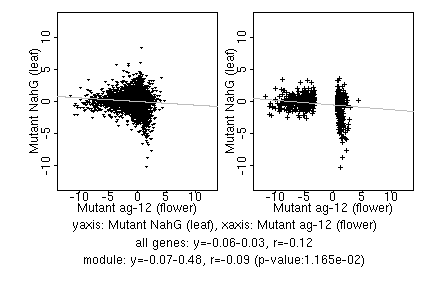
marker gene list
| Mutant ag-12(flower) |
| Mutant NahG(leaf) |
| probeID | AGICode | Annotation | gene expression |
|
|---|---|---|---|---|
| 1 | 261045_at | AT1G01310 | allergen V5/Tpx-1-related family protein | -5.78 |
| 2 | 261623_at | AT1G01980 | ATSEC1A; electron carrier | -5.31 |
| 3 | 259441_at | AT1G02300 | cathepsin B-like cysteine protease, putative | 1.26 |
| 4 | 260904_at | AT1G02450 | NIMIN-1/NIMIN1; protein binding | 1.58 |
| 5 | 262122_at | AT1G02790 | PGA4 (POLYGALACTURONASE 4); polygalacturonase | -10.40 |
| 6 | 262118_at | AT1G02850 | glycosyl hydrolase family 1 protein | 1.26 |
| 7 | 262131_at | AT1G02900 | RALFL1 (RALF-LIKE 1) | -3.17 |
| 8 | 263166_at | AT1G03050 | epsin N-terminal homology (ENTH) domain-containing protein / clathrin assembly protein-related | -4.89 |
| 9 | 265083_at | AT1G03820 | similar to E6-4 [Gossypium hirsutum] (GB:AAY43793.1); similar to E6-3 [Gossypium hirsutum] (GB:AAY43792.1); similar to E6 (GB:AAB03081.1) | 1.07 |
| 10 | 264319_at | AT1G04110 | SDD1 (STOMATAL DENSITY AND DISTRIBUTION); subtilase | 1.02 |
| 11 | 263656_at | AT1G04240 | SHY2 (SHORT HYPOCOTYL 2); transcription factor | 1.00 |
| 12 | 264603_at | AT1G04670 | similar to pollen-preferential protein [Brassica rapa subsp. chinensis] (GB:ABH09777.1) | -7.65 |
| 13 | 265215_at | AT1G05040 | contains InterPro domain UBA-like (InterPro:IPR009060) | -3.88 |
| 14 | 260791_at | AT1G06250 | lipase class 3 family protein | -3.89 |
| 15 | 260788_at | AT1G06260 | cysteine proteinase, putative | -5.44 |
| 16 | 262629_at | AT1G06460 | ACD32.1 (ALPHA-CRYSTALLIN DOMAIN 31.2) | 1.06 |
| 17 | 256059_at | AT1G06990 | GDSL-motif lipase/hydrolase family protein | -6.81 |
| 18 | 256050_at | AT1G07000 | ATEXO70B2 (exocyst subunit EXO70 family protein B2); protein binding | 0.93 |
| 19 | 256060_at | AT1G07050 | CONSTANS-like protein-related | 0.97 |
| 20 | 256054_at | AT1G07120 | similar to proline-rich family protein [Arabidopsis thaliana] (TAIR:AT4G18570.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO21423.1) | -3.33 |
| 21 | 261077_at | AT1G07430 | protein phosphatase 2C, putative / PP2C, putative | 1.93 |
| 22 | 261061_at | AT1G07540 | TRFL2 (TRF-LIKE 2); DNA binding | -4.30 |
| 23 | 261462_at | AT1G07850 | fringe-related protein | -4.05 |
| 24 | 261692_at | AT1G08450 | CRT3 (CALRETICULIN 3); calcium ion binding | 1.44 |
| 25 | 264645_at | AT1G08940 | phosphoglycerate/bisphosphoglycerate mutase family protein | 1.18 |
| 26 | 264648_at | AT1G09080 | BIP3; ATP binding | 1.27 |
| 27 | 264434_at | AT1G10340 | ankyrin repeat family protein | 1.32 |
| 28 | 262760_at | AT1G10770 | invertase/pectin methylesterase inhibitor family protein | -7.25 |
| 29 | 262479_at | AT1G11130 | SUB (STRUBBELIG); protein binding | 1.20 |
| 30 | 262507_at | AT1G11330 | S-locus lectin protein kinase family protein | 1.72 |
| 31 | 261873_at | AT1G11350 | S-locus lectin protein kinase family protein | 0.96 |
| 32 | 262817_at | AT1G11770 | electron carrier | -3.33 |
| 33 | 260991_at | AT1G12160 | flavin-containing monooxygenase family protein / FMO family protein | 1.08 |
| 34 | 259526_at | AT1G12570 | glucose-methanol-choline (GMC) oxidoreductase family protein | 1.05 |
| 35 | 261203_at | AT1G12845 | similar to hypothetical protein MtrDRAFT_AC149131g9v2 [Medicago truncatula] (GB:ABD32556.1) | 1.26 |
| 36 | 262795_at | AT1G13130 | glycosyl hydrolase family 5 protein / cellulase family protein | -5.92 |
| 37 | 262764_at | AT1G13140 | CYP86C3 (cytochrome P450, family 86, subfamily C, polypeptide 3); oxygen binding | -3.67 |
| 38 | 256098_at | AT1G13700 | glucosamine/galactosamine-6-phosphate isomerase family protein | 1.34 |
| 39 | 256100_at | AT1G13750 | calcineurin-like phosphoesterase family protein | 0.95 |
| 40 | 259451_at | AT1G13890 | SNAP30 (synaptosomal-associated protein 30) | -5.96 |
| 41 | 262662_at | AT1G13920 | remorin family protein | 0.97 |
| 42 | 262664_at | AT1G13970 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G29180.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO15029.1); contains InterPro domain Protein of unknown function DUF1336 (InterPro:IPR009769) | -4.06 |
| 43 | 261528_at | AT1G14420 | AT59 (Arabidopsis homolog of tomato LAT59); lyase/ pectate lyase | -5.28 |
| 44 | 261502_at | AT1G14440 | ATHB31 (ARABIDOPSIS THALIANA HOMEOBOX PROTEIN 31); transcription factor | 1.46 |
| 45 | 262585_at | AT1G15460 | ATBOR4/BOR4; anion exchanger | -3.39 |
| 46 | 261763_at | AT1G15520 | ATPDR12/PDR12 (PLEIOTROPIC DRUG RESISTANCE 12); ATPase, coupled to transmembrane movement of substances | 1.15 |
| 47 | 262755_at | AT1G16360 | LEM3 (ligand-effect modulator 3) family protein / CDC50 family protein | -3.13 |
| 48 | 261673_at | AT1G18280 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | -7.43 |
| 49 | 261670_at | AT1G18520 | TET11 (TETRASPANIN11) | -3.40 |
| 50 | 261151_at | AT1G19650 | SEC14 cytosolic factor, putative / phosphoglyceride transfer protein, putative | 1.03 |
| 51 | 261142_at | AT1G19780 | ATCNGC8 (CYCLIC NUCLEOTIDE GATED CHANNEL 8); calmodulin binding / cyclic nucleotide binding / ion channel | -5.61 |
| 52 | 255815_at | AT1G19890 | ATMGH3/MGH3 (MALE-GAMETE-SPECIFIC HISTONE H3); DNA binding | -6.24 |
| 53 | 261221_at | AT1G19960 | similar to transmembrane receptor [Arabidopsis thaliana] (TAIR:AT2G32140.1) | 1.89 |
| 54 | 261222_at | AT1G20120 | family II extracellular lipase, putative | -4.97 |
| 55 | 261245_at | AT1G20135 | hydrolase, acting on ester bonds / lipase | -5.40 |
| 56 | 259544_at | AT1G20620 | CAT3 (CATALASE 3); catalase | 1.00 |
| 57 | 259541_at | AT1G20650 | protein kinase | 1.15 |
| 58 | 259561_at | AT1G21250 | WAK1 (CELL WALL-ASSOCIATED KINASE); kinase | 1.55 |
| 59 | 259560_at | AT1G21270 | WAK2 (wall-associated kinase 2); protein serine/threonine kinase | 1.26 |
| 60 | 255956_at | AT1G22015 | DD46; transferase, transferring hexosyl groups | -4.63 |
| 61 | 255950_at | AT1G22110 | structural constituent of ribosome | -3.56 |
| 62 | 255952_at | AT1G22130 | AGL104; DNA binding / transcription factor | -5.95 |
| 63 | 255958_at | AT1G22150 | SULTR1;3 (sulfate transporter); sulfate transmembrane transporter | -4.77 |
| 64 | 255957_at | AT1G22160 | senescence-associated protein-related | 1.40 |
| 65 | 264208_at | AT1G22760 | PAB3 (POLY(A) BINDING PROTEIN 3); RNA binding | -5.16 |
| 66 | 264899_at | AT1G23130 | Bet v I allergen family protein | 2.02 |
| 67 | 262987_at | AT1G23240 | caleosin-related family protein | -6.92 |
| 68 | 263011_at | AT1G23250 | caleosin-related | -4.29 |
| 69 | 263043_at | AT1G23350 | invertase/pectin methylesterase inhibitor family protein | -4.22 |
| 70 | 263013_at | AT1G23380 | KNAT6 (Knotted-like Arabidopsis thaliana 6); DNA binding / transcription factor | 1.02 |
| 71 | 265176_at | AT1G23520 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G23600.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO66084.1); contains InterPro domain Protein of unknown function DUF220 (InterPro:IPR003863) | -4.37 |
| 72 | 265186_at | AT1G23560 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G70480.2); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G70480.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO66084.1); contains InterPro domain Protein of unknown function DUF220 (InterPro:IPR003863) | -3.80 |
| 73 | 257402_at | AT1G23570 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G23580.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO66084.1); contains InterPro domain Protein of unknown function DUF220 (InterPro:IPR003863) | -7.84 |
| 74 | 265163_at | AT1G23580 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G23600.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO66084.1); contains InterPro domain Protein of unknown function DUF220 (InterPro:IPR003863) | -3.89 |
| 75 | 265180_at | AT1G23590 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G23600.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO66084.1); contains InterPro domain Protein of unknown function DUF220 (InterPro:IPR003863) | -4.72 |
| 76 | 265177_at | AT1G23630 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G23660.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO66084.1); contains InterPro domain Protein of unknown function DUF220 (InterPro:IPR003863) | -4.62 |
| 77 | 265166_at | AT1G23640 | pseudogene, hypothetical protein, contains Pfam profile PF02713: Domain of unknown function DUF220 | -4.09 |
| 78 | 265179_at | AT1G23650 | unknown protein | -4.71 |
| 79 | 265185_at | AT1G23670 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G23600.2); similar to unnamed protein product [Vitis vinifera] (GB:CAO66084.1); contains InterPro domain Protein of unknown function DUF220 (InterPro:IPR003863) | -4.18 |
| 80 | 257399_at | AT1G23690 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G23600.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO66084.1); contains InterPro domain Protein of unknown function DUF220 (InterPro:IPR003863) | -4.01 |
| 81 | 265188_at | AT1G23800 | ALDH2B7 (Aldehyde dehydrogenase 2B7); 3-chloroallyl aldehyde dehydrogenase/ aldehyde dehydrogenase (NAD) | -8.73 |
| 82 | 263031_at | AT1G24070 | ATCSLA10 (Cellulose synthase-like A10); transferase, transferring glycosyl groups | 1.08 |
| 83 | 265002_at | AT1G24400 | LHT2 (LYSINE HISTIDINE TRANSPORTER 2); amino acid transmembrane transporter | -4.80 |
| 84 | 265022_at | AT1G24520 | BCP1 (Brassica campestris pollen protein 1) | -4.84 |
| 85 | 265028_at | AT1G24530 | transducin family protein / WD-40 repeat family protein | 0.94 |
| 86 | 257405_at | AT1G24620 | polcalcin, putative / calcium-binding pollen allergen, putative | -6.93 |
| 87 | 245640_at | AT1G25330 | basic helix-loop-helix (bHLH) family protein | -3.23 |
| 88 | 255727_at | AT1G25510 | aspartyl protease family protein | 1.31 |
| 89 | 245820_at | AT1G26320 | NADP-dependent oxidoreductase, putative | -4.21 |
| 90 | 261020_at | AT1G26390 | FAD-binding domain-containing protein | 1.78 |
| 91 | 261015_at | AT1G26480 | GRF12 (GENERAL REGULATORY FACTOR 12); protein phosphorylated amino acid binding | -7.63 |
| 92 | 263689_at | AT1G26820 | RNS3 (RIBONUCLEASE 3); endoribonuclease | -3.13 |
| 93 | 245658_at | AT1G28270 | RALFL4 (RALF-LIKE 4) | -9.16 |
| 94 | 261442_at | AT1G28375 | unknown protein | -6.24 |
| 95 | 261499_at | AT1G28430 | CYP705A24 (cytochrome P450, family 705, subfamily A, polypeptide 24); oxygen binding | -6.50 |
| 96 | 262742_at | AT1G28550 | AtRABA1i (Arabidopsis Rab GTPase homolog A1i); GTP binding | -5.30 |
| 97 | 262736_at | AT1G28570 | GDSL-motif lipase, putative | 1.42 |
| 98 | 262748_at | AT1G28610 | GDSL-motif lipase, putative | 1.29 |
| 99 | 260888_at | AT1G29140 | pollen Ole e 1 allergen and extensin family protein | -6.71 |
| 100 | 256307_at | AT1G30350 | pectate lyase family protein | -4.83 |
| 101 | 263219_at | AT1G30600 | subtilase family protein | 0.94 |
| 102 | 264499_at | AT1G30795 | hydroxyproline-rich glycoprotein family protein | -4.54 |
| 103 | 260702_at | AT1G32250 | calmodulin, putative | -5.58 |
