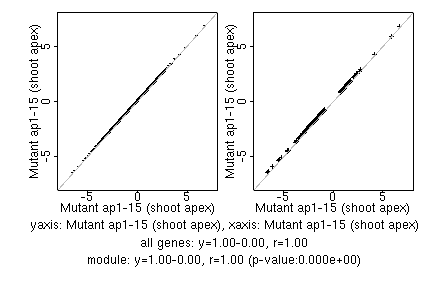
marker gene list
| Mutant ap1-15(shoot apex) |
| Mutant ap1-15(shoot apex) |
| probeID | AGICode | Annotation | gene expression |
|
|---|---|---|---|---|
| 1 | 261056_at | AT1G01360 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G01026.1); similar to hypothetical protein [Arabidopsis suecica] (GB:BAF63139.1); contains domain SSF55961 (SSF55961) | 0.85 |
| 2 | 259429_at | AT1G01600 | CYP86A4 (cytochrome P450, family 86, subfamily A, polypeptide 4); oxygen binding | -2.42 |
| 3 | 264151_at | AT1G02070 | similar to zinc ion binding [Arabidopsis thaliana] (TAIR:AT3G60520.1); similar to unknown [Populus trichocarpa] (GB:ABK95332.1) | 1.00 |
| 4 | 264363_at | AT1G03170 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G02810.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO22484.1) | 0.94 |
| 5 | 264604_at | AT1G04650 | similar to unnamed protein product [Vitis vinifera] (GB:CAO71563.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN72207.1) | 0.86 |
| 6 | 262637_at | AT1G06640 | 2-oxoglutarate-dependent dioxygenase, putative | -0.93 |
| 7 | 264624_at | AT1G08930 | ERD6 (EARLY RESPONSE TO DEHYDRATION 6); carbohydrate transmembrane transporter/ sugar:hydrogen ion symporter | 0.80 |
| 8 | 264507_at | AT1G09415 | NIMIN-3 (NIM1-INTERACTING 3) | -1.22 |
| 9 | 264525_at | AT1G10060 | ATBCAT-1; branched-chain-amino-acid transaminase/ catalytic | -1.18 |
| 10 | 264436_at | AT1G10370 | ATGSTU17/ERD9/GST30/GST30B (EARLY-RESPONSIVE TO DEHYDRATION 9); glutathione transferase | 1.50 |
| 11 | 262788_at | AT1G10690 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G60783.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN59885.1) | 1.81 |
| 12 | 262479_at | AT1G11130 | SUB (STRUBBELIG); protein binding | 1.06 |
| 13 | 264343_at | AT1G11850 | unknown protein | -6.39 |
| 14 | 264342_at | AT1G12080 | contains domain PTHR22683 (PTHR22683) | -2.17 |
| 15 | 260969_at | AT1G12240 | ATBETAFRUCT4/VAC-INV (VACUOLAR INVERTASE); beta-fructofuranosidase/ hydrolase, hydrolyzing O-glycosyl compounds | -0.79 |
| 16 | 259526_at | AT1G12570 | glucose-methanol-choline (GMC) oxidoreductase family protein | -2.61 |
| 17 | 262774_at | AT1G13230 | leucine-rich repeat family protein | -0.99 |
| 18 | 259364_at | AT1G13260 | RAV1 (Related to ABI3/VP1 1); DNA binding / transcription factor | 0.88 |
| 19 | 256102_at | AT1G13680 | phospholipase C | -0.81 |
| 20 | 262612_at | AT1G14150 | oxygen evolving enhancer 3 (PsbQ) family protein | -0.83 |
| 21 | 262842_at | AT1G14720 | XTR2 (XYLOGLUCAN ENDOTRANSGLYCOSYLASE RELATED 2); hydrolase, acting on glycosyl bonds | 1.14 |
| 22 | 262569_at | AT1G15180 | MATE efflux family protein | -1.33 |
| 23 | 262575_at | AT1G15210 | ATPDR7/PDR7 (PLEIOTROPIC DRUG RESISTANCE 7); ATPase, coupled to transmembrane movement of substances | -1.30 |
| 24 | 262598_at | AT1G15260 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G16070.1) | -2.20 |
| 25 | 259502_at | AT1G15670 | kelch repeat-containing F-box family protein | 1.05 |
| 26 | 261788_at | AT1G15980 | similar to unnamed protein product [Vitis vinifera] (GB:CAO49411.1); contains domain G3DSA:3.40.50.2000 (G3DSA:3.40.50.2000); contains domain SSF53756 (SSF53756) | -1.08 |
| 27 | 255768_at | AT1G16705 | p300/CBP acetyltransferase-related protein-related | -0.94 |
| 28 | 262519_at | AT1G17160 | pfkB-type carbohydrate kinase family protein | -0.87 |
| 29 | 256128_at | AT1G18140 | LAC1 (Laccase 1); copper ion binding / oxidoreductase | -2.09 |
| 30 | 255786_at | AT1G19670 | ATCLH1 (CORONATINE-INDUCED PROTEIN 1) | -0.83 |
| 31 | 261247_at | AT1G20070 | unknown protein | -0.84 |
| 32 | 259570_at | AT1G20440 | COR47 (cold regulated 47) | -0.83 |
| 33 | 256093_at | AT1G20823 | zinc finger (C3HC4-type RING finger) family protein | 1.40 |
| 34 | 255957_at | AT1G22160 | senescence-associated protein-related | 0.87 |
| 35 | 255962_at | AT1G22330 | RNA binding | -0.96 |
| 36 | 261929_at | AT1G22460 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G64600.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO61677.1); contains InterPro domain Protein of unknown function DUF246, plant (InterPro:IPR004348) | -1.41 |
| 37 | 263013_at | AT1G23380 | KNAT6 (Knotted-like Arabidopsis thaliana 6); DNA binding / transcription factor | 1.27 |
| 38 | 265025_at | AT1G24575 | unknown protein | 1.11 |
| 39 | 245870_at | AT1G26300 | BSD domain-containing protein | 0.92 |
| 40 | 245819_at | AT1G26310 | CAL (CAULIFLOWER); DNA binding / transcription factor | 1.76 |
| 41 | 261004_at | AT1G26450 | beta-1,3-glucanase-related | -0.86 |
| 42 | 261265_at | AT1G26800 | zinc finger (C3HC4-type RING finger) family protein | 0.82 |
| 43 | 259596_at | AT1G28130 | GH3.17; indole-3-acetic acid amido synthetase | 1.12 |
| 44 | 261467_at | AT1G28520 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G42400.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO66378.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO69825.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN71043.1) | 0.99 |
| 45 | 259783_at | AT1G29510 | SAUR68 (SMALL AUXIN UPREGULATED 68) | -1.08 |
| 46 | 260023_at | AT1G30040 | ATGA2OX2; gibberellin 2-beta-dioxygenase | 1.00 |
| 47 | 264497_at | AT1G30840 | ATPUP4 (Arabidopsis thaliana purine permease 4); purine transmembrane transporter | 1.33 |
| 48 | 257467_at | AT1G31320 | LBD4 (LOB DOMAIN-CONTAINING PROTEIN 4) | 0.84 |
| 49 | 246261_at | AT1G31810 | actin binding | -1.29 |
| 50 | 261567_at | AT1G33055 | unknown protein | 2.46 |
| 51 | 261594_at | AT1G33240 | AT-GTL1 (Arabidopsis thaliana GT2-like 1); transcription factor | -0.81 |
| 52 | 262543_at | AT1G34245 | similar to EPF1 (EPIDERMAL PATTERNING FACTOR 1) [Arabidopsis thaliana] (TAIR:AT2G20875.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO64636.1); contains domain PROKAR_LIPOPROTEIN (PS51257) | -2.36 |
| 53 | 261339_at | AT1G35710 | leucine-rich repeat transmembrane protein kinase, putative | 0.97 |
| 54 | 261285_at | AT1G35720 | ANNAT1 (ANNEXIN ARABIDOPSIS 1); calcium ion binding / calcium-dependent phospholipid binding | -0.81 |
| 55 | 261292_at | AT1G36940 | unknown protein | 1.04 |
| 56 | 261979_at | AT1G37130 | NIA2 (NITRATE REDUCTASE 2) | -2.97 |
| 57 | 261363_at | AT1G41830 | SKS6 (SKU5 Similar 6); pectinesterase | -0.94 |
| 58 | 262719_at | AT1G43590 | transposable element gene | 1.69 |
| 59 | 260869_at | AT1G43800 | acyl-(acyl-carrier-protein) desaturase, putative / stearoyl-ACP desaturase, putative | 1.08 |
| 60 | 260943_at | AT1G45145 | ATTRX5 (thioredoxin H-type 5); thiol-disulfide exchange intermediate | 1.07 |
| 61 | 262399_at | AT1G49500 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G19030.1) | -1.79 |
| 62 | 260203_at | AT1G52890 | ANAC019 (Arabidopsis NAC domain containing protein 19); transcription factor | -2.06 |
| 63 | 261369_at | AT1G53060 | legume lectin family protein | -1.26 |
