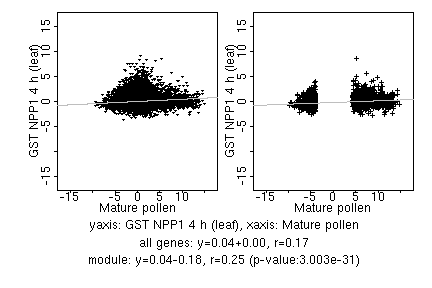
marker gene list
| Mature pollen |
| GST NPP1 4 h (leaf) |
| probeID | AGICode | Annotation | gene expression |
|
|---|---|---|---|---|
| 1 | 261570_at | AT1G01120 | KCS1 (3-KETOACYL-COA SYNTHASE 1); acyltransferase | -4.69 |
| 2 | 261572_at | AT1G01170 | ozone-responsive stress-related protein, putative | -4.95 |
| 3 | 261055_at | AT1G01300 | aspartyl protease family protein | -5.48 |
| 4 | 261045_at | AT1G01310 | allergen V5/Tpx-1-related family protein | 13.67 |
| 5 | 259425_at | AT1G01460 | ATPIPK11; 1-phosphatidylinositol-4-phosphate 5-kinase | 12.73 |
| 6 | 259426_at | AT1G01470 | LEA14 (LATE EMBRYOGENESIS ABUNDANT 14) | -4.15 |
| 7 | 259436_at | AT1G01500 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G19400.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G19400.2); similar to unknown [Picea sitchensis] (GB:ABK24253.1); contains domain PTHR10996:SF1 (PTHR10996:SF1); contains domain PTHR10996 (PTHR10996) | 5.25 |
| 8 | 259431_at | AT1G01620 | PIP1C (PLASMA MEMBRANE INTRINSIC PROTEIN 1;3); water channel | -7.09 |
| 9 | 261559_at | AT1G01780 | LIM domain-containing protein | 7.92 |
| 10 | 261537_at | AT1G01800 | short-chain dehydrogenase/reductase (SDR) family protein | -4.36 |
| 11 | 261623_at | AT1G01980 | ATSEC1A; electron carrier | 13.82 |
| 12 | 261624_at | AT1G02000 | GAE2 (UDP-D-GLUCURONATE 4-EPIMERASE 2); catalytic | 8.06 |
| 13 | 264147_at | AT1G02205 | CER1 (ECERIFERUM 1) | -4.35 |
| 14 | 259441_at | AT1G02300 | cathepsin B-like cysteine protease, putative | -4.19 |
| 15 | 262122_at | AT1G02790 | PGA4 (POLYGALACTURONASE 4); polygalacturonase | 11.18 |
| 16 | 263166_at | AT1G03050 | epsin N-terminal homology (ENTH) domain-containing protein / clathrin assembly protein-related | 12.86 |
| 17 | 263164_at | AT1G03070 | glutamate binding / | 5.37 |
| 18 | 263114_at | AT1G03130 | PSAD-2 (photosystem I subunit D-2) | -4.89 |
| 19 | 264822_at | AT1G03457 | RNA-binding protein, putative | 6.75 |
| 20 | 264837_at | AT1G03600 | photosystem II family protein | -5.87 |
| 21 | 264815_at | AT1G03620 | phagocytosis and cell motility protein ELMO1-related | 5.98 |
| 22 | 264839_at | AT1G03630 | POR C (PROTOCHLOROPHYLLIDE OXIDOREDUCTASE); NADPH dehydrogenase/ oxidoreductase/ protochlorophyllide reductase | -5.60 |
| 23 | 264830_at | AT1G03710 | cysteine protease inhibitor | 4.71 |
| 24 | 265066_at | AT1G03870 | FLA9 | -3.73 |
| 25 | 265038_at | AT1G03920 | protein kinase, putative | 7.00 |
| 26 | 265042_at | AT1G04040 | acid phosphatase class B family protein | -5.61 |
| 27 | 264324_at | AT1G04160 | XIB (Myosin-like protein XIB) | 5.54 |
| 28 | 263663_at | AT1G04410 | malate dehydrogenase, cytosolic, putative | -3.77 |
| 29 | 263655_at | AT1G04500 | zinc finger CONSTANS-related | 5.74 |
| 30 | 264601_at | AT1G04540 | C2 domain-containing protein | 10.90 |
| 31 | 264599_at | AT1G04600 | XIA (Myosin-like protein XIA); motor/ protein binding | 5.73 |
| 32 | 264613_at | AT1G04640 | LIP2 (LIPOYLTRANSFERASE 2) | -4.50 |
| 33 | 264603_at | AT1G04670 | similar to pollen-preferential protein [Brassica rapa subsp. chinensis] (GB:ABH09777.1) | 8.29 |
| 34 | 264602_at | AT1G04700 | protein kinase family protein | 7.84 |
| 35 | 261178_at | AT1G04760 | ATVAMP726 (VESICLE-ASSOCIATED MEMBRANE PROTEIN) | 4.64 |
| 36 | 261171_at | AT1G04880 | high mobility group (HMG1/2) family protein / ARID/BRIGHT DNA-binding domain-containing protein | 6.58 |
| 37 | 261179_at | AT1G04985 | similar to unnamed protein product [Vitis vinifera] (GB:CAO24071.1) | 4.66 |
| 38 | 265194_at | AT1G05010 | EFE (ETHYLENE FORMING ENZYME) | -6.53 |
| 39 | 265213_at | AT1G05020 | epsin N-terminal homology (ENTH) domain-containing protein / clathrin assembly protein-related | 6.83 |
| 40 | 265215_at | AT1G05040 | contains InterPro domain UBA-like (InterPro:IPR009060) | 5.93 |
| 41 | 264584_at | AT1G05140 | membrane-associated zinc metalloprotease, putative | -5.25 |
| 42 | 264575_at | AT1G05190 | EMB2394 (EMBRYO DEFECTIVE 2394); structural constituent of ribosome | -6.96 |
| 43 | 263233_at | AT1G05577 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G46110.2); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G59790.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G46110.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO67167.1); contains InterPro domain Protein of unknown function DUF966 (InterPro:IPR010369) | 7.65 |
| 44 | 260961_at | AT1G05960 | binding | 5.64 |
| 45 | 260955_at | AT1G06000 | UDP-glucoronosyl/UDP-glucosyl transferase family protein | -6.59 |
| 46 | 259395_at | AT1G06400 | ARA2 (Arabidopsis Rab GTPase homolog A1a); GTP binding | -3.94 |
| 47 | 262626_at | AT1G06430 | FTSH8 (FtsH protease 8); ATP-dependent peptidase/ ATPase/ metallopeptidase/ zinc ion binding | -6.05 |
| 48 | 262631_at | AT1G06500 | similar to unknown [Populus trichocarpa x Populus deltoides] (GB:ABK96557.1) | -4.93 |
| 49 | 262620_at | AT1G06540 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G30930.1) | 8.46 |
| 50 | 262635_at | AT1G06570 | PDS1 (PHYTOENE DESATURATION 1) | -5.22 |
| 51 | 262637_at | AT1G06640 | 2-oxoglutarate-dependent dioxygenase, putative | -4.38 |
| 52 | 262632_at | AT1G06680 | PSBP-1 (OXYGEN-EVOLVING ENHANCER PROTEIN 2); poly(U) binding | -5.32 |
| 53 | 260835_at | AT1G06700 | serine/threonine protein kinase, putative | 5.10 |
| 54 | 260828_at | AT1G06750 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G30630.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G30630.2); similar to hypothetical protein [Vitis vinifera] (GB:CAN76299.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO22658.1); contains domain no description (G3DSA:3.40.50.300); contains domain P-loop containing nucleoside triphosphate hydrolases (SSF52540) | 9.76 |
| 55 | 260823_at | AT1G06770 | zinc finger (C3HC4-type RING finger) family protein | 4.70 |
| 56 | 260818_at | AT1G06890 | transporter-related | 4.63 |
| 57 | 260815_at | AT1G06950 | ATTIC110/TIC110 (TRANSLOCON AT THE INNER ENVELOPE MEMBRANE OF CHLOROPLASTS 110) | -3.88 |
| 58 | 256051_at | AT1G06970 | ATCHX14/CHX14 (CATION/HYDROGEN EXCHANGER 14); monovalent cation:proton antiporter/ sodium:hydrogen antiporter | 7.65 |
| 59 | 256044_at | AT1G07160 | protein phosphatase 2C, putative / PP2C, putative | 4.61 |
| 60 | 261078_at | AT1G07320 | RPL4 (ribosomal protein L4); poly(U) binding / structural constituent of ribosome | -5.75 |
| 61 | 257489_at | AT1G07330 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G29620.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN81231.1) | 7.70 |
| 62 | 261072_at | AT1G07340 | ATSTP2 (SUGAR TRANSPORTER 2); carbohydrate transmembrane transporter/ sugar:hydrogen ion symporter | 5.81 |
| 63 | 261069_at | AT1G07410 | AtRABA2b (Arabidopsis Rab GTPase homolog A2b); GTP binding | 4.69 |
| 64 | 261061_at | AT1G07540 | TRFL2 (TRF-LIKE 2); DNA binding | 7.32 |
| 65 | 261410_at | AT1G07610 | MT1C (metallothionein 1C) | -7.12 |
| 66 | 261462_at | AT1G07850 | fringe-related protein | 5.50 |
| 67 | 260647_at | AT1G08030 | similar to hypothetical protein OsI_021001 [Oryza sativa (indica cultivar-group)] (GB:EAY99768.1); similar to hypothetical protein OsJ_019377 [Oryza sativa (japonica cultivar-group)] (GB:EAZ35894.1) | 4.79 |
| 68 | 261816_at | AT1G08135 | ATCHX6B/CHX6B (CATION/H+ EXCHANGER 6B); monovalent cation:proton antiporter | 6.69 |
| 69 | 261746_at | AT1G08380 | PSAO (photosystem I subunit O) | -9.12 |
| 70 | 261692_at | AT1G08450 | CRT3 (CALRETICULIN 3); calcium ion binding | -4.40 |
| 71 | 264781_at | AT1G08540 | SIGB (SIGMA FACTOR B); DNA binding / DNA-directed RNA polymerase/ transcription factor | -5.12 |
| 72 | 264803_at | AT1G08580 | similar to Pm52 [Prunus mume] (GB:BAE48663.1) | -4.43 |
| 73 | 264798_at | AT1G08730 | XIC (Myosin-like protein XIC); motor/ protein binding | 5.00 |
| 74 | 264646_at | AT1G08860 | BON3 (BONZAI 3) | 7.86 |
| 75 | 264656_at | AT1G09010 | glycoside hydrolase family 2 protein | -5.91 |
| 76 | 264253_at | AT1G09170 | kinesin motor protein-related | 4.74 |
| 77 | 263709_at | AT1G09310 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G56580.1); similar to unknown [Populus trichocarpa] (GB:ABK94207.1); contains InterPro domain Protein of unknown function DUF538 (InterPro:IPR007493) | -9.46 |
| 78 | 263676_at | AT1G09340 | CRB; binding / catalytic/ coenzyme binding | -7.99 |
| 79 | 264504_at | AT1G09430 | ACLA-3 (ATP-citrate lyase A-3) | -5.55 |
| 80 | 264508_at | AT1G09570 | PHYA (PHYTOCHROME A); G-protein coupled photoreceptor/ signal transducer | -5.68 |
| 81 | 264558_at | AT1G09600 | protein kinase family protein | 6.77 |
| 82 | 264672_at | AT1G09750 | chloroplast nucleoid DNA-binding protein-related | -3.83 |
| 83 | 264659_at | AT1G09930 | ATOPT2 (oligopeptide transporter 2); oligopeptide transporter | 5.41 |
| 84 | 264521_at | AT1G10020 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G29310.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO61535.1); contains InterPro domain Protein of unknown function DUF1005 (InterPro:IPR010410) | -5.38 |
| 85 | 264524_at | AT1G10070 | ATBCAT-2; branched-chain-amino-acid transaminase/ catalytic | 4.66 |
| 86 | 264462_at | AT1G10200 | WLIM1; transcription factor | -4.49 |
| 87 | 261830_at | AT1G10620 | protein kinase family protein | 12.00 |
| 88 | 257477_at | AT1G10660 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G62960.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO64308.1) | 5.83 |
| 89 | 261829_at | AT1G10680 | PGP10 (P-GLYCOPROTEIN 10); ATPase, coupled to transmembrane movement of substances | 8.91 |
| 90 | 262786_at | AT1G10740 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G23330.1); similar to Os05g0408300 [Oryza sativa (japonica cultivar-group)] (GB:NP_001055523.1); similar to hypothetical protein OsI_019255 [Oryza sativa (indica cultivar-group)] (GB:EAY98022.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO42648.1); contains domain SSF53474 (SSF53474); contains domain G3DSA:3.40.50.1820 (G3DSA:3.40.50.1820) | 5.54 |
| 91 | 262760_at | AT1G10770 | invertase/pectin methylesterase inhibitor family protein | 10.76 |
| 92 | 260481_at | AT1G10960 | ATFD1 (FERREDOXIN 1); 2 iron, 2 sulfur cluster binding / electron carrier/ iron-sulfur cluster binding | -5.47 |
| 93 | 260478_at | AT1G11040 | DNAJ chaperone C-terminal domain-containing protein | 6.66 |
| 94 | 262511_at | AT1G11250 | SYP125 (syntaxin 125); SNAP receptor | 7.77 |
| 95 | 262456_at | AT1G11260 | STP1 (SUGAR TRANSPORTER 1); carbohydrate transmembrane transporter/ sugar:hydrogen ion symporter | -4.02 |
| 96 | 262455_at | AT1G11310 | MLO2 (MILDEW RESISTANCE LOCUS O 2); calmodulin binding | -4.84 |
| 97 | 261873_at | AT1G11350 | S-locus lectin protein kinase family protein | -4.82 |
| 98 | 262811_at | AT1G11700 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G61930.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO17934.1); contains InterPro domain Protein of unknown function DUF584 (InterPro:IPR007608) | -3.78 |
| 99 | 262817_at | AT1G11770 | electron carrier | 11.05 |
| 100 | 264343_at | AT1G11850 | unknown protein | -5.84 |
| 101 | 264387_at | AT1G11990 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G62330.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO22725.1); contains InterPro domain Protein of unknown function DUF246, plant (InterPro:IPR004348) | 9.05 |
| 102 | 264396_at | AT1G12050 | fumarylacetoacetase, putative | -5.47 |
| 103 | 264395_at | AT1G12070 | Rho GDP-dissociation inhibitor family protein | 8.05 |
| 104 | 264371_at | AT1G12090 | ELP (EXTENSIN-LIKE PROTEIN); lipid binding | -6.64 |
| 105 | 264348_at | AT1G12110 | NRT1.1 (NITRATE TRANSPORTER 1.1); transporter | -5.57 |
| 106 | 260992_at | AT1G12150 | similar to myosin heavy chain-related [Arabidopsis thaliana] (TAIR:AT4G17210.1); similar to Prefoldin [Medicago truncatula] (GB:ABD28587.1); contains InterPro domain Protein of unknown function DUF827, plant (InterPro:IPR008545) | 6.91 |
| 107 | 260967_at | AT1G12230 | transaldolase, putative | -5.65 |
| 108 | 259537_at | AT1G12370 | PHR1 (PHOTOLYASE 1) | 4.71 |
| 109 | 259530_at | AT1G12450 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G22850.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO63604.1); contains InterPro domain SNARE associated Golgi protein (InterPro:IPR015414) | 6.59 |
| 110 | 261197_at | AT1G12900 | GAPA-2; glyceraldehyde-3-phosphate dehydrogenase | -5.69 |
| 111 | 262780_at | AT1G13090 | CYP71B28 (cytochrome P450, family 71, subfamily B, polypeptide 28); oxygen binding | -4.07 |
| 112 | 259363_at | AT1G13270 | MAP1C (METHIONINE AMINOPEPTIDASE 1B); metalloexopeptidase | -4.26 |
| 113 | 259451_at | AT1G13890 | SNAP30 (synaptosomal-associated protein 30) | 11.84 |
| 114 | 262609_at | AT1G13930 | similar to nodulin-related [Arabidopsis thaliana] (TAIR:AT2G03440.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN63832.1) | -7.63 |
| 115 | 262664_at | AT1G13970 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G29180.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO15029.1); contains InterPro domain Protein of unknown function DUF1336 (InterPro:IPR009769) | 9.90 |
| 116 | 261480_at | AT1G14280 | PKS2 (PHYTOCHROME KINASE SUBSTRATE 2) | -4.13 |
| 117 | 261492_at | AT1G14290 | acid phosphatase, putative | 5.29 |
| 118 | 261488_at | AT1G14345 | oxidoreductase | -6.32 |
| 119 | 261528_at | AT1G14420 | AT59 (Arabidopsis homolog of tomato LAT59); lyase/ pectate lyase | 12.48 |
| 120 | 261482_at | AT1G14530 | (TOM THREE HOMOLOG); virion binding | 4.66 |
| 121 | 262842_at | AT1G14720 | XTR2 (XYLOGLUCAN ENDOTRANSGLYCOSYLASE RELATED 2); hydrolase, acting on glycosyl bonds | -4.12 |
| 122 | 262844_at | AT1G14890 | pectinesterase inhibitor | -4.96 |
| 123 | 262829_at | AT1G14970 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G01480.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN77011.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO42288.1); contains InterPro domain Protein of unknown function DUF246, plant (InterPro:IPR004348) | 4.77 |
| 124 | 260741_at | AT1G15040 | glutamine amidotransferase-related | 6.53 |
| 125 | 262590_at | AT1G15100 | RHA2A (RING-H2 finger A2A); protein binding / zinc ion binding | -3.91 |
| 126 | 262599_at | AT1G15350 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G25360.1); similar to hypothetical protein [Glycine max] (GB:AAN03467.1) | 6.17 |
| 127 | 262603_at | AT1G15380 | lactoylglutathione lyase family protein / glyoxalase I family protein | 5.30 |
| 128 | 262571_at | AT1G15430 | similar to zinc ion binding [Arabidopsis thaliana] (TAIR:AT1G80220.1); similar to Os01g0612600 [Oryza sativa (japonica cultivar-group)] (GB:NP_001043557.1); similar to hypothetical protein OsI_002789 [Oryza sativa (indica cultivar-group)] (GB:EAY74942.1); contains InterPro domain Protein of unknown function DUF1644 (InterPro:IPR012866) | 5.05 |
| 129 | 262597_at | AT1G15470 | transducin family protein / WD-40 repeat family protein | 5.06 |
| 130 | 259499_at | AT1G15730 | PRLI-interacting factor L, putative | -4.03 |
| 131 | 259491_at | AT1G15820 | LHCB6 (LIGHT HARVESTING COMPLEX PSII); chlorophyll binding | -6.57 |
| 132 | 259498_at | AT1G15880 | GOS11 (GOLGI SNARE 11); SNARE binding | 4.57 |
| 133 | 261841_at | AT1G15920 | CCR4-NOT transcription complex protein, putative | 5.00 |
| 134 | 261788_at | AT1G15980 | similar to unnamed protein product [Vitis vinifera] (GB:CAO49411.1); contains domain G3DSA:3.40.50.2000 (G3DSA:3.40.50.2000); contains domain SSF53756 (SSF53756) | -3.80 |
| 135 | 261786_at | AT1G15990 | ATCNGC7 (CYCLIC NUCLEOTIDE GATED CHANNEL 7); calmodulin binding / cyclic nucleotide binding / ion channel | 5.54 |
| 136 | 262707_at | AT1G16290 | similar to predicted protein [Physcomitrella patens subsp. patens] (GB:EDQ78585.1); similar to Os02g0170900 [Oryza sativa (japonica cultivar-group)] (GB:NP_001046030.1); contains InterPro domain Lytic transglycosylase-like, catalytic (InterPro:IPR008258) | 5.81 |
| 137 | 262755_at | AT1G16360 | LEM3 (ligand-effect modulator 3) family protein / CDC50 family protein | 6.76 |
| 138 | 262757_at | AT1G16380 | ATCHX1 (CATION/H+ EXCHANGER 1); monovalent cation:proton antiporter | 7.07 |
| 139 | 255765_at | AT1G16760 | protein kinase family protein | 8.28 |
| 140 | 256115_at | AT1G16880 | uridylyltransferase-related | -7.52 |
| 141 | 262483_at | AT1G17220 | FUG1 (FU-GAERI1); translation initiation factor | -6.43 |
| 142 | 260737_at | AT1G17540 | kinase | 9.45 |
| 143 | 259399_at | AT1G17710 | phosphoric monoester hydrolase | 5.03 |
| 144 | 255900_at | AT1G17830 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G73210.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO45300.1); contains InterPro domain Protein of unknown function DUF789 (InterPro:IPR008507) | 4.98 |
| 145 | 256075_at | AT1G18150 | ATMPK8 (ARABIDOPSIS THALIANA MAP KINASE 8); MAP kinase | 4.67 |
| 146 | 256122_at | AT1G18180 | oxidoreductase, acting on the CH-CH group of donors | 4.71 |
| 147 | 256129_at | AT1G18210 | calcium-binding protein, putative | -4.96 |
| 148 | 261673_at | AT1G18280 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | 7.58 |
| 149 | 261716_at | AT1G18410 | kinesin motor protein-related | 9.00 |
| 150 | 261670_at | AT1G18520 | TET11 (TETRASPANIN11) | 5.66 |
| 151 | 255772_at | AT1G18530 | calmodulin, putative | 8.96 |
| 152 | 255773_at | AT1G18590 | sulfotransferase family protein | -5.18 |
| 153 | 255774_at | AT1G18620 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G74160.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO49165.1) | -5.52 |
| 154 | 261422_at | AT1G18730 | similar to unknown [Populus trichocarpa] (GB:ABK95263.1) | -6.67 |
| 155 | 261423_at | AT1G18750 | AGL65; DNA binding / transcription factor | 4.63 |
| 156 | 259464_at | AT1G18990 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G74830.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO62468.1); contains InterPro domain Protein of unknown function DUF593 (InterPro:IPR007656) | 10.87 |
| 157 | 259466_at | AT1G19050 | ARR7 (RESPONSE REGULATOR 7); transcription regulator/ two-component response regulator | -4.25 |
| 158 | 259482_at | AT1G19090 | RKF2 (RECEPTOR-LIKE SERINE/THREONINE KINASE 2); kinase | 6.78 |
| 159 | 256015_at | AT1G19150 | LHCA6 (Photosystem I light harvesting complex gene 6); chlorophyll binding | -5.61 |
| 160 | 256017_at | AT1G19180 | JAZ1/TIFY10A (JASMONATE-ZIM-DOMAIN PROTEIN 1); protein binding | -5.08 |
| 161 | 260666_at | AT1G19300 | GATL1/GLZ1/PARVUS (GALACTURONOSYLTRANSFERASE-LIKE 1); polygalacturonate 4-alpha-galacturonosyltransferase/ transferase, transferring glycosyl groups / transferase, transferring hexosyl groups | -4.54 |
| 162 | 260661_at | AT1G19500 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G35765.1) | 12.49 |
| 163 | 261150_at | AT1G19640 | JMT (JASMONIC ACID CARBOXYL METHYLTRANSFERASE); jasmonate O-methyltransferase | 7.21 |
| 164 | 255786_at | AT1G19670 | ATCLH1 (CORONATINE-INDUCED PROTEIN 1) | -5.56 |
| 165 | 261142_at | AT1G19780 | ATCNGC8 (CYCLIC NUCLEOTIDE GATED CHANNEL 8); calmodulin binding / cyclic nucleotide binding / ion channel | 7.94 |
| 166 | 261131_at | AT1G19835 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G47900.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO48232.1); similar to hypothetical protein OsI_010472 [Oryza sativa (indica cultivar-group)] (GB:EAY89239.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN60525.1); contains InterPro domain Protein of unknown function DUF869, plant (InterPro:IPR008587) | -4.75 |
| 167 | 255815_at | AT1G19890 | ATMGH3/MGH3 (MALE-GAMETE-SPECIFIC HISTONE H3); DNA binding | 9.52 |
| 168 | 255756_at | AT1G19940 | ATGH9B5 (ARABIDOPSIS THALIANA GLYCOSYL HYDROLASE 9B5); hydrolase, hydrolyzing O-glycosyl compounds | 5.04 |
| 169 | 261221_at | AT1G19960 | similar to transmembrane receptor [Arabidopsis thaliana] (TAIR:AT2G32140.1) | -5.95 |
| 170 | 261218_at | AT1G20020 | ATLFNR2 (LEAF FNR 2); NADPH dehydrogenase/ oxidoreductase/ poly(U) binding | -6.99 |
| 171 | 261246_at | AT1G20080 | ATSYTB/NTMC2T1.2/NTMC2TYPE1.2/SYTB | 6.91 |
| 172 | 261245_at | AT1G20135 | hydrolase, acting on ester bonds / lipase | 7.13 |
| 173 | 255885_at | AT1G20330 | SMT2 (STEROL METHYLTRANSFERASE 2) | -3.93 |
| 174 | 255886_at | AT1G20340 | DRT112 (DNA-damage-repair/toleration protein 112); copper ion binding / electron carrier | -5.27 |
| 175 | 259516_at | AT1G20450 | ERD10/LTI45 (EARLY RESPONSIVE TO DEHYDRATION 10) | -4.33 |
| 176 | 259545_at | AT1G20560 | AMP-dependent synthetase and ligase family protein | -4.78 |
| 177 | 259517_at | AT1G20630 | CAT1 (CATALASE 1); catalase | 5.40 |
| 178 | 262806_at | AT1G20950 | pyrophosphate--fructose-6-phosphate 1-phosphotransferase-related / pyrophosphate-dependent 6-phosphofructose-1-kinase-related | -5.45 |
| 179 | 262798_at | AT1G20980 | SPL14 (SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 14); DNA binding / transcription factor | -3.86 |
| 180 | 261457_at | AT1G21065 | similar to unnamed protein product [Vitis vinifera] (GB:CAO66320.1); contains InterPro domain Protein of unknown function UPF0047 (InterPro:IPR001602) | -3.76 |
| 181 | 261453_at | AT1G21130 | O-methyltransferase, putative | -4.70 |
| 182 | 259561_at | AT1G21250 | WAK1 (CELL WALL-ASSOCIATED KINASE); kinase | -3.79 |
| 183 | 260875_at | AT1G21410 | SKP2A; protein binding | -4.27 |
| 184 | 260877_at | AT1G21500 | similar to hypothetical protein OsI_030994 [Oryza sativa (indica cultivar-group)] (GB:EAZ09762.1); similar to Os09g0517000 [Oryza sativa (japonica cultivar-group)] (GB:NP_001063677.1); similar to unknown protein [Oryza sativa (japonica cultivar-group)] (GB:BAD33829.1) | -3.95 |