| 104 | 260693_at | AT1G32450 | proton-dependent oligopeptide transport (POT) family protein | -5.24 |
| 105 | 261240_at | AT1G32940 | ATSBT3.5; subtilase | 1.12 |
| 106 | 261567_at | AT1G33055 | unknown protein | -3.72 |
| 107 | 256481_at | AT1G33430 | galactosyltransferase family protein | -4.36 |
| 108 | 260116_at | AT1G33960 | AIG1 (AVRRPT2-INDUCED GENE 1); GTP binding | 2.01 |
| 109 | 262408_at | AT1G34750 | protein phosphatase 2C, putative / PP2C, putative | 0.93 |
| 110 | 259550_at | AT1G35230 | AGP5 (ARABINOGALACTAN-PROTEIN 5) | 1.69 |
| 111 | 259549_at | AT1G35290 | thioesterase family protein | 1.48 |
| 112 | 262022_at | AT1G35490 | bZIP family transcription factor | -6.57 |
| 113 | 262028_at | AT1G35560 | TCP family transcription factor, putative | 1.16 |
| 114 | 261339_at | AT1G35710 | leucine-rich repeat transmembrane protein kinase, putative | 1.55 |
| 115 | 245246_at | AT1G44224 | Encodes a ECA1 gametogenesis related family protein | -5.33 |
| 116 | 260941_at | AT1G44970 | peroxidase, putative | -4.96 |
| 117 | 261687_at | AT1G47280 | unknown protein | -4.47 |
| 118 | 262434_at | AT1G47670 | amino acid transporter family protein | 1.19 |
| 119 | 259615_at | AT1G47980 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G62730.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO44946.1) | -5.89 |
| 120 | 260727_at | AT1G48100 | glycoside hydrolase family 28 protein / polygalacturonase (pectinase) family protein | 0.94 |
| 121 | 262236_at | AT1G48330 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G17580.1); similar to unknown [Populus trichocarpa] (GB:ABK96144.1) | 1.85 |
| 122 | 261305_at | AT1G48470 | GLN1;5 (GLUTAMINE SYNTHETASE 1;5); glutamate-ammonia ligase | -3.60 |
| 123 | 257469_at | AT1G49290 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G13620.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN72326.1) | -5.80 |
| 124 | 262393_at | AT1G49490 | leucine-rich repeat family protein / extensin family protein | -3.59 |
| 125 | 262463_at | AT1G50310 | monosaccharide transporter (STP9) | -4.43 |
| 126 | 261876_at | AT1G50590 | pirin, putative | 1.05 |
| 127 | 246371_at | AT1G51940 | protein kinase family protein / peptidoglycan-binding LysM domain-containing protein | 0.99 |
| 128 | 259836_at | AT1G52240 | ATROPGEF11/ROPGEF11 (KINASE PARTNER PROTEIN-LIKE); Rho guanyl-nucleotide exchange factor/ | -4.56 |
| 129 | 262147_at | AT1G52570 | PLDALPHA2 (PHOSPHLIPASE D ALPHA 2); phospholipase D | -3.23 |
| 130 | 262146_at | AT1G52580 | rhomboid family protein | -3.55 |
| 131 | 262156_at | AT1G52680 | late embryogenesis abundant protein-related / LEA protein-related | -8.39 |
| 132 | 262128_at | AT1G52690 | late embryogenesis abundant protein, putative / LEA protein, putative | 1.37 |
| 133 | 261368_at | AT1G53070 | legume lectin family protein | 1.16 |
| 134 | 261375_at | AT1G53160 | SPL4 (SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 4); DNA binding / transcription factor | 1.61 |
| 135 | 260618_at | AT1G53230 | TCP3 (TEOSINTE BRANCHED1, CYCLOIDEA AND PCF TRANSCRIPTION FACTOR 3); transcription factor | 1.37 |
| 136 | 263144_at | AT1G54070 | dormancy/auxin associated protein-related | -4.97 |
| 137 | 264235_at | AT1G54560 | XIE (Myosin-like protein XIE); motor/ protein binding | -3.93 |
| 138 | 264240_at | AT1G54820 | protein kinase family protein | 1.20 |
| 139 | 264187_at | AT1G54860 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G19250.1); similar to GPI-anchored protein-like protein II [Cucumis melo] (GB:ABR67421.1) | -7.56 |
| 140 | 256354_at | AT1G54870 | oxidoreductase | -4.39 |
| 141 | 256149_at | AT1G55110 | ATIDD7 (ARABIDOPSIS THALIANA INDETERMINATE(ID)-DOMAIN 7); nucleic acid binding / transcription factor/ zinc ion binding | 1.00 |
| 142 | 265127_at | AT1G55560 | SKS14 (SKU5 Similar 14); copper ion binding / oxidoreductase | -4.96 |
| 143 | 265080_at | AT1G55570 | SKS12 (SKU5 Similar 12); copper ion binding / oxidoreductase | -10.46 |
| 144 | 264548_at | AT1G55680 | WD-40 repeat family protein | 1.00 |
| 145 | 256221_at | AT1G56300 | DNAJ heat shock N-terminal domain-containing protein | 1.27 |
| 146 | 246377_at | AT1G57550 | hydrophobic protein, putative / low temperature and salt responsive protein, putative | -3.31 |
| 147 | 246405_at | AT1G57630 | disease resistance protein (TIR class), putative | 2.86 |
| 148 | 245842_at | AT1G58430 | RXF26; carboxylesterase | 0.98 |
| 149 | 262899_at | AT1G59870 | PDR8/PEN3 (PLEIOTROPIC DRUG RESISTANCE8); ATPase, coupled to transmembrane movement of substances / cadmium ion transmembrane transporter | 1.00 |
| 150 | 264882_at | AT1G61110 | ANAC025 (Arabidopsis NAC domain containing protein 25); transcription factor | -7.31 |
| 151 | 265031_at | AT1G61590 | protein kinase, putative | -5.20 |
| 152 | 264400_at | AT1G61800 | GPT2 (glucose-6-phosphate/phosphate translocator 2); antiporter/ glucose-6-phosphate transmembrane transporter | 2.33 |
| 153 | 264287_at | AT1G61930 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G11700.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO17934.1); contains InterPro domain Protein of unknown function DUF584 (InterPro:IPR007608) | 1.35 |
| 154 | 264746_at | AT1G62300 | WRKY6 (WRKY DNA-binding protein 6); transcription factor | 1.73 |
| 155 | 265115_at | AT1G62450 | Rho GDP-dissociation inhibitor family protein | -3.47 |
| 156 | 262694_at | AT1G62790 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | 1.22 |
| 157 | 259693_at | AT1G63060 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G07330.1); similar to unknown [Xerophyta humilis] (GB:AAT45004.1) | -8.85 |
| 158 | 259694_at | AT1G63180 | UGE3 (UDP-D-GLUCOSE/UDP-D-GALACTOSE 4-EPIMERASE 3); UDP-glucose 4-epimerase/ protein dimerization | -3.15 |
| 159 | 260241_at | AT1G63710 | CYP86A7 (cytochrome P450, family 86, subfamily A, polypeptide 7); oxygen binding | 1.13 |
| 160 | 259764_at | AT1G64280 | NPR1 (NONEXPRESSER OF PR GENES 1); protein binding | 1.00 |
| 161 | 259766_at | AT1G64360 | unknown protein | 2.63 |
| 162 | 259765_at | AT1G64370 | unknown protein | 1.23 |
| 163 | 264163_at | AT1G65445 | transferase-related | 1.39 |
| 164 | 264638_at | AT1G65480 | FT (FLOWERING LOCUS T) | 1.09 |
| 165 | 264635_at | AT1G65500 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G65486.1) | 1.07 |
| 166 | 256381_at | AT1G66850 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | -8.28 |
| 167 | 256366_at | AT1G66880 | serine/threonine protein kinase family protein | 1.33 |
| 168 | 255913_at | AT1G66980 | protein kinase family protein / glycerophosphoryl diester phosphodiesterase family protein | 0.93 |
| 169 | 264993_at | AT1G67290 | glyoxal oxidase-related | -3.44 |
| 170 | 260195_at | AT1G67540 | similar to hypothetical protein [Vitis vinifera] (GB:CAN81893.1) | -3.30 |
| 171 | 260004_at | AT1G67860 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G67865.1) | 1.13 |
| 172 | 260012_at | AT1G67865 | unknown protein | 1.14 |
| 173 | 260011_at | AT1G68110 | epsin N-terminal homology (ENTH) domain-containing protein / clathrin assembly protein-related | -3.76 |
| 174 | 260484_at | AT1G68360 | zinc finger protein-related | 1.24 |
| 175 | 262282_at | AT1G68610 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G14870.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO42115.1); contains InterPro domain Protein of unknown function Cys-rich (InterPro:IPR006461) | -3.39 |
| 176 | 260032_at | AT1G68750 | ATPPC4 (Arabidopsis thaliana phosphoenolpyruvate carboxylase 4); phosphoenolpyruvate carboxylase | -4.15 |
| 177 | 260034_at | AT1G68810 | basic helix-loop-helix (bHLH) family protein | 0.95 |
| 178 | 260038_at | AT1G68875 | unknown protein | -7.92 |
| 179 | 259372_at | AT1G69120 | AP1 (APETALA1); DNA binding / transcription factor | 1.14 |
| 180 | 256300_at | AT1G69490 | NAP (NAC-LIKE, ACTIVATED BY AP3/PI); transcription factor | 1.50 |
| 181 | 260405_at | AT1G69930 | ATGSTU11 (Arabidopsis thaliana Glutathione S-transferase (class tau) 11); glutathione transferase | 1.02 |
| 182 | 264701_at | AT1G70160 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G54870.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G27020.1); similar to unknown [Picea sitchensis] (GB:ABK24649.1); similar to hypothetical protein OsI_030218 [Oryza sativa (indica cultivar-group)] (GB:EAZ08986.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO40967.1) | 0.97 |
| 183 | 260334_at | AT1G70510 | KNAT2 (KNOTTED-LIKE FROM ARABIDOPSIS THALIANA 2); transcription factor | 1.00 |
| 184 | 261532_at | AT1G71680 | amino acid transmembrane transporter | -7.69 |
| 185 | 261511_at | AT1G71770 | PAB5 (POLY(A)-BINDING PROTEIN); RNA binding | -4.89 |
| 186 | 259802_at | AT1G72260 | THI2.1 (THIONIN 2.1); toxin receptor binding | -3.62 |
| 187 | 259799_at | AT1G72290 | trypsin and protease inhibitor family protein / Kunitz family protein | -6.42 |
| 188 | 262378_at | AT1G72830 | HAP2C (Heme activator protein (yeast) homolog 2C); transcription factor | 1.88 |
| 189 | 262383_at | AT1G72940 | disease resistance protein (TIR-NBS class), putative | 0.96 |
| 190 | 262385_at | AT1G72960 | root hair defective 3 GTP-binding (RHD3) family protein | -6.45 |
| 191 | 260097_at | AT1G73220 | ATOCT1 (ARABIDOPSIS THALIANA ORGANIC CATION/CARNITINE TRANSPORTER1); carbohydrate transmembrane transporter/ carnitine transporter/ transporter | -6.48 |
| 192 | 260046_at | AT1G73805 | calmodulin binding | 1.77 |
| 193 | 260255_at | AT1G74330 | ATP binding / protein kinase | 1.05 |
| 194 | 260228_at | AT1G74540 | CYP98A8 (cytochrome P450, family 98, subfamily A, polypeptide 8); oxygen binding | -5.86 |
| 195 | 262177_at | AT1G74710 | ICS1 (ISOCHORISMATE SYNTHASEI); isochorismate synthase | 1.13 |
| 196 | 259925_at | AT1G75040 | PR5 (PATHOGENESIS-RELATED GENE 5) | 2.40 |
| 197 | 256452_at | AT1G75240 | ATHB33 (ARABIDOPSIS THALIANA HOMEOBOX PROTEIN 33); DNA binding / transcription factor | 1.72 |
| 198 | 261120_at | AT1G75410 | BLH3; DNA binding / transcription factor | 1.03 |
| 199 | 262969_at | AT1G75710 | zinc finger (C2H2 type) family protein | 0.95 |
| 200 | 262949_at | AT1G75790 | SKS18 (SKU5 Similar 18); copper ion binding / pectinesterase | -4.33 |
| 201 | 262674_at | AT1G75910 | EXL4 (extracellular lipase 4); acyltransferase/ carboxylesterase/ lipase | -6.72 |
| 202 | 262683_at | AT1G75920 | family II extracellular lipase 5 (EXL5) | -6.35 |
| 203 | 262675_at | AT1G75930 | EXL6 (extracellular lipase 6); acyltransferase/ carboxylesterase/ lipase | -9.92 |
| 204 | 262697_at | AT1G75940 | ATA27 (Arabidopsis thaliana anther 27); hydrolase, hydrolyzing O-glycosyl compounds | -10.34 |
| 205 | 261749_at | AT1G76180 | ERD14 (EARLY RESPONSE TO DEHYDRATION 14) | 0.96 |
| 206 | 261772_at | AT1G76240 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G17080.1); similar to Os06g0725500 [Oryza sativa (japonica cultivar-group)] (GB:NP_001058623.1); similar to hypothetical protein OsI_023643 [Oryza sativa (indica cultivar-group)] (GB:EAZ02411.1); contains InterPro domain Protein of unknown function DUF241, plant (InterPro:IPR004320) | -3.25 |