| 64 | 263174_at | AT1G54040 | ESP (EPITHIOSPECIFIER PROTEIN) | -3.29 |
| 65 | 259658_at | AT1G55370 | carbohydrate binding / catalytic | -1.20 |
| 66 | 260592_at | AT1G55850 | ATCSLE1 (Cellulose synthase-like E1); cellulose synthase/ transferase, transferring glycosyl groups | -1.48 |
| 67 | 256024_at | AT1G58340 | ZF14; transporter | 1.65 |
| 68 | 245842_at | AT1G58430 | RXF26; carboxylesterase | -4.49 |
| 69 | 264216_at | AT1G60180 | pseudogene of F-box family protein | 5.92 |
| 70 | 264920_at | AT1G60550 | naphthoate synthase, putative / dihydroxynaphthoic acid synthetase, putative / DHNA synthetase, putative | -0.89 |
| 71 | 264433_at | AT1G61810 | BGLU45; hydrolase, hydrolyzing O-glycosyl compounds | -1.34 |
| 72 | 260632_at | AT1G62360 | STM (SHOOT MERISTEMLESS); transcription factor | 0.80 |
| 73 | 261095_at | AT1G62930 | binding | -1.30 |
| 74 | 260241_at | AT1G63710 | CYP86A7 (cytochrome P450, family 86, subfamily A, polypeptide 7); oxygen binding | -6.00 |
| 75 | 262354_at | AT1G64200 | VHA-E3 (VACUOLAR H+-ATPASE SUBUNIT E ISOFORM 3); hydrogen ion transporting ATPase, rotational mechanism | -1.59 |
| 76 | 261950_at | AT1G64620 | Dof-type zinc finger domain-containing protein | 0.88 |
| 77 | 256324_at | AT1G66760 | MATE efflux family protein | -0.90 |
| 78 | 256383_at | AT1G66820 | glycine-rich protein | -0.80 |
| 79 | 256366_at | AT1G66880 | serine/threonine protein kinase family protein | 0.83 |
| 80 | 260012_at | AT1G67865 | unknown protein | 1.08 |
| 81 | 260265_at | AT1G68510 | LBD42 (LOB DOMAIN-CONTAINING PROTEIN 42) | 0.88 |
| 82 | 260266_at | AT1G68520 | zinc finger (B-box type) family protein | -0.78 |
| 83 | 260041_at | AT1G68780 | leucine-rich repeat family protein | -1.09 |
| 84 | 259372_at | AT1G69120 | AP1 (APETALA1); DNA binding / transcription factor | -1.32 |
| 85 | 260355_at | AT1G69180 | CRC (CRABS CLAW); transcription factor | 0.94 |
| 86 | 260353_at | AT1G69230 | SP1L2 | -1.43 |
| 87 | 256299_at | AT1G69530 | ATEXPA1 (ARABIDOPSIS THALIANA EXPANSIN A1) | -1.31 |
| 88 | 260414_at | AT1G69850 | ATNRT1:2 (NITRATE TRANSPORTER 1:2); calcium ion binding / transporter | 0.94 |
| 89 | 260334_at | AT1G70510 | KNAT2 (KNOTTED-LIKE FROM ARABIDOPSIS THALIANA 2); transcription factor | 0.89 |
| 90 | 262288_at | AT1G70760 | inorganic carbon transport protein-related | -0.91 |
| 91 | 262309_at | AT1G70820 | phosphoglucomutase, putative / glucose phosphomutase, putative | -0.96 |
| 92 | 259753_at | AT1G71050 | heavy-metal-associated domain-containing protein / copper chaperone (CCH)-related | -3.12 |
| 93 | 259892_at | AT1G72610 | GLP1 (GERMIN-LIKE PROTEIN 1); manganese ion binding / metal ion binding / nutrient reservoir | -1.91 |
| 94 | 260389_at | AT1G74055 | unknown protein | -0.94 |
| 95 | 260221_at | AT1G74670 | gibberellin-responsive protein, putative | -1.44 |
| 96 | 262216_at | AT1G74780 | nodulin family protein | 0.86 |
| 97 | 262175_at | AT1G74880 | NDH-O (NAD(P)H:PLASTOQUINONE DEHYDROGENASE COMPLEX SUBUNIT O) | -0.82 |
| 98 | 262976_at | AT1G75520 | SRS5 (SHI-RELATED SEQUENCE 5) | 1.43 |
| 99 | 261727_at | AT1G76090 | SMT3 (S-adenosyl-methionine-sterol-C-methyltransferase 3); S-adenosylmethionine-dependent methyltransferase | -0.96 |
| 100 | 261749_at | AT1G76180 | ERD14 (EARLY RESPONSE TO DEHYDRATION 14) | 0.95 |
| 101 | 259878_at | AT1G76790 | O-methyltransferase family 2 protein | -1.31 |
| 102 | 264953_at | AT1G77120 | ADH1 (ALCOHOL DEHYDROGENASE 1) | 0.96 |
| 103 | 259680_at | AT1G77690 | amino acid permease, putative | -0.77 |
| 104 | 262196_at | AT1G77870 | MUB5 (MEMBRANE-ANCHORED UBIQUITIN-FOLD PROTEIN 5 PRECURSOR) | -0.96 |
| 105 | 264137_at | AT1G78960 | ATLUP2 (Arabidopsis thaliana lupeol synthase 2); lupeol synthase | -1.10 |
| 106 | 264124_at | AT1G79360 | ATOCT2 (ARABIDOPSIS THALIANA ORGANIC CATION/CARNITINE TRANSPORTER2); carbohydrate transmembrane transporter/ sugar:hydrogen ion symporter | -1.23 |
| 107 | 260166_at | AT1G79840 | GL2 (GLABRA 2); DNA binding / transcription factor | 0.95 |
| 108 | 262040_at | AT1G80080 | TMM (TOO MANY MOUTHS); protein binding | -1.14 |
| 109 | 262041_at | AT1G80100 | AHP6 (ARABIDOPSIS HISTIDINE PHOSPHOTRANSFER PROTEIN 6); transferase, transferring phosphorus-containing groups | 1.25 |
| 110 | 266353_at | AT2G01520 | MLP328 (MLP-LIKE PROTEIN 328) | -3.21 |
| 111 | 263595_at | AT2G01890 | PAP8 (PURPLE ACID PHOSPHATASE PRECURSOR); acid phosphatase/ protein serine/threonine phosphatase | -0.95 |
| 112 | 263322_at | AT2G04270 | glycoside hydrolase starch-binding domain-containing protein | -0.77 |
| 113 | 263809_at | AT2G04570 | GDSL-motif lipase/hydrolase family protein | -2.93 |
| 114 | 263046_at | AT2G05380 | GRP3S (GLYCINE-RICH PROTEIN 3 SHORT ISOFORM) | -2.53 |
| 115 | 265511_at | AT2G05540 | glycine-rich protein | -2.57 |
| 116 | 266032_x_at | AT2G05890 | transposable element gene | -0.88 |
| 117 | 263819_x_at | AT2G10140 | transposable element gene | -1.33 |
| 118 | 265400_at | AT2G10940 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | -1.41 |
| 119 | 263350_at | AT2G13360 | AGT (ALANINE:GLYOXYLATE AMINOTRANSFERASE) | -2.25 |
| 120 | 263295_at | AT2G14210 | ANR1; DNA binding / transcription factor | 1.32 |
| 121 | 265475_at | AT2G15620 | NIR1 (NITRITE REDUCTASE); ferredoxin-nitrate reductase | -1.10 |
| 122 | 265414_at | AT2G16660 | nodulin family protein | -0.83 |
| 123 | 264909_at | AT2G17300 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G35320.1) | -0.75 |
| 124 | 263071_at | AT2G17490 | transposable element gene | 1.50 |
| 125 | 263073_at | AT2G17500 | auxin efflux carrier family protein | -1.40 |
| 126 | 263067_at | AT2G17550 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G02390.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO14481.1) | -1.08 |
| 127 | 264620_at | AT2G17770 | ATBZIP27/FDP (FD PARALOG); transcription factor | 1.01 |
| 128 | 264788_at | AT2G17880 | DNAJ heat shock protein, putative | -0.76 |
| 129 | 265983_at | AT2G18550 | ATHB21/HB-2 (homeobox-2); DNA binding / transcription factor | 1.86 |
| 130 | 266070_at | AT2G18660 | EXLB3 (EXPANSIN-LIKE B3 PRECURSOR) | 2.73 |
| 131 | 266019_at | AT2G18750 | calmodulin-binding protein | 0.82 |
| 132 | 265387_at | AT2G20670 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G32480.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO69754.1); contains InterPro domain Protein of unknown function DUF506, plant (InterPro:IPR006502) | -2.68 |
| 133 | 265441_at | AT2G20870 | cell wall protein precursor, putative | -5.43 |
| 134 | 263876_at | AT2G21880 | AtRABG2/AtRab7A (Arabidopsis Rab GTPase homolog G2); GTP binding | -1.21 |
| 135 | 263875_at | AT2G21970 | SEP2 (STRESS ENHANCED PROTEIN 2) | 1.06 |
| 136 | 264054_at | AT2G22540 | SVP (SHORT VEGETATIVE PHASE); transcription factor | 1.54 |
| 137 | 267265_at | AT2G22980 | SCPL13; serine carboxypeptidase | -1.24 |