| 185 | 262499_at | AT1G21770 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G77540.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO61877.1); contains InterPro domain Acyl-CoA N-acyltransferase (InterPro:IPR016181) | -4.63 |
| 186 | 262495_at | AT1G21780 | BTB/POZ domain-containing protein | -5.08 |
| 187 | 255976_at | AT1G22010 | unknown protein | 5.09 |
| 188 | 255950_at | AT1G22110 | structural constituent of ribosome | 7.54 |
| 189 | 255952_at | AT1G22130 | AGL104; DNA binding / transcription factor | 7.60 |
| 190 | 255958_at | AT1G22150 | SULTR1;3 (sulfate transporter); sulfate transmembrane transporter | 6.65 |
| 191 | 255925_at | AT1G22200 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G36050.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO61738.1); contains InterPro domain Protein of unknown function DUF1692 (InterPro:IPR012936) | 5.09 |
| 192 | 261929_at | AT1G22460 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G64600.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO61677.1); contains InterPro domain Protein of unknown function DUF246, plant (InterPro:IPR004348) | 5.82 |
| 193 | 264199_at | AT1G22700 | tetratricopeptide repeat (TPR)-containing protein | -5.91 |
| 194 | 264206_at | AT1G22730 | MA3 domain-containing protein | 4.57 |
| 195 | 264209_at | AT1G22740 | RAB7 (Ras-related protein 7); GTP binding | -3.78 |
| 196 | 264208_at | AT1G22760 | PAB3 (POLY(A) BINDING PROTEIN 3); RNA binding | 6.50 |
| 197 | 264722_at | AT1G22970 | protein binding | -4.00 |
| 198 | 264900_at | AT1G23080 | PIN7 (PIN-FORMED 7); auxin:hydrogen symporter/ transporter | -4.27 |
| 199 | 264897_at | AT1G23220 | dynein light chain type 1 family protein | -5.44 |
| 200 | 263043_at | AT1G23350 | invertase/pectin methylesterase inhibitor family protein | 11.40 |
| 201 | 262986_at | AT1G23390 | kelch repeat-containing F-box family protein | -4.36 |
| 202 | 265178_at | AT1G23540 | protein kinase family protein | 9.26 |
| 203 | 265170_at | AT1G23730 | carbonic anhydrase, putative / carbonate dehydratase, putative | 4.66 |
| 204 | 265182_at | AT1G23740 | oxidoreductase, zinc-binding dehydrogenase family protein | -6.87 |
| 205 | 263034_at | AT1G24020 | MLP423 (MLP-LIKE PROTEIN 423) | -6.52 |
| 206 | 264863_at | AT1G24110 | peroxidase, putative | 8.28 |
| 207 | 264862_at | AT1G24330 | armadillo/beta-catenin repeat family protein / U-box domain-containing family protein | 5.18 |
| 208 | 265002_at | AT1G24400 | LHT2 (LYSINE HISTIDINE TRANSPORTER 2); amino acid transmembrane transporter | 5.37 |
| 209 | 265022_at | AT1G24520 | BCP1 (Brassica campestris pollen protein 1) | 9.47 |
| 210 | 257405_at | AT1G24620 | polcalcin, putative / calcium-binding pollen allergen, putative | 12.31 |
| 211 | 245645_at | AT1G24764 | ATMAP70-2 (microtubule-associated proteins 70-2); microtubule binding | -4.55 |
| 212 | 245636_at | AT1G25240 | epsin N-terminal homology (ENTH) domain-containing protein / clathrin assembly protein-related | 10.29 |
| 213 | 245642_at | AT1G25275 | similar to unnamed protein product [Vitis vinifera] (GB:CAO43403.1) | -4.35 |
| 214 | 245846_at | AT1G26130 | haloacid dehalogenase-like hydrolase family protein | 4.75 |
| 215 | 245877_at | AT1G26220 | GCN5-related N-acetyltransferase (GNAT) family protein | -4.36 |
| 216 | 245820_at | AT1G26320 | NADP-dependent oxidoreductase, putative | 5.68 |
| 217 | 261015_at | AT1G26480 | GRF12 (GENERAL REGULATORY FACTOR 12); protein phosphorylated amino acid binding | 11.14 |
| 218 | 263681_at | AT1G26840 | ATORC6/ORC6 (Origin recognition complex protein 6); DNA binding | 7.24 |
| 219 | 264987_at | AT1G27030 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G27020.1); similar to unknown protein [Oryza sativa (japonica cultivar-group)] (GB:AAN05517.1); similar to Os10g0463800 [Oryza sativa (japonica cultivar-group)] (GB:NP_001064789.1) | -4.25 |
| 220 | 261643_at | AT1G27720 | TAF4b (TBP-ASSOCIATED FACTOR 4b); transcription initiation factor | 5.95 |
| 221 | 261650_at | AT1G27770 | ACA1 (autoinhibited Ca2+ -ATPase 1); calcium-transporting ATPase/ calmodulin binding | -5.31 |
| 222 | 259592_at | AT1G27950 | lipid transfer protein-related | -3.91 |
| 223 | 259591_at | AT1G28150 | Identical to UPF0426 protein At1g28150, chloroplast precursor [Arabidopsis Thaliana] (GB:Q9FZ89;GB:Q8LET5); similar to unknown [Populus trichocarpa] (GB:ABK94413.1) | -4.14 |
| 224 | 245658_at | AT1G28270 | RALFL4 (RALF-LIKE 4) | 13.56 |
| 225 | 245688_at | AT1G28290 | AGP31 (ARABINOGALACTAN-PROTEIN 31); structural constituent of cell wall | -6.37 |
| 226 | 245668_at | AT1G28330 | DRM1 (DORMANCY-ASSOCIATED PROTEIN 1) | -5.55 |
| 227 | 262742_at | AT1G28550 | AtRABA1i (Arabidopsis Rab GTPase homolog A1i); GTP binding | 12.50 |
| 228 | 262750_at | AT1G28710 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G28700.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN64421.1); contains domain PTHR10483:SF6 (PTHR10483:SF6); contains domain PTHR10483 (PTHR10483) | 5.09 |
| 229 | 262746_at | AT1G28960 | ATNUDT15 (ARABIDOPSIS THALIANA NUDIX HYDROLASE HOMOLOG 15); hydrolase | 5.02 |
| 230 | 260898_at | AT1G29070 | ribosomal protein L34 family protein | -4.13 |
| 231 | 260888_at | AT1G29140 | pollen Ole e 1 allergen and extensin family protein | 11.43 |
| 232 | 257506_at | AT1G29440 | auxin-responsive family protein | -4.56 |
| 233 | 259773_at | AT1G29500 | auxin-responsive protein, putative | -3.97 |
| 234 | 259780_at | AT1G29630 | Identical to Exonuclease 1 (EXO1) [Arabidopsis Thaliana] (GB:Q8L6Z7;GB:Q9C7N8); similar to exonuclease, putative [Arabidopsis thaliana] (TAIR:AT1G18090.2); similar to exonuclease, putative [Arabidopsis thaliana] (TAIR:AT1G18090.1); similar to Exonuclease 1 (OsEXO-1) (GB:Q60GC1); contains InterPro domain XPG N-terminal; (InterPro:IPR006085); contains InterPro domain XPG I; (InterPro:IPR006086); contains InterPro domain Helix-hairpin-helix motif, class 2; (InterPro:IPR008918); contains InterPro domain DNA repair protein (XPGC)/yeast Rad; (InterPro:IPR006084) | 5.98 |
| 235 | 259786_at | AT1G29660 | GDSL-motif lipase/hydrolase family protein | -5.71 |
| 236 | 259788_at | AT1G29670 | GDSL-motif lipase/hydrolase family protein | -8.97 |
| 237 | 245776_at | AT1G30260 | AGL79 (AGAMOUS-LIKE 79) | -4.18 |
| 238 | 256309_at | AT1G30380 | PSAK (PHOTOSYSTEM I SUBUNIT K) | -7.93 |
| 239 | 261804_at | AT1G30530 | UDP-glucoronosyl/UDP-glucosyl transferase family protein | -5.50 |
| 240 | 263226_at | AT1G30690 | SEC14 cytosolic factor family protein / phosphoglyceride transfer family protein | -4.59 |
| 241 | 263215_at | AT1G30710 | FAD-binding domain-containing protein | 12.97 |
| 242 | 263217_at | AT1G30740 | FAD-binding domain-containing protein | 6.76 |
| 243 | 262557_at | AT1G31330 | PSAF (photosystem I subunit F) | -4.77 |
| 244 | 256497_at | AT1G31580 | ECS1 | -5.42 |
| 245 | 246582_at | AT1G31750 | proline-rich family protein | 7.90 |
| 246 | 246288_at | AT1G31850 | dehydration-responsive protein, putative | -4.49 |
| 247 | 246313_at | AT1G31920 | pentatricopeptide (PPR) repeat-containing protein | -4.01 |
| 248 | 255720_at | AT1G32060 | PRK (PHOSPHORIBULOKINASE); ATP binding / phosphoribulokinase/ protein binding | -5.02 |
| 249 | 255718_at | AT1G32070 | ATNSI (NUCLEAR SHUTTLE INTERACTING); N-acetyltransferase | -4.60 |
| 250 | 245795_at | AT1G32160 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G17800.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G17800.2); similar to unnamed protein product [Vitis vinifera] (GB:CAO63331.1); contains InterPro domain Protein of unknown function DUF760 (InterPro:IPR008479) | -4.29 |
| 251 | 260702_at | AT1G32250 | calmodulin, putative | 7.27 |
| 252 | 260704_at | AT1G32470 | glycine cleavage system H protein, mitochondrial, putative | -4.50 |
| 253 | 256468_at | AT1G32550 | ferredoxin family protein | -4.88 |
| 254 | 261713_at | AT1G32640 | ATMYC2 (JASMONATE INSENSITIVE 1); DNA binding / transcription factor | -4.40 |
| 255 | 261709_at | AT1G32790 | CID11; RNA binding / protein binding | -6.69 |
| 256 | 261190_at | AT1G32990 | PRPL11 (PLASTID RIBOSOMAL PROTEIN L11); structural constituent of ribosome | -4.51 |
| 257 | 245768_at | AT1G33590 | disease resistance protein-related / LRR protein-related | -4.71 |
| 258 | 245765_at | AT1G33600 | leucine-rich repeat family protein | -4.92 |
| 259 | 261992_at | AT1G33680 | nucleic acid binding | -3.96 |
| 260 | 261983_at | AT1G33770 | protein kinase family protein | 7.34 |
| 261 | 255980_at | AT1G33970 | avirulence-responsive protein, putative / avirulence induced gene protein, putative / AIG protein, putative | -4.33 |
| 262 | 262022_at | AT1G35490 | bZIP family transcription factor | 12.33 |
| 263 | 262038_at | AT1G35580 | CINV1 (CYTOSOLIC INVERTASE 1); beta-fructofuranosidase | -5.01 |
| 264 | 262029_at | AT1G35680 | 50S ribosomal protein L21, chloroplast / CL21 (RPL21) | -4.33 |
| 265 | 263193_at | AT1G36050 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G22200.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO61738.1); contains InterPro domain Protein of unknown function DUF1692 (InterPro:IPR012936) | 4.80 |
| 266 | 263190_at | AT1G36105 | transposable element gene | 8.21 |
| 267 | 260127_at | AT1G36320 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G37920.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO61774.1) | -4.36 |
| 268 | 261979_at | AT1G37130 | NIA2 (NITRATE REDUCTASE 2) | -4.22 |
| 269 | 256542_at | AT1G42550 | PMI1 (PLASTID MOVEMENT IMPAIRED1) | -4.05 |
| 270 | 256544_at | AT1G42560 | ATMLO9/MLO9 (MILDEW RESISTANCE LOCUS O 9); calmodulin binding | 5.65 |
| 271 | 259625_at | AT1G42970 | GAPB (GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE B SUBUNIT); glyceraldehyde-3-phosphate dehydrogenase | -7.27 |
| 272 | 262725_at | AT1G43580 | similar to unnamed protein product [Vitis vinifera] (GB:CAO43621.1); contains domain PTHR21290 (PTHR21290) | -4.35 |
| 273 | 262723_at | AT1G43630 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G18740.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO43630.1); contains InterPro domain Protein of unknown function DUF793 (InterPro:IPR008511) | 6.43 |
| 274 | 260813_at | AT1G43700 | VIP1 (VIRE2-INTERACTING PROTEIN 1); transcription factor | -4.29 |
| 275 | 245741_at | AT1G44120 | C2 domain-containing protein / armadillo/beta-catenin repeat family protein | 7.22 |
| 276 | 245737_at | AT1G44160 | DNAJ chaperone C-terminal domain-containing protein | 9.06 |
| 277 | 245213_at | AT1G44575 | NPQ4 (NONPHOTOCHEMICAL QUENCHING) | -4.56 |
| 278 | 260944_at | AT1G45130 | BGAL5 (beta-galactosidase 5); beta-galactosidase | -4.46 |
| 279 | 245780_at | AT1G45688 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G42860.1); similar to H0814G11.12 [Oryza sativa (indica cultivar-group)] (GB:CAJ86345.1); similar to CAA30379.1 protein [Oryza sativa] (GB:CAB53482.1) | -7.10 |
| 280 | 245803_at | AT1G47128 | RD21 (RESPONSIVE TO DEHYDRATION 21); cysteine-type peptidase | -5.11 |
| 281 | 260502_at | AT1G47270 | AtTLP6 (TUBBY LIKE PROTEIN 6); phosphoric diester hydrolase/ transcription factor | 7.30 |
| 282 | 261687_at | AT1G47280 | unknown protein | 12.45 |
| 283 | 261688_at | AT1G47380 | protein phosphatase 2C-related / PP2C-related | 5.05 |
| 284 | 262432_at | AT1G47530 | ripening-responsive protein, putative | -4.70 |
| 285 | 261740_at | AT1G47740 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G25170.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO61202.1); contains InterPro domain Protein of unknown function DUF862, eukaryotic (InterPro:IPR008580) | -3.96 |
| 286 | 259615_at | AT1G47980 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G62730.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO44946.1) | 6.91 |
| 287 | 260725_at | AT1G48170 | similar to expressed protein [Oryza sativa (japonica cultivar-group)] (GB:ABA95965.1); similar to hypothetical protein OsI_036449 [Oryza sativa (indica cultivar-group)] (GB:EAY82490.1); similar to Os12g0182800 [Oryza sativa (japonica cultivar-group)] (GB:NP_001066323.1) | -3.73 |
| 288 | 261305_at | AT1G48470 | GLN1;5 (GLUTAMINE SYNTHETASE 1;5); glutamate-ammonia ligase | 5.64 |
| 289 | 261306_at | AT1G48610 | AT hook motif-containing protein | -3.84 |
| 290 | 260757_at | AT1G48940 | plastocyanin-like domain-containing protein | 4.89 |
| 291 | 262389_at | AT1G49270 | protein kinase family protein | 10.70 |
| 292 | 257469_at | AT1G49290 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G13620.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN72326.1) | 10.77 |
| 293 | 262393_at | AT1G49490 | leucine-rich repeat family protein / extensin family protein | 11.63 |
| 294 | 262399_at | AT1G49500 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G19030.1) | -4.55 |
| 295 | 261608_at | AT1G49650 | cell death associated protein-related | -3.96 |
| 296 | 261598_at | AT1G49750 | leucine-rich repeat family protein | -5.38 |
| 297 | 259819_at | AT1G49820 | ATMTK; S-methyl-5-thioribose kinase | -3.74 |
| 298 | 261638_at | AT1G49975 | similar to hypothetical protein [Vitis vinifera] (GB:CAN66219.1) | -4.19 |
| 299 | 262473_at | AT1G50250 | FTSH1 (FtsH protease 1); ATP-dependent peptidase/ ATPase/ metallopeptidase | -3.87 |
| 300 | 262421_at | AT1G50290 | unknown protein | 5.28 |
| 301 | 262463_at | AT1G50310 | monosaccharide transporter (STP9) | 6.67 |
| 302 | 262418_at | AT1G50320 | ATHX (THIOREDOXIN X); thiol-disulfide exchange intermediate | -7.04 |
| 303 | 261860_at | AT1G50600 | SCL5; transcription factor | -5.87 |
| 304 | 261875_at | AT1G50610 | leucine-rich repeat transmembrane protein kinase, putative | 9.04 |
| 305 | 256215_at | AT1G50900 | similar to hypothetical protein [Vitis vinifera] (GB:CAN65357.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO38995.1); contains InterPro domain Ankyrin (InterPro:IPR002110) | -6.67 |
| 306 | 256213_at | AT1G50990 | protein kinase-related | 5.59 |
| 307 | 245745_at | AT1G51110 | plastid-lipid associated protein PAP / fibrillin family protein | -7.97 |
| 308 | 265134_at | AT1G51260 | LPAT3 (Lysophosphatidyl acyltransferase 3); 1-acylglycerol-3-phosphate O-acyltransferase | 7.38 |
| 309 | 265149_at | AT1G51400 | photosystem II 5 kD protein | -4.57 |
| 310 | 260518_at | AT1G51410 | cinnamyl-alcohol dehydrogenase, putative (CAD) | 8.15 |
| 311 | 260513_at | AT1G51490 | glycosyl hydrolase family 1 protein | 7.18 |
| 312 | 256186_at | AT1G51680 | 4CL1 (4-COUMARATE:COA LIGASE 1); 4-coumarate-CoA ligase | -4.07 |
| 313 | 246349_at | AT1G51915 | cryptdin protein-related | 6.82 |
| 314 | 246371_at | AT1G51940 | protein kinase family protein / peptidoglycan-binding LysM domain-containing protein | -4.68 |
| 315 | 265053_at | AT1G52000 | jacalin lectin family protein | -5.48 |
| 316 | 259839_at | AT1G52190 | proton-dependent oligopeptide transport (POT) family protein | -6.22 |
| 317 | 259838_at | AT1G52220 | similar to PSAP/PSI-P/PTAC8/TMP14 (THYLAKOID MEMBRANE PHOSPHOPROTEIN OF 14 KDA) [Arabidopsis thaliana] (TAIR:AT2G46820.2); similar to PSAP/PSI-P/PTAC8/TMP14 (THYLAKOID MEMBRANE PHOSPHOPROTEIN OF 14 KDA), DNA binding [Arabidopsis thaliana] (TAIR:AT2G46820.1); similar to unknown [Populus trichocarpa] (GB:ABK95860.1) | -4.29 |
| 318 | 259840_at | AT1G52230 | PSAH-2/PSAH2/PSI-H (PHOTOSYSTEM I SUBUNIT H-2) | -7.84 |
| 319 | 259836_at | AT1G52240 | ATROPGEF11/ROPGEF11 (KINASE PARTNER PROTEIN-LIKE); Rho guanyl-nucleotide exchange factor/ | 11.51 |
| 320 | 259637_at | AT1G52260 | ATPDIL1-5 (PDI-LIKE 1-5); thiol-disulfide exchange intermediate | -3.87 |
| 321 | 259670_at | AT1G52310 | protein kinase family protein / C-type lectin domain-containing protein | -5.30 |
| 322 | 259669_at | AT1G52340 | ABA2 (ABA DEFICIENT 2); oxidoreductase | -5.13 |
| 323 | 259640_at | AT1G52400 | BGL1 (BETA-GLUCOSIDASE HOMOLOG 1); hydrolase, hydrolyzing O-glycosyl compounds | -6.11 |
| 324 | 262151_at | AT1G52510 | hydrolase, alpha/beta fold family protein | -3.85 |
| 325 | 262147_at | AT1G52570 | PLDALPHA2 (PHOSPHLIPASE D ALPHA 2); phospholipase D | 5.96 |
| 326 | 262146_at | AT1G52580 | rhomboid family protein | 9.64 |
| 327 | 262156_at | AT1G52680 | late embryogenesis abundant protein-related / LEA protein-related | 9.71 |
| 328 | 260153_at | AT1G52760 | esterase/lipase/thioesterase family protein | -5.30 |
| 329 | 261372_at | AT1G53010 | zinc finger (C3HC4-type RING finger) family protein | 4.63 |
| 330 | 260588_at | AT1G53320 | AtTLP7 (TUBBY LIKE PROTEIN 7); phosphoric diester hydrolase/ transcription factor | 4.97 |
| 331 | 260589_at | AT1G53400 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G45740.1); similar to unknown [Brassica rapa] (GB:ABL97984.1); contains domain PTHR13609 (PTHR13609) | 4.60 |
| 332 | 260975_at | AT1G53430 | leucine-rich repeat family protein / protein kinase family protein | -4.07 |
| 333 | 260978_at | AT1G53540 | 17.6 kDa class I small heat shock protein (HSP17.6C-CI) (AA 1-156) | 5.24 |
| 334 | 262225_at | AT1G53840 | ATPME1 (Arabidopsis thaliana pectin methylesterase 1); pectinesterase | -5.04 |
| 335 | 263156_at | AT1G54030 | GDSL-motif lipase, putative | -4.03 |
| 336 | 263174_at | AT1G54040 | ESP (EPITHIOSPECIFIER PROTEIN) | -3.87 |
| 337 | 263144_at | AT1G54070 | dormancy/auxin associated protein-related | 13.49 |
| 338 | 263006_at | AT1G54240 | DNA binding | 7.83 |
| 339 | 262998_at | AT1G54280 | haloacid dehalogenase-like hydrolase family protein | 7.99 |
| 340 | 262959_at | AT1G54290 | eukaryotic translation initiation factor SUI1, putative | 4.98 |
| 341 | 262954_at | AT1G54500 | rubredoxin family protein | -4.57 |
| 342 | 264235_at | AT1G54560 | XIE (Myosin-like protein XIE); motor/ protein binding | 6.01 |
| 343 | 264185_at | AT1G54780 | thylakoid lumen 18.3 kDa protein | -8.05 |
| 344 | 264187_at | AT1G54860 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G19250.1); similar to GPI-anchored protein-like protein II [Cucumis melo] (GB:ABR67421.1) | 6.37 |
| 345 | 256354_at | AT1G54870 | oxidoreductase | 5.41 |
| 346 | 256323_at | AT1G54920 | similar to hypothetical protein [Vitis vinifera] (GB:CAN77027.1) | 5.37 |
| 347 | 259646_at | AT1G55350 | DEK1 (DEFECTIVE KERNEL 1); calcium-dependent cysteine-type endopeptidase/ cysteine-type endopeptidase | -3.82 |
| 348 | 265125_at | AT1G55410 | pseudogene, CHP-rich zinc finger protein, putative, similar to putative CHP-rich zinc finger protein GB:CAB77744 GI:7268217 from (Arabidopsis thaliana) | 6.47 |
| 349 | 265073_at | AT1G55480 | binding / protein binding | -4.36 |
| 350 | 265076_at | AT1G55490 | CPN60B (CHAPERONIN 60 BETA); ATP binding / protein binding / unfolded protein binding | -4.84 |
| 351 | 265127_at | AT1G55560 | SKS14 (SKU5 Similar 14); copper ion binding / oxidoreductase | 12.94 |
| 352 | 265080_at | AT1G55570 | SKS12 (SKU5 Similar 12); copper ion binding / oxidoreductase | 13.59 |
| 353 | 264545_at | AT1G55670 | PSAG | -7.91 |
| 354 | 260598_at | AT1G55930 | CBS domain-containing protein / transporter associated domain-containing protein | -4.04 |
| 355 | 262086_at | AT1G56050 | GTP-binding protein-related | -4.04 |
| 356 | 256225_at | AT1G56220 | dormancy/auxin associated family protein | -7.04 |
| 357 | 259635_at | AT1G56360 | ATPAP6/PAP6 (purple acid phosphatase 6); acid phosphatase/ protein serine/threonine phosphatase | 5.27 |
| 358 | 246377_at | AT1G57550 | hydrophobic protein, putative / low temperature and salt responsive protein, putative | 6.03 |
| 359 | 245868_at | AT1G58030 | CAT2 (CATIONIC AMINO ACID TRANSPORTER 2); amino acid transmembrane transporter | 4.93 |
| 360 | 246396_at | AT1G58180 | carbonic anhydrase family protein / carbonate dehydratase family protein | -3.94 |
| 361 | 256200_at | AT1G58210 | EMB1674 (EMBRYO DEFECTIVE 1674) | 9.09 |
| 362 | 256022_at | AT1G58360 | AAP1 (AMINO ACID PERMEASE 1); amino acid transmembrane transporter | -4.46 |
| 363 | 264247_at | AT1G60160 | potassium transporter family protein | -4.16 |
| 364 | 264269_at | AT1G60240 | apical meristem formation protein-related | 11.74 |
| 365 | 259727_at | AT1G60950 | FED A (FERREDOXIN 2); 2 iron, 2 sulfur cluster binding / electron carrier/ iron-sulfur cluster binding | -5.79 |
| 366 | 259715_at | AT1G60990 | aminomethyltransferase | -3.86 |
| 367 | 259720_at | AT1G61080 | proline-rich family protein | 8.96 |
| 368 | 264760_at | AT1G61290 | SYP124 (syntaxin 124); SNAP receptor | 11.23 |
| 369 | 265033_at | AT1G61520 | LHCA3 (Photosystem I light harvesting complex gene 3); chlorophyll binding | -6.70 |
| 370 | 264424_at | AT1G61740 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G11540.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO38814.1); contains InterPro domain Protein of unknown function DUF81; (InterPro:IPR002781) | -4.75 |
| 371 | 264284_at | AT1G61860 | protein kinase, putative | 10.78 |
| 372 | 260637_at | AT1G62380 | ACO2 (ACC OXIDASE 2) | -6.05 |
| 373 | 265115_at | AT1G62450 | Rho GDP-dissociation inhibitor family protein | 7.06 |
| 374 | 265116_at | AT1G62480 | vacuolar calcium-binding protein-related | -7.75 |
| 375 | 259693_at | AT1G63060 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G07330.1); similar to unknown [Xerophyta humilis] (GB:AAT45004.1) | 11.86 |
| 376 | 257509_at | AT1G63190 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G63200.1); contains InterPro domain Cystatin-related, plant (InterPro:IPR006525); contains InterPro domain Oxidoreductase, molybdopterin binding; (InterPro:IPR000572) | 5.60 |