| 207 | 259886_at | AT1G76370 | protein kinase, putative | -5.14 |
| 208 | 264958_at | AT1G76960 | unknown protein | 4.61 |
| 209 | 264953_at | AT1G77120 | ADH1 (ALCOHOL DEHYDROGENASE 1) | 1.17 |
| 210 | 264483_at | AT1G77230 | tetratricopeptide repeat (TPR)-containing protein | 0.99 |
| 211 | 262179_at | AT1G77980 | AGL66; transcription factor | -4.09 |
| 212 | 264096_at | AT1G78995 | similar to unnamed protein product [Vitis vinifera] (GB:CAO41778.1) | 1.25 |
| 213 | 264127_at | AT1G79250 | protein kinase, putative | -3.27 |
| 214 | 262936_at | AT1G79400 | ATCHX2 (CATION/H+ EXCHANGER 2); monovalent cation:proton antiporter | -3.24 |
| 215 | 260158_at | AT1G79910 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G52315.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO45916.1); contains InterPro domain Protein of unknown function DUF292, eukaryotic (InterPro:IPR005061) | -3.92 |
| 216 | 262050_at | AT1G80130 | binding | 1.77 |
| 217 | 261943_at | AT1G80660 | AHA9 (Arabidopsis H(+)-ATPase 9); hydrogen-exporting ATPase, phosphorylative mechanism | -7.99 |
| 218 | 265740_at | AT2G01150 | RHA2B (RING-H2 FINGER PROTEIN 2B); protein binding / zinc ion binding | 1.24 |
| 219 | 266353_at | AT2G01520 | MLP328 (MLP-LIKE PROTEIN 328) | 1.30 |
| 220 | 265872_at | AT2G01670 | ATNUDT17 (Arabidopsis thaliana Nudix hydrolase homolog 17); hydrolase | 1.03 |
| 221 | 265871_at | AT2G01680 | ankyrin repeat family protein | 1.13 |
| 222 | 266115_at | AT2G02140 | LCR72/PDF2.6 (Low-molecular-weight cysteine-rich 72); protease inhibitor | -5.42 |
| 223 | 267476_at | AT2G02720 | pectate lyase family protein | -7.08 |
| 224 | 266770_at | AT2G03090 | ATEXPA15 (ARABIDOPSIS THALIANA EXPANSIN A15) | 1.04 |
| 225 | 264039_at | AT2G03740 | late embryogenesis abundant domain-containing protein / LEA domain-containing protein | -6.86 |
| 226 | 263363_at | AT2G03850 | late embryogenesis abundant domain-containing protein / LEA domain-containing protein | -8.71 |
| 227 | 263325_at | AT2G04240 | XERICO; protein binding / zinc ion binding | 1.24 |
| 228 | 263852_at | AT2G04450 | ATNUDT6 (Arabidopsis thaliana Nudix hydrolase homolog 6); ADP-ribose diphosphatase/ NAD binding / hydrolase | 1.28 |
| 229 | 263627_at | AT2G04675 | unknown protein | -5.19 |
| 230 | 265560_at | AT2G05520 | GRP-3 (GLYCINE-RICH PROTEIN 3) | 1.00 |
| 231 | 266037_at | AT2G05940 | protein kinase, putative | 1.08 |
| 232 | 266500_at | AT2G06925 | ATSPLA2-ALPHA/PLA2-ALPHA (PHOSPHOLIPASE A2-ALPHA); phospholipase A2 | 1.35 |
| 233 | 266494_at | AT2G07040 | ATPRK2A; ATP binding / protein serine/threonine kinase | -7.32 |
| 234 | 265552_at | AT2G07560 | AHA6 (ARABIDOPSIS H(+)-ATPASE 6); ATPase | -9.06 |
| 235 | 264112_at | AT2G13680 | CALS5 (CALLOSE SYNTHASE 5); 1,3-beta-glucan synthase | -3.91 |
| 236 | 265837_at | AT2G14560 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G33840.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO48825.1); contains InterPro domain Protein of unknown function DUF567 (InterPro:IPR007612) | 1.68 |
| 237 | 266385_at | AT2G14610 | PR1 (PATHOGENESIS-RELATED GENE 1) | 3.06 |
| 238 | 266376_at | AT2G14620 | xyloglucan:xyloglucosyl transferase, putative / xyloglucan endotransglycosylase, putative / endo-xyloglucan transferase, putative | 1.35 |
| 239 | 263566_at | AT2G15340 | glycine-rich protein | -4.22 |
| 240 | 265473_at | AT2G15535 | LCR10 (Low-molecular-weight cysteine-rich 10) | -5.39 |
| 241 | 265480_at | AT2G15970 | COR413-PM1 (cold regulated 413 plasma membrane 1) | 0.95 |
| 242 | 263608_at | AT2G16260 | pseudogene of glycine-rich RNA-binding protein | 1.00 |
| 243 | 263238_at | AT2G16580 | auxin-responsive protein, putative | 0.96 |
| 244 | 265404_at | AT2G16730 | BGAL13 (beta-galactosidase 13); beta-galactosidase | -4.66 |
| 245 | 265405_at | AT2G16750 | protein kinase family protein | -5.80 |
| 246 | 263584_at | AT2G17040 | ANAC036 (Arabidopsis NAC domain containing protein 36); transcription factor | 1.89 |
| 247 | 265821_at | AT2G17950 | WUS (WUSCHEL); DNA binding / transcription factor | -4.80 |
| 248 | 265379_at | AT2G18340 | late embryogenesis abundant domain-containing protein / LEA domain-containing protein | -5.61 |
| 249 | 265331_at | AT2G18420 | Encodes a Gibberellin-regulated GASA/GAST/Snakin family protein | -5.06 |
| 250 | 265922_at | AT2G18480 | mannitol transporter, putative | -3.42 |
| 251 | 266070_at | AT2G18660 | EXLB3 (EXPANSIN-LIKE B3 PRECURSOR) | 2.21 |
| 252 | 266017_at | AT2G18690 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G18680.1); similar to hypothetical protein OsI_029427 [Oryza sativa (indica cultivar-group)] (GB:EAZ08195.1) | 1.60 |
| 253 | 267443_at | AT2G19000 | similar to glycine-rich protein [Arabidopsis thaliana] (TAIR:AT4G21620.1); similar to hypothetical protein OsI_037979 [Oryza sativa (indica cultivar-group)] (GB:EAY84020.1); similar to expressed protein [Oryza sativa (japonica cultivar-group)] (GB:ABA99958.1); contains InterPro domain 4-disulphide core (InterPro:IPR015874) | -5.43 |
| 254 | 267440_at | AT2G19070 | transferase family protein | -3.56 |
| 255 | 267336_at | AT2G19310 | similar to HSP18.2 (HEAT SHOCK PROTEIN 18.2) [Arabidopsis thaliana] (TAIR:AT5G59720.1); similar to 18.5 kDa class I heat shock protein (HSP 18.5) (GB:P05478); contains InterPro domain HSP20-like chaperone (InterPro:IPR008978) | 0.94 |
| 256 | 266697_at | AT2G19770 | PRF5 (PROFILIN5); actin binding / actin monomer binding | -7.42 |
| 257 | 266695_at | AT2G19810 | zinc finger (CCCH-type) family protein | 1.45 |
| 258 | 263373_at | AT2G20515 | similar to unnamed protein product [Vitis vinifera] (GB:CAO40634.1) | 0.97 |
| 259 | 263715_at | AT2G20570 | GPRI1 (GOLDEN2-LIKE 1); transcription factor | 1.12 |
| 260 | 263711_at | AT2G20630 | protein phosphatase 2C, putative / PP2C, putative | 1.09 |
| 261 | 265443_at | AT2G20750 | ATEXPB1 (ARABIDOPSIS THALIANA EXPANSIN B1) | 1.01 |
| 262 | 265440_at | AT2G20960 | pEARLI4 | 0.94 |
| 263 | 264025_at | AT2G21050 | amino acid permease, putative | 1.22 |
| 264 | 263753_at | AT2G21490 | LEA (DEHYDRIN LEA) | -7.50 |
| 265 | 263517_at | AT2G21620 | RD2 (RESPONSIVE TO DESSICATION 2) | 1.01 |
| 266 | 263549_at | AT2G21650 | MEE3 (maternal effect embryo arrest 3); DNA binding / transcription factor | -3.55 |
| 267 | 263453_at | AT2G22180 | hydroxyproline-rich glycoprotein family protein | -3.45 |
| 268 | 263433_at | AT2G22240 | inositol-3-phosphate synthase isozyme 2 / myo-inositol-1-phosphate synthase 2 / MI-1-P synthase 2 / IPS 2 | 0.94 |
| 269 | 264001_at | AT2G22420 | peroxidase 17 (PER17) (P17) | 1.42 |
| 270 | 265344_at | AT2G22660 | similar to glycine-rich protein [Arabidopsis thaliana] (TAIR:AT4G37900.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO44607.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO17179.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO38708.1); contains InterPro domain Protein of unknown function DUF1399 (InterPro:IPR009836) | 0.99 |
| 271 | 267255_at | AT2G22950 | calcium-transporting ATPase, plasma membrane-type, putative / Ca2+-ATPase, putative (ACA7) | -3.88 |
| 272 | 267253_at | AT2G22960 | serine carboxypeptidase S10 family protein | 1.08 |
| 273 | 267150_at | AT2G23510 | transferase family protein | -4.35 |
| 274 | 267295_at | AT2G23800 | GGPS2 (GERANYLGERANYL PYROPHOSPHATE SYNTHASE 2); farnesyltranstransferase | -6.70 |
| 275 | 266572_at | AT2G23840 | HNH endonuclease domain-containing protein | 1.10 |
| 276 | 266558_at | AT2G23900 | glycoside hydrolase family 28 protein / polygalacturonase (pectinase) family protein | -5.18 |
| 277 | 266578_at | AT2G23910 | cinnamoyl-CoA reductase-related | 1.05 |
| 278 | 265993_at | AT2G24160 | pseudogene, leucine rich repeat protein family, contains leucine rich-repeat domains Pfam:PF00560, INTERPRO:IPR001611; contains some similarity to Cf-4 (Lycopersicon hirsutum) gi|2808683|emb|CAA05268; blastp match of 37% identity and 8.4e-98 P-value to GP|2808683|emb|CAA05268.1||AJ002235 Cf-4 {Lycopersicon hirsutum} | 0.97 |
| 279 | 257392_at | AT2G24450 | FLA3 (FASCICLIN-LIKE ARABINOGALACTAN PROTEIN 3 PRECURSOR) | -7.29 |
| 280 | 265609_at | AT2G25420 | similar to TPR4/WSIP2 (TOPLESS-RELATED 4) [Arabidopsis thaliana] (TAIR:AT3G15880.2); similar to TPR4/WSIP2 (TOPLESS-RELATED 4), protein binding [Arabidopsis thaliana] (TAIR:AT3G15880.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN64663.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO62716.1); contains InterPro domain WD40 repeat-like (InterPro:IPR011046); contains InterPro domain WD40/YVTN repeat-like (InterPro:IPR015943); contains InterPro domain WD40 repeat (InterPro:IPR001680); contains InterPro domain LisH dimerisation motif (InterPro:IPR006594); contains InterPro domain CTLH, C-terminal to LisH motif (InterPro:IPR006595) | 1.11 |
| 281 | 266654_at | AT2G25890 | glycine-rich protein / oleosin | -7.45 |
| 282 | 267381_at | AT2G26190 | calmodulin-binding family protein | 0.96 |
| 283 | 245035_at | AT2G26400 | ARD/ATARD3 (ACIREDUCTONE DIOXYGENASE); acireductone dioxygenase [iron(II)-requiring]/ heteroglycan binding / metal ion binding | 1.78 |
| 284 | 245036_at | AT2G26410 | IQD4 (IQ-domain 4); calmodulin binding | -5.85 |
| 285 | 245053_at | AT2G26450 | pectinesterase family protein | -3.76 |
| 286 | 245057_at | AT2G26490 | transducin family protein / WD-40 repeat family protein | -3.64 |
| 287 | 263084_at | AT2G27180 | unknown protein | -3.62 |
| 288 | 265545_at | AT2G28250 | protein kinase family protein. Binds AtRop4. | 1.23 |
| 289 | 265280_at | AT2G28355 | LCR5 (Low-molecular-weight cysteine-rich 5) | -7.97 |
| 290 | 263434_at | AT2G28610 | PRS (PRESSED FLOWER); transcription factor | 1.86 |
| 291 | 257442_at | AT2G28680 | cupin family protein | -4.29 |
| 292 | 266225_at | AT2G28900 | OEP16 (OUTER ENVELOPE PROTEIN 16); P-P-bond-hydrolysis-driven protein transmembrane transporter | 1.25 |
| 293 | 266292_at | AT2G29350 | SAG13 (Senescence-associated gene 13); oxidoreductase | 1.77 |
| 294 | 266267_at | AT2G29460 | ATGSTU4 (GLUTATHIONE S-TRANSFERASE 22); glutathione transferase | 1.88 |
| 295 | 266665_at | AT2G29790 | Encodes a Maternally expressed gene (MEG) family protein [pseudogene] | -7.90 |
| 296 | 266866_at | AT2G29940 | ATPDR3/PDR3 (PLEIOTROPIC DRUG RESISTANCE 3); ATPase, coupled to transmembrane movement of substances | -4.68 |
| 297 | 267275_at | AT2G30240 | ATCHX13 (cation/H+ exchanger 13); monovalent cation:proton antiporter | -4.19 |
| 298 | 257349_at | AT2G30630 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G06750.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN76299.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO22658.1); contains domain SSF52540 (SSF52540); contains domain G3DSA:3.40.50.300 (G3DSA:3.40.50.300) | -3.19 |
| 299 | 267567_at | AT2G30770 | CYP71A13 (CYTOCHROME P450, FAMILY 71, SUBFAMILY A, POLYPEPTIDE 13); indoleacetaldoxime dehydratase/ oxygen binding | 1.09 |
| 300 | 263450_at | AT2G31500 | CPK24 (calcium-dependent protein kinase 24); calmodulin-dependent protein kinase/ kinase | -7.41 |