| 138 | 267250_at | AT2G23060 | GCN5-related N-acetyltransferase (GNAT) family protein | 1.46 |
| 139 | 267298_at | AT2G23760 | BLH4 (SAWTOOTH 2); DNA binding / transcription factor | -0.89 |
| 140 | 266015_at | AT2G24190 | short-chain dehydrogenase/reductase (SDR) family protein | 1.27 |
| 141 | 265255_at | AT2G28420 | lactoylglutathione lyase family protein / glyoxalase I family protein | -2.04 |
| 142 | 264081_at | AT2G28540 | nucleic acid binding / nucleotide binding / protein binding / zinc ion binding | 0.87 |
| 143 | 267209_at | AT2G30930 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G06540.1) | -0.83 |
| 144 | 266481_at | AT2G31070 | TCP10 (TCP DOMAIN PROTEIN 10); transcription factor | -1.22 |
| 145 | 266474_at | AT2G31110 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G42570.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO69853.1); contains InterPro domain Protein of unknown function DUF231, plant (InterPro:IPR004253) | -0.80 |
| 146 | 266479_at | AT2G31160 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G23290.2); similar to unknown [Populus trichocarpa] (GB:ABK94898.1); contains InterPro domain Protein of unknown function DUF640 (InterPro:IPR006936) | 1.09 |
| 147 | 263478_at | AT2G31880 | leucine-rich repeat transmembrane protein kinase, putative | 1.14 |
| 148 | 265680_at | AT2G32150 | haloacid dehalogenase-like hydrolase family protein | -0.90 |
| 149 | 265698_at | AT2G32160 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G32170.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO64660.1); contains InterPro domain N2227-like (InterPro:IPR012901) | 0.90 |
| 150 | 267461_at | AT2G33830 | dormancy/auxin associated family protein | -1.10 |
| 151 | 266996_at | AT2G34490 | CYP710A2 (cytochrome P450, family 710, subfamily A, polypeptide 2); C-22 sterol desaturase/ oxygen binding | -1.02 |
| 152 | 267425_at | AT2G34810 | FAD-binding domain-containing protein | -2.23 |
| 153 | 265823_at | AT2G35760 | integral membrane family protein | 0.86 |
| 154 | 263931_at | AT2G36220 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G52710.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO15901.1) | 1.42 |
| 155 | 266097_at | AT2G37970 | SOUL-1; binding | 0.89 |
| 156 | 267144_at | AT2G38110 | ATGPAT6/GPAT6 (GLYCEROL-3-PHOSPHATE ACYLTRANSFERASE 6); 1-acylglycerol-3-phosphate O-acyltransferase/ acyltransferase | -1.54 |
| 157 | 266167_at | AT2G38860 | YLS5 (yellow-leaf-specific gene 5) | -0.80 |
| 158 | 266172_at | AT2G39010 | PIP2;6/PIP2E (plasma membrane intrinsic protein 2;6); water channel | -0.95 |
| 159 | 266990_at | AT2G39190 | ATATH8 (ABC2 homolog 8) | 0.83 |
| 160 | 266989_at | AT2G39330 | jacalin lectin family protein | -1.79 |
| 161 | 267361_at | AT2G39920 | acid phosphatase class B family protein | -1.38 |
| 162 | 267364_at | AT2G40080 | ELF4 (EARLY FLOWERING 4) | 0.96 |
| 163 | 263384_at | AT2G40130 | heat shock protein-related | 1.68 |
| 164 | 255825_at | AT2G40475 | unknown protein | -1.52 |
| 165 | 267077_at | AT2G40970 | myb family transcription factor | 2.10 |
| 166 | 257357_at | AT2G41050 | PQ-loop repeat family protein / transmembrane family protein | 1.01 |
| 167 | 267083_at | AT2G41100 | TCH3 (TOUCH 3) | 1.72 |
| 168 | 266391_at | AT2G41290 | strictosidine synthase family protein | -1.16 |
| 169 | 265261_at | AT2G42990 | GDSL-motif lipase/hydrolase family protein | -2.21 |
| 170 | 260551_at | AT2G43510 | ATTI1 (ARABIDOPSIS THALIANA TRYPSIN INHIBITOR PROTEIN 1) | -0.90 |
| 171 | 260557_at | AT2G43610 | glycoside hydrolase family 19 protein | -2.25 |
| 172 | 266123_at | AT2G45180 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | -2.08 |
| 173 | 267509_at | AT2G45660 | AGL20 (AGAMOUS-LIKE 20); transcription factor | 1.52 |
| 174 | 266326_at | AT2G46650 | B5 #1 (cytochrome b5 family protein #1); heme binding / transition metal ion binding | -1.65 |
| 175 | 266719_at | AT2G46830 | CCA1 (CIRCADIAN CLOCK ASSOCIATED 1); transcription factor | 0.83 |
| 176 | 266760_at | AT2G46870 | NGA1 (NGATHA1); transcription factor | -1.19 |
| 177 | 266502_at | AT2G47720 | similar to AtATG18a (Arabidopsis thaliana homolog of yeast autophagy 18 (ATG18) a) [Arabidopsis thaliana] (TAIR:AT3G62770.2); contains domain PTHR11227 (PTHR11227); contains domain PTHR11227:SF1 (PTHR11227:SF1) | 0.86 |
| 178 | 259269_at | AT3G01270 | pectate lyase family protein | -1.07 |
| 179 | 259161_at | AT3G01500 | CA1 (CARBONIC ANHYDRASE 1); carbonate dehydratase/ zinc ion binding | -3.77 |
| 180 | 258494_at | AT3G02450 | cell division protein ftsH, putative | -0.76 |
| 181 | 258487_at | AT3G02550 | LBD41 (LOB DOMAIN-CONTAINING PROTEIN 41) | 1.23 |
| 182 | 258483_at | AT3G02570 | MEE31 (maternal effect embryo arrest 31); mannose-6-phosphate isomerase | -0.77 |
| 183 | 258866_at | AT3G03180 | Got1-like family protein | 0.82 |
| 184 | 258879_at | AT3G03270 | universal stress protein (USP) family protein / early nodulin ENOD18 family protein | 0.99 |
| 185 | 257529_at | AT3G03290 | universal stress protein (USP) family protein | 6.79 |
| 186 | 258863_at | AT3G03300 | DCL2 (DICER-LIKE 2); ATP-dependent helicase/ ribonuclease III | 1.16 |
| 187 | 259057_at | AT3G03310 | lecithin:cholesterol acyltransferase family protein / LACT family protein | 1.33 |
| 188 | 259051_at | AT3G03330 | short-chain dehydrogenase/reductase (SDR) family protein | 0.92 |
| 189 | 259055_at | AT3G03340 | UNE6 (unfertilized embryo sac 6) | 0.91 |
| 190 | 259050_at | AT3G03360 | F-box family protein | 1.00 |
| 191 | 259049_at | AT3G03370 | similar to dehydration-responsive protein-related [Arabidopsis thaliana] (TAIR:AT5G06050.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO69144.1); contains InterPro domain Protein of unknown function DUF248, methyltransferase putative (InterPro:IPR004159) | 1.23 |
| 192 | 259047_at | AT3G03380 | DEGP7 (DEGP PROTEASE 7); serine-type peptidase/ trypsin | 0.82 |
| 193 | 259048_at | AT3G03380 | DEGP7 (DEGP PROTEASE 7); serine-type peptidase/ trypsin | 1.31 |
| 194 | 259219_at | AT3G03560 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G23490.1); similar to Os03g0707300 [Oryza sativa (japonica cultivar-group)] (GB:NP_001051031.1); similar to expressed protein [Oryza sativa (japonica cultivar-group)] (GB:ABF98471.1) | 1.10 |
| 195 | 259168_at | AT3G03570 | similar to signal transducer [Arabidopsis thaliana] (TAIR:AT4G40050.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO48140.1); contains InterPro domain Uncharacterised conserved protein UCP013022 (InterPro:IPR016607) | 1.32 |
| 196 | 259171_at | AT3G03590 | SWIB complex BAF60b domain-containing protein | 1.34 |
| 197 | 259196_at | AT3G03600 | RPS2 (RIBOSOMAL PROTEIN S2); structural constituent of ribosome | 0.87 |
| 198 | 259198_at | AT3G03610 | phagocytosis and cell motility protein ELMO1-related | 1.02 |