| 377 | 260083_at | AT1G63220 | C2 domain-containing protein | 5.30 |
| 378 | 261557_at | AT1G63640 | kinesin motor protein-related | 5.72 |
| 379 | 260317_at | AT1G63800 | UBC5 (UBIQUITIN-CONJUGATING ENZYME 5); ubiquitin-protein ligase | -4.13 |
| 380 | 260320_at | AT1G63930 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G23530.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN64338.1); contains InterPro domain Protein of unknown function DUF793 (InterPro:IPR008511) | 10.11 |
| 381 | 262347_at | AT1G64110 | AAA-type ATPase family protein | 4.70 |
| 382 | 259740_at | AT1G64300 | protein kinase family protein | 6.93 |
| 383 | 257508_at | AT1G64320 | myosin heavy chain-related | 7.25 |
| 384 | 259794_at | AT1G64330 | myosin heavy chain-related | -4.81 |
| 385 | 259765_at | AT1G64370 | unknown protein | -4.63 |
| 386 | 261951_at | AT1G64490 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G42060.1); contains domain DEK C-terminal domain (SSF109715) | -4.14 |
| 387 | 261954_at | AT1G64510 | ribosomal protein S6 family protein | -5.36 |
| 388 | 261973_at | AT1G64610 | WD-40 repeat family protein | 5.32 |
| 389 | 262885_at | AT1G64740 | TUA1 (ALPHA-1 TUBULIN) | 7.13 |
| 390 | 262883_at | AT1G64780 | ATAMT1;2 (AMMONIUM TRANSPORTER 1;2); ammonium transmembrane transporter | -4.08 |
| 391 | 262917_at | AT1G64800 | unknown protein | 6.01 |
| 392 | 262879_at | AT1G64860 | SIGA (SIGMA FACTOR A); DNA binding / DNA-directed RNA polymerase/ transcription factor | -5.17 |
| 393 | 263142_at | AT1G65230 | similar to hypothetical protein [Vitis vinifera] (GB:CAN76393.1) | -4.87 |
| 394 | 263108_at | AT1G65240 | aspartyl protease family protein | 7.66 |
| 395 | 262933_at | AT1G65840 | ATPAO4 (POLYAMINE OXIDASE 4); amine oxidase | 4.68 |
| 396 | 261970_at | AT1G65960 | GAD2 (GLUTAMATE DECARBOXYLASE 2); calmodulin binding | -4.60 |
| 397 | 256528_at | AT1G66140 | ZFP4 (ZINC FINGER PROTEIN 4); nucleic acid binding / transcription factor/ zinc ion binding | -4.01 |
| 398 | 256516_at | AT1G66150 | TMK1 (TRANSMEMBRANE KINASE 1) | -4.22 |
| 399 | 256524_at | AT1G66200 | ATGSR2 (Arabidopsis thaliana glutamine synthase clone R2); glutamate-ammonia ligase | -5.25 |
| 400 | 259820_at | AT1G66210 | subtilase family protein | 10.33 |
| 401 | 259823_at | AT1G66250 | glycosyl hydrolase family 17 protein | -4.48 |
| 402 | 260136_at | AT1G66360 | C2 domain-containing protein | 6.39 |
| 403 | 260107_at | AT1G66430 | pfkB-type carbohydrate kinase family protein | -3.91 |
| 404 | 256366_at | AT1G66880 | serine/threonine protein kinase family protein | -4.70 |
| 405 | 264993_at | AT1G67290 | glyoxal oxidase-related | 9.02 |
| 406 | 264229_at | AT1G67480 | kelch repeat-containing F-box family protein | 5.80 |
| 407 | 260195_at | AT1G67540 | similar to hypothetical protein [Vitis vinifera] (GB:CAN81893.1) | 6.54 |
| 408 | 260197_at | AT1G67623 | F-box family protein | 8.13 |
| 409 | 260193_at | AT1G67640 | lysine and histidine specific transporter, putative | 6.96 |
| 410 | 245189_at | AT1G67670 | unknown protein | 6.58 |
| 411 | 245195_at | AT1G67740 | PSBY (photosystem II BY) | -5.47 |
| 412 | 245193_at | AT1G67810 | Fe-S metabolism associated domain-containing protein | 6.86 |
| 413 | 245215_at | AT1G67830 | ATFXG1 (ALPHA-FUCOSIDASE 1); alpha-L-fucosidase/ carboxylesterase | -4.13 |
| 414 | 260004_at | AT1G67860 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G67865.1) | -4.72 |
| 415 | 260007_at | AT1G67870 | glycine-rich protein | -5.86 |
| 416 | 260014_at | AT1G68010 | HPR (HYDROXYPYRUVATE REDUCTASE); glycerate dehydrogenase/ poly(U) binding | -5.02 |
| 417 | 260013_at | AT1G68090 | ANN5/ANNAT5 (ANNEXIN ARABIDOPSIS 5); calcium ion binding / calcium-dependent phospholipid binding | 8.02 |
| 418 | 260011_at | AT1G68110 | epsin N-terminal homology (ENTH) domain-containing protein / clathrin assembly protein-related | 9.60 |
| 419 | 260442_at | AT1G68220 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G13380.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN76984.1); contains InterPro domain Protein of unknown function DUF1218 (InterPro:IPR009606) | -4.91 |
| 420 | 260268_at | AT1G68490 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G13390.2); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G13390.1); similar to unknown [Paeonia suffruticosa] (GB:ABW74476.1) | -5.13 |
| 421 | 260266_at | AT1G68520 | zinc finger (B-box type) family protein | -4.38 |
| 422 | 260267_at | AT1G68530 | CUT1 (CUTICULAR 1); acyltransferase/ catalytic | -3.83 |
| 423 | 262230_at | AT1G68560 | ATXYL1/XYL1 (ALPHA-XYLOSIDASE 1); alpha-N-arabinofuranosidase/ hydrolase, hydrolyzing O-glycosyl compounds / xylan 1,4-beta-xylosidase | -6.43 |
| 424 | 262283_at | AT1G68590 | plastid-specific 30S ribosomal protein 3, putative / PSRP-3, putative | -5.00 |
| 425 | 262282_at | AT1G68610 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G14870.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO42115.1); contains InterPro domain Protein of unknown function Cys-rich (InterPro:IPR006461) | 9.79 |
| 426 | 262279_at | AT1G68630 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G58320.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO42113.1); contains InterPro domain Protein of unknown function Cys-rich (InterPro:IPR006461) | 5.37 |
| 427 | 260032_at | AT1G68750 | ATPPC4 (Arabidopsis thaliana phosphoenolpyruvate carboxylase 4); phosphoenolpyruvate carboxylase | 6.70 |
| 428 | 259370_at | AT1G69050 | unknown protein | 7.69 |
| 429 | 256299_at | AT1G69530 | ATEXPA1 (ARABIDOPSIS THALIANA EXPANSIN A1) | -4.60 |
| 430 | 260395_at | AT1G69780 | ATHB13; DNA binding / transcription factor | -4.02 |
| 431 | 264716_at | AT1G70170 | MMP (MATRIX METALLOPROTEINASE); metalloendopeptidase | 6.44 |
| 432 | 264313_at | AT1G70410 | carbonic anhydrase, putative / carbonate dehydratase, putative | -3.98 |
| 433 | 264369_at | AT1G70430 | protein kinase family protein | 5.65 |
| 434 | 260303_at | AT1G70520 | protein kinase family protein | -4.35 |
| 435 | 260306_at | AT1G70540 | EDA24 (embryo sac development arrest 24); pectinesterase inhibitor | 12.84 |
| 436 | 260180_at | AT1G70660 | MMZ2/UEV1B (MMS ZWEI HOMOLOGE 2); protein binding / ubiquitin-protein ligase | -4.04 |
| 437 | 262291_at | AT1G70790 | C2 domain-containing protein | 8.20 |
| 438 | 262309_at | AT1G70820 | phosphoglucomutase, putative / glucose phosphomutase, putative | -4.74 |
| 439 | 262312_at | AT1G70830 | MLP28 (MLP-LIKE PROTEIN 28) | -6.68 |
| 440 | 262304_at | AT1G70890 | MLP43 (MLP-LIKE PROTEIN 43) | -4.96 |
| 441 | 259897_at | AT1G71380 | ATCEL3/ATGH9B3 (ARABIDOPSIS THALIANA GLYCOSYL HYDROLASE 9B3); hydrolase, hydrolyzing O-glycosyl compounds | 6.23 |
| 442 | 261532_at | AT1G71680 | amino acid transmembrane transporter | 8.54 |
| 443 | 261518_at | AT1G71695 | peroxidase 12 (PER12) (P12) (PRXR6) | -5.58 |
| 444 | 261506_at | AT1G71697 | ATCK1 (CHOLINE KINASE) | 6.21 |
| 445 | 261511_at | AT1G71770 | PAB5 (POLY(A)-BINDING PROTEIN); RNA binding | 8.08 |
| 446 | 260144_at | AT1G71960 | ABC transporter family protein | -4.74 |
| 447 | 260167_at | AT1G71970 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G22680.1); similar to unknown [Populus trichocarpa] (GB:ABK94498.1) | -4.42 |
| 448 | 256341_at | AT1G72040 | deoxynucleoside kinase family | -5.03 |
| 449 | 259803_at | AT1G72150 | PATL1 (PATELLIN 1); transporter | -4.45 |
| 450 | 259804_at | AT1G72160 | SEC14 cytosolic factor family protein / phosphoglyceride transfer family protein | -4.39 |
| 451 | 259801_at | AT1G72230 | plastocyanin-like domain-containing protein | -4.05 |
| 452 | 260429_at | AT1G72450 | JAZ6/TIFY11B (JASMONATE-ZIM-DOMAIN PROTEIN 6) | -3.92 |
| 453 | 260424_at | AT1G72460 | leucine-rich repeat transmembrane protein kinase, putative | 5.19 |
| 454 | 259892_at | AT1G72610 | GLP1 (GERMIN-LIKE PROTEIN 1); manganese ion binding / metal ion binding / nutrient reservoir | -4.73 |
| 455 | 262366_at | AT1G72890 | disease resistance protein (TIR-NBS class), putative | -4.14 |
| 456 | 262385_at | AT1G72960 | root hair defective 3 GTP-binding (RHD3) family protein | 7.74 |
| 457 | 262369_at | AT1G73010 | phosphoric monoester hydrolase | 6.19 |
| 458 | 262359_at | AT1G73066 | protein binding | 4.90 |
| 459 | 262360_at | AT1G73080 | PEPR1 (PEP1 RECEPTOR 1); ATP binding / kinase/ protein binding / protein serine/threonine kinase | -4.44 |
| 460 | 262377_at | AT1G73110 | ribulose bisphosphate carboxylase/oxygenase activase, putative / RuBisCO activase, putative | -5.05 |
| 461 | 245734_at | AT1G73480 | hydrolase, alpha/beta fold family protein | 4.80 |
| 462 | 260076_at | AT1G73630 | calcium-binding protein, putative | 5.61 |
| 463 | 260381_at | AT1G73860 | microtubule motor | 7.74 |
| 464 | 260335_at | AT1G74000 | SS3 (STRICTOSIDINE SYNTHASE 3) | 9.12 |
| 465 | 260386_at | AT1G74010 | strictosidine synthase family protein | 10.34 |
| 466 | 260246_at | AT1G74220 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G03630.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN71403.1) | 7.21 |
| 467 | 260244_at | AT1G74320 | choline kinase, putative | 5.06 |
| 468 | 260236_at | AT1G74470 | geranylgeranyl reductase | -5.67 |
| 469 | 260213_at | AT1G74490 | protein kinase, putative | 9.11 |
| 470 | 260214_at | AT1G74510 | kelch repeat-containing F-box family protein | -3.92 |
| 471 | 260238_at | AT1G74520 | ATHVA22A (Arabidopsis thaliana HVA22 homologue A) | -5.38 |
| 472 | 260221_at | AT1G74670 | gibberellin-responsive protein, putative | -5.57 |
| 473 | 262168_at | AT1G74730 | similar to unknown [Populus trichocarpa x Populus deltoides] (GB:ABK96654.1); contains InterPro domain Protein of unknown function DUF1118 (InterPro:IPR009500) | -3.94 |
| 474 | 262216_at | AT1G74780 | nodulin family protein | 5.28 |
| 475 | 262213_at | AT1G74870 | protein binding / zinc ion binding | 5.12 |
| 476 | 262175_at | AT1G74880 | NDH-O (NAD(P)H:PLASTOQUINONE DEHYDROGENASE COMPLEX SUBUNIT O) | -4.15 |
| 477 | 262172_at | AT1G74970 | RPS9 (RIBOSOMAL PROTEIN S9); structural constituent of ribosome | -4.15 |
| 478 | 259986_at | AT1G75050 | similar to ATLP-3 (Arabidopsis thaumatin-like protein 3) [Arabidopsis thaliana] (TAIR:AT1G75030.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN70296.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO61219.1); contains InterPro domain Thaumatin, pathogenesis-related (InterPro:IPR001938) | 4.95 |
| 479 | 259954_at | AT1G75130 | CYP721A1 (cytochrome P450, family 721, subfamily A, polypeptide 1); oxygen binding | -4.15 |
| 480 | 256506_at | AT1G75160 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G05840.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO61175.1); contains InterPro domain Protein of unknown function DUF620 (InterPro:IPR006873) | 7.72 |
| 481 | 256505_at | AT1G75200 | flavodoxin family protein / radical SAM domain-containing protein | -4.09 |
| 482 | 256454_at | AT1G75280 | isoflavone reductase, putative | -4.75 |
| 483 | 261120_at | AT1G75410 | BLH3; DNA binding / transcription factor | -4.05 |
| 484 | 261110_at | AT1G75440 | UBC16 (UBIQUITIN-CONJUGATING ENZYME 16); ubiquitin-protein ligase | -4.31 |
| 485 | 261118_at | AT1G75460 | ATP-dependent protease La (LON) domain-containing protein | -3.88 |
| 486 | 262951_at | AT1G75500 | nodulin MtN21 family protein | -3.74 |
| 487 | 262950_at | AT1G75510 | transcription initiation factor IIF beta subunit (TFIIF-beta) family protein | -4.22 |
| 488 | 262947_at | AT1G75750 | GASA1 (GAST1 PROTEIN HOMOLOG 1) | -4.54 |
| 489 | 262727_at | AT1G75800 | pathogenesis-related thaumatin family protein | -4.45 |
| 490 | 262728_at | AT1G75820 | CLV1 (CLAVATA 1); ATP binding / kinase/ protein serine/threonine kinase | -4.07 |
| 491 | 262729_at | AT1G75840 | ATGP3/ATROP4 (RHO-LIKE GTP BINDING PROTEIN 4); GTP binding / GTPase | -5.02 |
| 492 | 262696_at | AT1G75870 | unknown protein | 10.59 |
| 493 | 261751_at | AT1G76080 | ATCDSP32/CDSP32 (CHLOROPLASTIC DROUGHT-INDUCED STRESS PROTEIN OF 32 KD); thiol-disulfide exchange intermediate | -4.19 |
| 494 | 261769_at | AT1G76100 | plastocyanin | -7.54 |
| 495 | 259886_at | AT1G76370 | protein kinase, putative | 13.38 |
| 496 | 259980_at | AT1G76520 | auxin efflux carrier family protein | -4.63 |
| 497 | 259866_at | AT1G76640 | calmodulin-related protein, putative | 11.52 |
| 498 | 256332_at | AT1G76890 | GT2; transcription factor | -5.52 |
| 499 | 259676_at | AT1G77730 | pleckstrin homology (PH) domain-containing protein | 7.43 |
| 500 | 262179_at | AT1G77980 | AGL66; transcription factor | 7.07 |
| 501 | 262162_at | AT1G78020 | senescence-associated protein-related | -5.39 |
| 502 | 262195_at | AT1G78040 | pollen Ole e 1 allergen and extensin family protein | -4.26 |
| 503 | 262181_at | AT1G78060 | glycosyl hydrolase family 3 protein | -4.73 |
| 504 | 260051_at | AT1G78210 | hydrolase, alpha/beta fold family protein | -5.29 |
| 505 | 260799_at | AT1G78270 | ATUGT85A4 (UDP-GLUCOSYL TRANSFERASE 85A4); UDP-glycosyltransferase/ glucuronosyltransferase/ transferase, transferring hexosyl groups | -3.90 |
| 506 | 260745_at | AT1G78370 | ATGSTU20 (Arabidopsis thaliana Glutathione S-transferase (class tau) 20); glutathione transferase | -6.97 |
| 507 | 260746_at | AT1G78380 | ATGSTU19 (GLUTATHIONE TRANSFERASE 8); glutathione transferase | -3.90 |
| 508 | 260801_at | AT1G78430 | Identical to Uncharacterized protein At1g78430 [Arabidopsis Thaliana] (GB:Q9M9F9); similar to tropomyosin-related [Arabidopsis thaliana] (TAIR:AT1G17140.1); similar to tropomyosin-related [Arabidopsis thaliana] (TAIR:AT1G17140.2); similar to hypothetical protein [Vitis vinifera] (GB:CAN73256.1); contains InterPro domain Prefoldin (InterPro:IPR009053) | -3.92 |
| 509 | 263126_at | AT1G78460 | SOUL heme-binding family protein | 7.40 |
| 510 | 263131_at | AT1G78630 | EMB1473 (EMBRYO DEFECTIVE 1473); structural constituent of ribosome | -6.21 |
| 511 | 264139_at | AT1G78940 | protein kinase family protein | 7.19 |
| 512 | 257428_at | AT1G78990 | transferase family protein | 5.24 |
| 513 | 264092_at | AT1G79040 | PSBR (photosystem II subunit R) | -5.13 |
| 514 | 264099_at | AT1G79050 | DNA repair protein recA | -3.79 |
| 515 | 264127_at | AT1G79250 | protein kinase, putative | 6.53 |
| 516 | 264124_at | AT1G79360 | ATOCT2 (ARABIDOPSIS THALIANA ORGANIC CATION/CARNITINE TRANSPORTER2); carbohydrate transmembrane transporter/ sugar:hydrogen ion symporter | 4.67 |
| 517 | 262919_at | AT1G79380 | copine-related | -5.07 |
| 518 | 262936_at | AT1G79400 | ATCHX2 (CATION/H+ EXCHANGER 2); monovalent cation:proton antiporter | 7.09 |
| 519 | 262921_at | AT1G79430 | APL (ALTERED PHLOEM DEVELOPMENT); transcription factor | -4.12 |
| 520 | 262942_at | AT1G79450 | LEM3 (ligand-effect modulator 3) family protein / CDC50 family protein | 5.95 |
| 521 | 261353_at | AT1G79600 | ABC1 family protein | -5.81 |
| 522 | 261401_at | AT1G79640 | kinase | 5.88 |
| 523 | 261356_at | AT1G79660 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G16170.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO23158.1) | 5.05 |
| 524 | 260161_at | AT1G79860 | ATROPGEF12/MEE64/ROPGEF12 (MATERNAL EFFECT EMBRYO ARREST 64); Rho guanyl-nucleotide exchange factor/ | 9.43 |
| 525 | 260158_at | AT1G79910 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G52315.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO45916.1); contains InterPro domain Protein of unknown function DUF292, eukaryotic (InterPro:IPR005061) | 8.59 |
| 526 | 262047_at | AT1G80160 | lactoylglutathione lyase family protein / glyoxalase I family protein | 4.93 |
| 527 | 260297_at | AT1G80280 | hydrolase, alpha/beta fold family protein | 5.77 |
| 528 | 260285_at | AT1G80560 | 3-isopropylmalate dehydrogenase, chloroplast, putative | -4.24 |
| 529 | 260286_at | AT1G80600 | acetylornithine aminotransferase, mitochondrial, putative / acetylornithine transaminase, putative / AOTA, putative / ACOAT, putative | -5.21 |
| 530 | 261943_at | AT1G80660 | AHA9 (Arabidopsis H(+)-ATPase 9); hydrogen-exporting ATPase, phosphorylative mechanism | 10.97 |
| 531 | 262203_at | AT2G01060 | myb family transcription factor | -4.26 |
| 532 | 265761_at | AT2G01330 | transducin family protein / WD-40 repeat family protein | 4.78 |
| 533 | 265738_at | AT2G01350 | QPT (QUINOLINATE PHOSHORIBOSYLTRANSFERASE); nicotinate-nucleotide diphosphorylase (carboxylating) | -4.89 |
| 534 | 266348_at | AT2G01450 | ATMPK17 (Arabidopsis thaliana MAP kinase 17); MAP kinase | 4.71 |
| 535 | 263590_at | AT2G01820 | leucine-rich repeat protein kinase, putative | -4.88 |
| 536 | 263598_at | AT2G01850 | EXGT-A3 (endo-xyloglucan transferase A3); hydrolase, acting on glycosyl bonds / xyloglucan:xyloglucosyl transferase | -4.86 |
| 537 | 263304_at | AT2G01920 | epsin N-terminal homology (ENTH) domain-containing protein / clathrin assembly protein-related | 6.72 |
| 538 | 266119_at | AT2G02100 | LCR69/PDF2.2 (Low-molecular-weight cysteine-rich 69); protease inhibitor | -4.32 |
| 539 | 266118_at | AT2G02130 | LCR68/PDF2.3 (Low-molecular-weight cysteine-rich 68); protease inhibitor | -5.29 |
| 540 | 266115_at | AT2G02140 | LCR72/PDF2.6 (Low-molecular-weight cysteine-rich 72); protease inhibitor | 11.15 |
| 541 | 266181_at | AT2G02390 | ATGSTZ1 (GLUTATHIONE S-TRANSFERASE 18); glutathione transferase | -4.31 |
| 542 | 266204_at | AT2G02410 | similar to unnamed protein product [Vitis vinifera] (GB:CAO46936.1); contains InterPro domain Protein of unknown function DUF901 (InterPro:IPR010298) | -4.28 |
| 543 | 267243_at | AT2G02660 | similar to F-box family protein [Arabidopsis thaliana] (TAIR:AT2G02890.1); similar to Cyclin-like F-box; F-box protein interaction domain [Medicago truncatula] (GB:ABD33119.1); contains InterPro domain F-box associated type 3 (InterPro:IPR013187); contains InterPro domain F-box associated type 1 (InterPro:IPR006527) | 5.25 |
| 544 | 267476_at | AT2G02720 | pectate lyase family protein | 12.39 |
| 545 | 266744_at | AT2G02970 | nucleoside phosphatase family protein / GDA1/CD39 family protein | 6.43 |
| 546 | 266793_at | AT2G03060 | AGL30; DNA binding / transcription factor | 6.41 |
| 547 | 266725_at | AT2G03170 | ASK14 (ARABIDOPSIS SKP1-LIKE 14); protein binding / ubiquitin-protein ligase | 4.62 |
| 548 | 265705_at | AT2G03410 | Mo25 family protein | 10.95 |
| 549 | 265714_at | AT2G03520 | ATUPS4 (ARABIDOPSIS THALIANA UREIDE PERMEASE 4) | 5.84 |
| 550 | 264035_at | AT2G03630 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G74220.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN77667.1) | 6.47 |
| 551 | 263361_at | AT2G03840 | TET13 (TETRASPANIN13) | 7.43 |
| 552 | 263482_at | AT2G03980 | GDSL-motif lipase/hydrolase family protein | 5.59 |
| 553 | 263483_at | AT2G04030 | CR88 (EMBRYO DEFECTIVE 1956); ATP binding | -4.20 |
| 554 | 263410_at | AT2G04039 | similar to unnamed protein product [Vitis vinifera] (GB:CAO41070.1) | -4.77 |
| 555 | 263324_at | AT2G04220 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G12690.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO41124.1); contains InterPro domain Protein of unknown function DUF868, plant (InterPro:IPR008586) | 8.55 |
| 556 | 263325_at | AT2G04240 | XERICO; protein binding / zinc ion binding | -4.56 |
| 557 | 263627_at | AT2G04675 | unknown protein | 5.75 |
| 558 | 263628_at | AT2G04780 | FLA7 | -4.16 |
| 559 | 263046_at | AT2G05380 | GRP3S (GLYCINE-RICH PROTEIN 3 SHORT ISOFORM) | -4.65 |
| 560 | 265560_at | AT2G05520 | GRP-3 (GLYCINE-RICH PROTEIN 3) | -4.96 |
| 561 | 265377_at | AT2G05790 | glycosyl hydrolase family 17 protein | -3.83 |
| 562 | 266029_at | AT2G05850 | SCPL38 (serine carboxypeptidase-like 38); serine carboxypeptidase | 13.34 |
| 563 | 265518_at | AT2G06040 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G21900.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO65254.1); contains InterPro domain Leucine-rich repeat, cysteine-containing subtype (InterPro:IPR006553) | 4.85 |
| 564 | 265519_at | AT2G06040 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G21900.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO65254.1); contains InterPro domain Leucine-rich repeat, cysteine-containing subtype (InterPro:IPR006553) | 4.70 |
| 565 | 265530_at | AT2G06050 | OPR3 (OPDA-REDUCTASE 3); 12-oxophytodienoate reductase | -4.92 |
| 566 | 265374_at | AT2G06520 | PSBX (photosystem II subunit X) | -6.41 |
| 567 | 266215_at | AT2G06850 | EXGT-A1 (ENDOXYLOGLUCAN TRANSFERASE); hydrolase, acting on glycosyl bonds | -4.47 |
| 568 | 266494_at | AT2G07040 | ATPRK2A; ATP binding / protein serine/threonine kinase | 12.76 |
| 569 | 266428_at | AT2G07180 | protein kinase, putative | 6.34 |
| 570 | 265552_at | AT2G07560 | AHA6 (ARABIDOPSIS H(+)-ATPASE 6); ATPase | 9.94 |
| 571 | 265400_at | AT2G10940 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | -5.16 |
| 572 | 265401_at | AT2G10970 | invertase/pectin methylesterase inhibitor family protein | 7.39 |
| 573 | 263637_at | AT2G11890 | adenylate cyclase | -4.53 |
| 574 | 265368_at | AT2G13350 | C2 domain-containing protein | 9.55 |
| 575 | 263350_at | AT2G13360 | AGT (ALANINE:GLYOXYLATE AMINOTRANSFERASE) | -4.44 |
| 576 | 263718_at | AT2G13570 | CCAAT-box binding transcription factor, putative | 10.01 |
| 577 | 263720_at | AT2G13620 | ATCHX15 (cation/hydrogen exchanger 15); monovalent cation:proton antiporter | 8.97 |
| 578 | 264108_at | AT2G13680 | CALS5 (CALLOSE SYNTHASE 5); 1,3-beta-glucan synthase | 7.68 |
| 579 | 264112_at | AT2G13680 | CALS5 (CALLOSE SYNTHASE 5); 1,3-beta-glucan synthase | 7.28 |
| 580 | 265656_at | AT2G13820 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | -5.25 |