| 301 | 263478_at | AT2G31880 | leucine-rich repeat transmembrane protein kinase, putative | 1.27 |
| 302 | 265672_at | AT2G31980 | cysteine proteinase inhibitor-related | -5.16 |
| 303 | 267546_at | AT2G32680 | disease resistance family protein | 1.94 |
| 304 | 267643_at | AT2G32890 | RALFL17 (RALF-LIKE 17) | -5.17 |
| 305 | 255835_at | AT2G33420 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G25800.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G20010.2); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G04470.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO71451.1); similar to hypothetical protein LOC_Os03g04560 [Oryza sativa (japonica cultivar-group)] (GB:ABF93881.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN63332.1); contains InterPro domain Munc13 homology 2 (InterPro:IPR014772); contains InterPro domain Protein of unknown function DUF810 (InterPro:IPR008528); contains InterPro domain Munc13 homology 1 (InterPro:IPR014770) | -3.33 |
| 306 | 267449_at | AT2G33690 | late embryogenesis abundant protein, putative / LEA protein, putative | -5.54 |
| 307 | 267317_at | AT2G34700 | pollen Ole e 1 allergen and extensin family protein | -3.92 |
| 308 | 267414_at | AT2G34790 | EDA28/MEE23 (MATERNAL EFFECT EMBRYO ARREST 23); electron carrier | -3.41 |
| 309 | 267431_at | AT2G34870 | MEE26 (maternal effect embryo arrest 26) | -3.17 |
| 310 | 263950_at | AT2G36020 | HVA22J (HVA22-LIKE PROTEIN J) | -6.21 |
| 311 | 263905_at | AT2G36190 | ATCWINV4 (ARABIDOPSIS THALIANA CELL WALL INVERTASE 4); hydrolase, hydrolyzing O-glycosyl compounds | -4.64 |
| 312 | 263931_at | AT2G36220 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G52710.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO15901.1) | 1.19 |
| 313 | 263847_at | AT2G36970 | UDP-glucoronosyl/UDP-glucosyl transferase family protein | 1.53 |
| 314 | 267178_at | AT2G37750 | unknown protein | -4.34 |
| 315 | 267094_at | AT2G38080 | IRX12/LAC4 (laccase 4); copper ion binding / oxidoreductase | -3.19 |
| 316 | 267028_at | AT2G38470 | WRKY33 (WRKY DNA-binding protein 33); transcription factor | 1.13 |
| 317 | 266167_at | AT2G38860 | YLS5 (yellow-leaf-specific gene 5) | 0.93 |
| 318 | 266143_at | AT2G38905 | hydrophobic protein, putative / low temperature and salt responsive protein, putative | -3.76 |
| 319 | 266142_at | AT2G39030 | GCN5-related N-acetyltransferase (GNAT) family protein | 1.70 |
| 320 | 245087_at | AT2G39830 | zinc ion binding | 0.95 |
| 321 | 267361_at | AT2G39920 | acid phosphatase class B family protein | 1.55 |
| 322 | 267364_at | AT2G40080 | ELF4 (EARLY FLOWERING 4) | 1.42 |
| 323 | 263829_at | AT2G40435 | transcription regulator | 2.03 |
| 324 | 257382_at | AT2G40750 | WRKY54 (WRKY DNA-binding protein 54); transcription factor | 1.71 |
| 325 | 245096_at | AT2G40880 | FL3-27; cysteine protease inhibitor | 1.00 |
| 326 | 267076_at | AT2G41090 | calmodulin-like calcium-binding protein, 22 kDa (CaBP-22) | 1.15 |
| 327 | 267582_at | AT2G41970 | protein kinase, putative | -4.42 |
| 328 | 267639_at | AT2G42200 | squamosa promoter-binding protein-like 9 (SPL9) | 1.04 |
| 329 | 260551_at | AT2G43510 | ATTI1 (ARABIDOPSIS THALIANA TRYPSIN INHIBITOR PROTEIN 1) | 2.48 |
| 330 | 260568_at | AT2G43570 | chitinase, putative | 2.98 |
| 331 | 260557_at | AT2G43610 | glycoside hydrolase family 19 protein | -3.27 |
| 332 | 266126_at | AT2G45040 | matrix metalloproteinase | -3.25 |
| 333 | 267527_at | AT2G45610 | hydrolase | -4.41 |
| 334 | 266920_at | AT2G45750 | dehydration-responsive family protein | -3.51 |
| 335 | 266918_at | AT2G45800 | LIM domain-containing protein | -5.21 |
| 336 | 263783_at | AT2G46400 | WRKY46 (WRKY DNA-binding protein 46); transcription factor | 1.53 |
| 337 | 266765_at | AT2G46860 | ATPPA3 (ARABIDOPSIS THALIANA PYROPHOSPHORYLASE 3); inorganic diphosphatase/ pyrophosphatase | -8.73 |
| 338 | 266764_at | AT2G47050 | invertase/pectin methylesterase inhibitor family protein | -3.88 |
| 339 | 263319_at | AT2G47160 | BOR1 (REQUIRES HIGH BORON 1); anion exchanger | 1.24 |
| 340 | 245126_at | AT2G47460 | ATMYB12/MYB12 (MYB DOMAIN PROTEIN 12); DNA binding / transcription activator/ transcription factor | 1.01 |
| 341 | 245151_at | AT2G47550 | pectinesterase family protein | -3.55 |
| 342 | 266488_at | AT2G47670 | invertase/pectin methylesterase inhibitor family protein | -4.41 |
| 343 | 266462_at | AT2G47770 | benzodiazepine receptor-related | -3.55 |
| 344 | 266516_at | AT2G47880 | glutaredoxin family protein | 1.06 |
| 345 | 259262_at | AT3G01020 | ATISU2/ISU2 (IscU-like 2); structural molecule | -5.70 |
| 346 | 259281_at | AT3G01140 | MYB106 (myb domain protein 106); DNA binding / transcription factor | 1.38 |
| 347 | 259266_at | AT3G01240 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G01230.1) | -8.27 |
| 348 | 259265_at | AT3G01250 | unknown protein | -9.01 |
| 349 | 259269_at | AT3G01270 | pectate lyase family protein | -7.17 |
| 350 | 259116_at | AT3G01350 | proton-dependent oligopeptide transport (POT) family protein | -3.13 |
| 351 | 259189_at | AT3G01700 | AGP11 (ARABINOGALACTAN PROTEIN 11) | -8.23 |
| 352 | 258947_at | AT3G01830 | calmodulin-related protein, putative | 0.99 |
| 353 | 258857_at | AT3G02110 | SCPL25 (serine carboxypeptidase-like 25); serine carboxypeptidase | 0.95 |
| 354 | 259124_at | AT3G02310 | SEP2 (SEPALLATA2); DNA binding / transcription factor | 1.03 |
| 355 | 258498_at | AT3G02480 | ABA-responsive protein-related | -8.41 |
| 356 | 258600_at | AT3G02810 | protein kinase family protein | -6.05 |
| 357 | 258605_at | AT3G02970 | phosphate-responsive 1 family protein | -3.39 |
| 358 | 258870_at | AT3G03080 | NADP-dependent oxidoreductase, putative | -3.15 |
| 359 | 259044_at | AT3G03430 | polcalcin, putative / calcium-binding pollen allergen, putative | -6.33 |
| 360 | 259338_at | AT3G03800 | SYP131 (syntaxin 131); SNAP receptor | -3.85 |
| 361 | 259346_at | AT3G03910 | glutamate dehydrogenase, putative | -3.45 |
| 362 | 258537_at | AT3G04210 | disease resistance protein (TIR-NBS class), putative | 1.25 |
| 363 | 258840_at | AT3G04620 | nucleic acid binding | -3.49 |
| 364 | 258843_at | AT3G04690 | protein kinase family protein | -4.55 |
| 365 | 259351_at | AT3G05150 | sugar transporter family protein | -4.39 |
| 366 | 258889_at | AT3G05610 | pectinesterase family protein | -7.23 |
| 367 | 258894_at | AT3G05650 | disease resistance family protein | 1.31 |
| 368 | 258742_at | AT3G05800 | transcription factor | 1.64 |
| 369 | 258748_at | AT3G05930 | GLP8 (GERMIN-LIKE PROTEIN 8); manganese ion binding / metal ion binding / nutrient reservoir | -4.63 |
| 370 | 258561_at | AT3G05960 | sugar transporter, putative | -3.88 |
| 371 | 258523_at | AT3G06830 | pectinesterase family protein | -3.73 |
| 372 | 258639_at | AT3G07820 | polygalacturonase 3 (PGA3) / pectinase | -10.82 |
| 373 | 258685_at | AT3G07830 | polygalacturonase, putative / pectinase, putative | -5.43 |
| 374 | 258686_at | AT3G07840 | polygalacturonase, putative / pectinase, putative | -6.03 |
| 375 | 258671_at | AT3G08560 | VHA-E2 (VACUOLAR H+-ATPASE SUBUNIT E ISOFORM 2); hydrogen-exporting ATPase, phosphorylative mechanism | -7.44 |
| 376 | 259213_at | AT3G09010 | protein kinase family protein | 0.95 |
| 377 | 258753_at | AT3G09530 | ATEXO70H3 (exocyst subunit EXO70 family protein H3); protein binding | -6.38 |
| 378 | 258650_at | AT3G09830 | protein kinase, putative | 1.08 |
| 379 | 258940_at | AT3G09930 | GDSL-motif lipase/hydrolase family protein | -3.46 |
| 380 | 258938_at | AT3G10080 | germin-like protein, putative | 0.98 |
| 381 | 258968_at | AT3G10460 | self-incompatibility protein-related | -5.02 |
| 382 | 258962_at | AT3G10570 | CYP77A6 (cytochrome P450, family 77, subfamily A, polypeptide 6); oxygen binding | 1.00 |
| 383 | 258767_at | AT3G10890 | (1-4)-beta-mannan endohydrolase, putative | -3.64 |
| 384 | 256255_at | AT3G11280 | myb family transcription factor | 1.35 |
| 385 | 259282_at | AT3G11430 | ATGPAT5/GPAT5 (GLYCEROL-3-PHOSPHATE ACYLTRANSFERASE 5); 1-acylglycerol-3-phosphate O-acyltransferase/ acyltransferase/ organic anion transmembrane transporter | -3.25 |
| 386 | 256261_at | AT3G12160 | AtRABA4d (Arabidopsis Rab GTPase homolog A4d); GTP binding | -5.36 |
| 387 | 257691_at | AT3G12660 | FLA14 (FASCICLIN-LIKE ARABINOGALACTAN PROTEIN 14 PRECURSOR) | -3.71 |
| 388 | 257702_at | AT3G12670 | EMB2742 (EMBRYO DEFECTIVE 2742); CTP synthase | 1.10 |
| 389 | 257850_at | AT3G13065 | SRF4 (STRUBBELIG-RECEPTOR FAMILY 4); ATP binding / kinase/ protein serine/threonine kinase | -5.20 |
| 390 | 257181_at | AT3G13190 | myosin heavy chain-related | 0.94 |
| 391 | 256955_at | AT3G13390 | SKS11 (SKU5 Similar 11); copper ion binding / oxidoreductase | -5.74 |
| 392 | 256966_at | AT3G13400 | SKS13 (SKU5 Similar 13); copper ion binding / oxidoreductase | -7.59 |
| 393 | 256647_at | AT3G13610 | oxidoreductase, 2OG-Fe(II) oxygenase family protein | 1.10 |
| 394 | 257610_at | AT3G13810 | ATIDD11 (ARABIDOPSIS THALIANA INDETERMINATE(ID)-DOMAIN 11); nucleic acid binding / transcription factor/ zinc ion binding | 1.08 |
| 395 | 257605_at | AT3G13840 | scarecrow transcription factor family protein | 1.50 |
| 396 | 258203_at | AT3G13950 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G13266.1); similar to Ankyrin [Medicago truncatula] (GB:ABN08906.1) | 1.70 |
| 397 | 258204_at | AT3G13960 | AtGRF5 (GROWTH-REGULATING FACTOR 5) | 0.99 |
| 398 | 256999_at | AT3G14200 | DNAJ heat shock N-terminal domain-containing protein | 1.30 |
| 399 | 257008_at | AT3G14210 | ESM1 (EPITHIOSPECIFIER MODIFIER 1); carboxylesterase | 1.33 |
| 400 | 258357_at | AT3G14350 | SRF7 (STRUBBELIG-RECEPTOR FAMILY 7); ATP binding / protein serine/threonine kinase | 0.93 |
| 401 | 257267_at | AT3G15030 | TCP4 (TCP FAMILY TRANSCRIPTION FACTOR 4); transcription factor | 1.67 |
| 402 | 257059_at | AT3G15280 | similar to unnamed protein product [Vitis vinifera] (GB:CAO66421.1) | -4.31 |
| 403 | 257057_at | AT3G15310 | transposable element gene | 1.32 |
| 404 | 258392_at | AT3G15400 | ATA20 (Arabidopsis thaliana anther 20) | -7.29 |
| 405 | 258337_at | AT3G16040 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G15270.1); contains InterPro domain Translation machinery associated TMA7 (InterPro:IPR015157) | -3.35 |
| 406 | 258338_at | AT3G16150 | L-asparaginase, putative / L-asparagine amidohydrolase, putative | 0.94 |
| 407 | 257886_at | AT3G17060 | pectinesterase family protein | -6.94 |
| 408 | 258465_at | AT3G17220 | invertase/pectin methylesterase inhibitor family protein | -4.85 |
| 409 | 258408_at | AT3G17630 | ATCHX19 (CATION/H+ EXCHANGER 19); monovalent cation:proton antiporter | -3.90 |
| 410 | 257065_at | AT3G18220 | phosphatidic acid phosphatase family protein / PAP2 family protein | -6.29 |
| 411 | 257061_at | AT3G18250 | contains domain PROKAR_LIPOPROTEIN (PS51257) | 1.31 |
| 412 | 257066_at | AT3G18280 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | 1.15 |