| 199 | 259173_at | AT3G03640 | GLUC (Beta-glucosidase homolog); hydrolase, hydrolyzing O-glycosyl compounds | 0.83 |
| 200 | 259222_at | AT3G03680 | C2 domain-containing protein | 0.97 |
| 201 | 259344_at | AT3G03710 | RIF10 (RESISTANT TO INHIBITION WITH FSM 10); 3'-5'-exoribonuclease/ RNA binding / nucleic acid binding | 0.90 |
| 202 | 259341_at | AT3G03740 | ATBPM4 (BTB-POZ AND MATH DOMAIN 4); protein binding | 1.00 |
| 203 | 259348_at | AT3G03770 | leucine-rich repeat transmembrane protein kinase, putative | 1.53 |
| 204 | 259334_at | AT3G03790 | ankyrin repeat family protein / regulator of chromosome condensation (RCC1) family protein | 1.52 |
| 205 | 259333_at | AT3G03810 | EDA30 (embryo sac development arrest 30) | 0.92 |
| 206 | 259349_at | AT3G03860 | ATAPRL5 (APR-LIKE 5) | 1.19 |
| 207 | 257518_at | AT3G03880 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G55340.1); similar to unknown [Populus trichocarpa] (GB:ABK95741.1); contains InterPro domain Protein of unknown function DUF1639 (InterPro:IPR012438) | 1.19 |
| 208 | 259342_at | AT3G03890 | FMN binding | 1.03 |
| 209 | 259347_at | AT3G03920 | Gar1 RNA-binding region family protein | 0.90 |
| 210 | 258812_at | AT3G03950 | ECT1; protein binding | 1.21 |
| 211 | 258816_at | AT3G03960 | chaperonin, putative | 1.02 |
| 212 | 258810_at | AT3G03970 | binding | 1.05 |
| 213 | 258814_at | AT3G03980 | short-chain dehydrogenase/reductase (SDR) family protein | 0.88 |
| 214 | 258811_at | AT3G03990 | esterase/lipase/thioesterase family protein | 1.36 |
| 215 | 258567_at | AT3G04080 | ATAPY1 (APYRASE 1); calmodulin binding | 0.80 |
| 216 | 258584_at | AT3G04090 | SIP1;1 (SMALL AND BASIC INTRINSIC PROTEIN 1A) | 1.09 |
| 217 | 258588_s_at | AT3G04120 | GAPC (GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE C SUBUNIT); glyceraldehyde-3-phosphate dehydrogenase | 0.91 |
| 218 | 258592_at | AT3G04130 | pentatricopeptide (PPR) repeat-containing protein | 1.10 |
| 219 | 258581_at | AT3G04160 | similar to hypothetical protein [Vitis vinifera] (GB:CAN82741.1) | 1.13 |
| 220 | 258575_at | AT3G04240 | SEC (SECRET AGENT); transferase, transferring glycosyl groups | 1.13 |
| 221 | 258587_at | AT3G04310 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G33250.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO38748.1) | 1.09 |
| 222 | 258585_at | AT3G04340 | EMB2458 (EMBRYO DEFECTIVE 2458); ATPase | 1.05 |
| 223 | 258565_at | AT3G04350 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G18490.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G43950.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G04090.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN77840.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN61465.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO40557.1); contains InterPro domain Protein of unknown function DUF946, plant (InterPro:IPR009291) | 0.99 |
| 224 | 258626_at | AT3G04450 | transcription factor | 0.81 |
| 225 | 258568_at | AT3G04470 | similar to ankyrin repeat family protein [Arabidopsis thaliana] (TAIR:AT1G04780.1); similar to hypothetical protein OsI_009190 [Oryza sativa (indica cultivar-group)] (GB:EAY87957.1); similar to Os02g0810100 [Oryza sativa (japonica cultivar-group)] (GB:NP_001048467.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO44660.1); contains InterPro domain Ankyrin (InterPro:IPR002110) | 0.94 |
| 226 | 258593_at | AT3G04480 | endoribonuclease | 0.87 |
| 227 | 258595_at | AT3G04500 | RNA recognition motif (RRM)-containing protein | 1.04 |
| 228 | 258599_at | AT3G04520 | THA2 (THREONINE ALDOLASE 2); threonine aldolase | 1.38 |
| 229 | 258801_at | AT3G04560 | similar to unnamed protein product [Vitis vinifera] (GB:CAO65029.1) | 1.01 |
| 230 | 258818_at | AT3G04580 | EIN4 (ETHYLENE INSENSITIVE 4); receptor | 1.16 |
| 231 | 258819_at | AT3G04590 | DNA-binding family protein | 1.46 |
| 232 | 258820_at | AT3G04600 | tRNA synthetase class I (W and Y) family protein | 0.99 |
| 233 | 258840_at | AT3G04620 | nucleic acid binding | 1.60 |
| 234 | 258796_at | AT3G04630 | WDL1 (WVD2-LIKE 1) | 0.91 |
| 235 | 258803_at | AT3G04670 | WRKY39 (WRKY DNA-binding protein 39); transcription factor | 1.23 |
| 236 | 258842_at | AT3G04680 | pre-mRNA cleavage complex family protein | 1.05 |
| 237 | 258794_at | AT3G04710 | ankyrin repeat family protein | 1.06 |
| 238 | 258797_at | AT3G04730 | IAA16 (indoleacetic acid-induced protein 16); transcription factor | 1.34 |
| 239 | 258844_at | AT3G04740 | SWP (STRUWWELPETER) | 0.93 |
| 240 | 258817_at | AT3G04750 | pentatricopeptide (PPR) repeat-containing protein | 1.07 |
| 241 | 258793_at | AT3G04780 | Encodes a protein with little sequence identity with any other protein of known structure or function. Part of this protein shows a 42% sequence identity with the C-terminal domain of the 32-kD human thioredoxin-like protein. | 1.20 |
| 242 | 259096_at | AT3G04840 | 40S ribosomal protein S3A (RPS3aA) | 0.85 |
| 243 | 259093_at | AT3G04860 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G28150.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN64151.1); contains InterPro domain Protein of unknown function DUF868, plant (InterPro:IPR008586) | 1.13 |
| 244 | 259092_at | AT3G04870 | ZDS (ZETA-CAROTENE DESATURASE); carotene 7,8-desaturase | 0.87 |
| 245 | 259100_at | AT3G04880 | DRT102 (DNA-DAMAGE-REPAIR/TOLERATION 2) | 0.91 |
| 246 | 259094_at | AT3G04940 | ATCYSD1 (Arabidopsis thaliana cysteine synthase D1); cysteine synthase | 0.89 |
| 247 | 259088_at | AT3G04970 | zinc finger (DHHC type) family protein | 1.11 |
| 248 | 259087_at | AT3G04980 | DNAJ heat shock N-terminal domain-containing protein | 1.17 |
| 249 | 259085_at | AT3G05000 | transport protein particle (TRAPP) component Bet3 family protein | 1.08 |
| 250 | 259099_at | AT3G05010 | transmembrane protein, putative | 1.33 |
| 251 | 259081_at | AT3G05030 | NHX2 (sodium proton exchanger 2); sodium:hydrogen antiporter | 1.34 |
| 252 | 259309_at | AT3G05050 | protein kinase family protein | 1.19 |
| 253 | 259297_at | AT3G05360 | disease resistance family protein / LRR family protein | 1.14 |
| 254 | 259133_at | AT3G05400 | sugar transporter, putative | -1.31 |
| 255 | 258894_at | AT3G05650 | disease resistance family protein | 0.83 |
| 256 | 258513_at | AT3G06630 | protein kinase family protein | -3.20 |
| 257 | 259244_at | AT3G07650 | COL9 (CONSTANS-LIKE 9); transcription factor/ zinc ion binding | 0.83 |
| 258 | 258639_at | AT3G07820 | polygalacturonase 3 (PGA3) / pectinase | -4.54 |
| 259 | 258942_at | AT3G09960 | calcineurin-like phosphoesterase family protein | -1.63 |
| 260 | 258930_at | AT3G10040 | transcription factor | 1.56 |
| 261 | 258920_at | AT3G10520 | AHB2 (NON-SYMBIOTIC HAEMOGLOBIN 2) | -0.96 |
| 262 | 258962_at | AT3G10570 | CYP77A6 (cytochrome P450, family 77, subfamily A, polypeptide 6); oxygen binding | -2.43 |