| 581 | 265831_at | AT2G14460 | unknown protein | -4.45 |
| 582 | 266587_at | AT2G14880 | SWIB complex BAF60b domain-containing protein | -6.00 |
| 583 | 266588_at | AT2G14890 | AGP9 (ARABINOGALACTAN PROTEIN 9) | -4.45 |
| 584 | 265894_at | AT2G15050 | LTP; lipid binding | -4.37 |
| 585 | 263566_at | AT2G15340 | glycine-rich protein | 7.29 |
| 586 | 265473_at | AT2G15535 | LCR10 (Low-molecular-weight cysteine-rich 10) | 9.63 |
| 587 | 265475_at | AT2G15620 | NIR1 (NITRITE REDUCTASE); ferredoxin-nitrate reductase | -4.71 |
| 588 | 265489_at | AT2G15640 | F-box family protein | 5.55 |
| 589 | 265484_at | AT2G15820 | pentatricopeptide (PPR) repeat-containing protein | -3.93 |
| 590 | 265536_at | AT2G15880 | leucine-rich repeat family protein / extensin family protein | 8.85 |
| 591 | 265478_at | AT2G15890 | MEE14 (maternal effect embryo arrest 14) | -6.23 |
| 592 | 265481_at | AT2G15960 | unknown protein | -4.71 |
| 593 | 263095_at | AT2G16040 | hAT dimerisation domain-containing protein / transposase-related | 8.90 |
| 594 | 263611_at | AT2G16450 | F-box family protein | 5.28 |
| 595 | 265404_at | AT2G16730 | BGAL13 (beta-galactosidase 13); beta-galactosidase | 10.48 |
| 596 | 265405_at | AT2G16750 | protein kinase family protein | 5.78 |
| 597 | 266526_at | AT2G16980 | tetracycline transporter | 4.57 |
| 598 | 263417_at | AT2G17180 | zinc finger (C2H2 type) family protein | 5.88 |
| 599 | 264850_at | AT2G17340 | pantothenate kinase-related | -4.94 |
| 600 | 264856_at | AT2G17370 | HMG2 (3-HYDROXY-3-METHYLGLUTARYL-COA REDUCTASE 2) | 4.57 |
| 601 | 264854_at | AT2G17450 | RHA3A (RING-H2 finger A3A); protein binding / zinc ion binding | -4.01 |
| 602 | 263073_at | AT2G17500 | auxin efflux carrier family protein | 6.59 |
| 603 | 264617_at | AT2G17660 | nitrate-responsive NOI protein, putative | 6.97 |
| 604 | 264619_at | AT2G17760 | aspartyl protease family protein | -4.00 |
| 605 | 264813_at | AT2G17890 | CPK16 (calcium-dependent protein kinase 16); calmodulin-dependent protein kinase | 9.45 |
| 606 | 265811_at | AT2G18080 | EDA2 (embryo sac development arrest 2); serine-type peptidase | 7.38 |
| 607 | 263062_at | AT2G18180 | SEC14 cytosolic factor, putative / phosphoglyceride transfer protein, putative | 9.70 |
| 608 | 265339_at | AT2G18230 | ATPPA2 (ARABIDOPSIS THALIANA PYROPHOSPHORYLASE 2); inorganic diphosphatase/ pyrophosphatase | -4.85 |
| 609 | 265323_at | AT2G18260 | SYP112 (syntaxin 112); SNAP receptor | 4.68 |
| 610 | 265321_at | AT2G18280 | AtTLP2 (TUBBY LIKE PROTEIN 2); phosphoric diester hydrolase/ transcription factor | 4.82 |
| 611 | 265379_at | AT2G18340 | late embryogenesis abundant domain-containing protein / LEA domain-containing protein | 6.68 |
| 612 | 265923_at | AT2G18470 | protein kinase family protein | 10.52 |
| 613 | 267443_at | AT2G19000 | similar to glycine-rich protein [Arabidopsis thaliana] (TAIR:AT4G21620.1); similar to hypothetical protein OsI_037979 [Oryza sativa (indica cultivar-group)] (GB:EAY84020.1); similar to expressed protein [Oryza sativa (japonica cultivar-group)] (GB:ABA99958.1); contains InterPro domain 4-disulphide core (InterPro:IPR015874) | 9.85 |
| 614 | 267466_at | AT2G19010 | GDSL-motif lipase/hydrolase family protein | 6.45 |
| 615 | 267438_at | AT2G19050 | GDSL-motif lipase/hydrolase family protein | 8.30 |
| 616 | 267322_at | AT2G19330 | leucine-rich repeat family protein | 6.19 |
| 617 | 267281_at | AT2G19400 | protein kinase, putative | 6.32 |
| 618 | 267280_at | AT2G19450 | TAG1 (TRIACYLGLYCEROL BIOSYNTHESIS DEFECT 1); diacylglycerol O-acyltransferase | 5.37 |
| 619 | 266697_at | AT2G19770 | PRF5 (PROFILIN5); actin binding / actin monomer binding | 10.98 |
| 620 | 266693_at | AT2G19800 | MIOX2 (MYO-INOSITOL OXYGENASE 2) | 5.49 |
| 621 | 266703_at | AT2G19880 | ceramide glucosyltransferase, putative | 5.62 |
| 622 | 265587_at | AT2G19980 | allergen V5/Tpx-1-related family protein | 10.23 |
| 623 | 265576_at | AT2G20190 | ATCLASP/CLASP; binding | -4.07 |
| 624 | 265287_at | AT2G20260 | PSAE-2 (photosystem I subunit E-2); catalytic | -4.84 |
| 625 | 265306_at | AT2G20320 | DENN (AEX-3) domain-containing protein | 7.29 |
| 626 | 257397_at | AT2G20430 | RIC6 (ROP-INTERACTIVE CRIB MOTIF-CONTAINING PROTEIN 6) | 6.71 |
| 627 | 263371_at | AT2G20490 | EDA27/NOP10 (embryo sac development arrest 27); RNA binding | -3.85 |
| 628 | 263714_at | AT2G20610 | SUR1 (SUPERROOT 1); transaminase | -6.84 |
| 629 | 263711_at | AT2G20630 | protein phosphatase 2C, putative / PP2C, putative | -7.37 |
| 630 | 265430_at | AT2G20700 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G28280.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO69731.1) | 8.57 |
| 631 | 264018_at | AT2G21170 | TIM (TRIOSEPHOSPHATE ISOMERASE); triose-phosphate isomerase | -6.11 |
| 632 | 264014_at | AT2G21210 | auxin-responsive protein, putative | -3.81 |
| 633 | 263760_at | AT2G21280 | GC1 (GIANT CHLOROPLAST 1); binding / catalytic/ coenzyme binding | -5.70 |
| 634 | 263739_at | AT2G21320 | zinc finger (B-box type) family protein | -4.63 |
| 635 | 263761_at | AT2G21330 | fructose-bisphosphate aldolase, putative | -7.16 |
| 636 | 263763_at | AT2G21385 | similar to Os09g0531100 [Oryza sativa (japonica cultivar-group)] (GB:NP_001063753.1); similar to unknown [Picea sitchensis] (GB:ABK27019.1) | -5.36 |
| 637 | 263766_at | AT2G21440 | RNA recognition motif (RRM)-containing protein | -3.74 |
| 638 | 263748_at | AT2G21480 | protein kinase family protein | 8.66 |
| 639 | 263765_at | AT2G21540 | SEC14 cytosolic factor, putative / phosphoglyceride transfer protein, putative | 8.06 |
| 640 | 263876_at | AT2G21880 | AtRABG2/AtRab7A (Arabidopsis Rab GTPase homolog G2); GTP binding | 5.37 |
| 641 | 263880_at | AT2G21960 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G56180.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO71767.1) | -4.23 |
| 642 | 257433_at | AT2G21990 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G39610.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO71771.1); contains InterPro domain Protein of unknown function DUF617, plant (InterPro:IPR006460) | 9.02 |
| 643 | 263453_at | AT2G22180 | hydroxyproline-rich glycoprotein family protein | 9.79 |
| 644 | 263458_at | AT2G22290 | AtRABH1d (Arabidopsis Rab GTPase homolog H1d); GTP binding | 8.45 |
| 645 | 264051_at | AT2G22340 | unknown protein | 9.06 |
| 646 | 266799_at | AT2G22860 | ATPSK2 (PHYTOSULFOKINE 2 PRECURSOR); growth factor | 5.61 |
| 647 | 266801_at | AT2G22870 | EMB2001 (EMBRYO DEFECTIVE 2001); GTP binding | -4.02 |
| 648 | 267255_at | AT2G22950 | calcium-transporting ATPase, plasma membrane-type, putative / Ca2+-ATPase, putative (ACA7) | 6.63 |
| 649 | 267264_at | AT2G22970 | SCPL11; serine carboxypeptidase | -5.50 |
| 650 | 267134_at | AT2G23450 | protein kinase family protein | -4.38 |
| 651 | 267296_at | AT2G23660 | LBD10 (LOB DOMAIN-CONTAINING PROTEIN 10) | 5.10 |
| 652 | 267294_at | AT2G23670 | YCF37 (Arabidopsis homolog of Synechocystis YCF37) | -6.36 |
| 653 | 266558_at | AT2G23900 | glycoside hydrolase family 28 protein / polygalacturonase (pectinase) family protein | 10.99 |
| 654 | 266521_at | AT2G24020 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G30620.1); similar to unknown [Picea sitchensis] (GB:ABK26000.1); similar to Os02g0180200 [Oryza sativa (japonica cultivar-group)] (GB:NP_001046090.1); contains InterPro domain Conserved hypothetical protein CHP00103 (InterPro:IPR004401) | -4.59 |
| 655 | 266566_at | AT2G24040 | hydrophobic protein, putative / low temperature and salt responsive protein, putative | -5.30 |
| 656 | 265999_at | AT2G24100 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G30780.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO66612.1) | 5.01 |
| 657 | 265998_at | AT2G24270 | ALDH11A3 (Aldehyde dehydrogenase 11A3); 3-chloroallyl aldehyde dehydrogenase/ glyceraldehyde-3-phosphate dehydrogenase (NADP+) | -5.00 |
| 658 | 265690_at | AT2G24320 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G31020.2); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G31020.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO22751.1); contains domain PTHR12277:SF8 (PTHR12277:SF8); contains domain PTHR12277 (PTHR12277); contains domain SSF53474 (SSF53474); contains domain G3DSA:3.40.50.1820 (G3DSA:3.40.50.1820) | 6.21 |
| 659 | 265681_at | AT2G24370 | kinase | 9.56 |
| 660 | 257392_at | AT2G24450 | FLA3 (FASCICLIN-LIKE ARABINOGALACTAN PROTEIN 3 PRECURSOR) | 12.43 |
| 661 | 263318_at | AT2G24762 | ATGDU4 (ARABIDOPSIS THALIANA GLUTAMINE DUMPER 4) | -3.99 |
| 662 | 263537_at | AT2G24790 | COL3 (CONSTANS-LIKE 3); protein binding / transcription factor/ zinc ion binding | -5.59 |
| 663 | 264376_at | AT2G25070 | protein phosphatase 2C, putative / PP2C, putative | -6.53 |
| 664 | 263640_at | AT2G25270 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G12400.1); similar to Os02g0799300 [Oryza sativa (japonica cultivar-group)] (GB:NP_001048408.1); similar to hypothetical protein OsJ_008445 [Oryza sativa (japonica cultivar-group)] (GB:EAZ24962.1) | -4.08 |
| 665 | 263638_at | AT2G25310 | carbohydrate binding | -4.10 |
| 666 | 265615_at | AT2G25450 | 2-oxoglutarate-dependent dioxygenase, putative | -4.17 |
| 667 | 265618_at | AT2G25460 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G04860.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN68625.1); contains InterPro domain C2 calcium-dependent membrane targeting (InterPro:IPR000008) | 5.44 |
| 668 | 265611_at | AT2G25510 | unknown protein | -4.54 |
| 669 | 265903_at | AT2G25600 | SPIK (SHAKER POLLEN INWARD K+ CHANNEL); cyclic nucleotide binding / inward rectifier potassium channel/ potassium channel | 10.57 |
| 670 | 265904_at | AT2G25630 | glycosyl hydrolase family 1 protein | 7.37 |
| 671 | 266662_at | AT2G25830 | YebC-related | -3.95 |
| 672 | 266655_at | AT2G25880 | ATAUR2 (ATAURORA2); histone serine kinase(H3-S10 specific) / kinase | 6.37 |
| 673 | 266654_at | AT2G25890 | glycine-rich protein / oleosin | 8.48 |
| 674 | 267377_at | AT2G26250 | FDH (FIDDLEHEAD); acyltransferase | -3.92 |
| 675 | 267375_at | AT2G26300 | GPA1 (G PROTEIN ALPHA SUBUNIT 1); signal transducer | -5.79 |
| 676 | 267376_at | AT2G26330 | ER (ERECTA) | -4.51 |
| 677 | 245036_at | AT2G26410 | IQD4 (IQ-domain 4); calmodulin binding | 11.06 |
| 678 | 245053_at | AT2G26450 | pectinesterase family protein | 11.08 |
| 679 | 245057_at | AT2G26490 | transducin family protein / WD-40 repeat family protein | 10.47 |
| 680 | 245044_at | AT2G26500 | cytochrome b6f complex subunit (petM), putative | -5.21 |
| 681 | 245038_at | AT2G26560 | PLP2 (PHOSPHOLIPASE A 2A); nutrient reservoir | -3.72 |
| 682 | 267617_at | AT2G26670 | HY1 (HEME OXYGENASE 1) | -4.00 |
| 683 | 267612_at | AT2G26690 | nitrate transporter (NTP2) | -4.62 |
| 684 | 267619_at | AT2G26730 | leucine-rich repeat transmembrane protein kinase, putative | -4.16 |
| 685 | 266850_at | AT2G26850 | F-box family protein | 10.13 |
| 686 | 266313_at | AT2G26980 | CIPK3 (CBL-INTERACTING PROTEIN KINASE 3); kinase/ protein kinase | -3.88 |
| 687 | 266305_at | AT2G27120 | POL2B/TIL2 (TILTED2); DNA-directed DNA polymerase | 5.11 |
| 688 | 263084_at | AT2G27180 | unknown protein | 10.79 |
| 689 | 263083_at | AT2G27190 | PAP1 (PURPLE ACID PHOSPHATASE 1); acid phosphatase/ protein serine/threonine phosphatase | -4.98 |
| 690 | 265646_at | AT2G27360 | lipase, putative | -4.78 |
| 691 | 266160_at | AT2G28180 | ATCHX8 (cation/hydrogen exchanger 8); monovalent cation:proton antiporter | 9.55 |
| 692 | 265573_at | AT2G28200 | nucleic acid binding / transcription factor/ zinc ion binding | -3.75 |
| 693 | 265280_at | AT2G28355 | LCR5 (Low-molecular-weight cysteine-rich 5) | 8.27 |
| 694 | 265275_at | AT2G28440 | proline-rich family protein | 9.40 |
| 695 | 263440_at | AT2G28640 | ATEXO70H5 (exocyst subunit EXO70 family protein H5); protein binding | 7.74 |
| 696 | 257442_at | AT2G28680 | cupin family protein | 6.82 |
| 697 | 263412_at | AT2G28720 | histone H2B, putative | -5.52 |
| 698 | 266773_at | AT2G29040 | exostosin family protein | 6.34 |
| 699 | 266774_at | AT2G29050 | ATRBL1 (ARABIDOPSIS THALIANA RHOMBOID-LIKE 1) | 7.34 |
| 700 | 266273_at | AT2G29410 | MTPB1; efflux transmembrane transporter/ zinc ion transmembrane transporter | 6.72 |
| 701 | 266299_at | AT2G29450 | ATGSTU5 (Arabidopsis thaliana Glutathione S-transferase (class tau) 5); glutathione transferase | -5.51 |
| 702 | 266294_at | AT2G29500 | 17.6 kDa class I small heat shock protein (HSP17.6B-CI) | 5.00 |
| 703 | 266674_at | AT2G29620 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G07330.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN81231.1) | 6.76 |
| 704 | 266665_at | AT2G29790 | Encodes a Maternally expressed gene (MEG) family protein [pseudogene] | 9.27 |
| 705 | 266645_at | AT2G29880 | Identical to Uncharacterized protein At2g29880 [Arabidopsis Thaliana] (GB:O82368;GB:Q1PEY9); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G27260.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN74698.1) | 6.00 |
| 706 | 266866_at | AT2G29940 | ATPDR3/PDR3 (PLEIOTROPIC DRUG RESISTANCE 3); ATPase, coupled to transmembrane movement of substances | 4.69 |
| 707 | 266805_at | AT2G30010 | similar to PMR5 (POWDERY MILDEW RESISTANT 5) [Arabidopsis thaliana] (TAIR:AT5G58600.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO21434.1); contains InterPro domain Protein of unknown function DUF231, plant (InterPro:IPR004253) | -3.98 |
| 708 | 267275_at | AT2G30240 | ATCHX13 (cation/H+ exchanger 13); monovalent cation:proton antiporter | 6.28 |
| 709 | 255871_at | AT2G30260 | U2B'' (U2 small nuclear ribonucleoprotein B); RNA binding | -4.93 |
| 710 | 255861_at | AT2G30290 | vacuolar sorting receptor, putative | 5.56 |
| 711 | 267470_at | AT2G30490 | ATC4H/C4H/CYP73A5 (CINNAMATE 4-HYDROXYLASE, CINNAMATE-4-HYDROXYLASE); trans-cinnamate 4-monooxygenase | -5.13 |
| 712 | 267517_at | AT2G30520 | RPT2 (ROOT PHOTOTROPISM 2); protein binding | -4.38 |
| 713 | 267526_at | AT2G30570 | PSBW (PHOTOSYSTEM II REACTION CENTER W) | -5.22 |
| 714 | 267492_at | AT2G30620 | histone H1.2 | -4.80 |
| 715 | 257349_at | AT2G30630 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G06750.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN76299.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO22658.1); contains domain SSF52540 (SSF52540); contains domain G3DSA:3.40.50.300 (G3DSA:3.40.50.300) | 5.16 |
| 716 | 267575_at | AT2G30690 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G08800.2); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G08800.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO70635.1); contains InterPro domain Protein of unknown function DUF593 (InterPro:IPR007656) | 5.78 |
| 717 | 267153_at | AT2G30860 | ATGSTF9 (Arabidopsis thaliana Glutathione S-transferase (class phi) 9); glutathione transferase | -4.67 |
| 718 | 267209_at | AT2G30930 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G06540.1) | -4.55 |
| 719 | 267196_at | AT2G30950 | VAR2 (VARIEGATED 2); ATP-dependent peptidase/ ATPase/ metallopeptidase/ zinc ion binding | -3.95 |
| 720 | 267199_at | AT2G30990 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G29240.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN81695.1); contains InterPro domain Protein of unknown function DUF688 (InterPro:IPR007789) | -4.17 |
| 721 | 263450_at | AT2G31500 | CPK24 (calcium-dependent protein kinase 24); calmodulin-dependent protein kinase/ kinase | 12.41 |
| 722 | 263426_at | AT2G31570 | ATGPX2 (GLUTATHIONE PEROXIDASE 2); glutathione peroxidase | -4.06 |
| 723 | 263477_at | AT2G31790 | UDP-glucoronosyl/UDP-glucosyl transferase family protein | -3.77 |
| 724 | 263489_at | AT2G31830 | endonuclease/exonuclease/phosphatase family protein | 6.55 |
| 725 | 263478_at | AT2G31880 | leucine-rich repeat transmembrane protein kinase, putative | -3.82 |
| 726 | 267087_at | AT2G32460 | AtM1/AtMYB101/MYB101 (myb domain protein 101); DNA binding / transcription factor | 8.51 |
| 727 | 267547_at | AT2G32670 | ATVAMP725 (Arabidopsis thaliana vesicle-associated membrane protein 725) | 4.64 |
| 728 | 267643_at | AT2G32890 | RALFL17 (RALF-LIKE 17) | 6.13 |
| 729 | 245159_at | AT2G33100 | ATCSLD1 (Cellulose synthase-like D1); cellulose synthase/ transferase, transferring glycosyl groups | 10.46 |
| 730 | 255847_at | AT2G33270 | thioredoxin family protein | 9.24 |
| 731 | 255849_at | AT2G33320 | C2 domain-containing protein | 8.92 |
| 732 | 255831_at | AT2G33350 | similar to zinc finger CONSTANS-related [Arabidopsis thaliana] (TAIR:AT1G04500.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN83004.1); contains InterPro domain CCT (InterPro:IPR010402) | 5.24 |
| 733 | 255835_at | AT2G33420 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G25800.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G20010.2); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G04470.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO71451.1); similar to hypothetical protein LOC_Os03g04560 [Oryza sativa (japonica cultivar-group)] (GB:ABF93881.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN63332.1); contains InterPro domain Munc13 homology 2 (InterPro:IPR014772); contains InterPro domain Protein of unknown function DUF810 (InterPro:IPR008528); contains InterPro domain Munc13 homology 1 (InterPro:IPR014770) | 10.47 |
| 734 | 255837_at | AT2G33460 | RIC1 (ROP-INTERACTIVE CRIB MOTIF-CONTAINING PROTEIN 1) | 8.92 |
| 735 | 255818_at | AT2G33570 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G44670.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO66312.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN76802.1); contains InterPro domain Protein of unknown function DUF23 (InterPro:IPR008166) | -5.40 |
| 736 | 255846_at | AT2G33610 | ATSWI3B (Arabidopsis thaliana switching protein 3B); DNA binding | -4.04 |
| 737 | 267458_at | AT2G33670 | MLO5 (MILDEW RESISTANCE LOCUS O 5); calmodulin binding | 6.55 |
| 738 | 267449_at | AT2G33690 | late embryogenesis abundant protein, putative / LEA protein, putative | 9.91 |
| 739 | 267461_at | AT2G33830 | dormancy/auxin associated family protein | -3.98 |
| 740 | 267447_at | AT2G33870 | ArRABA1h (Arabidopsis Rab GTPase homolog A1h); GTP binding | 10.62 |
| 741 | 256724_at | AT2G34040 | apoptosis inhibitory 5 (API5) family protein | -3.84 |
| 742 | 267044_at | AT2G34357 | binding | -4.02 |
| 743 | 266994_at | AT2G34440 | AGL29; transcription factor | 7.59 |
| 744 | 266956_at | AT2G34510 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G29980.1); similar to unknown [Populus trichocarpa] (GB:ABK95079.1); contains InterPro domain Protein of unknown function DUF642 (InterPro:IPR006946); contains InterPro domain Galactose-binding like (InterPro:IPR008979) | -4.87 |
| 745 | 266905_at | AT2G34560 | katanin, putative | -6.72 |
| 746 | 267318_at | AT2G34770 | FAH1 (FATTY ACID HYDROXYLASE 1); catalytic | 4.93 |
| 747 | 267430_at | AT2G34860 | EDA3 (embryo sac development arrest 3); heat shock protein binding / unfolded protein binding | -3.97 |
| 748 | 266548_at | AT2G35210 | AGD10/MEE28/RPA (MATERNAL EFFECT EMBRYO ARREST 28); DNA binding | 8.72 |
| 749 | 266551_at | AT2G35260 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G17840.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN74782.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO63413.1) | -5.32 |
| 750 | 266636_at | AT2G35370 | GDCH (Glycine decarboxylase complex H) | -6.22 |
| 751 | 263942_at | AT2G35860 | FLA16 (FASCICLIN-LIKE ARABINOGALACTAN PROTEIN 16 PRECURSOR) | -4.92 |
| 752 | 263940_at | AT2G35890 | CPK25 (calcium-dependent protein kinase 25); calmodulin-dependent protein kinase/ kinase | 6.73 |
| 753 | 263950_at | AT2G36020 | HVA22J (HVA22-LIKE PROTEIN J) | 10.89 |
| 754 | 263287_at | AT2G36145 | similar to unnamed protein product [Vitis vinifera] (GB:CAO15876.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN74693.1) | -5.34 |
| 755 | 263905_at | AT2G36190 | ATCWINV4 (ARABIDOPSIS THALIANA CELL WALL INVERTASE 4); hydrolase, hydrolyzing O-glycosyl compounds | 4.82 |
| 756 | 263901_at | AT2G36320 | zinc finger (AN1-like) family protein | 5.07 |
| 757 | 263920_at | AT2G36410 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G52920.1); similar to unknown [Populus trichocarpa] (GB:ABK93998.1); contains InterPro domain Protein of unknown function DUF662 (InterPro:IPR007033) | -4.28 |
| 758 | 265205_at | AT2G36660 | PAB7 (POLY(A) BINDING PROTEIN 7); RNA binding / translation initiation factor | 6.78 |
| 759 | 263867_at | AT2G36830 | GAMMA-TIP (Tonoplast intrinsic protein (TIP) gamma); water channel | -4.21 |
| 760 | 263865_at | AT2G36910 | ATPGP1 (ARABIDOPSIS THALIANA P GLYCOPROTEIN1); calmodulin binding | -5.15 |
| 761 | 263889_at | AT2G37010 | ATNAP12 (Arabidopsis thaliana non-intrinsic ABC protein 12) | 6.34 |
| 762 | 263845_at | AT2G37040 | PAL1 (PHE AMMONIA LYASE 1); phenylalanine ammonia-lyase | -3.85 |
| 763 | 265467_at | AT2G37050 | kinase | -5.42 |
| 764 | 265465_at | AT2G37070 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G53320.1) | 6.73 |
| 765 | 265464_at | AT2G37080 | myosin heavy chain-related | -4.76 |
| 766 | 265961_at | AT2G37400 | chloroplast lumen common family protein | -4.17 |
| 767 | 267156_at | AT2G37610 | unknown protein | 5.92 |
| 768 | 267172_at | AT2G37660 | binding / catalytic/ coenzyme binding | -4.73 |
| 769 | 267160_at | AT2G37670 | WD-40 repeat family protein | 6.95 |