| 413 | 257726_at | AT3G18360 | VQ motif-containing protein | -3.82 |
| 414 | 257734_at | AT3G18370 | ATSYTF/NTMC2T3/NTMC2TYPE3/SYTF | 1.02 |
| 415 | 256827_at | AT3G18570 | glycine-rich protein / oleosin | -4.78 |
| 416 | 256945_at | AT3G19020 | leucine-rich repeat family protein / extensin family protein | -4.04 |
| 417 | 256638_at | AT3G19090 | RNA-binding protein, putative | -3.56 |
| 418 | 258016_at | AT3G19350 | polyadenylate-binding protein-related / PABP-related | 1.60 |
| 419 | 258005_at | AT3G19390 | cysteine proteinase, putative / thiol protease, putative | -6.44 |
| 420 | 257017_at | AT3G19620 | glycosyl hydrolase family 3 protein | 1.20 |
| 421 | 257129_at | AT3G20100 | CYP705A19 (cytochrome P450, family 705, subfamily A, polypeptide 19); oxygen binding | 0.95 |
| 422 | 257119_at | AT3G20190 | leucine-rich repeat transmembrane protein kinase, putative | -4.93 |
| 423 | 257130_at | AT3G20210 | DELTA-VPE (delta vacuolar processing enzyme); cysteine-type endopeptidase | -4.35 |
| 424 | 257121_at | AT3G20220 | auxin-responsive protein, putative | -5.50 |
| 425 | 257089_at | AT3G20520 | glycerophosphoryl diester phosphodiesterase family protein | -3.64 |
| 426 | 257090_at | AT3G20530 | protein kinase family protein | -4.92 |
| 427 | 257082_at | AT3G20580 | COBL10 (COBRA-LIKE PROTEIN 10 PRECURSOR) | -3.72 |
| 428 | 257986_at | AT3G20865 | AGP40 (ARABINOGALACTAN-PROTEIN 40) | -6.26 |
| 429 | 258035_at | AT3G21180 | ACA9 (autoinhibited Ca2+ -ATPase 9); calcium-transporting ATPase/ calmodulin binding | -4.70 |
| 430 | 257255_at | AT3G21960 | receptor-like protein kinase-related | -3.62 |
| 431 | 257256_at | AT3G21970 | receptor-like protein kinase-related | -5.49 |
| 432 | 258451_at | AT3G22360 | AOX1B (alternative oxidase 1B); alternative oxidase | -4.58 |
| 433 | 256933_at | AT3G22600 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | 1.81 |
| 434 | 256621_at | AT3G24450 | copper-binding family protein | 0.94 |
| 435 | 258129_at | AT3G24620 | ATROPGEF8/ROPGEF8 (KINASE PARTNER PROTEIN-LIKE); Rho guanyl-nucleotide exchange factor/ | -3.73 |
| 436 | 257100_at | AT3G25010 | disease resistance family protein | 1.85 |
| 437 | 257101_at | AT3G25020 | disease resistance family protein | 1.80 |
| 438 | 257102_at | AT3G25050 | XTH3 (XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 3); hydrolase, acting on glycosyl bonds | -7.02 |
| 439 | 257819_at | AT3G25165 | RALFL25 (RALF-LIKE 25) | -4.90 |
| 440 | 257821_at | AT3G25170 | RALFL26 (RALF-LIKE 26) | -5.17 |
| 441 | 257637_at | AT3G25810 | myrcene/ocimene synthase, putative | -4.27 |
| 442 | 258077_at | AT3G26110 | similar to BCP1 (Brassica campestris pollen protein 1) [Arabidopsis thaliana] (TAIR:AT1G24520.1); contains domain PD015210 (PD015210) | -5.23 |
| 443 | 257633_at | AT3G26125 | CYP86C2 (cytochrome P450, family 86, subfamily C, polypeptide 2); oxygen binding | -4.44 |
| 444 | 257629_at | AT3G26140 | glycosyl hydrolase family 5 protein / cellulase family protein | -3.91 |
| 445 | 257623_at | AT3G26210 | CYP71B23 (cytochrome P450, family 71, subfamily B, polypeptide 23); oxygen binding | 1.58 |
| 446 | 257628_at | AT3G26290 | CYP71B26 (cytochrome P450, family 71, subfamily B, polypeptide 26); oxygen binding | 1.71 |
| 447 | 258277_at | AT3G26830 | PAD3 (PHYTOALEXIN DEFICIENT 3); oxygen binding | 1.29 |
| 448 | 258278_at | AT3G26860 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G27331.1); contains InterPro domain Plant self-incompatibility S1 (InterPro:IPR010264) | -7.79 |
| 449 | 256980_at | AT3G26932 | DRB3 (DSRNA-BINDING PROTEIN 3); double-stranded RNA binding | 0.93 |
| 450 | 257737_at | AT3G27440 | uracil phosphoribosyltransferase, putative / UMP pyrophosphorylase, putative / UPRTase, putative | -3.19 |
| 451 | 256596_at | AT3G28540 | AAA-type ATPase family protein | 1.57 |
| 452 | 256584_at | AT3G28750 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G39880.1) | -7.96 |
| 453 | 256587_at | AT3G28780 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G28840.1); contains InterPro domain Protein of unknown function DUF1216 (InterPro:IPR009605) | -9.80 |
| 454 | 256588_at | AT3G28790 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G28830.1); contains InterPro domain Protein of unknown function DUF1216 (InterPro:IPR009605) | -9.05 |
| 455 | 256581_at | AT3G28830 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G28790.1); contains InterPro domain Protein of unknown function DUF1216 (InterPro:IPR009605) | -10.76 |
| 456 | 256582_at | AT3G28840 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G28780.1); contains InterPro domain Protein of unknown function DUF1216 (InterPro:IPR009605) | -8.55 |
| 457 | 257543_at | AT3G28960 | amino acid transporter family protein | -3.35 |
| 458 | 258058_at | AT3G28980 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G28810.1); contains InterPro domain Protein of unknown function DUF1216 (InterPro:IPR009605) | -8.01 |
| 459 | 257106_at | AT3G29060 | similar to EXS family protein / ERD1/XPR1/SYG1 family protein [Arabidopsis thaliana] (TAIR:AT2G03260.1); similar to ATP binding / ATPase, coupled to transmembrane movement of substances [Arabidopsis thaliana] (TAIR:AT1G14040.1); similar to EXS family protein / ERD1/XPR1/SYG1 family protein [Arabidopsis thaliana] (TAIR:AT2G03240.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO68453.1); similar to SPX, N-terminal; EXS, C-terminal [Medicago truncatula] (GB:ABD32751.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO23292.1); contains InterPro domain SPX, N-terminal (InterPro:IPR004331); contains InterPro domain EXS, C-terminal; (InterPro:IPR004342) | -3.35 |
| 460 | 257107_at | AT3G29070 | protein transmembrane transporter | -4.06 |
| 461 | 256736_at | AT3G29410 | terpene synthase/cyclase family protein | 2.17 |
| 462 | 252820_at | AT3G42640 | AHA8 (ARABIDOPSIS H(+)-ATPASE 8); ATPase | -3.53 |
| 463 | 252769_at | AT3G42850 | galactokinase, putative | -4.05 |
| 464 | 252733_at | AT3G43120 | auxin-responsive protein-related | -4.50 |
| 465 | 252698_at | AT3G43670 | copper amine oxidase, putative | 1.31 |
| 466 | 252712_at | AT3G43800 | ATGSTU27 (Arabidopsis thaliana Glutathione S-transferase (class tau) 27); glutathione transferase | 0.96 |
| 467 | 252710_at | AT3G43860 | ATGH9A4 (ARABIDOPSIS THALIANA GLYCOSYL HYDROLASE 9A4); hydrolase, hydrolyzing O-glycosyl compounds | -3.86 |
| 468 | 252592_at | AT3G45640 | ATMPK3 (MITOGEN-ACTIVATED PROTEIN KINASE 3); MAP kinase/ kinase/ protein kinase | 0.98 |
| 469 | 252549_at | AT3G45860 | receptor-like protein kinase, putative | 1.73 |
| 470 | 252531_at | AT3G46520 | ACT12 (ACTIN-12); structural constituent of cytoskeleton | -7.16 |
| 471 | 252478_at | AT3G46540 | epsin N-terminal homology (ENTH) domain-containing protein / clathrin assembly protein-related | 1.01 |
| 472 | 252493_at | AT3G46750 | similar to hypothetical protein [Vitis vinifera] (GB:CAN62428.1) | -3.65 |
| 473 | 252468_at | AT3G46970 | ATPHS2/PHS2 (ALPHA-GLUCAN PHOSPHORYLASE 2); phosphorylase/ transferase, transferring glycosyl groups | 1.03 |
| 474 | 252415_at | AT3G47340 | ASN1 (DARK INDUCIBLE 6) | -3.24 |
| 475 | 252440_at | AT3G47440 | TIP5;1 (tonoplast intrinsic protein 5;1); water channel | -4.98 |
| 476 | 252417_at | AT3G47480 | calcium-binding EF hand family protein | 1.86 |
| 477 | 252429_at | AT3G47500 | CDF3 (CYCLING DOF FACTOR 3); DNA binding / protein binding / transcription factor | 1.18 |
| 478 | 252395_at | AT3G47950 | AHA4 (Arabidopsis H(+)-ATPase 4); ATPase | 1.13 |
| 479 | 252373_at | AT3G48090 | EDS1 (ENHANCED DISEASE SUSCEPTIBILITY 1); signal transducer/ triacylglycerol lipase | 1.16 |
| 480 | 252345_at | AT3G48640 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G66670.2); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G66670.1) | 1.52 |
| 481 | 252346_at | AT3G48650 | pseudogene, At14a-related protein, similar to At14a (GI:11994571 and GI:11994573) (Arabidopsis thaliana) | 1.74 |
| 482 | 252296_at | AT3G48970 | copper-binding family protein | 0.97 |
| 483 | 252280_at | AT3G49260 | IQD21 (IQ-DOMAIN 21, IQ-domain 21); calmodulin binding | 1.01 |
| 484 | 252309_at | AT3G49340 | cysteine proteinase, putative | 1.72 |
| 485 | 252227_at | AT3G49900 | BTB/POZ domain-containing protein | 1.13 |
| 486 | 252205_at | AT3G50350 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G33985.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN61779.1); contains InterPro domain Protein of unknown function DUF1685 (InterPro:IPR012881) | 1.16 |
| 487 | 252175_at | AT3G50700 | ATIDD2 (ARABIDOPSIS THALIANA INDETERMINATE(ID)-DOMAIN 2); nucleic acid binding / transcription factor/ zinc ion binding | 0.99 |
| 488 | 252131_at | AT3G50930 | AAA-type ATPase family protein | 1.36 |
| 489 | 252140_at | AT3G51070 | dehydration-responsive protein-related | -4.05 |
| 490 | 252103_at | AT3G51410 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G35680.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO23714.1); contains InterPro domain Protein of unknown function DUF241, plant (InterPro:IPR004320) | 1.36 |
| 491 | 252063_at | AT3G51590 | LTP12 (LIPID TRANSFER PROTEIN 12); lipid binding | -8.24 |
| 492 | 252037_at | AT3G51920 | CAM9 (CALMODULIN 9); calcium ion binding | 0.95 |
| 493 | 256676_at | AT3G52180 | ATPTPKIS1/DSP4/SEX4 (STARCH-EXCESS 4); polysaccharide binding / protein tyrosine/serine/threonine phosphatase | 0.97 |
| 494 | 252060_at | AT3G52430 | PAD4 (PHYTOALEXIN DEFICIENT 4); triacylglycerol lipase | 1.41 |
| 495 | 252052_at | AT3G52600 | ATCWINV2 (ARABIDOPSIS THALIANA CELL WALL INVERTASE 2); hydrolase, hydrolyzing O-glycosyl compounds | -5.50 |
| 496 | 252061_at | AT3G52620 | unknown protein | -6.01 |
| 497 | 252004_at | AT3G52780 | ATPAP20/PAP20; acid phosphatase/ protein serine/threonine phosphatase | -4.41 |
| 498 | 252005_at | AT3G52810 | ATPAP21/PAP21 (purple acid phosphatase 21); acid phosphatase/ protein serine/threonine phosphatase | -7.28 |
| 499 | 251981_at | AT3G53080 | galactose-binding lectin family protein | -5.42 |
| 500 | 251944_at | AT3G53510 | ABC transporter family protein | -3.75 |
| 501 | 251928_at | AT3G53980 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | 2.33 |
| 502 | 251892_at | AT3G54320 | WRI1 (WRINKLED 1); DNA binding / transcription factor | 0.93 |
| 503 | 251898_at | AT3G54340 | AP3 (APETALA 3); DNA binding / transcription factor | 0.95 |
| 504 | 251854_at | AT3G54800 | pleckstrin homology (PH) domain-containing protein / lipid-binding START domain-containing protein | -4.50 |
| 505 | 251839_at | AT3G54950 | PLA IIIA/PLP7 (Patatin-like protein 7) | 1.06 |
| 506 | 251825_at | AT3G55100 | ABC transporter family protein | -4.31 |
| 507 | 251753_at | AT3G55760 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G42430.1); similar to hypothetical protein [Trifolium pratense] (GB:BAE71279.1) | 0.94 |
| 508 | 251725_at | AT3G56260 | unknown protein | 1.31 |
| 509 | 251705_at | AT3G56400 | WRKY70 (WRKY DNA-binding protein 70); transcription factor | 1.54 |
| 510 | 246293_at | AT3G56710 | SIB1 (SIGMA FACTOR BINDING PROTEIN 1); binding | 1.41 |