| 263 | 259293_at | AT3G11580 | DNA-binding protein, putative | 1.08 |
| 264 | 256237_at | AT3G12610 | DRT100 (DNA-DAMAGE REPAIR/TOLERATION 100); protein binding | -0.96 |
| 265 | 257706_at | AT3G12685 | similar to catalytic [Arabidopsis thaliana] (TAIR:AT1G24350.1); similar to unknown [Populus trichocarpa] (GB:ABK94924.1); contains InterPro domain Acid phosphatase/vanadium-dependent haloperoxidase related (InterPro:IPR003832) | -0.91 |
| 266 | 257654_at | AT3G13310 | DNAJ heat shock N-terminal domain-containing protein | 0.82 |
| 267 | 256965_at | AT3G13450 | DIN4 (DARK INDUCIBLE 4); 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) | -0.92 |
| 268 | 256960_at | AT3G13510 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G55360.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G56530.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO22094.1); similar to unknown [Populus trichocarpa] (GB:ABK93966.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN72570.1); contains InterPro domain Protein of unknown function DUF239, plant (InterPro:IPR004314) | 0.81 |
| 269 | 258209_at | AT3G14060 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G54120.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO45609.1) | -1.51 |
| 270 | 257008_at | AT3G14210 | ESM1 (EPITHIOSPECIFIER MODIFIER 1); carboxylesterase | -3.67 |
| 271 | 257213_at | AT3G15020 | malate dehydrogenase (NAD), mitochondrial, putative | -1.00 |
| 272 | 257054_at | AT3G15353 | MT3 (METALLOTHIONEIN 3) | 0.80 |
| 273 | 258270_at | AT3G15650 | phospholipase/carboxylesterase family protein | 1.08 |
| 274 | 257798_at | AT3G15950 | (TSA1-LIKE); protein binding | -1.67 |
| 275 | 258338_at | AT3G16150 | L-asparaginase, putative / L-asparagine amidohydrolase, putative | 1.15 |
| 276 | 258349_at | AT3G17609 | HYH (HY5-HOMOLOG); DNA binding / transcription factor | 1.18 |
| 277 | 257026_at | AT3G19200 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G34420.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN61148.1) | -0.79 |
| 278 | 257964_at | AT3G19850 | phototropic-responsive NPH3 family protein | 0.82 |
| 279 | 257679_at | AT3G20470 | encodes a glycine-rich protein that is expressed more abundantly in immature seed pods than in stems and leaves. Expression is not detected in roots or flowers. | -1.39 |
| 280 | 256976_at | AT3G21020 | transposable element gene | -1.20 |
| 281 | 258181_at | AT3G21670 | nitrate transporter (NTP3) | -1.58 |
| 282 | 258452_at | AT3G22370 | AOX1A (alternative oxidase 1A); alternative oxidase | 0.94 |
| 283 | 256926_at | AT3G22540 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G14819.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO71190.1); contains InterPro domain Protein of unknown function DUF1677, plant (InterPro:IPR012876) | 0.95 |
| 284 | 258321_at | AT3G22840 | ELIP1 (EARLY LIGHT-INDUCABLE PROTEIN); chlorophyll binding | 2.28 |
| 285 | 257772_at | AT3G23080 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G14500.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO71306.1); contains InterPro domain Lipid-binding START (InterPro:IPR002913) | -1.21 |
| 286 | 258095_at | AT3G23610 | dual specificity protein phosphatase (DsPTP1) | -0.91 |
| 287 | 257173_at | AT3G23810 | SAHH2 (S-ADENOSYL-L-HOMOCYSTEINE (SAH) HYDROLASE 2); adenosylhomocysteinase | -1.16 |
| 288 | 257247_at | AT3G24140 | FMA (FAMA); DNA binding / transcription activator/ transcription factor | -1.34 |
| 289 | 258133_at | AT3G24500 | ATMBF1C/MBF1C (MULTIPROTEIN BRIDGING FACTOR 1C); DNA binding / transcription coactivator/ transcription factor | 1.06 |
| 290 | 257101_at | AT3G25020 | disease resistance family protein | 0.83 |
| 291 | 256759_at | AT3G25640 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G23100.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO42071.1); contains InterPro domain Protein of unknown function DUF617, plant (InterPro:IPR006460) | 1.69 |
| 292 | 257623_at | AT3G26210 | CYP71B23 (cytochrome P450, family 71, subfamily B, polypeptide 23); oxygen binding | 1.02 |
| 293 | 256880_at | AT3G26450 | major latex protein-related / MLP-related | -0.91 |
| 294 | 257793_at | AT3G26960 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G41050.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO49052.1) | -1.31 |
| 295 | 257153_at | AT3G27220 | kelch repeat-containing protein | 1.54 |
| 296 | 256848_at | AT3G27960 | kinesin light chain-related | -0.83 |
| 297 | 256577_at | AT3G28220 | meprin and TRAF homology domain-containing protein / MATH domain-containing protein | -1.59 |
| 298 | 256987_at | AT3G28560 | similar to AAA-type ATPase family protein [Arabidopsis thaliana] (TAIR:AT3G28510.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN79616.1); contains domain PTHR23070 (PTHR23070); contains domain PTHR23070:SF1 (PTHR23070:SF1) | 1.73 |
| 299 | 258003_at | AT3G29030 | ATEXPA5 (ARABIDOPSIS THALIANA EXPANSIN A5) | -1.25 |
| 300 | 256940_at | AT3G30720 | unknown protein | -5.29 |
| 301 | 252618_at | AT3G45140 | LOX2 (LIPOXYGENASE 2) | -3.65 |
| 302 | 252483_at | AT3G46600 | scarecrow transcription factor family protein | 0.93 |
| 303 | 252441_at | AT3G46780 | PTAC16 (PLASTID TRANSCRIPTIONALLY ACTIVE18); binding / catalytic | -0.87 |
| 304 | 252415_at | AT3G47340 | ASN1 (DARK INDUCIBLE 6) | -2.85 |
| 305 | 252414_at | AT3G47420 | glycerol-3-phosphate transporter, putative / glycerol 3-phosphate permease, putative | 0.81 |
| 306 | 252373_at | AT3G48090 | EDS1 (ENHANCED DISEASE SUSCEPTIBILITY 1); signal transducer/ triacylglycerol lipase | 1.01 |
| 307 | 252353_at | AT3G48200 | similar to hypothetical protein OsI_020499 [Oryza sativa (indica cultivar-group)] (GB:EAY99266.1); similar to hypothetical protein OsJ_018975 [Oryza sativa (japonica cultivar-group)] (GB:EAZ35492.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO64300.1) | -1.30 |
| 308 | 252363_at | AT3G48460 | GDSL-motif lipase/hydrolase family protein | -1.65 |
| 309 | 252168_at | AT3G50440 | hydrolase | -1.52 |
| 310 | 252181_at | AT3G50685 | similar to unnamed protein product [Vitis vinifera] (GB:CAO22868.1) | -0.77 |
| 311 | 252130_at | AT3G50820 | PSBO-2/PSBO2 (PHOTOSYSTEM II SUBUNIT O-2); oxygen evolving/ poly(U) binding | -1.35 |
| 312 | 252131_at | AT3G50930 | AAA-type ATPase family protein | 1.19 |
| 313 | 252060_at | AT3G52430 | PAD4 (PHYTOALEXIN DEFICIENT 4); triacylglycerol lipase | 0.90 |
| 314 | 252010_at | AT3G52740 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G44450.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO70571.1) | 2.49 |
| 315 | 251962_at | AT3G53420 | PIP2A (PLASMA MEMBRANE INTRINSIC PROTEIN 2A); water channel | -0.80 |
| 316 | 251940_at | AT3G53450 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G37210.1); similar to Conserved hypothetical protein 730 [Medicago truncatula] (GB:ABD32794.1); contains InterPro domain Conserved hypothetical protein CHP00730 (InterPro:IPR005269) | 1.18 |