| 770 | 266100_at | AT2G37980 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G01100.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G54100.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO15763.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN72579.1); contains InterPro domain Protein of unknown function DUF246, plant (InterPro:IPR004348) | 7.09 |
| 771 | 267088_at | AT2G38140 | PSRP4 (PLASTID-SPECIFIC RIBOSOMAL PROTEIN 4); structural constituent of ribosome | -6.37 |
| 772 | 267093_at | AT2G38170 | CAX1 (CATION EXCHANGER 1); calcium ion transmembrane transporter/ calcium:hydrogen antiporter | -4.35 |
| 773 | 267034_at | AT2G38310 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G05440.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO48777.1); contains InterPro domain Bet v I allergen; (InterPro:IPR000916); contains InterPro domain Streptomyces cyclase/dehydrase (InterPro:IPR005031) | -4.84 |
| 774 | 267035_at | AT2G38400 | AGT3 (ALANINE:GLYOXYLATE AMINOTRANSFERASE 3); alanine-glyoxylate transaminase | 6.21 |
| 775 | 267030_at | AT2G38440 | ITB1 (IRREGULAR TRICHOME BRANCH1) | -4.44 |
| 776 | 267051_at | AT2G38500 | similar to DTA4 (DOWNSTREAM TARGET OF AGL15-4) [Arabidopsis thaliana] (TAIR:AT1G79760.1); similar to hypothetical protein [Oryza sativa (japonica cultivar-group)] (GB:BAD87077.1); contains domain SSF51197 (SSF51197) | 11.08 |
| 777 | 266408_at | AT2G38520 | transposable element gene | 5.68 |
| 778 | 266421_at | AT2G38540 | LP1 (nonspecific lipid transfer protein 1) | -7.92 |
| 779 | 266420_at | AT2G38610 | KH domain-containing protein | -4.16 |
| 780 | 263296_at | AT2G38800 | calmodulin-binding protein-related | -5.03 |
| 781 | 266168_at | AT2G38870 | protease inhibitor, putative | -4.62 |
| 782 | 266169_at | AT2G38900 | serine protease inhibitor, potato inhibitor I-type family protein | 6.32 |
| 783 | 266143_at | AT2G38905 | hydrophobic protein, putative / low temperature and salt responsive protein, putative | 6.29 |
| 784 | 266199_at | AT2G38910 | CPK20 (calcium-dependent protein kinase 20); calmodulin-dependent protein kinase/ kinase | 6.46 |
| 785 | 266200_at | AT2G38920 | SPX (SYG1/Pho81/XPR1) domain-containing protein / zinc finger (C3HC4-type RING finger) protein-related | 7.26 |
| 786 | 266172_at | AT2G39010 | PIP2;6/PIP2E (plasma membrane intrinsic protein 2;6); water channel | -8.63 |
| 787 | 267012_at | AT2G39220 | PLA IIB/PLP6 (Patatin-like protein 6); nutrient reservoir | -4.30 |
| 788 | 266977_at | AT2G39420 | esterase/lipase/thioesterase family protein | 5.03 |
| 789 | 266963_at | AT2G39450 | ATMTP11/MTP11; cation transmembrane transporter/ manganese ion transmembrane transporter/ manganese:hydrogen antiporter | -4.39 |
| 790 | 267562_at | AT2G39670 | radical SAM domain-containing protein | -4.41 |
| 791 | 267590_at | AT2G39700 | ATEXPA4 (ARABIDOPSIS THALIANA EXPANSIN A4) | 6.62 |
| 792 | 245061_at | AT2G39730 | RCA (RUBISCO ACTIVASE) | -5.85 |
| 793 | 245063_at | AT2G39795 | mitochondrial glycoprotein family protein / MAM33 family protein | -3.83 |
| 794 | 245086_at | AT2G39820 | eukaryotic translation initiation factor 6, putative / eIF-6, putative | 5.90 |
| 795 | 267339_at | AT2G39870 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G55690.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO69095.1) | -3.74 |
| 796 | 267358_at | AT2G39890 | ProT1 (PROLINE TRANSPORTER 1); amino acid transmembrane transporter | 5.79 |
| 797 | 267360_at | AT2G40060 | protein binding / protein transporter/ structural molecule | -4.74 |
| 798 | 265721_at | AT2G40090 | ATATH9 (ABC2 homolog 9) | 5.71 |
| 799 | 265722_at | AT2G40100 | LHCB4.3 (LIGHT HARVESTING COMPLEX PSII); chlorophyll binding | -3.80 |
| 800 | 263378_at | AT2G40180 | ATHPP2C5; protein serine/threonine phosphatase | 8.83 |
| 801 | 263804_at | AT2G40270 | protein kinase family protein | -5.48 |
| 802 | 263836_at | AT2G40330 | Bet v I allergen family protein | -3.96 |
| 803 | 255826_at | AT2G40490 | HEME2; uroporphyrinogen decarboxylase | -3.80 |
| 804 | 255822_at | AT2G40610 | ATEXPA8 (ARABIDOPSIS THALIANA EXPANSIN A8) | -4.43 |
| 805 | 255878_at | AT2G40620 | bZIP transcription factor family protein | 5.88 |
| 806 | 257382_at | AT2G40750 | WRKY54 (WRKY DNA-binding protein 54); transcription factor | -4.26 |
| 807 | 266048_at | AT2G40790 | thioredoxin family protein | 8.28 |
| 808 | 257358_at | AT2G40990 | zinc finger (DHHC type) family protein | 9.33 |
| 809 | 267076_at | AT2G41090 | calmodulin-like calcium-binding protein, 22 kDa (CaBP-22) | -5.20 |
| 810 | 267083_at | AT2G41100 | TCH3 (TOUCH 3) | -4.21 |
| 811 | 267081_at | AT2G41210 | phosphatidylinositol-4-phosphate 5-kinase family protein | 6.12 |
| 812 | 267104_at | AT2G41430 | ERD15 (EARLY RESPONSIVE TO DEHYDRATION 15) | -6.52 |
| 813 | 260501_at | AT2G41770 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G57420.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO23590.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN82225.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO40788.1) | -4.09 |
| 814 | 260494_at | AT2G41820 | leucine-rich repeat transmembrane protein kinase, putative | -4.79 |
| 815 | 267531_at | AT2G41860 | CPK14 (calcium-dependent protein kinase 14); calmodulin-dependent protein kinase/ kinase | 11.33 |
| 816 | 267534_at | AT2G41900 | zinc finger (CCCH-type) family protein | -3.97 |
| 817 | 267582_at | AT2G41970 | protein kinase, putative | 7.76 |
| 818 | 267630_at | AT2G42130 | Identical to Probable plastid-lipid-associated protein 13, chloroplast precursor (PAP13) [Arabidopsis Thaliana] (GB:Q8S9M1;GB:O48521;GB:Q84X37;GB:Q84X38;GB:Q84X39;GB:Q8GY49); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G58010.1); similar to unknown [Populus trichocarpa] (GB:ABK96151.1) | -5.25 |
| 819 | 267635_at | AT2G42220 | rhodanese-like domain-containing protein | -5.42 |
| 820 | 265877_at | AT2G42380 | bZIP transcription factor family protein | 5.62 |
| 821 | 263495_at | AT2G42530 | COR15B | -4.71 |
| 822 | 263497_at | AT2G42540 | COR15A (COLD-REGULATED 15A) | -4.44 |
| 823 | 263491_at | AT2G42600 | ATPPC2 (PHOSPHOENOLPYRUVATE CARBOXYLASE 2); phosphoenolpyruvate carboxylase | -3.91 |
| 824 | 263987_at | AT2G42690 | lipase, putative | -3.73 |
| 825 | 265247_at | AT2G43030 | ribosomal protein L3 family protein | -6.39 |
| 826 | 265243_at | AT2G43040 | NPG1 (NO POLLEN GERMINATION 1); calmodulin binding | 5.33 |
| 827 | 266436_at | AT2G43160 | epsin N-terminal homology (ENTH) domain-containing protein | 4.57 |
| 828 | 266453_at | AT2G43230 | serine/threonine protein kinase, putative | 9.43 |
| 829 | 260547_at | AT2G43550 | trypsin inhibitor, putative | -4.90 |
| 830 | 260542_at | AT2G43560 | immunophilin / FKBP-type peptidyl-prolyl cis-trans isomerase family protein | -4.53 |
| 831 | 260610_at | AT2G43680 | IQD14; calmodulin binding | -4.32 |
| 832 | 260566_at | AT2G43750 | OASB (O-ACETYLSERINE (THIOL) LYASE B); cysteine synthase | -4.08 |
| 833 | 260567_at | AT2G43820 | GT/UGT74F2 (UDP-GLUCOSYLTRANSFERASE 74F2); UDP-glucosyltransferase/ UDP-glycosyltransferase/ transferase, transferring glycosyl groups / transferase, transferring hexosyl groups | -4.03 |
| 834 | 267214_at | AT2G43970 | La domain-containing protein | -5.26 |
| 835 | 267236_at | AT2G44100 | ATGDI1 (Arabidopsis thaliana guanosine diphosphate dissociation inhibitor 1) | -6.36 |
| 836 | 267367_at | AT2G44210 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G55360.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO67164.1); contains InterPro domain Protein of unknown function DUF239, plant (InterPro:IPR004314) | -4.25 |
| 837 | 267392_at | AT2G44490 | PEN2 (PENETRATION 2); hydrolase, hydrolyzing O-glycosyl compounds | -4.14 |
| 838 | 267398_at | AT2G44560 | ATGH9B11 (ARABIDOPSIS THALIANA GLYCOSYL HYDROLASE 9B11); hydrolase, hydrolyzing O-glycosyl compounds | 12.36 |
| 839 | 266882_at | AT2G44670 | senescence-associated protein-related | -4.32 |
| 840 | 266870_at | AT2G44710 | RNA recognition motif (RRM)-containing protein | 4.58 |
| 841 | 266123_at | AT2G45180 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | -5.17 |
| 842 | 245133_at | AT2G45310 | GAE4 (UDP-D-GLUCURONATE 4-EPIMERASE 4); catalytic | 7.17 |
| 843 | 245130_at | AT2G45340 | leucine-rich repeat transmembrane protein kinase, putative | -3.94 |
| 844 | 251395_at | AT2G45470 | FLA8 (Arabinogalactan protein 8) | -6.96 |
| 845 | 267527_at | AT2G45610 | hydrolase | 7.08 |
| 846 | 267509_at | AT2G45660 | AGL20 (AGAMOUS-LIKE 20); transcription factor | -3.98 |
| 847 | 266918_at | AT2G45800 | LIM domain-containing protein | 11.60 |
| 848 | 266927_at | AT2G45960 | PIP1B (plasma membrane intrinsic protein 1;2); water channel | -5.27 |
| 849 | 266603_at | AT2G46040 | ARID/BRIGHT DNA-binding domain-containing protein / ELM2 domain-containing protein | 5.34 |
| 850 | 266581_at | AT2G46140 | late embryogenesis abundant protein, putative / LEA protein, putative | 5.51 |
| 851 | 266592_at | AT2G46210 | delta-8 sphingolipid desaturase, putative | 9.27 |
| 852 | 266580_at | AT2G46260 | BTB/POZ domain-containing protein | 6.16 |
| 853 | 266607_at | AT2G46300 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G01453.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO39716.1); contains InterPro domain Harpin-induced 1 (InterPro:IPR010847) | 12.13 |
| 854 | 263781_at | AT2G46360 | unknown protein | 9.94 |
| 855 | 265451_at | AT2G46490 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G35110.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN80242.1) | 4.84 |
| 856 | 265461_at | AT2G46500 | phosphatidylinositol 3- and 4-kinase family protein / ubiquitin family protein | 5.25 |
| 857 | 266324_at | AT2G46710 | rac GTPase activating protein, putative | -5.20 |
| 858 | 266716_at | AT2G46820 | PSAP/PSI-P/PTAC8/TMP14 (THYLAKOID MEMBRANE PHOSPHOPROTEIN OF 14 KDA); DNA binding | -4.88 |
| 859 | 266765_at | AT2G46860 | ATPPA3 (ARABIDOPSIS THALIANA PYROPHOSPHORYLASE 3); inorganic diphosphatase/ pyrophosphatase | 11.76 |
| 860 | 266764_at | AT2G47050 | invertase/pectin methylesterase inhibitor family protein | 12.89 |
| 861 | 266763_at | AT2G47070 | SPL1 (SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 1); DNA binding / transcription factor | -3.82 |
| 862 | 266762_at | AT2G47120 | short-chain dehydrogenase/reductase (SDR) family protein | 8.08 |
| 863 | 263319_at | AT2G47160 | BOR1 (REQUIRES HIGH BORON 1); anion exchanger | -3.78 |
| 864 | 260577_at | AT2G47340 | invertase/pectin methylesterase inhibitor family protein | 10.65 |
| 865 | 260529_at | AT2G47400 | CP12-1 (CP12 domain-containing protein 1) | -5.39 |
| 866 | 245176_at | AT2G47440 | DNAJ heat shock N-terminal domain-containing protein | -4.00 |
| 867 | 245123_at | AT2G47450 | CAO (CHAOS); chromatin binding | -3.99 |
| 868 | 266458_at | AT2G47710 | universal stress protein (USP) family protein | -4.73 |
| 869 | 266462_at | AT2G47770 | benzodiazepine receptor-related | 4.98 |
| 870 | 266505_at | AT2G47830 | cation efflux family protein / metal tolerance protein, putative (MTPc1) | -4.02 |
| 871 | 266514_at | AT2G47890 | zinc finger (B-box type) family protein | -3.92 |
| 872 | 266483_at | AT2G47910 | CRR6 (CHLORORESPIRATORY REDUCTION 6) | -4.55 |
| 873 | 266460_at | AT2G47930 | AGP26/ATAGP26 (ARABINOGALACTAN PROTEINS 26) | -5.13 |
| 874 | 266509_at | AT2G47940 | DEGP2 (DEGP PROTEASE 2); serine-type peptidase/ trypsin | -4.19 |
| 875 | 262350_at | AT2G48150 | ATGPX4 (GLUTATHIONE PEROXIDASE 4); glutathione peroxidase | 9.90 |
| 876 | 258442_at | AT3G01015 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G15510.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO15061.1); contains InterPro domain Targeting for Xklp2 (InterPro:IPR009675) | 5.13 |
| 877 | 259262_at | AT3G01020 | ATISU2/ISU2 (IscU-like 2); structural molecule | 8.83 |
| 878 | 259321_at | AT3G01040 | GAUT13 (Galacturonosyltransferase 13); polygalacturonate 4-alpha-galacturonosyltransferase/ transferase, transferring glycosyl groups / transferase, transferring hexosyl groups | 4.96 |
| 879 | 259266_at | AT3G01240 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G01230.1) | 11.72 |
| 880 | 259265_at | AT3G01250 | unknown protein | 13.36 |
| 881 | 259269_at | AT3G01270 | pectate lyase family protein | 10.46 |
| 882 | 259272_at | AT3G01290 | band 7 family protein | -4.08 |
| 883 | 259161_at | AT3G01500 | CA1 (CARBONIC ANHYDRASE 1); carbonate dehydratase/ zinc ion binding | -7.53 |
| 884 | 257523_at | AT3G01620 | glycosyl transferase family 17 protein | 8.36 |
| 885 | 259177_at | AT3G01630 | similar to NFD4 (NUCLEAR FUSION DEFECTIVE 4) [Arabidopsis thaliana] (TAIR:AT1G31470.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO63775.1); contains InterPro domain MFS general substrate transporter (InterPro:IPR016196); contains InterPro domain Nodulin-like (InterPro:IPR010658) | 10.75 |
| 886 | 259181_at | AT3G01690 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G14390.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO14651.1); contains InterPro domain Alpha/beta hydrolase fold-1 (InterPro:IPR000073) | -6.00 |
| 887 | 259189_at | AT3G01700 | AGP11 (ARABINOGALACTAN PROTEIN 11) | 13.64 |
| 888 | 258999_at | AT3G01850 | ribulose-phosphate 3-epimerase, cytosolic, putative / pentose-5-phosphate 3-epimerase, putative | -3.98 |
| 889 | 258975_at | AT3G01970 | WRKY45 (WRKY DNA-binding protein 45); transcription factor | 5.26 |
| 890 | 258856_at | AT3G02040 | SRG3 (SENESCENCE-RELATED GENE 3); glycerophosphodiester phosphodiesterase | 5.07 |
| 891 | 259074_at | AT3G02130 | RPK2/TOAD2 (RECEPTOR-LIKE PROTEIN KINASE2); ATP binding / kinase/ protein serine/threonine kinase | -4.29 |
| 892 | 258479_at | AT3G02440 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G15900.1); similar to hypothetical protein [Cleome spinosa] (GB:ABD96951.1); contains InterPro domain Protein of unknown function DUF231, plant (InterPro:IPR004253) | 6.38 |
| 893 | 258498_at | AT3G02480 | ABA-responsive protein-related | 9.89 |
| 894 | 258493_at | AT3G02555 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G16110.1); similar to hypothetical protein [Cleome spinosa] (GB:ABD96883.1) | 7.21 |
| 895 | 258485_at | AT3G02630 | acyl-(acyl-carrier-protein) desaturase, putative / stearoyl-ACP desaturase, putative | -5.70 |
| 896 | 258495_at | AT3G02690 | integral membrane family protein | -3.76 |
| 897 | 258607_at | AT3G02730 | ATF1/TRXF1 (THIOREDOXIN F-TYPE 1); thiol-disulfide exchange intermediate | -5.13 |
| 898 | 258600_at | AT3G02810 | protein kinase family protein | 14.42 |
| 899 | 258629_at | AT3G02850 | SKOR (stelar K+ outward rectifier); cyclic nucleotide binding / outward rectifier potassium channel | 6.09 |
| 900 | 258605_at | AT3G02970 | phosphate-responsive 1 family protein | 10.55 |
| 901 | 258870_at | AT3G03080 | NADP-dependent oxidoreductase, putative | 6.26 |
| 902 | 258869_at | AT3G03090 | ATVGT1 (ARABIDOPSIS THALIANA VACUOLAR GLUCOSE TRANSPORTER 1); carbohydrate transmembrane transporter/ fructose transmembrane transporter/ glucose transmembrane transporter/ sugar:hydrogen ion symporter | 5.62 |
| 903 | 258845_at | AT3G03150 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G17165.1); similar to seed specific protein Bn15D1B [Brassica napus] (GB:AAP37967.1) | -5.16 |
| 904 | 259044_at | AT3G03430 | polcalcin, putative / calcium-binding pollen allergen, putative | 10.14 |
| 905 | 259338_at | AT3G03800 | SYP131 (syntaxin 131); SNAP receptor | 10.88 |
| 906 | 259339_at | AT3G03900 | adenylylsulfate kinase, putative | 4.72 |
| 907 | 258812_at | AT3G03950 | ECT1; protein binding | -3.79 |
| 908 | 258573_at | AT3G04260 | PTAC3 (PLASTID TRANSCRIPTIONALLY ACTIVE3); DNA binding | -4.44 |
| 909 | 258589_at | AT3G04290 | ATLTL1/LTL1 (LI-TOLERANT LIPASE 1); carboxylesterase | -3.84 |
| 910 | 258591_at | AT3G04360 | C2 domain-containing protein | 9.88 |
| 911 | 258840_at | AT3G04620 | nucleic acid binding | 8.30 |
| 912 | 258796_at | AT3G04630 | WDL1 (WVD2-LIKE 1) | 6.15 |
| 913 | 258843_at | AT3G04690 | protein kinase family protein | 11.33 |
| 914 | 257532_at | AT3G04700 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G28690.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO65016.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN62670.1); contains InterPro domain Protein of unknown function DUF1685 (InterPro:IPR012881) | 10.31 |
| 915 | 258804_at | AT3G04760 | pentatricopeptide (PPR) repeat-containing protein | -4.55 |
| 916 | 259094_at | AT3G04940 | ATCYSD1 (Arabidopsis thaliana cysteine synthase D1); cysteine synthase | -4.84 |
| 917 | 259350_at | AT3G05140 | RBK2 (ROP BINDING PROTEIN KINASES 2); kinase | 6.95 |
| 918 | 259351_at | AT3G05150 | sugar transporter family protein | 10.87 |
| 919 | 259312_at | AT3G05200 | ATL6 (Arabidopsis T?xicos en Levadura 6); protein binding / zinc ion binding | -5.11 |
| 920 | 259354_at | AT3G05220 | heavy-metal-associated domain-containing protein | -3.81 |
| 921 | 259106_at | AT3G05490 | RALFL22 (RALF-LIKE 22) | -4.79 |
| 922 | 259109_at | AT3G05580 | serine/threonine protein phosphatase, putative | 5.07 |
| 923 | 258889_at | AT3G05610 | pectinesterase family protein | 13.57 |
| 924 | 258886_at | AT3G05720 | importin alpha-1 subunit, putative | 4.88 |
| 925 | 258732_at | AT3G05820 | beta-fructofuranosidase, putative / invertase, putative / saccharase, putative / beta-fructosidase, putative | 7.80 |
| 926 | 258735_at | AT3G05880 | RCI2A (RARE-COLD-INDUCIBLE 2A) | -4.36 |
| 927 | 258750_at | AT3G05910 | pectinacetylesterase, putative | -6.85 |
| 928 | 258748_at | AT3G05930 | GLP8 (GERMIN-LIKE PROTEIN 8); manganese ion binding / metal ion binding / nutrient reservoir | 11.39 |
| 929 | 258561_at | AT3G05960 | sugar transporter, putative | 5.53 |
| 930 | 258557_at | AT3G05990 | leucine-rich repeat family protein | 5.74 |
| 931 | 256396_at | AT3G06150 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G19060.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO71094.1); contains InterPro domain Immunoglobulin E-set (InterPro:IPR014756) | 5.14 |
| 932 | 256392_at | AT3G06260 | GATL4 (Galacturonosyltransferase-like 4); polygalacturonate 4-alpha-galacturonosyltransferase/ transferase, transferring glycosyl groups / transferase, transferring hexosyl groups | 10.27 |
| 933 | 258564_at | AT3G06560 | polynucleotide adenylyltransferase/ protein binding | 6.48 |
| 934 | 258523_at | AT3G06830 | pectinesterase family protein | 8.90 |
| 935 | 258545_at | AT3G07050 | GTP-binding family protein | -3.90 |
| 936 | 258832_at | AT3G07070 | protein kinase family protein | 7.67 |
| 937 | 258825_at | AT3G07180 | GPI transamidase component PIG-S-related | -4.32 |
| 938 | 259010_at | AT3G07340 | basic helix-loop-helix (bHLH) family protein | -5.20 |
| 939 | 259012_at | AT3G07360 | armadillo/beta-catenin repeat family protein / U-box domain-containing protein | -4.84 |
| 940 | 259064_at | AT3G07490 | AGD11 (ARF-GAP DOMAIN 11); calcium ion binding | 8.07 |
| 941 | 259230_at | AT3G07780 | protein binding / zinc ion binding | -4.31 |
| 942 | 258639_at | AT3G07820 | polygalacturonase 3 (PGA3) / pectinase | 13.48 |
| 943 | 258685_at | AT3G07830 | polygalacturonase, putative / pectinase, putative | 9.54 |
| 944 | 258686_at | AT3G07840 | polygalacturonase, putative / pectinase, putative | 8.06 |
| 945 | 258637_at | AT3G07880 | Rho GDP-dissociation inhibitor family protein | 5.14 |
| 946 | 258690_at | AT3G07960 | phosphatidylinositol-4-phosphate 5-kinase family protein | 6.87 |
| 947 | 258643_at | AT3G08010 | ATAB2; RNA binding | -3.89 |
| 948 | 258666_at | AT3G08550 | ABI8/ELD1/KOB1 (KOBITO) | -4.44 |
| 949 | 258671_at | AT3G08560 | VHA-E2 (VACUOLAR H+-ATPASE SUBUNIT E ISOFORM 2); hydrogen-exporting ATPase, phosphorylative mechanism | 8.97 |
| 950 | 258677_at | AT3G08730 | ATPK1 (P70 RIBOSOMAL S6 KINASE); kinase/ protein binding | 6.08 |
| 951 | 258993_at | AT3G08940 | LHCB4.2 (LIGHT HARVESTING COMPLEX PSII); chlorophyll binding | -9.01 |
| 952 | 258984_at | AT3G08970 | DNAJ heat shock N-terminal domain-containing protein | 5.26 |
| 953 | 257522_at | AT3G08990 | yippee family protein | 5.88 |
| 954 | 258707_at | AT3G09480 | histone H2B, putative | 4.64 |
| 955 | 258753_at | AT3G09530 | ATEXO70H3 (exocyst subunit EXO70 family protein H3); protein binding | 12.97 |
| 956 | 258720_at | AT3G09550 | ankyrin repeat family protein | 5.71 |
| 957 | 258723_at | AT3G09600 | myb family transcription factor | -3.98 |
| 958 | 258696_at | AT3G09650 | HCF152 (HIGH CHLOROPHYLL FLUORESCENCE 152) | -4.14 |
| 959 | 258717_at | AT3G09740 | SYP71 (SYNTAXIN OF PLANTS 71) | -5.09 |
| 960 | 258718_at | AT3G09760 | zinc finger (C3HC4-type RING finger) family protein | 6.15 |
| 961 | 258940_at | AT3G09930 | GDSL-motif lipase/hydrolase family protein | 5.74 |
| 962 | 258933_at | AT3G09980 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G03660.1); similar to unknown [Populus trichocarpa] (GB:ABK95581.1); similar to unknown [Populus trichocarpa] (GB:ABK93998.1); contains InterPro domain Protein of unknown function DUF662 (InterPro:IPR007033) | -5.46 |
| 963 | 259154_at | AT3G10260 | reticulon family protein | -5.98 |
| 964 | 258968_at | AT3G10460 | self-incompatibility protein-related | 7.86 |
| 965 | 258967_at | AT3G10470 | zinc finger (C2H2 type) family protein | 6.20 |
| 966 | 258964_at | AT3G10540 | 3-phosphoinositide-dependent protein kinase, putative | 4.88 |
| 967 | 258945_at | AT3G10660 | CPK2 (CALMODULIN-DOMAIN PROTEIN KINASE CDPK ISOFORM 2); calmodulin-dependent protein kinase/ kinase | 4.64 |
| 968 | 258764_at | AT3G10720 | pectinesterase, putative | -5.61 |
| 969 | 258760_at | AT3G10780 | emp24/gp25L/p24 family protein | 5.18 |