| 511 | 251625_at | AT3G57260 | BGL2 (PATHOGENESIS-RELATED PROTEIN 2); glucan 1,3-beta-glucosidase/ hydrolase, hydrolyzing O-glycosyl compounds | 2.75 |
| 512 | 251633_at | AT3G57460 | catalytic/ metal ion binding / metalloendopeptidase/ zinc ion binding | 1.08 |
| 513 | 251590_at | AT3G57690 | AGP23/ATAGP23 (ARABINOGALACTAN-PROTEIN 23) | -5.40 |
| 514 | 251560_at | AT3G57920 | squamosa promoter-binding protein, putative | 1.23 |
| 515 | 251497_at | AT3G59060 | PIL6 (PHYTOCHROME-INTERACTING FACTOR 5); DNA binding / transcription factor | 1.27 |
| 516 | 251400_at | AT3G60420 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G60450.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO70569.1); contains InterPro domain Phosphoglycerate mutase (InterPro:IPR013078); contains InterPro domain PRIB5 (InterPro:IPR012398) | 1.11 |
| 517 | 251339_at | AT3G60780 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G45360.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO24498.1); contains InterPro domain Protein of unknown function DUF1442 (InterPro:IPR009902) | -6.28 |
| 518 | 251358_at | AT3G61160 | shaggy-related protein kinase beta / ASK-beta (ASK2) | -3.68 |
| 519 | 251361_at | AT3G61230 | LIM domain-containing protein | -5.22 |
| 520 | 251319_at | AT3G61610 | aldose 1-epimerase family protein | 1.16 |
| 521 | 251299_at | AT3G61950 | basic helix-loop-helix (bHLH) family protein | 1.18 |
| 522 | 251245_at | AT3G62090 | PIL2 (PHYTOCHROME INTERACTING FACTOR 3-LIKE 2); transcription factor | 1.36 |
| 523 | 251258_at | AT3G62170 | VGDH2 (VANGUARD 1 HOMOLOG 2); pectinesterase | -7.57 |
| 524 | 251250_at | AT3G62180 | invertase/pectin methylesterase inhibitor family protein | -5.86 |
| 525 | 251252_at | AT3G62230 | F-box family protein | -8.68 |
| 526 | 251223_at | AT3G62610 | AtMYB11 (myb domain protein 11); DNA binding / transcription factor | 1.42 |
| 527 | 251180_at | AT3G62640 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G47480.1); similar to unknown [Picea sitchensis] (GB:ABK21328.1) | -4.13 |
| 528 | 251228_at | AT3G62710 | glycosyl hydrolase family 3 protein | -4.21 |
| 529 | 251169_at | AT3G63210 | MARD1 (MEDIATOR OF ABA-REGULATED DORMANCY 1) | 1.05 |
| 530 | 255717_at | AT4G00350 | MATE efflux family protein | -4.74 |
| 531 | 255626_at | AT4G00780 | meprin and TRAF homology domain-containing protein / MATH domain-containing protein | 0.98 |
| 532 | 255644_at | AT4G00870 | basic helix-loop-helix (bHLH) family protein | 0.99 |
| 533 | 255607_at | AT4G01130 | acetylesterase, putative | 1.82 |
| 534 | 255576_at | AT4G01440 | nodulin MtN21 family protein | 0.96 |
| 535 | 255580_at | AT4G01470 | GAMMA-TIP3/TIP1;3 (tonoplast intrinsic protein 1;3); water channel | -4.28 |
| 536 | 255530_at | AT4G02140 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G02700.1) | -5.27 |
| 537 | 255515_at | AT4G02250 | invertase/pectin methylesterase inhibitor family protein | -7.11 |
| 538 | 255517_at | AT4G02290 | ATGH9B13 (ARABIDOPSIS THALIANA GLYCOSYL HYDROLASE 9B13); hydrolase, hydrolyzing O-glycosyl compounds | 1.08 |
| 539 | 255437_at | AT4G03060 | AOP2 (ALKENYL HYDROXALKYL PRODUCING 2); oxidoreductase, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors | 1.67 |
| 540 | 255423_at | AT4G03290 | calcium-binding protein, putative | -5.74 |
| 541 | 255386_at | AT4G03620 | myosin heavy chain-related | -4.93 |
| 542 | 255345_at | AT4G04460 | aspartyl protease family protein | -6.65 |
| 543 | 255341_at | AT4G04500 | protein kinase family protein | 1.79 |
| 544 | 255285_at | AT4G04630 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G21970.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO17943.1); contains InterPro domain Protein of unknown function DUF584 (InterPro:IPR007608) | 0.96 |
| 545 | 255294_at | AT4G04750 | carbohydrate transmembrane transporter/ sugar:hydrogen ion symporter | 1.48 |
| 546 | 255276_at | AT4G04930 | DES-1-LIKE (fatty acid desaturase 1-like); oxidoreductase | -4.52 |
| 547 | 255101_at | AT4G08670 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | -7.13 |
| 548 | 255070_at | AT4G09020 | ATISA3/ISA3 (ISOAMYLASE 3); alpha-amylase | 1.14 |
| 549 | 254998_at | AT4G09760 | choline kinase, putative | 1.41 |
| 550 | 255805_at | AT4G10240 | zinc finger (B-box type) family protein | 0.96 |
| 551 | 254996_at | AT4G10390 | protein kinase family protein | 2.03 |
| 552 | 254972_at | AT4G10440 | dehydration-responsive family protein | -3.94 |
| 553 | 254975_at | AT4G10500 | oxidoreductase, 2OG-Fe(II) oxygenase family protein | 1.38 |
| 554 | 254956_at | AT4G10850 | nodulin MtN3 family protein | -4.68 |
| 555 | 254961_at | AT4G11030 | long-chain-fatty-acid--CoA ligase, putative / long-chain acyl-CoA synthetase, putative | -4.62 |
| 556 | 254886_at | AT4G11760 | LCR17 (Low-molecular-weight cysteine-rich 17) | -9.81 |
| 557 | 254869_at | AT4G11890 | protein kinase family protein | 1.16 |
| 558 | 254809_at | AT4G12410 | auxin-responsive family protein | -4.69 |
| 559 | 254786_at | AT4G12890 | gamma interferon responsive lysosomal thiol reductase family protein / GILT family protein | -4.35 |
| 560 | 254777_at | AT4G12960 | gamma interferon responsive lysosomal thiol reductase family protein / GILT family protein | -4.87 |
| 561 | 254754_at | AT4G13210 | pectate lyase family protein | 1.07 |
| 562 | 254762_at | AT4G13230 | late embryogenesis abundant domain-containing protein / LEA domain-containing protein | -6.57 |
| 563 | 254716_at | AT4G13560 | UNE15 (unfertilized embryo sac 15) | -9.46 |
| 564 | 254689_at | AT4G13710 | pectate lyase family protein | 0.98 |
| 565 | 245329_at | AT4G14365 | zinc finger (C3HC4-type RING finger) family protein / ankyrin repeat family protein | 1.94 |
| 566 | 245609_at | AT4G14370 | disease resistance protein (TIR-NBS-LRR class), putative | -3.25 |
| 567 | 245265_at | AT4G14400 | ACD6 (ACCELERATED CELL DEATH 6); protein binding | 1.38 |
| 568 | 245577_at | AT4G14780 | protein kinase, putative | -3.61 |
| 569 | 245322_at | AT4G14815 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | -4.42 |
| 570 | 245501_at | AT4G15620 | integral membrane family protein | 1.41 |
| 571 | 245503_at | AT4G15650 | protein kinase-related | -4.31 |
| 572 | 245335_at | AT4G16160 | ATOEP16-2/ATOEP16-S; P-P-bond-hydrolysis-driven protein transmembrane transporter | -5.10 |
| 573 | 245488_at | AT4G16270 | peroxidase 40 (PER40) (P40) | 2.22 |
| 574 | 245465_at | AT4G16590 | ATCSLA01 (Cellulose synthase-like A1); glucosyltransferase/ transferase, transferring glycosyl groups | 1.24 |
| 575 | 245307_at | AT4G16770 | oxidoreductase, 2OG-Fe(II) oxygenase family protein | 0.97 |
| 576 | 245376_at | AT4G17690 | peroxidase, putative | -3.22 |
| 577 | 254704_at | AT4G18020 | APRR2 (PSEUDO-RESPONSE REGULATOR 2); transcription factor | 0.98 |
| 578 | 254627_at | AT4G18395 | unknown protein | -4.23 |
| 579 | 254572_at | AT4G19380 | alcohol oxidase-related | 1.93 |
| 580 | 254550_at | AT4G19690 | IRT1 (IRON-REGULATED TRANSPORTER 1); cadmium ion transmembrane transporter/ iron ion transmembrane transporter/ manganese ion transmembrane transporter/ zinc ion transmembrane transporter | -3.70 |
| 581 | 254494_at | AT4G20050 | QRT3 (QUARTET 3) | -5.94 |
| 582 | 254490_at | AT4G20320 | CTP synthase | 0.97 |
| 583 | 254468_at | AT4G20460 | NAD-dependent epimerase/dehydratase family protein | -3.12 |
| 584 | 254416_at | AT4G21380 | ARK3 (Arabidopsis Receptor Kinase 3); kinase | 1.35 |
| 585 | 254394_at | AT4G21630 | subtilase family protein | -4.68 |
| 586 | 254386_at | AT4G21960 | PRXR1 (peroxidase 42); peroxidase | 0.93 |
| 587 | 254271_at | AT4G23150 | protein kinase family protein | 2.62 |
| 588 | 254255_at | AT4G23220 | protein kinase family protein | 1.17 |
| 589 | 254231_at | AT4G23810 | WRKY53 (WRKY DNA-binding protein 53); DNA binding / protein binding / transcription activator/ transcription factor | 1.01 |
| 590 | 254178_at | AT4G23880 | unknown protein | 1.11 |
| 591 | 254202_at | AT4G24140 | hydrolase, alpha/beta fold family protein | 1.18 |
| 592 | 254123_at | AT4G24640 | APPB1; pectinesterase inhibitor | -4.69 |
| 593 | 254112_at | AT4G24970 | ATP-binding region, ATPase-like domain-containing protein | 1.15 |
| 594 | 254101_at | AT4G25000 | AMY1/ATAMY1 (ALPHA-AMYLASE-LIKE); alpha-amylase | 1.11 |
| 595 | 254090_at | AT4G25010 | nodulin MtN3 family protein | -3.38 |
| 596 | 254104_at | AT4G25040 | integral membrane family protein | -6.28 |
| 597 | 245232_at | AT4G25590 | ADF7 (ACTIN DEPOLYMERIZING FACTOR 7); actin binding | -8.21 |
| 598 | 254024_at | AT4G25780 | pathogenesis-related protein, putative | -3.59 |
| 599 | 254045_at | AT4G25880 | APUM6 (ARABIDOPSIS PUMILIO 6); RNA binding | 0.98 |
| 600 | 254033_at | AT4G25950 | VATG3 (VACUOLAR ATP SYNTHASE G3) | -5.94 |
| 601 | 254043_at | AT4G25990 | CIL | 1.10 |
| 602 | 253993_at | AT4G26070 | MEK1 (mitogen-activated protein kinase kinase 1); MAP kinase kinase/ kinase | 0.93 |
| 603 | 254001_at | AT4G26260 | MIOX4 (MYO-INOSITOL OXYGENASE 4) | -3.52 |
| 604 | 253934_at | AT4G26830 | hydrolase, hydrolyzing O-glycosyl compounds | -3.50 |
| 605 | 253831_at | AT4G27580 | unknown protein | -5.26 |
| 606 | 253846_at | AT4G28000 | AAA-type ATPase family protein | -4.75 |
| 607 | 253850_at | AT4G28090 | SKS10 (SKU5 Similar 10); copper ion binding / oxidoreductase | -4.24 |
| 608 | 253815_at | AT4G28250 | ATEXPB3 (ARABIDOPSIS THALIANA EXPANSIN B3) | 1.29 |
| 609 | 253807_at | AT4G28280 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G20700.1); similar to hypothetical protein OsJ_007917 [Oryza sativa (japonica cultivar-group)] (GB:EAZ24434.1) | -3.79 |
| 610 | 253779_at | AT4G28490 | HAESA (RECEPTOR-LIKE PROTEIN KINASE 5); ATP binding / kinase/ protein serine/threonine kinase | 1.11 |
| 611 | 253736_at | AT4G28780 | GDSL-motif lipase/hydrolase family protein | 1.25 |
| 612 | 253749_at | AT4G29080 | PAP2 (PHYTOCHROME-ASSOCIATED PROTEIN 2); transcription factor | 1.14 |
| 613 | 253722_at | AT4G29190 | zinc finger (CCCH-type) family protein | 1.07 |
| 614 | 253725_at | AT4G29340 | PRF4 (PROFILIN 4); actin binding | -6.73 |
| 615 | 253650_at | AT4G30020 | subtilase family protein | 0.94 |
| 616 | 253661_at | AT4G30130 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G19090.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN72045.1); contains InterPro domain Protein of unknown function DUF632 (InterPro:IPR006867); contains InterPro domain Protein of unknown function DUF630 (InterPro:IPR006868) | 1.10 |
| 617 | 253660_at | AT4G30140 | GDSL-motif lipase/hydrolase family protein | -5.60 |
| 618 | 253614_at | AT4G30350 | heat shock protein-related | 0.99 |
| 619 | 253627_at | AT4G30650 | hydrophobic protein, putative / low temperature and salt responsive protein, putative | 1.86 |
| 620 | 253581_at | AT4G30660 | hydrophobic protein, putative / low temperature and salt responsive protein, putative | 1.35 |
| 621 | 253525_at | AT4G31330 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G10580.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN79714.1); contains InterPro domain Protein of unknown function DUF599 (InterPro:IPR006747) | -3.60 |