| 317 | 251858_at | AT3G54820 | PIP2;5/PIP2D (plasma membrane intrinsic protein 2;5); water channel | 0.81 |
| 318 | 251839_at | AT3G54950 | PLA IIIA/PLP7 (Patatin-like protein 7) | -0.80 |
| 319 | 251827_at | AT3G55120 | A11/CFI/TT5 (TRANSPARENT TESTA 5); chalcone isomerase | 0.93 |
| 320 | 251759_at | AT3G55630 | ATDFD (A. THALIANA DHFS-FPGS HOMOLOG D); tetrahydrofolylpolyglutamate synthase | -1.47 |
| 321 | 251705_at | AT3G56400 | WRKY70 (WRKY DNA-binding protein 70); transcription factor | 1.04 |
| 322 | 251598_at | AT3G57600 | AP2 domain-containing transcription factor, putative | -2.01 |
| 323 | 251590_at | AT3G57690 | AGP23/ATAGP23 (ARABINOGALACTAN-PROTEIN 23) | -1.17 |
| 324 | 251617_at | AT3G58000 | VQ motif-containing protein | -1.58 |
| 325 | 251575_at | AT3G58120 | bZIP transcription factor family protein | -0.90 |
| 326 | 251374_at | AT3G60390 | HAT3 (homeobox-leucine zipper protein 3); transcription factor | 0.85 |
| 327 | 251363_at | AT3G61250 | AtMYB17 (myb domain protein 17); DNA binding / transcription factor | 0.97 |
| 328 | 251223_at | AT3G62610 | AtMYB11 (myb domain protein 11); DNA binding / transcription factor | 0.97 |
| 329 | 251181_at | AT3G62820 | invertase/pectin methylesterase inhibitor family protein | -1.01 |
| 330 | 251203_at | AT3G63070 | PWWP domain-containing protein | -4.47 |
| 331 | 251157_at | AT3G63140 | mRNA-binding protein, putative | -0.95 |
| 332 | 251174_at | AT3G63200 | PLA IIIB/PLP9 (Patatin-like protein 9); nutrient reservoir | 0.83 |
| 333 | 251178_at | AT3G63440 | ATCKX6/ATCKX7/CKX6 (CYTOKININ OXIDASE/DEHYDROGENASE 6); cytokinin dehydrogenase | 0.84 |
| 334 | 255675_at | AT4G00480 | ATMYC1 (Arabidopsis thaliana myc-related transcription factor 1); DNA binding / transcription factor | -0.77 |
| 335 | 255576_at | AT4G01440 | nodulin MtN21 family protein | -1.32 |
| 336 | 255527_at | AT4G02360 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G02813.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO40037.1); contains InterPro domain Protein of unknown function DUF538 (InterPro:IPR007493) | -1.19 |
| 337 | 255448_at | AT4G02810 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G03170.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN75990.1) | 1.23 |
| 338 | 255403_at | AT4G03400 | DFL2 (DWARF IN LIGHT 2) | -1.46 |
| 339 | 255365_at | AT4G04040 | MEE51 (maternal effect embryo arrest 51); diphosphate-fructose-6-phosphate 1-phosphotransferase | -1.08 |
| 340 | 255302_at | AT4G04830 | methionine sulfoxide reductase domain-containing protein / SeIR domain-containing protein | -1.88 |
| 341 | 255310_at | AT4G04955 | ATALN (ARABIDOPSIS ALLANTOINASE); allantoinase/ hydrolase | 1.34 |
| 342 | 255080_at | AT4G09030 | AGP10 (Arabinogalactan protein 10) | -0.79 |
| 343 | 255807_at | AT4G10270 | wound-responsive family protein | 1.26 |
| 344 | 254907_at | AT4G11190 | disease resistance-responsive family protein / dirigent family protein | -2.23 |
| 345 | 254786_at | AT4G12890 | gamma interferon responsive lysosomal thiol reductase family protein / GILT family protein | 1.33 |
| 346 | 254777_at | AT4G12960 | gamma interferon responsive lysosomal thiol reductase family protein / GILT family protein | -2.63 |
| 347 | 254754_at | AT4G13210 | pectate lyase family protein | -1.10 |
| 348 | 245329_at | AT4G14365 | zinc finger (C3HC4-type RING finger) family protein / ankyrin repeat family protein | 1.61 |
| 349 | 245275_at | AT4G15210 | ATBETA-AMY (BETA-AMYLASE); beta-amylase | -0.80 |
| 350 | 245253_at | AT4G15440 | HPL1 (HYDROPEROXIDE LYASE 1); heme binding / iron ion binding / monooxygenase | -1.49 |
| 351 | 245515_at | AT4G15810 | chloroplast outer membrane protein, putative | 0.93 |
| 352 | 245488_at | AT4G16270 | peroxidase 40 (PER40) (P40) | -1.84 |
| 353 | 245349_at | AT4G16690 | esterase/lipase/thioesterase family protein | 0.98 |
| 354 | 245422_at | AT4G17470 | palmitoyl protein thioesterase family protein | -2.77 |
| 355 | 245401_at | AT4G17670 | senescence-associated protein-related | 1.49 |
| 356 | 254663_at | AT4G18290 | KAT2 (K+ ATPase 2); cyclic nucleotide binding / inward rectifier potassium channel | -2.04 |
| 357 | 254665_at | AT4G18340 | glycosyl hydrolase family 17 protein | -1.06 |
| 358 | 254609_at | AT4G18970 | GDSL-motif lipase/hydrolase family protein | -1.09 |
| 359 | 254563_at | AT4G19120 | ERD3 (EARLY-RESPONSIVE TO DEHYDRATION 3) | -0.78 |
| 360 | 254496_at | AT4G20070 | ATAAH (ARABIDOPSIS THALIANA ALLANTOATE AMIDOHYDROLASE); allantoate deiminase/ metallopeptidase | 0.88 |
| 361 | 254492_at | AT4G20260 | DREPP plasma membrane polypeptide family protein | -0.86 |
| 362 | 254416_at | AT4G21380 | ARK3 (Arabidopsis Receptor Kinase 3); kinase | 0.82 |
| 363 | 254386_at | AT4G21960 | PRXR1 (peroxidase 42); peroxidase | 0.83 |
| 364 | 254327_at | AT4G22490 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | 1.63 |
| 365 | 254329_at | AT4G22540 | oxysterol-binding family protein | 0.87 |
| 366 | 254232_at | AT4G23600 | CORI3 (CORONATINE INDUCED 1, JASMONIC ACID RESPONSIVE 2); transaminase | -2.26 |
| 367 | 254234_at | AT4G23680 | major latex protein-related / MLP-related | -3.14 |
| 368 | 254200_at | AT4G24110 | similar to unnamed protein product [Vitis vinifera] (GB:CAO62924.1) | 1.24 |
| 369 | 254122_at | AT4G24510 | CER2 (ECERIFERUM 2); transferase | -1.04 |
| 370 | 254130_at | AT4G24540 | AGL24 (AGAMOUS-LIKE 24); protein binding / protein heterodimerization/ protein homodimerization/ transcription factor | 0.97 |
| 371 | 254022_at | AT4G25750 | ABC transporter family protein | 0.91 |
| 372 | 254001_at | AT4G26260 | MIOX4 (MYO-INOSITOL OXYGENASE 4) | -2.10 |
| 373 | 253907_at | AT4G27250 | dihydroflavonol 4-reductase family / dihydrokaempferol 4-reductase family | -0.98 |
| 374 | 253908_at | AT4G27260 | GH3.5/WES1; indole-3-acetic acid amido synthetase | 0.81 |
| 375 | 253915_at | AT4G27280 | calcium-binding EF hand family protein | 1.01 |
| 376 | 253874_at | AT4G27450 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G15450.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO39242.1); contains domain G3DSA:3.60.20.10 (G3DSA:3.60.20.10); contains domain SSF56235 (SSF56235) | 0.93 |
| 377 | 253880_at | AT4G27590 | copper-binding protein-related | -1.71 |
| 378 | 253886_at | AT4G27710 | CYP709B3 (cytochrome P450, family 709, subfamily B, polypeptide 3); oxygen binding | -0.96 |
| 379 | 253702_at | AT4G29900 | ACA10 (autoinhibited Ca2+ -ATPase 10); calcium-transporting ATPase/ calmodulin binding | 0.80 |
| 380 | 253614_at | AT4G30350 | heat shock protein-related | -1.31 |
| 381 | 253534_at | AT4G31500 | CYP83B1 (CYTOCHROME P450 MONOOXYGENASE 83B1); oxygen binding | -1.32 |
| 382 | 253514_at | AT4G31805 | WRKY family transcription factor | -1.11 |
| 383 | 253493_at | AT4G31820 | ENP (ENHANCER OF PINOID); signal transducer | 0.95 |