| 970 | 256435_at | AT3G11180 | oxidoreductase, 2OG-Fe(II) oxygenase family protein | 6.73 |
| 971 | 259288_at | AT3G11500 | small nuclear ribonucleoprotein G, putative / snRNP-G, putative / Sm protein G, putative | -4.94 |
| 972 | 259237_at | AT3G11630 | 2-cys peroxiredoxin, chloroplast (BAS1) | -4.26 |
| 973 | 259103_at | AT3G11690 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G06380.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO67416.1) | 7.15 |
| 974 | 258781_at | AT3G11740 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G01750.2); similar to unnamed protein product [Vitis vinifera] (GB:CAO48825.1); contains InterPro domain Protein of unknown function DUF567 (InterPro:IPR007612) | 8.38 |
| 975 | 258788_at | AT3G11780 | MD-2-related lipid recognition domain-containing protein / ML domain-containing protein | -4.04 |
| 976 | 258777_at | AT3G11850 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G06560.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO39886.1); contains InterPro domain Protein of unknown function DUF593 (InterPro:IPR007656) | 6.35 |
| 977 | 258727_at | AT3G11930 | universal stress protein (USP) family protein | -5.51 |
| 978 | 256261_at | AT3G12160 | AtRABA4d (Arabidopsis Rab GTPase homolog A4d); GTP binding | 10.65 |
| 979 | 256291_at | AT3G12200 | ATNEK7 (NIMA-RELATED KINASE7); kinase | -4.66 |
| 980 | 256233_at | AT3G12360 | ankyrin repeat family protein | -6.01 |
| 981 | 256241_at | AT3G12390 | nascent polypeptide associated complex alpha chain protein, putative / alpha-NAC, putative | -4.48 |
| 982 | 256285_at | AT3G12510 | similar to At3g12510-like protein [Boechera stricta] (GB:ABB89768.1); contains domain PTHR11945 (PTHR11945); contains domain PTHR11945:SF13 (PTHR11945:SF13) | 7.24 |
| 983 | 256240_at | AT3G12600 | ATNUDT16 (Arabidopsis thaliana Nudix hydrolase homolog 16); hydrolase | 4.72 |
| 984 | 256279_at | AT3G12620 | protein phosphatase 2C family protein / PP2C family protein | 5.90 |
| 985 | 257691_at | AT3G12660 | FLA14 (FASCICLIN-LIKE ARABINOGALACTAN PROTEIN 14 PRECURSOR) | 4.99 |
| 986 | 257696_at | AT3G12690 | protein kinase, putative | 6.24 |
| 987 | 257699_at | AT3G12780 | PGK1 (PHOSPHOGLYCERATE KINASE 1); phosphoglycerate kinase | -3.78 |
| 988 | 257856_at | AT3G12930 | similar to unknown [Populus trichocarpa] (GB:ABK94112.1); contains InterPro domain Iojap-related protein (InterPro:IPR004394) | -4.37 |
| 989 | 257855_at | AT3G13040 | myb family transcription factor | -4.13 |
| 990 | 257861_at | AT3G13050 | transporter-related | 4.68 |
| 991 | 257850_at | AT3G13065 | SRF4 (STRUBBELIG-RECEPTOR FAMILY 4); ATP binding / kinase/ protein serine/threonine kinase | 10.79 |
| 992 | 256955_at | AT3G13390 | SKS11 (SKU5 Similar 11); copper ion binding / oxidoreductase | 10.27 |
| 993 | 256966_at | AT3G13400 | SKS13 (SKU5 Similar 13); copper ion binding / oxidoreductase | 10.98 |
| 994 | 256964_at | AT3G13520 | AGP12 (ARABINOGALACTAN PROTEIN 12) | -4.68 |
| 995 | 256781_at | AT3G13650 | disease resistance response | -3.91 |
| 996 | 256782_at | AT3G13660 | disease resistance response | 7.56 |
| 997 | 256767_at | AT3G13680 | F-box family protein | 4.63 |
| 998 | 256772_at | AT3G13750 | BGAL1 (BETA GALACTOSIDASE 1); beta-galactosidase | -5.91 |
| 999 | 256778_at | AT3G13782 | NAP1;4 (NUCLEOSOME ASSEMBLY PROTEIN1;4); DNA binding | 5.22 |
| 1000 | 257610_at | AT3G13810 | ATIDD11 (ARABIDOPSIS THALIANA INDETERMINATE(ID)-DOMAIN 11); nucleic acid binding / transcription factor/ zinc ion binding | -3.85 |
| 1001 | 258200_at | AT3G13900 | haloacid dehalogenase-like hydrolase family protein | 6.32 |
| 1002 | 258205_at | AT3G13965 | pseudogene, hypothetical protein | 5.29 |
| 1003 | 257010_at | AT3G14090 | ATEXO70D3 (exocyst subunit EXO70 family protein D3); protein binding | -4.47 |
| 1004 | 257008_at | AT3G14210 | ESM1 (EPITHIOSPECIFIER MODIFIER 1); carboxylesterase | -4.56 |
| 1005 | 258369_at | AT3G14310 | ATPME3 (Arabidopsis thaliana pectin methylesterase 3) | -5.44 |
| 1006 | 258092_at | AT3G14595 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G17080.1); similar to unknown [Populus trichocarpa] (GB:ABK95219.1); contains domain PTHR10052 (PTHR10052); contains domain PTHR10052:SF2 (PTHR10052:SF2) | 5.01 |
| 1007 | 257216_at | AT3G14990 | 4-methyl-5(b-hydroxyethyl)-thiazole monophosphate biosynthesis protein, putative | -4.52 |
| 1008 | 256856_at | AT3G15110 | similar to unnamed protein product [Vitis vinifera] (GB:CAO39343.1) | -5.39 |
| 1009 | 256855_at | AT3G15190 | chloroplast 30S ribosomal protein S20, putative | -3.91 |
| 1010 | 257059_at | AT3G15280 | similar to unnamed protein product [Vitis vinifera] (GB:CAO66421.1) | 5.54 |
| 1011 | 257054_at | AT3G15353 | MT3 (METALLOTHIONEIN 3) | -5.11 |
| 1012 | 258398_at | AT3G15360 | ATHM4 (Arabidopsis thioredoxin M-type 4); thiol-disulfide exchange intermediate | -4.65 |
| 1013 | 258222_at | AT3G15680 | zinc finger (Ran-binding) family protein | -4.92 |
| 1014 | 258269_at | AT3G15690 | biotin carboxyl carrier protein of acetyl-CoA carboxylase-related | -4.52 |
| 1015 | 258251_at | AT3G15810 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G80120.1); similar to unknown [Populus trichocarpa] (GB:ABK95691.1); contains InterPro domain Protein of unknown function DUF567 (InterPro:IPR007612) | -5.05 |
| 1016 | 258223_at | AT3G15840 | PIFI (POST-ILLUMINATION CHLOROPHYLL FLUORESCENCE INCREASE) | -4.12 |
| 1017 | 258333_at | AT3G16000 | MFP1 (MAR BINDING FILAMENT-LIKE PROTEIN 1) | -4.65 |
| 1018 | 258337_at | AT3G16040 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G15270.1); contains InterPro domain Translation machinery associated TMA7 (InterPro:IPR015157) | 11.09 |
| 1019 | 258336_at | AT3G16050 | A37 (PYRIDOXINE BIOSYNTHESIS 1.2); protein heterodimerization | -3.73 |
| 1020 | 258329_at | AT3G16110 | ATPDIL1-6 (PDI-LIKE 1-6); thiol-disulfide exchange intermediate | -5.69 |
| 1021 | 258330_at | AT3G16130 | ATROPGEF13/ROPGEF13 (KINASE PARTNER PROTEIN-LIKE); Rho guanyl-nucleotide exchange factor/ | 5.18 |
| 1022 | 258285_at | AT3G16140 | PSAH-1 (photosystem I subunit H-1) | -5.58 |
| 1023 | 258338_at | AT3G16150 | L-asparaginase, putative / L-asparagine amidohydrolase, putative | 5.09 |
| 1024 | 258049_at | AT3G16220 | similar to RNA binding / catalytic [Arabidopsis thaliana] (TAIR:AT3G16230.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO45862.1); contains InterPro domain Predicted eukaryotic LigT; (InterPro:IPR009210) | -3.87 |
| 1025 | 258054_at | AT3G16240 | DELTA-TIP (delta tonoplast integral protein); water channel | -7.66 |
| 1026 | 258055_at | AT3G16250 | ferredoxin-related | -3.80 |
| 1027 | 258048_at | AT3G16290 | EMB2083 (EMBRYO DEFECTIVE 2083); ATPase/ metallopeptidase | -5.29 |
| 1028 | 259379_at | AT3G16350 | myb family transcription factor | 5.50 |
| 1029 | 259375_at | AT3G16370 | GDSL-motif lipase/hydrolase family protein | -4.34 |
| 1030 | 259377_at | AT3G16380 | PAB6 (POLY(A) BINDING PROTEIN 6); RNA binding / translation initiation factor | 6.38 |
| 1031 | 259383_at | AT3G16470 | JR1 (Jacalin lectin family protein) | -4.34 |
| 1032 | 257206_at | AT3G16530 | legume lectin family protein | -3.96 |
| 1033 | 257895_at | AT3G16950 | LPD1 (LIPOAMIDE DEHYDROGENASE 1) | -4.28 |
| 1034 | 257886_at | AT3G17060 | pectinesterase family protein | 13.83 |
| 1035 | 258412_at | AT3G17210 | stable protein 1-related | -4.07 |
| 1036 | 258465_at | AT3G17220 | invertase/pectin methylesterase inhibitor family protein | 7.77 |
| 1037 | 258349_at | AT3G17609 | HYH (HY5-HOMOLOG); DNA binding / transcription factor | -4.27 |
| 1038 | 258408_at | AT3G17630 | ATCHX19 (CATION/H+ EXCHANGER 19); monovalent cation:proton antiporter | 5.25 |
| 1039 | 258158_at | AT3G17790 | ATACP5 (acid phosphatase 5); acid phosphatase/ protein serine/threonine phosphatase | 5.00 |
| 1040 | 258161_at | AT3G17930 | similar to unknown [Populus trichocarpa] (GB:ABK96142.1) | -7.01 |
| 1041 | 258216_at | AT3G17980 | C2 domain-containing protein | 9.82 |
| 1042 | 258151_at | AT3G18080 | glycosyl hydrolase family 1 protein | -5.41 |
| 1043 | 257065_at | AT3G18220 | phosphatidic acid phosphatase family protein / PAP2 family protein | 13.52 |
| 1044 | 257726_at | AT3G18360 | VQ motif-containing protein | 6.72 |
| 1045 | 257717_at | AT3G18390 | EMB1865 (EMBRYO DEFECTIVE 1865) | -4.14 |
| 1046 | 257722_at | AT3G18490 | aspartyl protease family protein | -5.21 |
| 1047 | 256827_at | AT3G18570 | glycine-rich protein / oleosin | 6.82 |
| 1048 | 256853_at | AT3G18640 | zinc finger protein-related | -4.29 |
| 1049 | 257801_at | AT3G18750 | WNK6 (Arabidopsis WNK kinase 6); kinase | 6.05 |
| 1050 | 257804_at | AT3G18810 | protein kinase family protein | 8.57 |
| 1051 | 256945_at | AT3G19020 | leucine-rich repeat family protein / extensin family protein | 11.09 |
| 1052 | 256891_at | AT3G19030 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G49500.1) | -4.69 |
| 1053 | 256638_at | AT3G19090 | RNA-binding protein, putative | 5.49 |
| 1054 | 257033_at | AT3G19170 | ATPREP1/ATZNMP (PRESEQUENCE PROTEASE 1); metalloendopeptidase | -5.05 |
| 1055 | 258012_at | AT3G19310 | phospholipase C | 9.57 |
| 1056 | 258006_at | AT3G19400 | cysteine proteinase, putative | -5.20 |
| 1057 | 258023_at | AT3G19450 | CAD4 (CINNAMYL ALCOHOL DEHYDROGENASE 4); cinnamyl-alcohol dehydrogenase | -4.40 |
| 1058 | 258025_at | AT3G19480 | D-3-phosphoglycerate dehydrogenase, putative / 3-PGDH, putative | -3.84 |
| 1059 | 257563_at | AT3G19610 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G08760.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN75715.1); similar to predicted protein [Physcomitrella patens subsp. patens] (GB:EDQ63015.1); similar to predicted protein [Physcomitrella patens subsp. patens] (GB:EDQ49909.1); contains InterPro domain Protein of unknown function DUF936, plant (InterPro:IPR010341) | 6.16 |
| 1060 | 257077_at | AT3G19690 | pathogenesis-related protein, putative | 7.86 |
| 1061 | 257938_at | AT3G19820 | DWF1 (DIMINUTO 1); catalytic | -7.64 |
| 1062 | 256627_at | AT3G19970 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G18245.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO66476.1); contains InterPro domain Protein of unknown function DUF829, eukaryotic (InterPro:IPR008547) | 7.14 |
| 1063 | 257129_at | AT3G20100 | CYP705A19 (cytochrome P450, family 705, subfamily A, polypeptide 19); oxygen binding | -5.15 |
| 1064 | 257119_at | AT3G20190 | leucine-rich repeat transmembrane protein kinase, putative | 12.89 |
| 1065 | 257120_at | AT3G20200 | protein kinase family protein | 7.64 |
| 1066 | 257121_at | AT3G20220 | auxin-responsive protein, putative | 11.24 |
| 1067 | 257672_at | AT3G20300 | extracellular ligand-gated ion channel | 7.91 |
| 1068 | 257090_at | AT3G20530 | protein kinase family protein | 13.22 |
| 1069 | 257082_at | AT3G20580 | COBL10 (COBRA-LIKE PROTEIN 10 PRECURSOR) | 10.60 |
| 1070 | 256697_at | AT3G20660 | ATOCT4 (ARABIDOPSIS THALIANA ORGANIC CATION/CARNITINE TRANSPORTER4); carbohydrate transmembrane transporter/ sugar:hydrogen ion symporter | 5.32 |
| 1071 | 257574_at | AT3G20710 | F-box protein-related | 5.82 |
| 1072 | 257974_at | AT3G20820 | leucine-rich repeat family protein | -8.43 |
| 1073 | 257986_at | AT3G20865 | AGP40 (ARABINOGALACTAN-PROTEIN 40) | 10.12 |
| 1074 | 256974_at | AT3G20990 | transposable element gene | 5.02 |
| 1075 | 256975_at | AT3G21000 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G20980.1); similar to putative polyprotein [Oryza sativa (japonica cultivar-group)] (GB:AAP46257.1); contains domain PTHR11439 (PTHR11439) | 5.05 |
| 1076 | 256979_at | AT3G21055 | PSBTN (photosystem II subunit T) | -7.84 |
| 1077 | 258035_at | AT3G21180 | ACA9 (autoinhibited Ca2+ -ATPase 9); calcium-transporting ATPase/ calmodulin binding | 9.78 |
| 1078 | 258040_at | AT3G21190 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G51630.1); similar to unknown [Populus trichocarpa] (GB:ABK93128.1); contains InterPro domain Protein of unknown function DUF246, plant (InterPro:IPR004348) | -6.45 |
| 1079 | 258183_at | AT3G21550 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G21520.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO71058.1); contains InterPro domain Protein of unknown function DUF679 (InterPro:IPR007770) | -3.94 |
| 1080 | 258168_at | AT3G21570 | unknown protein | 9.86 |
| 1081 | 258181_at | AT3G21670 | nitrate transporter (NTP3) | -5.86 |
| 1082 | 257951_at | AT3G21700 | GTP binding | 8.54 |
| 1083 | 257256_at | AT3G21970 | receptor-like protein kinase-related | 6.14 |
| 1084 | 256825_at | AT3G22120 | CWLP (CELL WALL-PLASMA MEMBRANE LINKER PROTEIN); lipid binding | -7.22 |
| 1085 | 256796_at | AT3G22210 | unknown protein | -4.83 |
| 1086 | 256766_at | AT3G22231 | PCC1 (PATHOGEN AND CIRCADIAN CONTROLLED 1) | -5.02 |
| 1087 | 256617_at | AT3G22240 | unknown protein | -5.05 |
| 1088 | 258448_at | AT3G22290 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G36050.1); similar to unknown [Populus trichocarpa] (GB:ABK93267.1); contains InterPro domain Protein of unknown function DUF1692 (InterPro:IPR012936) | 4.61 |
| 1089 | 258451_at | AT3G22360 | AOX1B (alternative oxidase 1B); alternative oxidase | 7.93 |
| 1090 | 258455_at | AT3G22440 | hydroxyproline-rich glycoprotein family protein | -4.72 |
| 1091 | 258344_at | AT3G22650 | F-box family protein | 7.97 |
| 1092 | 258323_at | AT3G22750 | protein kinase, putative | 6.73 |
| 1093 | 258326_at | AT3G22760 | SOL1 (TSO1-Like); transcription factor | 5.87 |
| 1094 | 256835_at | AT3G22890 | APS1 (ATP sulfurylase 3) | -5.23 |
| 1095 | 257771_at | AT3G23000 | CIPK7 (CBL-INTERACTING PROTEIN KINASE 7); kinase | -4.14 |
| 1096 | 257766_at | AT3G23030 | IAA2 (indoleacetic acid-induced protein 2); transcription factor | -4.41 |
| 1097 | 257925_at | AT3G23170 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G14450.1) | 7.56 |
| 1098 | 257921_at | AT3G23270 | regulator of chromosome condensation (RCC1) family protein | 6.54 |
| 1099 | 258294_at | AT3G23350 | similar to hypothetical protein [Vitis vinifera] (GB:CAN77915.1); contains InterPro domain ENTH/VHS (InterPro:IPR008942); contains InterPro domain Epsin, N-terminal (InterPro:IPR001026) | 5.88 |
| 1100 | 258295_at | AT3G23400 | plastid-lipid associated protein PAP / fibrillin family protein | -4.93 |
| 1101 | 258293_at | AT3G23430 | PHO1 (PHOSPHATE 1) | -4.13 |
| 1102 | 257203_at | AT3G23730 | xyloglucan:xyloglucosyl transferase, putative / xyloglucan endotransglycosylase, putative / endo-xyloglucan transferase, putative | -4.30 |
| 1103 | 257242_at | AT3G24220 | NCED6 (NINE-CIS-EPOXYCAROTENOID DIOXYGENASE 6) | 8.73 |
| 1104 | 257161_at | AT3G24280 | SMAP2 (SMALL ACIDIC PROTEIN 2) | 6.59 |
| 1105 | 257168_at | AT3G24430 | HCF101 (HIGH-CHLOROPHYLL-FLUORESCENCE 101); ATP binding | -4.33 |
| 1106 | 258134_at | AT3G24530 | AAA-type ATPase family protein / ankyrin repeat family protein | 5.24 |
| 1107 | 258129_at | AT3G24620 | ATROPGEF8/ROPGEF8 (KINASE PARTNER PROTEIN-LIKE); Rho guanyl-nucleotide exchange factor/ | 10.57 |
| 1108 | 257618_at | AT3G24715 | ATP binding / protein kinase/ protein-tyrosine kinase | 8.11 |
| 1109 | 257868_at | AT3G25070 | RIN4 (RPM1 INTERACTING PROTEIN 4); protein binding | -4.24 |
| 1110 | 257819_at | AT3G25165 | RALFL25 (RALF-LIKE 25) | 11.23 |
| 1111 | 257821_at | AT3G25170 | RALFL26 (RALF-LIKE 26) | 11.59 |
| 1112 | 256754_at | AT3G25690 | CHUP1 (CHLOROPLAST UNUSUAL POSITIONING 1) | -3.75 |
| 1113 | 258086_at | AT3G25860 | LTA2 (PLASTID E2 SUBUNIT OF PYRUVATE DECARBOXYLASE); dihydrolipoyllysine-residue acetyltransferase | -4.32 |
| 1114 | 258075_at | AT3G25900 | ATHMT-1/HMT-1; homocysteine S-methyltransferase | 6.56 |
| 1115 | 258060_at | AT3G26030 | ATB' DELTA (Arabidopsis thaliana serine/threonine protein phosphatase 2A 55 kDa regulatory subunit B prime delta); protein phosphatase type 2A regulator | -6.34 |
| 1116 | 258087_at | AT3G26060 | ATPRX Q; antioxidant/ peroxiredoxin | -4.45 |
| 1117 | 258077_at | AT3G26110 | similar to BCP1 (Brassica campestris pollen protein 1) [Arabidopsis thaliana] (TAIR:AT1G24520.1); contains domain PD015210 (PD015210) | 13.19 |
| 1118 | 257552_at | AT3G26130 | glycosyl hydrolase family 5 protein / cellulase family protein | 6.47 |
| 1119 | 256880_at | AT3G26450 | major latex protein-related / MLP-related | -5.07 |
| 1120 | 257313_at | AT3G26520 | TIP2 (TONOPLAST INTRINSIC PROTEIN 2); water channel | -8.61 |
| 1121 | 257611_at | AT3G26580 | binding | -4.44 |
| 1122 | 257807_at | AT3G26650 | GAPA (GLYCERALDEHYDE 3-PHOSPHATE DEHYDROGENASE A SUBUNIT); glyceraldehyde-3-phosphate dehydrogenase | -5.95 |
| 1123 | 257832_at | AT3G26740 | CCL (CCR-LIKE) | -5.67 |
| 1124 | 258278_at | AT3G26860 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G27331.1); contains InterPro domain Plant self-incompatibility S1 (InterPro:IPR010264) | 13.18 |
| 1125 | 258280_at | AT3G26880 | self-incompatibility protein-related | 7.56 |
| 1126 | 257151_at | AT3G27200 | plastocyanin-like domain-containing protein | -3.81 |
| 1127 | 257155_at | AT3G27290 | F-box family protein-related | 5.25 |
| 1128 | 257714_at | AT3G27360 | histone H3 | -4.70 |
| 1129 | 257736_at | AT3G27410 | unknown protein | 8.04 |
| 1130 | 257737_at | AT3G27440 | uracil phosphoribosyltransferase, putative / UMP pyrophosphorylase, putative / UPRTase, putative | 4.97 |
| 1131 | 257970_at | AT3G27570 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G40510.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO14698.1); contains InterPro domain Thioredoxin-like fold (InterPro:IPR012336); contains InterPro domain Thioredoxin fold (InterPro:IPR012335); contains InterPro domain Sucraseferredoxin-like (InterPro:IPR009737) | -5.36 |
| 1132 | 258239_at | AT3G27690 | LHCB2:4 (Photosystem II light harvesting complex gene 2.3); chlorophyll binding | -4.31 |
| 1133 | 257228_at | AT3G27890 | NQR (NADPH:QUINONE OXIDOREDUCTASE); FMN reductase | -5.26 |
| 1134 | 257309_at | AT3G28150 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G15900.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN82134.1); contains InterPro domain Protein of unknown function DUF231, plant (InterPro:IPR004253) | 8.13 |
| 1135 | 256576_at | AT3G28210 | PMZ; zinc ion binding | 7.07 |
| 1136 | 256577_at | AT3G28220 | meprin and TRAF homology domain-containing protein / MATH domain-containing protein | -3.72 |
| 1137 | 257870_at | AT3G28490 | oxidoreductase, 2OG-Fe(II) oxygenase family protein | 8.52 |
| 1138 | 256992_at | AT3G28630 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G59710.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO14885.1); contains InterPro domain Protein of unknown function DUF569 (InterPro:IPR007679); contains InterPro domain Actin-crosslinking proteins (InterPro:IPR008999) | 6.25 |
| 1139 | 256584_at | AT3G28750 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G39880.1) | 11.78 |
| 1140 | 256586_at | AT3G28770 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G28810.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G28820.1); similar to PREDICTED: similar to QUAhog (hedgehog related) family member (qua-1) [Rattus norvegicus] (GB:XP_001069720.1); contains InterPro domain Protein of unknown function DUF1216 (InterPro:IPR009605) | 6.96 |
| 1141 | 256587_at | AT3G28780 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G28840.1); contains InterPro domain Protein of unknown function DUF1216 (InterPro:IPR009605) | 12.52 |
| 1142 | 256588_at | AT3G28790 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G28830.1); contains InterPro domain Protein of unknown function DUF1216 (InterPro:IPR009605) | 10.80 |
| 1143 | 256581_at | AT3G28830 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G28790.1); contains InterPro domain Protein of unknown function DUF1216 (InterPro:IPR009605) | 14.06 |
| 1144 | 256582_at | AT3G28840 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G28780.1); contains InterPro domain Protein of unknown function DUF1216 (InterPro:IPR009605) | 9.94 |
| 1145 | 256583_at | AT3G28850 | glutaredoxin family protein | 6.39 |
| 1146 | 257137_at | AT3G28860 | ATMDR11/ATPGP19/MDR1/MDR11/PGP19 (P-GLYCOPROTEIN 19); ATPase, coupled to transmembrane movement of substances / auxin efflux transmembrane transporter | -3.74 |
| 1147 | 258000_at | AT3G28940 | avirulence-responsive protein, putative / avirulence induced gene (AIG) protein, putative | -5.81 |
| 1148 | 258058_at | AT3G28980 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G28810.1); contains InterPro domain Protein of unknown function DUF1216 (InterPro:IPR009605) | 11.07 |
| 1149 | 258003_at | AT3G29030 | ATEXPA5 (ARABIDOPSIS THALIANA EXPANSIN A5) | -4.53 |
| 1150 | 257106_at | AT3G29060 | similar to EXS family protein / ERD1/XPR1/SYG1 family protein [Arabidopsis thaliana] (TAIR:AT2G03260.1); similar to ATP binding / ATPase, coupled to transmembrane movement of substances [Arabidopsis thaliana] (TAIR:AT1G14040.1); similar to EXS family protein / ERD1/XPR1/SYG1 family protein [Arabidopsis thaliana] (TAIR:AT2G03240.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO68453.1); similar to SPX, N-terminal; EXS, C-terminal [Medicago truncatula] (GB:ABD32751.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO23292.1); contains InterPro domain SPX, N-terminal (InterPro:IPR004331); contains InterPro domain EXS, C-terminal; (InterPro:IPR004342) | 8.57 |
| 1151 | 257107_at | AT3G29070 | protein transmembrane transporter | 5.90 |
| 1152 | 256747_at | AT3G29180 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G39430.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO15029.1); contains InterPro domain Protein of unknown function DUF1336 (InterPro:IPR009769) | 5.57 |