| 622 | 253358_at | AT4G32940 | GAMMA-VPE (Vacuolar processing enzyme gamma); cysteine-type endopeptidase | 0.93 |
| 623 | 253414_at | AT4G33050 | EDA39 (embryo sac development arrest 39); calmodulin binding | 1.17 |
| 624 | 253346_at | AT4G33600 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G33590.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN80154.1) | -4.91 |
| 625 | 253285_at | AT4G34250 | fatty acid elongase, putative | -4.09 |
| 626 | 253258_at | AT4G34400 | DNA binding / transcription factor | 1.14 |
| 627 | 253240_at | AT4G34510 | KCS2 (3-ketoacyl-CoA synthase 2); acyltransferase | -5.35 |
| 628 | 253215_at | AT4G34950 | nodulin family protein | 1.13 |
| 629 | 253226_at | AT4G35010 | BGAL11 (beta-galactosidase 11); beta-galactosidase | -9.03 |
| 630 | 253182_at | AT4G35190 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G37210.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO47480.1); contains InterPro domain Conserved hypothetical protein CHP00730 (InterPro:IPR005269) | 2.12 |
| 631 | 253147_at | AT4G35600 | CONNEXIN 32; kinase | 0.97 |
| 632 | 253151_at | AT4G35670 | glycoside hydrolase family 28 protein / polygalacturonase (pectinase) family protein | -7.79 |
| 633 | 253153_at | AT4G35700 | zinc finger (C2H2 type) family protein | -5.00 |
| 634 | 246208_at | AT4G36490 | SEC14 cytosolic factor, putative / phosphoglyceride transfer protein, putative | -4.83 |
| 635 | 246242_at | AT4G36600 | late embryogenesis abundant domain-containing protein / LEA domain-containing protein | -3.79 |
| 636 | 246217_at | AT4G36920 | AP2 (APETALA 2); transcription factor | 1.07 |
| 637 | 253069_at | AT4G37840 | hexokinase, putative | -3.67 |
| 638 | 253012_at | AT4G37900 | glycine-rich protein | -4.43 |
| 639 | 252916_at | AT4G38950 | kinesin motor family protein | 0.96 |
| 640 | 252924_at | AT4G39070 | zinc finger (B-box type) family protein | 1.03 |
| 641 | 252933_at | AT4G39110 | protein kinase family protein | -3.25 |
| 642 | 252862_at | AT4G39830 | L-ascorbate oxidase, putative | 0.96 |
| 643 | 251132_at | AT5G01200 | myb family transcription factor | 1.17 |
| 644 | 251065_at | AT5G01870 | lipid transfer protein, putative | 2.52 |
| 645 | 251066_at | AT5G01880 | zinc finger (C3HC4-type RING finger) family protein | 1.58 |
| 646 | 250997_at | AT5G02570 | histone H2B, putative | -3.34 |
| 647 | 250971_at | AT5G02810 | PRR7 (PSEUDO-RESPONSE REGULATOR 7); transcription regulator | 0.98 |
| 648 | 250942_at | AT5G03350 | legume lectin family protein | 1.49 |
| 649 | 250904_at | AT5G03620 | subtilase family protein | -4.50 |
| 650 | 245700_at | AT5G04180 | carbonic anhydrase family protein | -4.24 |
| 651 | 250798_at | AT5G05340 | peroxidase, putative | 1.14 |
| 652 | 250744_at | AT5G05840 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G55720.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO69043.1); contains InterPro domain Protein of unknown function DUF620 (InterPro:IPR006873) | 0.98 |
| 653 | 250648_at | AT5G06760 | late embryogenesis abundant group 1 domain-containing protein / LEA group 1 domain-containing protein | -3.39 |
| 654 | 250613_at | AT5G07240 | IQD24 (IQ-domain 24); calmodulin binding | 1.08 |
| 655 | 250608_at | AT5G07420 | pectinesterase family protein | -9.41 |
| 656 | 250631_at | AT5G07430 | pectinesterase family protein | -7.64 |
| 657 | 250635_at | AT5G07510 | GRP14 (Glycine rich protein 14); nutrient reservoir | -6.44 |
| 658 | 250636_at | AT5G07520 | GRP18 (Glycine rich protein 18); nutrient reservoir | -6.29 |
| 659 | 250637_at | AT5G07530 | GRP17 (Glycine rich protein 17) | -7.35 |
| 660 | 250638_at | AT5G07540 | GRP16 (Glycine rich protein 16); nutrient reservoir | -6.38 |
| 661 | 250610_at | AT5G07550 | GRP19 (Glycine rich protein 19) | -9.09 |
| 662 | 250639_at | AT5G07560 | GRP20 (Glycine rich protein 20); nutrient reservoir | -8.76 |
| 663 | 250597_at | AT5G07680 | ANAC079/ANAC080/ATNAC4 (Arabidopsis NAC domain containing protein 79, Arabidopsis NAC domain containing protein 80); transcription factor | -3.19 |
| 664 | 250535_at | AT5G08480 | VQ motif-containing protein | -3.20 |
| 665 | 245883_at | AT5G09500 | 40S ribosomal protein S15 (RPS15C) | -3.19 |
| 666 | 250514_at | AT5G09550 | RAB GDP-dissociation inhibitor | -5.57 |
| 667 | 250515_at | AT5G09570 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G64400.1); similar to unknown [Populus trichocarpa] (GB:ABK94558.1); contains InterPro domain CHCH (InterPro:IPR010625) | -3.44 |
| 668 | 250499_at | AT5G09730 | BXL3 (BETA-XYLOSIDASE 3); hydrolase, hydrolyzing O-glycosyl compounds | -3.11 |
| 669 | 250481_at | AT5G10310 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G37810.1); similar to 80A08_28 [Brassica rapa subsp. pekinensis] (GB:AAZ67613.1) | 1.02 |
| 670 | 250482_at | AT5G10320 | similar to 80A08_29 [Brassica rapa subsp. pekinensis] (GB:AAZ67614.1) | 1.14 |
| 671 | 250445_at | AT5G10760 | aspartyl protease family protein | 2.68 |
| 672 | 245901_at | AT5G11060 | KNAT4 (KNOTTED1-LIKE HOMEOBOX GENE 4); transcription factor | 1.19 |
| 673 | 250349_at | AT5G12000 | kinase | -3.52 |
| 674 | 250304_at | AT5G12110 | elongation factor 1B alpha-subunit 1 (eEF1Balpha1) | 1.22 |
| 675 | 245979_at | AT5G13150 | ATEXO70C1 (exocyst subunit EXO70 family protein C1); protein binding | -4.42 |
| 676 | 250204_at | AT5G13990 | ATEXO70C2 (exocyst subunit EXO70 family protein C2); protein binding | -4.72 |
| 677 | 250201_at | AT5G14230 | ankyrin repeat family protein | 1.23 |
| 678 | 250174_at | AT5G14380 | AGP6 (ARABINOGALACTAN PROTEINS 6) | -6.83 |
| 679 | 246592_at | AT5G14890 | NHL repeat-containing protein | -3.88 |
| 680 | 246545_at | AT5G15110 | pectate lyase family protein | -5.76 |
| 681 | 250154_at | AT5G15140 | aldose 1-epimerase family protein | -3.78 |
| 682 | 250155_at | AT5G15160 | bHLH family protein | 1.57 |
| 683 | 246419_at | AT5G17030 | UDP-glucoronosyl/UDP-glucosyl transferase family protein | 1.12 |
| 684 | 250083_at | AT5G17220 | ATGSTF12 (GLUTATHIONE S-TRANSFERASE 26); glutathione transferase | 1.03 |
| 685 | 246431_at | AT5G17480 | polcalcin, putative / calcium-binding pollen allergen, putative | -8.64 |
| 686 | 250045_at | AT5G17700 | MATE efflux family protein | 0.97 |
| 687 | 250062_at | AT5G17760 | AAA-type ATPase family protein | 1.30 |
| 688 | 249983_at | AT5G18470 | curculin-like (mannose-binding) lectin family protein | 1.25 |
| 689 | 249950_at | AT5G18910 | protein kinase family protein | -3.95 |
| 690 | 245946_at | AT5G19580 | glyoxal oxidase-related | -8.12 |
| 691 | 245958_at | AT5G19610 | sec7 domain-containing protein | -5.57 |
| 692 | 245993_at | AT5G20700 | senescence-associated protein-related | 1.05 |
| 693 | 246001_at | AT5G20790 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G43110.1) | -4.70 |
| 694 | 249895_at | AT5G22500 | acyl CoA reductase, putative / male-sterility protein, putative | 0.98 |
| 695 | 249911_at | AT5G22740 | ATCSLA02 (Cellulose synthase-like A2); transferase, transferring glycosyl groups | 1.08 |
| 696 | 249850_at | AT5G23240 | DNAJ heat shock N-terminal domain-containing protein | 1.35 |
| 697 | 249760_at | AT5G23960 | terpene synthase/cyclase family protein | -5.19 |
| 698 | 249777_at | AT5G24210 | lipase class 3 family protein | 1.70 |
| 699 | 249780_at | AT5G24240 | phosphatidylinositol 3- and 4-kinase family protein / ubiquitin family protein | -4.92 |
| 700 | 249732_at | AT5G24420 | glucosamine/galactosamine-6-phosphate isomerase-related | -3.29 |
| 701 | 249754_at | AT5G24530 | oxidoreductase, 2OG-Fe(II) oxygenase family protein | 1.18 |
| 702 | 249750_at | AT5G24570 | unknown protein | 1.03 |
| 703 | 246939_at | AT5G25390 | SHN2 (SHINE2); DNA binding / transcription factor | 1.00 |
| 704 | 246896_at | AT5G25550 | leucine-rich repeat family protein / extensin family protein | -3.64 |
| 705 | 246878_at | AT5G26060 | S1 self-incompatibility protein-related | -6.98 |
| 706 | 246885_at | AT5G26230 | similar to hypothetical protein [Vitis vinifera] (GB:CAN75978.1) | 1.17 |
| 707 | 246841_at | AT5G26700 | germin-like protein, putative | -6.29 |
| 708 | 246797_at | AT5G26790 | unknown protein | 1.07 |
| 709 | 246849_at | AT5G26850 | binding | 1.03 |
| 710 | 246821_at | AT5G26920 | calmodulin binding | 1.87 |
| 711 | 246751_at | AT5G27870 | pectinesterase family protein | -4.46 |
| 712 | 246761_at | AT5G27980 | seed maturation family protein | -4.88 |
| 713 | 245861_at | AT5G28300 | trihelix DNA-binding protein, putative | 1.44 |
| 714 | 246106_at | AT5G28680 | protein kinase family protein | -4.30 |
| 715 | 246646_at | AT5G35090 | unknown protein | -5.88 |
| 716 | 249709_at | AT5G35670 | IQD33 (IQ-domain 33); calmodulin binding | 1.01 |
| 717 | 249688_at | AT5G36160 | aminotransferase-related | 1.01 |
| 718 | 246616_at | AT5G36260 | aspartyl protease family protein | -5.02 |
| 719 | 249619_at | AT5G37500 | GORK (GATED OUTWARDLY-RECTIFYING K+ CHANNEL); cyclic nucleotide binding / inward rectifier potassium channel/ outward rectifier potassium channel | 0.99 |
| 720 | 249626_at | AT5G37540 | aspartyl protease family protein | 1.15 |
| 721 | 249581_at | AT5G37600 | ATGSR1 (Arabidopsis thaliana glutamine synthase clone R1); glutamate-ammonia ligase | 1.17 |
| 722 | 249540_at | AT5G38120 | 4-coumarate--CoA ligase family protein / 4-coumaroyl-CoA synthase family protein | 1.12 |
| 723 | 249515_at | AT5G38530 | tryptophan synthase-related | 1.57 |
| 724 | 249536_at | AT5G38760 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G53820.1); similar to ABA-inducible protein-like protein [Brassica rapa] (GB:ABV89610.1); contains domain PTHR23241:SF11 (PTHR23241:SF11); contains domain PTHR23241 (PTHR23241) | -11.64 |
| 725 | 249497_at | AT5G39220 | hydrolase, alpha/beta fold family protein | 1.25 |
| 726 | 249503_at | AT5G39310 | ATEXPA24 (ARABIDOPSIS THALIANA EXPANSIN A24) | -4.26 |
| 727 | 249447_at | AT5G39400 | pollen specific phosphatase, putative / phosphatase and tensin, putative (PTEN1) | -3.64 |
| 728 | 249417_at | AT5G39670 | calcium-binding EF hand family protein | 1.34 |
| 729 | 249429_at | AT5G39880 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G28750.1) | -5.91 |
| 730 | 249381_at | AT5G40040 | 60S acidic ribosomal protein P2 (RPP2E) | -5.66 |
| 731 | 249401_at | AT5G40260 | nodulin MtN3 family protein | -4.08 |
| 732 | 249375_at | AT5G40730 | AGP24 (ARABINOGALACTAN PROTEIN 24) | -4.01 |
| 733 | 249346_at | AT5G40780 | LHT1 (LYSINE HISTIDINE TRANSPORTER 1); amino acid transmembrane transporter | 0.95 |
| 734 | 249288_at | AT5G41050 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G26960.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO49052.1) | 1.06 |
| 735 | 249336_at | AT5G41070 | DRB5 (DSRNA-BINDING PROTEIN 5); double-stranded RNA binding | 1.12 |
| 736 | 249303_at | AT5G41460 | fringe-related protein | 1.07 |
| 737 | 249279_at | AT5G41920 | scarecrow transcription factor family protein | 0.99 |