| 384 | 253483_at | AT4G31910 | transferase family protein | 1.26 |
| 385 | 253500_at | AT4G31920 | ARR10 (ARABIDOPSIS RESPONSE REGULATOR 10); transcription factor/ two-component response regulator | 0.94 |
| 386 | 253451_at | AT4G32730 | PC-MYB1 (myb domain protein 3R1); DNA binding / transcription coactivator/ transcription factor | 0.93 |
| 387 | 253358_at | AT4G32940 | GAMMA-VPE (Vacuolar processing enzyme gamma); cysteine-type endopeptidase | 1.01 |
| 388 | 253411_at | AT4G32980 | ATH1 (ARABIDOPSIS THALIANA HOMEOBOX GENE 1); transcription factor | -0.83 |
| 389 | 253298_at | AT4G33560 | unknown protein | 1.64 |
| 390 | 253203_at | AT4G34710 | ADC2 (ARGININE DECARBOXYLASE 2) | -1.08 |
| 391 | 253182_at | AT4G35190 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G37210.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO47480.1); contains InterPro domain Conserved hypothetical protein CHP00730 (InterPro:IPR005269) | 1.61 |
| 392 | 253161_at | AT4G35770 | SEN1 (DARK INDUCIBLE 1) | -2.69 |
| 393 | 253108_at | AT4G35900 | FD; DNA binding / transcription factor | 1.83 |
| 394 | 246247_at | AT4G36640 | SEC14 cytosolic factor family protein / phosphoglyceride transfer family protein | -0.78 |
| 395 | 246217_at | AT4G36920 | AP2 (APETALA 2); transcription factor | -0.78 |
| 396 | 253043_at | AT4G37540 | LBD39 (LOB DOMAIN-CONTAINING PROTEIN 39) | -1.38 |
| 397 | 253012_at | AT4G37900 | glycine-rich protein | 0.84 |
| 398 | 252971_at | AT4G38770 | PRP4 (PROLINE-RICH PROTEIN 4) | -6.54 |
| 399 | 252870_at | AT4G39940 | AKN2 (APS-KINASE 2); ATP binding / kinase/ transferase, transferring phosphorus-containing groups | -1.24 |
| 400 | 251109_at | AT5G01600 | ATFER1 (FERRETIN 1); ferric iron binding | -1.16 |
| 401 | 251065_at | AT5G01870 | lipid transfer protein, putative | -1.55 |
| 402 | 251020_at | AT5G02270 | ATNAP9 (Non-intrinsic ABC protein 9) | 0.89 |
| 403 | 251041_at | AT5G02310 | PRT6 (PROTEOLYSIS 6); ubiquitin-protein ligase | 1.26 |
| 404 | 250723_at | AT5G06300 | carboxy-lyase | 0.81 |
| 405 | 250669_at | AT5G06870 | PGIP2 (POLYGALACTURONASE INHIBITING PROTEIN 2); protein binding | -1.39 |
| 406 | 250620_at | AT5G07190 | ATS3 (ARABIDOPSIS THALIANA SEED GENE 3) | 1.06 |
| 407 | 250580_at | AT5G07440 | GDH2 (GLUTAMATE DEHYDROGENASE 2); oxidoreductase | -0.77 |
| 408 | 250570_at | AT5G08170 | ATAIH/EMB1873 (AGMATINE IMINOHYDROLASE); agmatine deiminase | 0.82 |
| 409 | 250517_at | AT5G08260 | SCPL35 (serine carboxypeptidase-like 35); serine carboxypeptidase | -2.06 |
| 410 | 245891_at | AT5G09220 | AAP2 (AMINO ACID PERMEASE 2); amino acid transmembrane transporter | 1.08 |
| 411 | 250435_at | AT5G10380 | zinc finger (C3HC4-type RING finger) family protein | 0.97 |
| 412 | 250445_at | AT5G10760 | aspartyl protease family protein | 1.76 |
| 413 | 245901_at | AT5G11060 | KNAT4 (KNOTTED1-LIKE HOMEOBOX GENE 4); transcription factor | -1.10 |
| 414 | 245906_at | AT5G11070 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G35090.1) | -1.00 |
| 415 | 245905_at | AT5G11090 | serine-rich protein-related | 1.01 |
| 416 | 250420_at | AT5G11260 | HY5 (ELONGATED HYPOCOTYL 5); DNA binding / transcription factor | 0.91 |
| 417 | 250255_at | AT5G13730 | SIG4 (SIGMA FACTOR 4); DNA binding / DNA-directed RNA polymerase/ transcription factor | -0.98 |
| 418 | 250207_at | AT5G13930 | ATCHS/CHS/TT4 (CHALCONE SYNTHASE); naringenin-chalcone synthase | 0.95 |
| 419 | 246583_at | AT5G14720 | protein kinase family protein | 0.92 |
| 420 | 250152_at | AT5G15120 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G39890.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO14912.1); contains InterPro domain Protein of unknown function DUF1637 (InterPro:IPR012864) | 0.80 |
| 421 | 250167_at | AT5G15310 | AtMIXTA/AtMYB16 (myb domain protein 16); DNA binding / transcription factor | -1.13 |
| 422 | 246487_at | AT5G16030 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G02500.1); similar to hypothetical protein [Cleome spinosa] (GB:ABD96956.1) | -1.08 |
| 423 | 246468_at | AT5G17050 | UDP-glucoronosyl/UDP-glucosyl transferase family protein | 0.87 |
| 424 | 246146_at | AT5G20050 | protein kinase family protein | -0.83 |
| 425 | 246071_at | AT5G20150 | SPX (SYG1/Pho81/XPR1) domain-containing protein | 0.90 |
| 426 | 246114_at | AT5G20250 | DIN10 (DARK INDUCIBLE 10); hydrolase, hydrolyzing O-glycosyl compounds | -0.83 |
| 427 | 246004_at | AT5G20630 | GLP3 (GERMIN-LIKE PROTEIN 3); manganese ion binding / metal ion binding / nutrient reservoir | -1.26 |
| 428 | 249939_at | AT5G22430 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G27385.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN68427.1) | -3.43 |
| 429 | 249817_at | AT5G23820 | MD-2-related lipid recognition domain-containing protein / ML domain-containing protein | -1.53 |
| 430 | 249761_at | AT5G23970 | transferase family protein | -2.45 |
| 431 | 246966_at | AT5G24850 | CRY3 (CRYPTOCHROME 3); DNA binding / DNA photolyase/ FMN binding | 0.85 |
| 432 | 246967_at | AT5G24860 | FPF1 (FLOWERING PROMOTING FACTOR 1) | 1.57 |
| 433 | 246978_at | AT5G24910 | CYP714A1 (cytochrome P450, family 714, subfamily A, polypeptide 1); oxygen binding | -1.06 |
| 434 | 246888_at | AT5G26270 | unknown protein | -3.63 |
| 435 | 246756_at | AT5G27930 | protein phosphatase 2C, putative / PP2C, putative | 1.06 |
| 436 | 246701_at | AT5G28020 | ATCYSD2 (Arabidopsis thaliana cysteine synthase D2); cysteine synthase | -1.07 |
| 437 | 246103_at | AT5G28640 | AN3 (ANGUSITFOLIA3) | 0.91 |
| 438 | 246168_at | AT5G32460 | pseudogene, transcriptional factor B3 family, low similarity to reproductive meristem protein 1 from (Brassica oleracea var. botrytis) GI:3170424, (Arabidopsis thaliana) GI:13604227; contains Pfam profile PF02362: B3 DNA binding domain; blastp match of 30% identity and 5.7e-22 P-value to GP|3170424|gb|AAC18082.1||AF051772 reproductive meristem gene 1 {Brassica oleracea var. botrytis} | 2.88 |
| 439 | 246687_at | AT5G33370 | GDSL-motif lipase/hydrolase family protein | -5.17 |
| 440 | 249716_at | AT5G35770 | SAP (STERILE APETALA); transcription factor | 1.00 |
| 441 | 249645_at | AT5G36910 | THI2.2 (THIONIN 2.2); toxin receptor binding | -1.53 |
| 442 | 249384_at | AT5G39890 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G15120.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO14912.1); contains InterPro domain Protein of unknown function DUF1637 (InterPro:IPR012864) | 1.23 |
| 443 | 249375_at | AT5G40730 | AGP24 (ARABINOGALACTAN PROTEIN 24) | -0.83 |
| 444 | 249327_at | AT5G40890 | ATCLC-A (CHLORIDE CHANNEL A); anion channel/ voltage-gated chloride channel | -1.13 |
| 445 | 249237_at | AT5G42050 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G27090.1); similar to unknown [Populus trichocarpa] (GB:ABK95892.1); contains InterPro domain Kelch related (InterPro:IPR013089); contains InterPro domain Development and cell death (InterPro:IPR013989) | 0.84 |