| 1153 | 257566_x_at | AT3G29610 | transposable element gene | 6.67 |
| 1154 | 252820_at | AT3G42640 | AHA8 (ARABIDOPSIS H(+)-ATPASE 8); ATPase | 8.87 |
| 1155 | 252769_at | AT3G42850 | galactokinase, putative | 4.67 |
| 1156 | 252771_at | AT3G42880 | leucine-rich repeat transmembrane protein kinase, putative | 10.41 |
| 1157 | 252733_at | AT3G43120 | auxin-responsive protein-related | 10.75 |
| 1158 | 252711_at | AT3G43720 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | -3.88 |
| 1159 | 252710_at | AT3G43860 | ATGH9A4 (ARABIDOPSIS THALIANA GLYCOSYL HYDROLASE 9A4); hydrolase, hydrolyzing O-glycosyl compounds | 10.88 |
| 1160 | 252663_at | AT3G44070 | beta-galactosidase | 7.54 |
| 1161 | 252669_at | AT3G44100 | MD-2-related lipid recognition domain-containing protein / ML domain-containing protein | -5.48 |
| 1162 | 252618_at | AT3G45140 | LOX2 (LIPOXYGENASE 2) | -7.09 |
| 1163 | 252612_at | AT3G45160 | contains domain PROKAR_LIPOPROTEIN (PS51257) | -4.03 |
| 1164 | 252571_at | AT3G45280 | SYP72 (SYNTAXIN OF PLANTS 72) | 9.59 |
| 1165 | 252622_at | AT3G45310 | cysteine proteinase, putative | -4.22 |
| 1166 | 252623_at | AT3G45320 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G60330.1) | 4.93 |
| 1167 | 252544_at | AT3G45810 | ferric reductase-like transmembrane component family protein | 6.25 |
| 1168 | 252563_at | AT3G45970 | ATEXLA1 (ARABIDOPSIS THALIANA EXPANSIN-LIKE A1) | -4.28 |
| 1169 | 252561_at | AT3G45980 | HTB9; DNA binding | -4.93 |
| 1170 | 252565_at | AT3G46000 | ADF2 (ACTIN DEPOLYMERIZING FACTOR 2); actin binding | -3.82 |
| 1171 | 252564_at | AT3G46010 | ADF1 (ACTIN DEPOLYMERIZING FACTOR 1) | -5.03 |
| 1172 | 252560_at | AT3G46030 | HTB11; DNA binding | -5.36 |
| 1173 | 252515_at | AT3G46230 | ATHSP17.4 (Arabidopsis thaliana heat shock protein 17.4) | 5.88 |
| 1174 | 252531_at | AT3G46520 | ACT12 (ACTIN-12); structural constituent of cytoskeleton | 10.35 |
| 1175 | 252485_at | AT3G46530 | RPP13 (RECOGNITION OF PERONOSPORA PARASITICA 13); ATP binding | -5.82 |
| 1176 | 252493_at | AT3G46750 | similar to hypothetical protein [Vitis vinifera] (GB:CAN62428.1) | 6.87 |
| 1177 | 252441_at | AT3G46780 | PTAC16 (PLASTID TRANSCRIPTIONALLY ACTIVE18); binding / catalytic | -6.18 |
| 1178 | 252463_at | AT3G47070 | similar to unknown [Populus trichocarpa] (GB:ABK95428.1) | -4.29 |
| 1179 | 252467_at | AT3G47080 | binding | 4.98 |
| 1180 | 252454_at | AT3G47130 | F-box family protein-related | 5.71 |
| 1181 | 252440_at | AT3G47440 | TIP5;1 (tonoplast intrinsic protein 5;1); water channel | 9.51 |
| 1182 | 252430_at | AT3G47470 | LHCA4 (Photosystem I light harvesting complex gene 4); chlorophyll binding | -6.81 |
| 1183 | 252433_at | AT3G47560 | esterase/lipase/thioesterase family protein | -3.88 |
| 1184 | 252425_at | AT3G47620 | TCP family transcription factor, putative | -5.62 |
| 1185 | 252409_at | AT3G47650 | bundle-sheath defective protein 2 family / bsd2 family | -4.76 |
| 1186 | 252428_at | AT3G47660 | regulator of chromosome condensation (RCC1) family protein | 5.91 |
| 1187 | 252383_at | AT3G47780 | ATATH6 (ABC2 homolog 6); ATPase, coupled to transmembrane movement of substances | 4.95 |
| 1188 | 252391_at | AT3G47860 | apolipoprotein D-related | -5.87 |
| 1189 | 252398_at | AT3G47990 | zinc finger (C3HC4-type RING finger) family protein | -4.30 |
| 1190 | 252399_at | AT3G48010 | ATCNGC16 (cyclic nucleotide gated channel 16); calmodulin binding / cyclic nucleotide binding / ion channel | 6.31 |
| 1191 | 252364_at | AT3G48450 | nitrate-responsive NOI protein, putative | 8.76 |
| 1192 | 252338_at | AT3G48890 | ATMP2 (MEMBRANE STEROID BINDING PROTEIN 2); heme binding / transition metal ion binding | -4.45 |
| 1193 | 252255_at | AT3G49220 | pectinesterase family protein | -4.08 |
| 1194 | 252270_at | AT3G49560 | mitochondrial import inner membrane translocase subunit Tim17/Tim22/Tim23 family protein | -3.96 |
| 1195 | 252212_at | AT3G50310 | MAPKKK20 (Mitogen-activated protein kinase kinase kinase 20); kinase | 9.12 |
| 1196 | 252168_at | AT3G50440 | hydrolase | -5.23 |
| 1197 | 252160_at | AT3G50570 | hydroxyproline-rich glycoprotein family protein | 4.95 |
| 1198 | 252161_at | AT3G50580 | histamine receptor | 6.21 |
| 1199 | 252130_at | AT3G50820 | PSBO-2/PSBO2 (PHOTOSYSTEM II SUBUNIT O-2); oxygen evolving/ poly(U) binding | -5.41 |
| 1200 | 252133_at | AT3G50900 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G66490.1); similar to hypothetical protein [Thellungiella halophila] (GB:ABB45855.1) | -4.54 |
| 1201 | 252134_at | AT3G50910 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G66480.1); similar to hypothetical protein [Thellungiella halophila] (GB:ABB45854.1) | 4.91 |
| 1202 | 252119_at | AT3G51030 | ATTRX1 (Arabidopsis thaliana thioredoxin H-type 1); thiol-disulfide exchange intermediate | 5.28 |
| 1203 | 252140_at | AT3G51070 | dehydration-responsive protein-related | 6.48 |
| 1204 | 252143_at | AT3G51150 | kinesin motor family protein | 5.41 |
| 1205 | 252101_at | AT3G51300 | ARAC11/ATRAC11/ATROP1/ROP1/ROP1AT (RHO-RELATED PROTEIN FROM PLANTS 1); GTP binding / GTPase/ protein binding | 7.82 |
| 1206 | 252066_at | AT3G51550 | FER (FERONIA); kinase/ protein kinase | -7.49 |
| 1207 | 252115_at | AT3G51600 | LTP5 (LIPID TRANSFER PROTEIN 5); lipid transporter | -4.09 |
| 1208 | 252080_at | AT3G51670 | SEC14 cytosolic factor family protein / phosphoglyceride transfer family protein | -4.63 |
| 1209 | 246298_at | AT3G51770 | ETO1 (ETHYLENE OVERPRODUCER 1) | 5.37 |
| 1210 | 252037_at | AT3G51920 | CAM9 (CALMODULIN 9); calcium ion binding | -3.95 |
| 1211 | 252087_at | AT3G52080 | CHX28 (cation/hydrogen exchanger 28); monovalent cation:proton antiporter | 8.51 |
| 1212 | 252032_at | AT3G52150 | RNA recognition motif (RRM)-containing protein | -6.31 |
| 1213 | 256727_at | AT3G52240 | similar to unnamed protein product [Vitis vinifera] (GB:CAO46543.1) | -4.28 |
| 1214 | 256674_at | AT3G52360 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G35850.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO39911.1) | 5.32 |
| 1215 | 256678_at | AT3G52380 | CP33 (PIGMENT DEFECTIVE 322); RNA binding | -4.79 |
| 1216 | 252046_at | AT3G52460 | hydroxyproline-rich glycoprotein family protein | 9.10 |
| 1217 | 252048_at | AT3G52500 | aspartyl protease family protein | -5.73 |
| 1218 | 252052_at | AT3G52600 | ATCWINV2 (ARABIDOPSIS THALIANA CELL WALL INVERTASE 2); hydrolase, hydrolyzing O-glycosyl compounds | 11.20 |
| 1219 | 252061_at | AT3G52620 | unknown protein | 10.21 |
| 1220 | 251981_at | AT3G53080 | galactose-binding lectin family protein | 7.26 |
| 1221 | 251984_at | AT3G53260 | PAL2 (phenylalanine ammonia-lyase 2); phenylalanine ammonia-lyase | -4.97 |
| 1222 | 251988_at | AT3G53300 | CYP71B31 (cytochrome P450, family 71, subfamily B, polypeptide 31); oxygen binding | 4.82 |
| 1223 | 251962_at | AT3G53420 | PIP2A (PLASMA MEMBRANE INTRINSIC PROTEIN 2A); water channel | -7.54 |
| 1224 | 251961_at | AT3G53620 | ATPPA4 (ARABIDOPSIS THALIANA PYROPHOSPHORYLASE 4); inorganic diphosphatase/ pyrophosphatase | -3.99 |
| 1225 | 251936_at | AT3G53700 | MEE40 (maternal effect embryo arrest 40) | -4.02 |
| 1226 | 251927_at | AT3G53990 | universal stress protein (USP) family protein | -5.97 |
| 1227 | 251918_at | AT3G54040 | photoassimilate-responsive protein-related | 7.34 |
| 1228 | 251885_at | AT3G54050 | fructose-1,6-bisphosphatase, putative / D-fructose-1,6-bisphosphate 1-phosphohydrolase, putative / FBPase, putative | -5.96 |
| 1229 | 251889_at | AT3G54080 | sugar binding | -4.29 |
| 1230 | 251935_at | AT3G54090 | pfkB-type carbohydrate kinase family protein | -5.62 |
| 1231 | 251874_at | AT3G54240 | hydrolase, alpha/beta fold family protein | 11.74 |
| 1232 | 251899_at | AT3G54400 | aspartyl protease family protein | -4.81 |
| 1233 | 251855_at | AT3G54690 | sugar isomerase (SIS) domain-containing protein / CBS domain-containing protein | 4.76 |
| 1234 | 251854_at | AT3G54800 | pleckstrin homology (PH) domain-containing protein / lipid-binding START domain-containing protein | 10.31 |
| 1235 | 251814_at | AT3G54890 | LHCA1; chlorophyll binding | -6.72 |
| 1236 | 251865_at | AT3G54930 | serine/threonine protein phosphatase 2A (PP2A) regulatory subunit B', putative | 4.99 |
| 1237 | 251821_at | AT3G55050 | serine/threonine protein phosphatase 2C (PP2C6) | 4.84 |
| 1238 | 251835_at | AT3G55180 | esterase/lipase/thioesterase family protein | 7.93 |
| 1239 | 251836_at | AT3G55190 | esterase/lipase/thioesterase family protein | 6.93 |
| 1240 | 251804_at | AT3G55430 | glycosyl hydrolase family 17 protein / beta-1,3-glucanase, putative | 4.75 |
| 1241 | 251791_at | AT3G55500 | ATEXPA16 (ARABIDOPSIS THALIANA EXPANSIN A16) | 4.76 |
| 1242 | 251762_at | AT3G55800 | SBPASE (SEDOHEPTULOSE-BISPHOSPHATASE); phosphoric ester hydrolase/ sedoheptulose-bisphosphatase | -5.77 |
| 1243 | 251742_at | AT3G56050 | protein kinase family protein | -4.22 |
| 1244 | 251690_at | AT3G56510 | TBP-binding protein, putative | -3.99 |
| 1245 | 251697_at | AT3G56600 | inositol or phosphatidylinositol kinase | 9.96 |
| 1246 | 246294_at | AT3G56910 | PSRP5 (PLASTID-SPECIFIC 50S RIBOSOMAL PROTEIN 5) | -5.96 |
| 1247 | 251664_at | AT3G56940 | AT103 (DICARBOXYLATE DIIRON 1) | -4.89 |
| 1248 | 251711_at | AT3G56960 | PIP5K4 (PHOSPHATIDYL INOSITOL MONOPHOSPHATE 5 KINASE 4); 1-phosphatidylinositol-4-phosphate 5-kinase | 6.12 |
| 1249 | 251654_at | AT3G57140 | SDP1-LIKE (SDP1-LIKE); GTP binding | 7.67 |
| 1250 | 251669_at | AT3G57180 | GTP binding | -4.00 |
| 1251 | 251626_at | AT3G57220 | UDP-GlcNAc:dolichol phosphate N-acetylglucosamine-1-phosphate transferase, putative | -3.94 |
| 1252 | 251623_at | AT3G57390 | AGL18 (AGAMOUS-LIKE 18); transcription factor | 7.24 |
| 1253 | 251634_at | AT3G57480 | zinc finger (C2H2 type, AN1-like) family protein | 5.90 |
| 1254 | 251590_at | AT3G57690 | AGP23/ATAGP23 (ARABINOGALACTAN-PROTEIN 23) | 8.38 |
| 1255 | 251497_at | AT3G59060 | PIL6 (PHYTOCHROME-INTERACTING FACTOR 5); DNA binding / transcription factor | -4.74 |
| 1256 | 251468_at | AT3G59290 | (EPSIN3); binding | 7.22 |
| 1257 | 251494_at | AT3G59350 | serine/threonine protein kinase, putative | 5.75 |
| 1258 | 251519_at | AT3G59400 | GUN4 (Genomes uncoupled 4) | -6.95 |
| 1259 | 251477_at | AT3G59680 | similar to hypothetical protein [Vitis vinifera] (GB:CAN69676.1) | 4.85 |
| 1260 | 251433_at | AT3G59830 | ankyrin protein kinase, putative | 8.64 |
| 1261 | 251441_at | AT3G60020 | ASK5 (ARABIDOPSIS SKP1-LIKE 5); ubiquitin-protein ligase | 6.31 |
| 1262 | 251455_at | AT3G60100 | CSY5 (CITRATE SYNTHASE 5); citrate (SI)-synthase | 4.65 |
| 1263 | 251413_at | AT3G60320 | DNA binding | -3.91 |
| 1264 | 251405_at | AT3G60330 | AHA7 (ARABIDOPSIS H(+)-ATPASE 7); hydrogen-exporting ATPase, phosphorylative mechanism | 5.96 |
| 1265 | 251400_at | AT3G60420 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G60450.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO70569.1); contains InterPro domain Phosphoglycerate mutase (InterPro:IPR013078); contains InterPro domain PRIB5 (InterPro:IPR012398) | -4.17 |
| 1266 | 251397_at | AT3G60570 | ATEXPB5 (ARABIDOPSIS THALIANA EXPANSIN B5) | 8.69 |
| 1267 | 251384_at | AT3G60760 | unknown protein | 5.82 |
| 1268 | 251339_at | AT3G60780 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G45360.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO24498.1); contains InterPro domain Protein of unknown function DUF1442 (InterPro:IPR009902) | 8.67 |
| 1269 | 251358_at | AT3G61160 | shaggy-related protein kinase beta / ASK-beta (ASK2) | 8.53 |
| 1270 | 251361_at | AT3G61230 | LIM domain-containing protein | 9.20 |
| 1271 | 251324_at | AT3G61430 | PIP1A (PLASMA MEMBRANE INTRINSIC PROTEIN 1A); water channel | -5.53 |
| 1272 | 251322_at | AT3G61440 | ATCYSC1 (BETA-SUBSTITUTED ALA SYNTHASE 3;1); L-3-cyanoalanine synthase/ cysteine synthase | -4.80 |
| 1273 | 251329_at | AT3G61450 | SYP73 (SYNTAXIN OF PLANTS 73) | 4.72 |
| 1274 | 251277_at | AT3G61760 | dynamin-like protein B (DL1B) | 5.38 |
| 1275 | 251287_at | AT3G61820 | aspartyl protease family protein | -4.58 |
| 1276 | 251243_at | AT3G61870 | similar to unknown [Populus trichocarpa] (GB:ABK95718.1) | -4.15 |
| 1277 | 251305_at | AT3G62030 | ROC4 (ROTAMASE CYP 4); peptidyl-prolyl cis-trans isomerase | -4.12 |
| 1278 | 251258_at | AT3G62170 | VGDH2 (VANGUARD 1 HOMOLOG 2); pectinesterase | 11.07 |
| 1279 | 251250_at | AT3G62180 | invertase/pectin methylesterase inhibitor family protein | 11.89 |
| 1280 | 251252_at | AT3G62230 | F-box family protein | 12.26 |
| 1281 | 251218_at | AT3G62410 | CP12-2 | -4.96 |
| 1282 | 251180_at | AT3G62640 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G47480.1); similar to unknown [Picea sitchensis] (GB:ABK21328.1) | 11.62 |
| 1283 | 251225_at | AT3G62660 | GATL7 (Galacturonosyltransferase-like 7); polygalacturonate 4-alpha-galacturonosyltransferase/ transferase, transferring glycosyl groups / transferase, transferring hexosyl groups | 5.58 |
| 1284 | 251228_at | AT3G62710 | glycosyl hydrolase family 3 protein | 11.30 |
| 1285 | 251193_at | AT3G62910 | APG3 (ALBINO AND PALE GREEN); translation release factor | -5.10 |
| 1286 | 251196_at | AT3G62950 | glutaredoxin family protein | -4.71 |
| 1287 | 251199_at | AT3G62980 | TIR1 (TRANSPORT INHIBITOR RESPONSE 1); ubiquitin-protein ligase | -3.86 |
| 1288 | 251155_at | AT3G63160 | unknown protein | -4.24 |
| 1289 | 251172_at | AT3G63190 | RRF (RIBOSOME RECYCLING FACTOR, CHLOROPLAST PRECURSOR) | -5.36 |
| 1290 | 251174_at | AT3G63200 | PLA IIIB/PLP9 (Patatin-like protein 9); nutrient reservoir | -4.57 |
| 1291 | 251118_at | AT3G63410 | APG1 (ALBINO OR PALE GREEN MUTANT 1); methyltransferase | -4.15 |
| 1292 | 251120_at | AT3G63490 | ribosomal protein L1 family protein | -6.36 |
| 1293 | 255696_at | AT4G00110 | GAE3 (UDP-D-GLUCURONATE 4-EPIMERASE 3); catalytic | 5.76 |
| 1294 | 255717_at | AT4G00350 | MATE efflux family protein | 10.03 |
| 1295 | 255691_at | AT4G00370 | ANTR2 (anion transporter 2); organic anion transmembrane transporter | -5.08 |
| 1296 | 255674_at | AT4G00430 | TMP-C (PLASMA MEMBRANE INTRINSIC PROTEIN 1;4); water channel | -7.29 |
| 1297 | 255632_at | AT4G00680 | actin-depolymerizing factor, putative | 4.75 |
| 1298 | 255631_at | AT4G00710 | protein kinase family protein | -3.83 |
| 1299 | 255645_at | AT4G00880 | auxin-responsive family protein | -4.63 |
| 1300 | 255572_at | AT4G01050 | hydroxyproline-rich glycoprotein family protein | -6.32 |
| 1301 | 255567_at | AT4G01150 | Identical to Uncharacterized protein At4g01150, chloroplast precursor [Arabidopsis Thaliana] (GB:O04616;GB:Q38835); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G38100.1); similar to unknown [Populus trichocarpa] (GB:ABK95729.1) | -5.28 |
| 1302 | 255623_at | AT4G01310 | ribosomal protein L5 family protein | -4.79 |
| 1303 | 255580_at | AT4G01470 | GAMMA-TIP3/TIP1;3 (tonoplast intrinsic protein 1;3); water channel | 7.05 |
| 1304 | 255536_at | AT4G01575 | serine protease inhibitor, Kazal-type family protein | 7.28 |
| 1305 | 255590_at | AT4G01610 | cathepsin B-like cysteine protease, putative | -4.49 |
| 1306 | 255595_at | AT4G01700 | chitinase, putative | -3.79 |
| 1307 | 255540_at | AT4G01800 | preprotein translocase secA subunit, putative | -4.51 |
| 1308 | 255553_at | AT4G01960 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G02380.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO40142.1) | 4.96 |
| 1309 | 255506_at | AT4G02130 | GATL6/LGT10; polygalacturonate 4-alpha-galacturonosyltransferase/ transferase, transferring glycosyl groups / transferase, transferring hexosyl groups | -4.61 |
| 1310 | 255530_at | AT4G02140 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G02700.1) | 7.25 |
| 1311 | 255504_at | AT4G02200 | drought-responsive family protein | -4.59 |
| 1312 | 255515_at | AT4G02250 | invertase/pectin methylesterase inhibitor family protein | 11.55 |
| 1313 | 255524_at | AT4G02330 | ATPMEPCRB; pectinesterase | -3.75 |
| 1314 | 255479_at | AT4G02380 | SAG21 (SENESCENCE-ASSOCIATED GENE 21) | 4.69 |
| 1315 | 255440_at | AT4G02530 | chloroplast thylakoid lumen protein | -4.36 |
| 1316 | 255497_at | AT4G02590 | UNE12 (unfertilized embryo sac 12); DNA binding / transcription factor | -3.94 |
| 1317 | 255490_at | AT4G02660 | WD-40 repeat family protein / beige-related | 4.84 |
| 1318 | 255492_at | AT4G02680 | EOL1 (ETO1-LIKE 1); binding / protein binding | -3.99 |
| 1319 | 255457_at | AT4G02770 | PSAD-1 (photosystem I subunit D-1) | -5.24 |
| 1320 | 255433_at | AT4G03210 | XTH9 (XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 9); hydrolase, acting on glycosyl bonds | -5.95 |
| 1321 | 255435_at | AT4G03280 | PETC (PHOTOSYNTHETIC ELECTRON TRANSFER C) | -5.81 |
| 1322 | 255423_at | AT4G03290 | calcium-binding protein, putative | 12.02 |
| 1323 | 255386_at | AT4G03620 | myosin heavy chain-related | 11.64 |
| 1324 | 255369_at | AT4G04080 | ATISU3/ISU3 (IscU-like 3); structural molecule | 7.15 |
| 1325 | 255290_at | AT4G04640 | ATPC1 (ATP synthase gamma chain 1) | -5.78 |
| 1326 | 255276_at | AT4G04930 | DES-1-LIKE (fatty acid desaturase 1-like); oxidoreductase | 10.46 |
| 1327 | 255251_at | AT4G04980 | similar to proline-rich family protein [Arabidopsis thaliana] (TAIR:AT1G61080.1); similar to Phosphoinositide-binding clathrin adaptor, N-terminal; Wiscott-Aldrich syndrome, C-terminal [Medicago truncatula] (GB:ABN08602.1); contains domain PTHR23213:SF18 (PTHR23213:SF18); contains domain PTHR23213 (PTHR23213) | 11.92 |
| 1328 | 255248_at | AT4G05180 | PSBQ/PSBQ-2/PSII-Q (PHOTOSYSTEM II SUBUNIT Q-2); calcium ion binding | -6.41 |
| 1329 | 255232_at | AT4G05330 | AGD13 (ARF-GAP DOMAIN 13); ARF GTPase activator/ zinc ion binding | 7.03 |
| 1330 | 255175_at | AT4G07960 | ATCSLC12 (Cellulose synthase-like C12); transferase, transferring glycosyl groups | 7.78 |
| 1331 | 255094_at | AT4G08590 | zinc finger (C3HC4-type RING finger) family protein | 5.39 |
| 1332 | 255101_at | AT4G08670 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | 8.48 |
| 1333 | 255068_at | AT4G08920 | CRY1 (CRYPTOCHROME 1) | -5.78 |
| 1334 | 255064_at | AT4G08950 | phosphate-responsive protein, putative (EXO) | -7.24 |
| 1335 | 255078_at | AT4G09010 | APX4 (ASCORBATE PEROXIDASE 4); peroxidase | -6.59 |
| 1336 | 255080_at | AT4G09030 | AGP10 (Arabinogalactan protein 10) | -4.30 |
| 1337 | 255046_at | AT4G09650 | ATP synthase delta chain, chloroplast, putative / H(+)-transporting two-sector ATPase, delta (OSCP) subunit, putative | -4.15 |
| 1338 | 255003_at | AT4G09950 | avirulence-responsive family protein / avirulence induced gene (AIG1) family protein | 6.73 |
| 1339 | 255006_at | AT4G10010 | protein kinase family protein | 9.29 |
| 1340 | 255008_at | AT4G10060 | catalytic | -4.99 |
| 1341 | 255016_at | AT4G10120 | ATSPS4F; sucrose-phosphate synthase/ transferase, transferring glycosyl groups | -3.75 |
| 1342 | 255806_at | AT4G10260 | pfkB-type carbohydrate kinase family protein | 6.36 |
| 1343 | 255809_at | AT4G10300 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G04300.1); similar to unknown [Populus trichocarpa] (GB:ABK95374.1); contains InterPro domain RmlC-like jelly roll fold (InterPro:IPR014710); contains InterPro domain Protein of unknown function DUF861, cupin-3 (InterPro:IPR008579); contains InterPro domain Cupin, RmlC-type (InterPro:IPR011051) | -4.03 |
| 1344 | 254970_at | AT4G10340 | LHCB5 (LIGHT HARVESTING COMPLEX OF PHOTOSYSTEM II 5); chlorophyll binding | -7.11 |
| 1345 | 254972_at | AT4G10440 | dehydration-responsive family protein | 6.14 |
| 1346 | 254961_at | AT4G11030 | long-chain-fatty-acid--CoA ligase, putative / long-chain acyl-CoA synthetase, putative | 5.48 |
| 1347 | 254886_at | AT4G11760 | LCR17 (Low-molecular-weight cysteine-rich 17) | 11.12 |
| 1348 | 254815_at | AT4G12420 | SKU5 (skewed 5); copper ion binding | -4.69 |
| 1349 | 254785_at | AT4G12730 | FLA2 | -6.00 |
| 1350 | 254790_at | AT4G12800 | PSAL (photosystem I subunit L) | -5.84 |
| 1351 | 254782_at | AT4G12860 | UNE14 (unfertilized embryo sac 14); calcium ion binding | 6.67 |
| 1352 | 254789_at | AT4G12880 | plastocyanin-like domain-containing protein | -5.12 |
| 1353 | 254793_at | AT4G12930 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G28930.1) | 7.26 |
| 1354 | 254777_at | AT4G12960 | gamma interferon responsive lysosomal thiol reductase family protein / GILT family protein | 5.57 |
| 1355 | 254753_at | AT4G13190 | kinase | 5.98 |
| 1356 | 254760_at | AT4G13200 | Identical to Uncharacterized protein At4g13200, chloroplast precursor [Arabidopsis Thaliana] (GB:Q8LDV3;GB:Q9SVQ7); similar to unknown [Picea sitchensis] (GB:ABK25970.1); similar to unknown [Picea sitchensis] (GB:ABK22042.1) | -3.76 |
| 1357 | 254755_at | AT4G13220 | similar to hypothetical protein [Vitis vinifera] (GB:CAN75300.1) | -5.66 |
| 1358 | 254762_at | AT4G13230 | late embryogenesis abundant domain-containing protein / LEA domain-containing protein | 8.70 |
| 1359 | 254757_at | AT4G13240 | ATROPGEF9/ROPGEF9 (KINASE PARTNER PROTEIN-LIKE); Rho guanyl-nucleotide exchange factor/ | 7.45 |
| 1360 | 254775_at | AT4G13450 | universal stress protein (USP) family protein | 7.22 |
| 1361 | 254722_at | AT4G13530 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G10080.1); similar to unknown protein [Oryza sativa (japonica cultivar-group)] (GB:BAD82228.1) | -4.67 |
| 1362 | 254716_at | AT4G13560 | UNE15 (unfertilized embryo sac 15) | 9.30 |