| 738 | 249226_at | AT5G42170 | carboxylesterase | -5.76 |
| 739 | 249250_at | AT5G42340 | binding / ubiquitin-protein ligase | -3.14 |
| 740 | 249128_at | AT5G43440 | 2-oxoglutarate-dependent dioxygenase, putative | 1.41 |
| 741 | 249087_at | AT5G44210 | ATERF-9/ATERF9/ERF9 (ERF domain protein 9); DNA binding / transcription factor/ transcription repressor | 1.14 |
| 742 | 249048_at | AT5G44300 | dormancy/auxin associated family protein | -6.43 |
| 743 | 249046_at | AT5G44400 | FAD-binding domain-containing protein | -3.25 |
| 744 | 248970_at | AT5G45380 | sodium:solute symporter family protein | 0.95 |
| 745 | 248961_at | AT5G45650 | subtilase family protein | 1.03 |
| 746 | 248926_at | AT5G45880 | pollen Ole e 1 allergen and extensin family protein | -9.33 |
| 747 | 248895_at | AT5G46330 | FLS2 (FLAGELLIN-SENSITIVE 2); ATP binding / kinase/ protein binding / protein serine/threonine kinase/ transmembrane receptor protein serine/threonine kinase | 1.01 |
| 748 | 248845_at | AT5G46470 | disease resistance protein (TIR-NBS-LRR class), putative | 0.95 |
| 749 | 248840_at | AT5G46770 | unknown protein | -4.90 |
| 750 | 248824_at | AT5G46940 | invertase/pectin methylesterase inhibitor family protein | -4.67 |
| 751 | 248822_at | AT5G47000 | peroxidase, putative | -6.39 |
| 752 | 248714_at | AT5G48140 | polygalacturonase, putative / pectinase, putative | -5.86 |
| 753 | 248741_at | AT5G48170 | SLY2 (SLEEPY2) | 1.19 |
| 754 | 248716_at | AT5G48210 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G42565.1) | -3.16 |
| 755 | 248593_at | AT5G49180 | pectinesterase family protein | -4.12 |
| 756 | 248596_at | AT5G49330 | AtMYB111 (myb domain protein 111); DNA binding / transcription factor | 1.27 |
| 757 | 248619_at | AT5G49630 | AAP6 (AMINO ACID PERMEASE 6); amino acid transmembrane transporter | 0.96 |
| 758 | 248534_at | AT5G50030 | invertase/pectin methylesterase inhibitor family protein | -4.96 |
| 759 | 248527_at | AT5G50740 | metal ion binding | 1.08 |
| 760 | 248470_at | AT5G50830 | unknown protein | -5.46 |
| 761 | 248484_at | AT5G51030 | short-chain dehydrogenase/reductase (SDR) family protein | -3.69 |
| 762 | 248355_at | AT5G52340 | ATEXO70A2 (exocyst subunit EXO70 family protein A2); protein binding | -5.21 |
| 763 | 248367_at | AT5G52360 | actin-depolymerizing factor, putative | -7.27 |
| 764 | 248333_at | AT5G52390 | photoassimilate-responsive protein, putative | 2.05 |
| 765 | 248327_at | AT5G52750 | heavy-metal-associated domain-containing protein | 1.34 |
| 766 | 248322_at | AT5G52760 | heavy-metal-associated domain-containing protein | 1.66 |
| 767 | 248245_at | AT5G53190 | nodulin MtN3 family protein | -3.95 |
| 768 | 248213_at | AT5G53660 | AtGRF7 (GROWTH-REGULATING FACTOR 7) | 1.75 |
| 769 | 248227_at | AT5G53820 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G38760.1); similar to pollen coat protein [Brassica oleracea] (GB:CAA63531.1); contains domain PTHR23241:SF11 (PTHR23241:SF11); contains domain PTHR23241 (PTHR23241) | -7.23 |
| 770 | 248194_at | AT5G54095 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G27580.1) | -7.61 |
| 771 | 248200_at | AT5G54160 | ATOMT1 (O-METHYLTRANSFERASE 1) | 0.94 |
| 772 | 248164_at | AT5G54490 | PBP1 (PINOID-BINDING PROTEIN 1); calcium ion binding | 1.09 |
| 773 | 248104_at | AT5G55250 | IAMT1 (IAA CARBOXYLMETHYLTRANSFERASE 1); S-adenosylmethionine-dependent methyltransferase | 1.34 |
| 774 | 248062_at | AT5G55450 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | 2.48 |
| 775 | 248028_at | AT5G55620 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G09950.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO39940.1) | 0.96 |
| 776 | 248073_at | AT5G55720 | pectate lyase family protein | 0.97 |
| 777 | 248003_at | AT5G56220 | nucleoside-triphosphatase/ nucleotide binding | 1.33 |
| 778 | 247906_at | AT5G57420 | IAA33 (indoleacetic acid-induced protein 33); transcription factor | 1.12 |
| 779 | 247897_at | AT5G57810 | TET15 (TETRASPANIN15) | -6.95 |
| 780 | 247843_at | AT5G58050 | glycerophosphoryl diester phosphodiesterase family protein | -5.73 |
| 781 | 247804_at | AT5G58170 | glycerophosphoryl diester phosphodiesterase family protein | -3.43 |
| 782 | 247812_at | AT5G58390 | peroxidase, putative | 1.09 |
| 783 | 247747_at | AT5G59000 | zinc finger (C3HC4-type RING finger) family protein | 1.33 |
| 784 | 247758_at | AT5G59120 | ATSBT4.13; subtilase | -6.99 |
| 785 | 247718_at | AT5G59310 | LTP4 (LIPID TRANSFER PROTEIN 4); lipid binding | 1.24 |
| 786 | 247736_at | AT5G59370 | ACT4 (ACTIN 4) | -4.85 |
| 787 | 247684_at | AT5G59670 | leucine-rich repeat protein kinase, putative | 2.27 |
| 788 | 247657_at | AT5G59845 | gibberellin-regulated family protein | -3.35 |
| 789 | 247626_at | AT5G60300 | lectin protein kinase family protein | 1.04 |
| 790 | 247602_at | AT5G60900 | RLK1 (RECEPTOR-LIKE PROTEIN KINASE 1); carbohydrate binding / kinase | 1.30 |
| 791 | 247604_at | AT5G60950 | COBL5 (COBRA-LIKE PROTEIN 5 PRECURSOR) | 1.01 |
| 792 | 247554_at | AT5G61010 | ATEXO70E2 (EXOCYST SUBUNIT EXO70 FAMILY PROTEIN E2); protein binding | 0.94 |
| 793 | 247519_at | AT5G61430 | ANAC100/ATNAC5 (Arabidopsis NAC domain containing protein 100); transcription factor | -4.49 |
| 794 | 247529_at | AT5G61520 | hexose transporter, putative | 1.06 |
| 795 | 247546_at | AT5G61605 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G16505.1) | -7.80 |
| 796 | 247512_at | AT5G61720 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G28830.1); contains InterPro domain Protein of unknown function DUF1216 (InterPro:IPR009605) | -10.84 |
| 797 | 247424_at | AT5G62850 | ATVEX1 (VEGETATIVE CELL EXPRESSED1) | -5.88 |
| 798 | 247253_at | AT5G64790 | glycosyl hydrolase family 17 protein | -4.75 |
| 799 | 247189_at | AT5G65390 | AGP7 (Arabinogalactan protein 7) | 1.02 |
| 800 | 247182_at | AT5G65410 | ATHB25/ZFHD2 (ZINC FINGER HOMEODOMAIN 2); transcription factor | 2.48 |
| 801 | 247122_at | AT5G66020 | ATSAC1B/IBS2 (IMPAIRED IN BABA-INDUCED STERILITY 2); phosphoinositide 5-phosphatase | -4.56 |
| 802 | 247068_at | AT5G66800 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G50640.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN61148.1) | 1.40 |
| 803 | 247066_at | AT5G66940 | Dof-type zinc finger domain-containing protein | 1.05 |
| 804 | 247003_at | AT5G67550 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G71110.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN65326.1) | -3.83 |
| 805 | 244939_at | ATCG00065 | chloroplast gene encoding ribosomal protein s12. The gene is located in three distinct loci on the chloroplast genome and is transpliced to make one transcript. | 1.17 |
| 806 | 245024_at | ATCG00120 | Encodes the ATPase alpha subunit, which is a subunit of ATP synthase and part of the CF1 portion which catalyzes the conversion of ADP to ATP using the proton motive force. This complex is located in the thylakoid membrane of the chloroplast. | 1.03 |
| 807 | 245025_at | ATCG00130 | ATPase F subunit. | 1.02 |
| 808 | 245026_at | ATCG00140 | ATPase III subunit | 1.08 |
| 809 | 244997_at | ATCG00170 | RNA polymerase beta' subunit-2 | 1.23 |
| 810 | 244998_at | ATCG00180 | RNA polymerase beta' subunit-1 | 1.04 |
| 811 | 244999_at | ATCG00190 | Chloroplast DNA-dependent RNA polymerase B subunit. The transcription of this gene is regulated by a nuclear encoded RNA polymerase. This gene has been transferred to mitochondrial genome during crucifer evolution. | 1.80 |
| 812 | 245002_at | ATCG00270 | PSII D2 protein | 1.79 |
| 813 | 245007_at | ATCG00350 | Encodes psaA protein comprising the reaction center for photosystem I along with psaB protein; hydrophobic protein encoded by the chloroplast genome. | 1.20 |
| 814 | 245008_at | ATCG00360 | Encodes a protein required for photosystem I assembly and stability. In Chlamydomonas reinhardtii, this protein seems to act as a PSI specific chaperone facilitating the assembly of the complex by interacting with PsaA and PsaD. A loss of function mutation in tobacco leads to a loss of photosystem I. | 1.04 |
| 815 | 245010_at | ATCG00420 | Encodes NADH dehydrogenase subunit J. Its transcription is increased upon sulfur depletion. | 1.33 |
| 816 | 245011_at | ATCG00430 | Encodes a protein which was originally thought to be part of photosystem II but its wheat homolog was later shown to encode for subunit K of NADH dehydrogenase. | 1.90 |
| 817 | 245015_at | ATCG00490 | large subunit of RUBISCO. | 0.95 |
| 818 | 245016_at | ATCG00500 | Encodes the carboxytransferase beta subunit of the Acetyl-CoA carboxylase (ACCase) complex in plastids. This complex catalyzes the carboxylation of acetyl-CoA to produce malonyl-CoA, the first committed step in fatty acid synthesis. | 1.11 |
| 819 | 245018_at | ATCG00520 | Encodes a protein required for photosystem I assembly and stability. In cyanobacteria, loss of function mutation in this gene increases PSII/PSI ratio without any influence on photoautotrophic growth. | 1.26 |
| 820 | 245019_at | ATCG00530 | hypothetical protein | 1.05 |
| 821 | 244964_at | ATCG00580 | PSII cytochrome b559. There have been many speculations about the function of Cyt b559, but the most favored at present is that it plays a protective role by acting as an electron acceptor or electron donor under conditions when electron flow through PSII is not optimized. | 1.00 |
| 822 | 244970_at | ATCG00660 | encodes a chloroplast ribosomal protein L20, a constituent of the large subunit of the ribosomal complex | 1.44 |
| 823 | 244972_at | ATCG00680 | encodes for CP47, subunit of the photosystem II reaction center. | 2.69 |
| 824 | 244973_at | ATCG00690 | Encodes photosystem II 5 kD protein subunit PSII-T. This is a plastid-encoded gene (PsbTc) which also has a nuclear-encoded paralog (PsbTn). | 1.32 |
| 825 | 244975_at | ATCG00710 | Encodes a 8 kD phosphoprotein that is a component of the photosystem II oxygen evolving core. Its exact molecular function has not been determined but it may play a role in mediating electron transfer between the secondary quinone acceptors, QA and QB, associated with the acceptor side of PSII. | 1.05 |
| 826 | 244962_at | ATCG01050 | Represents a plastid-encoded subunit of a NAD(P)H dehydrogenase complex. Its mRNA is edited at four positions. Translation data is not available for this gene. | 1.29 |
| 827 | 244932_at | ATCG01060 | Encodes the PsaC subunit of photosystem I. | 2.12 |
| 828 | 244933_at | ATCG01070 | NADH dehydrogenase ND4L | 2.00 |
| 829 | 244937_at | ATCG01110 | Encodes the 49KDa plastid NAD(P)H dehydrogenase subunit H protein. Its transcription is regulated by an ndhF-specific plastid sigma factor, SIG4. | 1.37 |
| 830 | 244938_at | ATCG01120 | encodes a chloroplast ribosomal protein S15, a constituent of the small subunit of the ribosomal complex | 1.25 |
| 831 | 244901_at | ATMG00640 | encodes a plant b subunit of mitochondrial ATP synthase based on structural similarity and the presence in the F(0) complex. | 0.95 |
| 832 | 257333_at | ATMG01360 | cytochrome c oxidase subunit 1 | 1.27 |