| 446 | 249245_at | AT5G42280 | DC1 domain-containing protein | -0.79 |
| 447 | 249073_at | AT5G44020 | acid phosphatase class B family protein | -2.04 |
| 448 | 248848_at | AT5G46520 | ATP binding / nucleoside-triphosphatase/ nucleotide binding / protein binding / transmembrane receptor | 1.17 |
| 449 | 248855_at | AT5G46590 | ANAC096 (Arabidopsis NAC domain containing protein 96); transcription factor | -0.80 |
| 450 | 248862_at | AT5G46730 | glycine-rich protein | -1.64 |
| 451 | 248819_at | AT5G47050 | ATP binding / protein binding / shikimate kinase/ zinc ion binding | -2.10 |
| 452 | 248820_at | AT5G47060 | senescence-associated protein-related | 0.85 |
| 453 | 248794_at | AT5G47220 | ATERF-2/ATERF2/ERF2 (ETHYLENE RESPONSE FACTOR 2); DNA binding / transcription activator/ transcription factor | 1.47 |
| 454 | 248793_at | AT5G47240 | ATNUDT8 (Arabidopsis thaliana Nudix hydrolase homolog 8); hydrolase | 0.96 |
| 455 | 248763_at | AT5G47550 | cysteine protease inhibitor, putative / cystatin, putative | -1.06 |
| 456 | 248719_at | AT5G47910 | RBOHD (RESPIRATORY BURST OXIDASE PROTEIN D) | 0.81 |
| 457 | 248683_at | AT5G48490 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | -1.10 |
| 458 | 248539_at | AT5G50130 | short-chain dehydrogenase/reductase (SDR) family protein | -0.95 |
| 459 | 248374_at | AT5G51870 | AGL71; transcription factor | 2.16 |
| 460 | 248333_at | AT5G52390 | photoassimilate-responsive protein, putative | -1.13 |
| 461 | 248327_at | AT5G52750 | heavy-metal-associated domain-containing protein | 1.40 |
| 462 | 248322_at | AT5G52760 | heavy-metal-associated domain-containing protein | 4.20 |
| 463 | 248246_at | AT5G53200 | TRY (TRIPTYCHON); DNA binding / transcription factor | -0.78 |
| 464 | 248160_at | AT5G54470 | zinc finger (B-box type) family protein | 1.73 |
| 465 | 248062_at | AT5G55450 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | 0.90 |
| 466 | 248073_at | AT5G55720 | pectate lyase family protein | -4.57 |
| 467 | 248011_at | AT5G56300 | GAMT2; S-adenosylmethionine-dependent methyltransferase/ gibberellin carboxyl-O-methyltransferase | 1.47 |
| 468 | 247925_at | AT5G57560 | TCH4 (TOUCH 4); hydrolase, acting on glycosyl bonds / xyloglucan:xyloglucosyl transferase | -0.82 |
| 469 | 247921_at | AT5G57660 | zinc finger (B-box type) family protein | -0.80 |
| 470 | 247880_at | AT5G57780 | transcription regulator | -1.04 |
| 471 | 247768_at | AT5G58900 | myb family transcription factor | 1.29 |
| 472 | 247718_at | AT5G59310 | LTP4 (LIPID TRANSFER PROTEIN 4); lipid binding | 1.04 |
| 473 | 247684_at | AT5G59670 | leucine-rich repeat protein kinase, putative | -1.13 |
| 474 | 247689_at | AT5G59770 | similar to PAS2 (PASTICCINO 2) [Arabidopsis thaliana] (TAIR:AT5G10480.1); similar to PAS2 (PASTICCINO 2) [Arabidopsis thaliana] (TAIR:AT5G10480.2); similar to unnamed protein product [Vitis vinifera] (GB:CAO21719.1); contains InterPro domain Protein-tyrosine phosphatase-like, PTPLA (InterPro:IPR007482) | 0.94 |
| 475 | 247553_at | AT5G60910 | AGL8 (AGAMOUS-LIKE 8); transcription factor | 0.88 |
| 476 | 247529_at | AT5G61520 | hexose transporter, putative | -0.81 |
| 477 | 247530_at | AT5G61540 | L-asparaginase, putative / L-asparagine amidohydrolase, putative | 2.31 |
| 478 | 247490_at | AT5G61850 | LFY (LEAFY); transcription factor | 0.96 |
| 479 | 247462_at | AT5G62080 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | 1.00 |
| 480 | 247486_at | AT5G62140 | similar to unknown [Populus trichocarpa] (GB:ABK94834.1) | -0.76 |
| 481 | 247469_at | AT5G62165 | AGL42 (AGAMOUS LIKE 42); transcription factor | 1.15 |
| 482 | 247463_at | AT5G62210 | embryo-specific protein-related | 0.99 |
| 483 | 247455_at | AT5G62470 | MYB96 (myb domain protein 96); DNA binding / transcription factor | 1.05 |
| 484 | 247447_at | AT5G62730 | proton-dependent oligopeptide transport (POT) family protein | -1.49 |
| 485 | 247337_at | AT5G63660 | LCR74/PDF2.5 (Low-molecular-weight cysteine-rich 74) | -1.41 |
| 486 | 247304_at | AT5G63850 | AAP4 (amino acid permease 4); amino acid transmembrane transporter | 0.99 |
| 487 | 247212_at | AT5G65040 | senescence-associated protein-related | -0.77 |
| 488 | 247170_at | AT5G65530 | protein kinase, putative | -0.80 |
| 489 | 247167_at | AT5G65850 | F-box family protein | -0.93 |
| 490 | 247107_at | AT5G66040 | STR16 (SULFURTRANSFERASE PROTEIN 16) | -1.16 |
| 491 | 247098_at | AT5G66470 | GTP binding / RNA binding | -0.80 |
| 492 | 247035_at | AT5G67110 | ALC (ALCATRAZ); DNA binding / transcription factor | 1.00 |
| 493 | 245024_at | ATCG00120 | Encodes the ATPase alpha subunit, which is a subunit of ATP synthase and part of the CF1 portion which catalyzes the conversion of ADP to ATP using the proton motive force. This complex is located in the thylakoid membrane of the chloroplast. | 0.85 |
| 494 | 245000_at | ATCG00210 | hypothetical protein | -1.94 |
| 495 | 245005_at | ATCG00330 | 30S chloroplast ribosomal protein S14 | -1.75 |
| 496 | 245007_at | ATCG00350 | Encodes psaA protein comprising the reaction center for photosystem I along with psaB protein; hydrophobic protein encoded by the chloroplast genome. | 0.87 |
| 497 | 245008_at | ATCG00360 | Encodes a protein required for photosystem I assembly and stability. In Chlamydomonas reinhardtii, this protein seems to act as a PSI specific chaperone facilitating the assembly of the complex by interacting with PsaA and PsaD. A loss of function mutation in tobacco leads to a loss of photosystem I. | -0.93 |
| 498 | 245015_at | ATCG00490 | large subunit of RUBISCO. | -0.75 |
| 499 | 244966_at | ATCG00600 | Cytochrome b6-f complex, subunit V. Disruption of homologous gene in Chlamydomonas results in disruption of cytochrome b6-f complex. | -1.04 |
| 500 | 244970_at | ATCG00660 | encodes a chloroplast ribosomal protein L20, a constituent of the large subunit of the ribosomal complex | -1.25 |
| 501 | 244977_at | ATCG00730 | A chloroplast gene encoding subunit IV of the cytochrome b6/f complex | -1.69 |
| 502 | 244994_at | ATCG01010 | Chloroplast encoded NADH dehydrogenase unit. | -3.34 |
| 503 | 244933_at | ATCG01070 | NADH dehydrogenase ND4L | -1.49 |
| 504 | 244934_at | ATCG01080 | NADH dehydrogenase ND6 | -1.50 |
| 505 | 244941_at | ATMG00010 | hypothetical protein | -1.01 |
| 506 | 244924_at | ATMG01040 | hypothetical protein | -1.06 |
| 507 | 257319_at | ATMG01100 | hypothetical protein | -1.00 |
| 508 | 257330_at | ATMG01290 | hypothetical protein | -1.43 |
| 509 | 257336_at | ATMG01410 | Identical to Uncharacterized mitochondrial protein AtMg01410 [Arabidopsis Thaliana] (GB:P92567;GB:Q1ZXV1); similar to hypothetical protein BrnapMp025 [Brassica napus] (GB:YP_717123.1); contains InterPro domain RNA-dependent RNA polymerase, mitoviral (InterPro:IPR008686) | -0.97 |