| 1363 | 254727_at | AT4G13670 | PTAC5 (PLASTID TRANSCRIPTIONALLY ACTIVE5); heat shock protein binding / unfolded protein binding | -4.36 |
| 1364 | 254687_at | AT4G13770 | CYP83A1 (CYTOCHROME P450 83A1); oxygen binding | -4.76 |
| 1365 | 245310_at | AT4G13950 | RALFL31 (RALF-LIKE 31) | -4.97 |
| 1366 | 245386_at | AT4G14010 | RALFL32 (RALF-LIKE 32) | -5.50 |
| 1367 | 245609_at | AT4G14370 | disease resistance protein (TIR-NBS-LRR class), putative | 6.28 |
| 1368 | 245591_at | AT4G14530 | similar to AGL97, DNA binding / transcription factor [Arabidopsis thaliana] (TAIR:AT1G46408.1) | 6.06 |
| 1369 | 245577_at | AT4G14780 | protein kinase, putative | 6.76 |
| 1370 | 245579_at | AT4G14810 | unknown protein | 9.54 |
| 1371 | 245398_at | AT4G14900 | hydroxyproline-rich glycoprotein family protein | -4.98 |
| 1372 | 245538_at | AT4G15200 | formin homology 2 domain-containing protein / FH2 domain-containing protein | 6.77 |
| 1373 | 245528_at | AT4G15530 | PPDK (PYRUVATE ORTHOPHOSPHATE DIKINASE); kinase/ pyruvate, phosphate dikinase | 5.75 |
| 1374 | 245501_at | AT4G15620 | integral membrane family protein | -4.94 |
| 1375 | 245503_at | AT4G15650 | protein kinase-related | 10.74 |
| 1376 | 245334_at | AT4G15800 | RALFL33 (RALF-LIKE 33) | -4.87 |
| 1377 | 245468_at | AT4G15980 | pectinesterase family protein | 7.24 |
| 1378 | 245472_at | AT4G16040 | unknown protein | 5.72 |
| 1379 | 245477_at | AT4G16110 | ARR2 (ARABIDOPSIS RESPONSE REGULATOR 2); transcription factor/ two-component response regulator | 4.88 |
| 1380 | 245335_at | AT4G16160 | ATOEP16-2/ATOEP16-S; P-P-bond-hydrolysis-driven protein transmembrane transporter | 7.90 |
| 1381 | 245483_at | AT4G16190 | cysteine proteinase, putative | -4.74 |
| 1382 | 245296_at | AT4G16370 | ATOPT3 (OLIGOPEPTIDE TRANSPORTER); oligopeptide transporter | -4.58 |
| 1383 | 245314_at | AT4G16745 | exostosin family protein | 8.33 |
| 1384 | 245318_at | AT4G16980 | arabinogalactan-protein family | -5.73 |
| 1385 | 245405_at | AT4G17150 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G14290.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO23689.1); contains domain PTHR12277:SF9 (PTHR12277:SF9); contains domain PTHR12277 (PTHR12277); contains domain SSF53474 (SSF53474); contains domain G3DSA:3.40.50.1820 (G3DSA:3.40.50.1820) | -4.34 |
| 1386 | 245362_at | AT4G17460 | HAT1 (homeobox-leucine zipper protein 1); DNA binding / transcription factor | -5.54 |
| 1387 | 245423_at | AT4G17483 | palmitoyl protein thioesterase family protein | 5.80 |
| 1388 | 245354_at | AT4G17600 | LIL3:1; transcription factor | -4.96 |
| 1389 | 245376_at | AT4G17690 | peroxidase, putative | 6.59 |
| 1390 | 245348_at | AT4G17770 | ATTPS5 (Arabidopsis thaliana trehalose phosphatase/synthase 5); transferase, transferring glycosyl groups / trehalose-phosphatase | -4.41 |
| 1391 | 245361_at | AT4G17790 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G71940.1); similar to hypothetical protein 23.t00026 [Brassica oleracea] (GB:ABD65614.1); contains InterPro domain SNARE associated Golgi protein (InterPro:IPR015414) | 5.89 |
| 1392 | 254710_at | AT4G18050 | PGP9 (P-GLYCOPROTEIN 9); ATPase, coupled to transmembrane movement of substances | 4.95 |
| 1393 | 254668_at | AT4G18350 | NCED2 (NINE-CIS-EPOXYCAROTENOID DIOXYGENASE 2) | 5.48 |
| 1394 | 254627_at | AT4G18395 | unknown protein | 9.20 |
| 1395 | 254623_at | AT4G18480 | CHLI1 (CHLORINA 42); magnesium chelatase | -4.57 |
| 1396 | 254633_at | AT4G18640 | MRH1 (morphogenesis of root hair 1); ATP binding / protein serine/threonine kinase | 7.17 |
| 1397 | 254636_at | AT4G18700 | CIPK12 (SNF1-RELATED PROTEIN KINASE 3.9); kinase | 6.19 |
| 1398 | 254640_at | AT4G18790 | NRAMP5 (NRAMP metal ion transporter 5); metal ion transmembrane transporter | 8.41 |
| 1399 | 254642_at | AT4G18810 | binding / catalytic/ transcription repressor | -4.96 |
| 1400 | 254598_at | AT4G18990 | xyloglucan:xyloglucosyl transferase, putative / xyloglucan endotransglycosylase, putative / endo-xyloglucan transferase, putative | 5.35 |
| 1401 | 254527_at | AT4G19610 | RNA binding | -3.86 |
| 1402 | 254532_at | AT4G19660 | NPR4 (NPR1-LIKE PROTEIN 4); protein binding | -5.43 |
| 1403 | 254551_at | AT4G19840 | ATPP2-A1 (Arabidopsis thaliana phloem protein 2-A1) | -6.45 |
| 1404 | 254504_at | AT4G20030 | RNA recognition motif (RRM)-containing protein | -5.44 |
| 1405 | 254507_at | AT4G20160 | similar to protein binding / zinc ion binding [Arabidopsis thaliana] (TAIR:AT1G30860.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO15310.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN63223.1); contains domain PTHR12429 (PTHR12429) | 7.53 |
| 1406 | 254508_at | AT4G20170 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G44670.1); similar to Os06g0328800 [Oryza sativa (japonica cultivar-group)] (GB:NP_001057533.1); similar to Os02g0712500 [Oryza sativa (japonica cultivar-group)] (GB:NP_001047907.1); similar to unknown protein [Oryza sativa (japonica cultivar-group)] (GB:BAD72474.1); contains InterPro domain Protein of unknown function DUF23 (InterPro:IPR008166) | -4.32 |
| 1407 | 254515_at | AT4G20270 | BAM3 (big apical meristem 3); ATP binding / protein serine/threonine kinase | -4.30 |
| 1408 | 254480_at | AT4G20360 | AtRABE1b/AtRab8D (Arabidopsis Rab GTPase homolog E1b); translation elongation factor | -4.63 |
| 1409 | 254477_at | AT4G20380 | LSD1 (LESION SIMULATING DISEASE) | 5.56 |
| 1410 | 254468_at | AT4G20460 | NAD-dependent epimerase/dehydratase family protein | 5.50 |
| 1411 | 254448_at | AT4G20870 | fatty acid hydroxylase, putative | 5.60 |
| 1412 | 254434_at | AT4G20880 | ethylene-responsive nuclear protein / ethylene-regulated nuclear protein (ERT2) | 5.46 |
| 1413 | 254446_at | AT4G20890 | TUB9 (tubulin beta-9 chain); structural molecule | 4.73 |
| 1414 | 254460_at | AT4G21210 | phosphoprotein phosphatase/ protein kinase | -4.50 |
| 1415 | 254398_at | AT4G21280 | PSBQ/PSBQ-1/PSBQA; calcium ion binding | -4.46 |
| 1416 | 254415_at | AT4G21326 | ATSBT3.12; subtilase | 6.25 |
| 1417 | 254372_at | AT4G21620 | glycine-rich protein | -5.28 |
| 1418 | 254386_at | AT4G21960 | PRXR1 (peroxidase 42); peroxidase | -5.25 |
| 1419 | 254362_at | AT4G22160 | unknown protein | -4.13 |
| 1420 | 254356_at | AT4G22190 | similar to unknown [Populus trichocarpa] (GB:ABK94177.1) | -6.66 |
| 1421 | 254305_at | AT4G22200 | AKT2 (Arabidopsis K+ transporter 2); cyclic nucleotide binding / inward rectifier potassium channel | -5.16 |
| 1422 | 254328_at | AT4G22570 | APT3 (ADENINE PHOSPHORIBOSYL TRANSFERASE 3); adenine phosphoribosyltransferase | -6.61 |
| 1423 | 254275_at | AT4G22670 | ATHIP1 (ARABIDOPSIS THALIANA HSP70-INTERACTING PROTEIN 1); binding | -3.98 |
| 1424 | 254273_at | AT4G22720 | glycoprotease M22 family protein | -4.48 |
| 1425 | 254298_at | AT4G22890 | PGR5-LIKE A | -7.79 |
| 1426 | 254256_at | AT4G23180 | CRK10 (CYSTEINE-RICH RLK10); kinase | -4.40 |
| 1427 | 254239_at | AT4G23400 | PIP1;5/PIP1D (plasma membrane intrinsic protein 1;5); water channel | -5.60 |
| 1428 | 254224_at | AT4G23650 | CDPK6 (CALCIUM-DEPENDENT PROTEIN KINASE 6); calmodulin-dependent protein kinase/ kinase | -4.34 |
| 1429 | 254225_at | AT4G23670 | major latex protein-related / MLP-related | -7.76 |
| 1430 | 254234_at | AT4G23680 | major latex protein-related / MLP-related | -3.96 |
| 1431 | 254221_at | AT4G23820 | glycoside hydrolase family 28 protein / polygalacturonase (pectinase) family protein | -3.90 |
| 1432 | 254187_at | AT4G23890 | similar to unnamed protein product [Vitis vinifera] (GB:CAO69542.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN73381.1) | -4.45 |
| 1433 | 254181_at | AT4G23940 | FtsH protease, putative | -4.44 |
| 1434 | 254183_at | AT4G23960 | F-box family protein | 4.76 |
| 1435 | 254205_at | AT4G24170 | kinesin motor family protein | 7.42 |
| 1436 | 254122_at | AT4G24510 | CER2 (ECERIFERUM 2); transferase | -5.96 |
| 1437 | 254173_at | AT4G24580 | pleckstrin homology (PH) domain-containing protein-related / RhoGAP domain-containing protein | 7.89 |
| 1438 | 254142_at | AT4G24630 | receptor/ zinc ion binding | 5.80 |
| 1439 | 254123_at | AT4G24640 | APPB1; pectinesterase inhibitor | 11.06 |
| 1440 | 254117_at | AT4G24750 | similar to rhodanese-like domain-containing protein [Arabidopsis thaliana] (TAIR:AT3G08920.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO64218.1); contains InterPro domain Rhodanese-like (InterPro:IPR001763) | -3.79 |
| 1441 | 254119_at | AT4G24780 | pectate lyase family protein | -4.74 |
| 1442 | 254104_at | AT4G25040 | integral membrane family protein | 4.77 |
| 1443 | 254102_at | AT4G25050 | ACP4 (ACYL CARRIER PROTEIN 4) | -5.78 |
| 1444 | 254105_at | AT4G25080 | CHLM (MAGNESIUM-PROTOPORPHYRIN IX METHYLTRANSFERASE); magnesium protoporphyrin IX methyltransferase | -4.34 |
| 1445 | 254098_at | AT4G25100 | FSD1 (FE SUPEROXIDE DISMUTASE 1); iron superoxide dismutase | -8.38 |
| 1446 | 254096_at | AT4G25150 | acid phosphatase, putative | 8.11 |
| 1447 | 254068_at | AT4G25450 | ATNAP8 (Arabidopsis thaliana non-intrinsic ABC protein 8); ATPase, coupled to transmembrane movement of substances | -3.83 |
| 1448 | 245232_at | AT4G25590 | ADF7 (ACTIN DEPOLYMERIZING FACTOR 7); actin binding | 12.89 |
| 1449 | 254024_at | AT4G25780 | pathogenesis-related protein, putative | 9.45 |
| 1450 | 254045_at | AT4G25880 | APUM6 (ARABIDOPSIS PUMILIO 6); RNA binding | -4.39 |
| 1451 | 254040_at | AT4G25900 | aldose 1-epimerase family protein | 5.73 |
| 1452 | 254033_at | AT4G25950 | VATG3 (VACUOLAR ATP SYNTHASE G3) | 10.12 |
| 1453 | 254034_at | AT4G25960 | PGP2 (P-GLYCOPROTEIN 2); ATPase, coupled to transmembrane movement of substances | -4.60 |
| 1454 | 253989_at | AT4G26130 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G56980.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN72576.1) | -4.08 |
| 1455 | 254001_at | AT4G26260 | MIOX4 (MYO-INOSITOL OXYGENASE 4) | 10.36 |
| 1456 | 254005_at | AT4G26330 | ATSBT3.18/UNE17 (UNFERTILIZED EMBRYO SAC 17); subtilase | 5.37 |
| 1457 | 254011_at | AT4G26370 | antitermination NusB domain-containing protein | -3.90 |
| 1458 | 254009_at | AT4G26390 | pyruvate kinase, putative | 5.91 |
| 1459 | 253958_at | AT4G26400 | zinc finger (C3HC4-type RING finger) family protein | 5.71 |
| 1460 | 253961_at | AT4G26440 | WRKY34 (MICROSPORE-SPECIFIC PROMOTER 3); transcription factor | 7.84 |
| 1461 | 253963_at | AT4G26470 | calcium ion binding | 5.92 |
| 1462 | 253964_at | AT4G26480 | KH domain-containing protein | -4.50 |
| 1463 | 253971_at | AT4G26530 | fructose-bisphosphate aldolase, putative | -4.43 |
| 1464 | 253977_at | AT4G26630 | GTP binding / RNA binding | -4.83 |
| 1465 | 253926_at | AT4G26650 | RNA recognition motif (RRM)-containing protein | 4.83 |
| 1466 | 253931_at | AT4G26770 | phosphatidate cytidylyltransferase, putative / CDP-diglyceride synthetase, putative | 6.00 |
| 1467 | 253946_at | AT4G26790 | GDSL-motif lipase/hydrolase family protein | -3.99 |
| 1468 | 253939_at | AT4G26930 | MYB97 (myb domain protein 97); DNA binding / transcription factor | 7.87 |
| 1469 | 253943_at | AT4G27030 | small conjugating protein ligase | -4.33 |
| 1470 | 253924_at | AT4G27110 | COBL11 (COBRA-LIKE PROTEIN 11 PRECURSOR) | 8.05 |
| 1471 | 253919_at | AT4G27350 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G54240.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO39196.1); contains InterPro domain Protein of unknown function DUF1223 (InterPro:IPR010634) | 7.08 |
| 1472 | 253913_at | AT4G27370 | VIIIB (Myosin-like protein VIIB); motor | 5.56 |
| 1473 | 253871_at | AT4G27440 | PORB (PROTOCHLOROPHYLLIDE OXIDOREDUCTASE B); oxidoreductase/ protochlorophyllide reductase | -3.82 |
| 1474 | 253831_at | AT4G27580 | unknown protein | 11.45 |
| 1475 | 253859_at | AT4G27657 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G27652.1) | -4.31 |
| 1476 | 253860_at | AT4G27700 | rhodanese-like domain-containing protein | -5.84 |
| 1477 | 253833_at | AT4G27790 | calcium-binding EF hand family protein | 9.76 |
| 1478 | 253834_at | AT4G27800 | protein phosphatase 2C PPH1 / PP2C PPH1 (PPH1) | -3.80 |
| 1479 | 253838_at | AT4G27880 | seven in absentia (SINA) family protein | 5.67 |
| 1480 | 253845_at | AT4G27980 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G04960.2); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G04960.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G04960.3); similar to unnamed protein product [Vitis vinifera] (GB:CAO71037.1); contains InterPro domain Molecular chaperone, heat shock protein, Hsp40, DnaJ (InterPro:IPR015609) | 5.67 |
| 1481 | 253846_at | AT4G28000 | AAA-type ATPase family protein | 9.07 |
| 1482 | 253807_at | AT4G28280 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G20700.1); similar to hypothetical protein OsJ_007917 [Oryza sativa (japonica cultivar-group)] (GB:EAZ24434.1) | 9.80 |
| 1483 | 253780_at | AT4G28400 | protein phosphatase 2C, putative / PP2C, putative | -6.43 |
| 1484 | 253775_at | AT4G28440 | DNA-binding protein-related | -4.68 |
| 1485 | 253790_at | AT4G28660 | PSB28 (PHOTOSYSTEM II REACTION CENTER PSB28 PROTEIN) | -4.28 |
| 1486 | 253787_at | AT4G28670 | protein kinase family protein | 8.06 |
| 1487 | 253788_at | AT4G28680 | tyrosine decarboxylase, putative | 4.91 |
| 1488 | 253792_at | AT4G28700 | ammonium transporter, putative | 6.42 |
| 1489 | 253765_at | AT4G28740 | similar to LPA1 (LOW PSII ACCUMULATION1), binding [Arabidopsis thaliana] (TAIR:AT1G02910.1); similar to OSJNBa0043L24.5 [Oryza sativa (japonica cultivar-group)] (GB:CAE04717.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO40509.1) | -4.77 |
| 1490 | 253738_at | AT4G28750 | PSAE-1 (PSA E1 KNOCKOUT); catalytic | -7.12 |
| 1491 | 253739_at | AT4G28760 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G20240.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN81514.1); contains domain PTHR21726:SF7 (PTHR21726:SF7); contains domain PTHR21726 (PTHR21726) | -4.91 |
| 1492 | 253758_at | AT4G29060 | EMB2726 (EMBRYO DEFECTIVE 2726); translation elongation factor | -5.40 |
| 1493 | 253749_at | AT4G29080 | PAP2 (PHYTOCHROME-ASSOCIATED PROTEIN 2); transcription factor | -5.13 |
| 1494 | 253725_at | AT4G29340 | PRF4 (PROFILIN 4); actin binding | 9.12 |
| 1495 | 253727_at | AT4G29350 | PFN2/PRF2/PRO2 (PROFILIN 2); actin binding / protein binding | -6.49 |
| 1496 | 253718_at | AT4G29450 | leucine-rich repeat protein kinase, putative | 4.91 |
| 1497 | 253687_at | AT4G29520 | similar to unnamed protein product [Vitis vinifera] (GB:CAO65389.1); contains InterPro domain Saposin B (InterPro:IPR008139) | -3.76 |
| 1498 | 253697_at | AT4G29700 | type I phosphodiesterase/nucleotide pyrophosphatase family protein | -3.77 |
| 1499 | 253650_at | AT4G30020 | subtilase family protein | -3.83 |
| 1500 | 253660_at | AT4G30140 | GDSL-motif lipase/hydrolase family protein | 9.46 |
| 1501 | 253627_at | AT4G30650 | hydrophobic protein, putative / low temperature and salt responsive protein, putative | -6.74 |
| 1502 | 253581_at | AT4G30660 | hydrophobic protein, putative / low temperature and salt responsive protein, putative | -4.86 |
| 1503 | 253587_at | AT4G30770 | similar to Os11g0195700 [Oryza sativa (japonica cultivar-group)] (GB:NP_001067426.1); similar to hypothetical protein OsJ_032844 [Oryza sativa (japonica cultivar-group)] (GB:EAZ18635.1); contains domain PROKAR_LIPOPROTEIN (PS51257) | 7.26 |
| 1504 | 253599_at | AT4G30860 | ASHR3 (ASH1-RELATED 3); protein binding / zinc ion binding | 6.69 |
| 1505 | 253547_at | AT4G30950 | FAD6 (FATTY ACID DESATURASE 6); omega-6 fatty acid desaturase | -3.88 |
| 1506 | 253550_at | AT4G30960 | CIPK6 (CBL-INTERACTING PROTEIN KINASE 6); kinase | -4.92 |
| 1507 | 253573_at | AT4G31020 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G24320.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO22751.1); contains domain PTHR12277:SF8 (PTHR12277:SF8); contains domain PTHR12277 (PTHR12277); contains domain SSF53474 (SSF53474); contains domain G3DSA:3.40.50.1820 (G3DSA:3.40.50.1820) | 6.11 |
| 1508 | 253558_at | AT4G31120 | ATPRMT5/PRMT5/SKB1 (SHK1 BINDING PROTEIN 1); protein methyltransferase | -4.89 |
| 1509 | 253559_at | AT4G31140 | glycosyl hydrolase family 17 protein | -4.09 |
| 1510 | 253564_at | AT4G31170 | protein kinase family protein | -3.93 |
| 1511 | 253567_at | AT4G31230 | protein kinase family protein | 8.27 |
| 1512 | 253534_at | AT4G31500 | CYP83B1 (CYTOCHROME P450 MONOOXYGENASE 83B1); oxygen binding | -7.71 |
| 1513 | 253531_at | AT4G31540 | ATEXO70G1 (exocyst subunit EXO70 family protein G1); protein binding | -4.08 |
| 1514 | 253420_at | AT4G32260 | ATP synthase family | -3.91 |
| 1515 | 253440_at | AT4G32570 | TIFY8 | -5.66 |
| 1516 | 253447_at | AT4G32630 | ARF GTPase activator/ zinc ion binding | 6.74 |
| 1517 | 253403_at | AT4G32830 | ATAUR1 (ATAURORA1); histone serine kinase(H3-S10 specific) / kinase/ protein serine/threonine kinase | 5.47 |
| 1518 | 253385_at | AT4G32930 | similar to unnamed protein product [Vitis vinifera] (GB:CAO43772.1); contains InterPro domain Protein of unknown function DUF866, eukaryotic (InterPro:IPR008584) | -4.23 |
| 1519 | 253370_at | AT4G33230 | pectinesterase family protein | 8.76 |
| 1520 | 253356_at | AT4G33390 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G26570.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO61337.1); contains InterPro domain Protein of unknown function DUF827, plant (InterPro:IPR008545) | 9.00 |
| 1521 | 253326_at | AT4G33440 | glycoside hydrolase family 28 protein / polygalacturonase (pectinase) family protein | 4.79 |
| 1522 | 253337_at | AT4G33470 | HDA14 (histone deacetylase 14); histone deacetylase | -4.06 |
| 1523 | 253302_at | AT4G33660 | unknown protein | -4.98 |
| 1524 | 253351_at | AT4G33700 | CBS domain-containing protein | -5.84 |
| 1525 | 253317_at | AT4G33960 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G15830.1) | -3.75 |
| 1526 | 253318_at | AT4G33970 | leucine-rich repeat family protein / extensin family protein | 9.97 |
| 1527 | 253276_at | AT4G34050 | caffeoyl-CoA 3-O-methyltransferase, putative | -6.61 |
| 1528 | 253237_at | AT4G34240 | ALDH3I1 (Aldehyde dehydrogenase 3I1); 3-chloroallyl aldehyde dehydrogenase/ aldehyde dehydrogenase (NAD) | -4.71 |
| 1529 | 253262_at | AT4G34440 | protein kinase family protein | 7.79 |
| 1530 | 253201_at | AT4G34620 | SSR16 (ribosomal protein S16); structural constituent of ribosome | -6.55 |
| 1531 | 253253_at | AT4G34750 | auxin-responsive protein, putative / small auxin up RNA (SAUR_E) | 5.76 |
| 1532 | 253255_at | AT4G34760 | auxin-responsive family protein | -5.35 |
| 1533 | 253214_at | AT4G34940 | armadillo/beta-catenin repeat family protein | 6.27 |
| 1534 | 253226_at | AT4G35010 | BGAL11 (beta-galactosidase 11); beta-galactosidase | 12.30 |
| 1535 | 253168_at | AT4G35070 | similar to SBP1 (S-RIBONUCLEASE BINDING PROTEIN 1), protein binding / zinc ion binding [Arabidopsis thaliana] (TAIR:AT1G45976.1); similar to unknown [Populus trichocarpa] (GB:ABK93023.1); contains InterPro domain S-ribonuclease binding protein, SBP1, pollen (InterPro:IPR017066) | 5.08 |
| 1536 | 253181_at | AT4G35180 | LHT7 (LYS/HIS TRANSPORTER 7); amino acid transmembrane transporter | 9.08 |
| 1537 | 253184_at | AT4G35230 | protein kinase family protein | -6.50 |
| 1538 | 253197_at | AT4G35250 | vestitone reductase-related | -8.02 |
| 1539 | 253188_at | AT4G35300 | TMT2 (TONOPLAST MONOSACCHARIDE TRANSPORTER2); carbohydrate transmembrane transporter/ nucleoside transmembrane transporter/ sugar:hydrogen ion symporter | -4.35 |
| 1540 | 253167_at | AT4G35310 | CPK5 (CALMODULIN-DOMAIN PROTEIN KINASE 5); calmodulin-dependent protein kinase/ kinase | -4.87 |
| 1541 | 253193_at | AT4G35380 | guanine nucleotide exchange family protein | 5.06 |
| 1542 | 253137_at | AT4G35500 | protein kinase family protein | 4.56 |
| 1543 | 253132_at | AT4G35580 | no apical meristem (NAM) family protein | -4.29 |
| 1544 | 253149_at | AT4G35650 | isocitrate dehydrogenase, putative / NAD+ isocitrate dehydrogenase, putative | 7.99 |
| 1545 | 253151_at | AT4G35670 | glycoside hydrolase family 28 protein / polygalacturonase (pectinase) family protein | 9.47 |
| 1546 | 253153_at | AT4G35700 | zinc finger (C2H2 type) family protein | 11.28 |
| 1547 | 253087_at | AT4G36350 | ATPAP25/PAP25 (purple acid phosphatase 25); acid phosphatase/ protein serine/threonine phosphatase | 5.17 |
| 1548 | 253090_at | AT4G36360 | BGAL3 (beta-galactosidase 3); beta-galactosidase | -4.64 |
| 1549 | 246208_at | AT4G36490 | SEC14 cytosolic factor, putative / phosphoglyceride transfer protein, putative | 10.90 |
| 1550 | 246275_at | AT4G36540 | BEE2 (BR ENHANCED EXPRESSION 2); DNA binding / transcription factor | -4.03 |
| 1551 | 246242_at | AT4G36600 | late embryogenesis abundant domain-containing protein / LEA domain-containing protein | 5.86 |
| 1552 | 246223_at | AT4G36890 | IRX14 (IRREGULAR XYLEM 14); transferase, transferring glycosyl groups / xylosyltransferase | -4.41 |
| 1553 | 246205_at | AT4G36970 | remorin family protein | -3.77 |
| 1554 | 246231_at | AT4G37080 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G42690.2); similar to hypothetical protein [Populus tremula] (GB:CAM84232.1); contains InterPro domain Protein of unknown function DUF547 (InterPro:IPR006869) | -4.19 |
| 1555 | 246253_at | AT4G37260 | AtMYB73/MYB73 (myb domain protein 73); DNA binding / transcription factor | -4.85 |
| 1556 | 246234_at | AT4G37280 | MRG family protein | 5.67 |
| 1557 | 253095_at | AT4G37510 | ribonuclease III family protein | -5.72 |
| 1558 | 253062_at | AT4G37590 | phototropic-responsive NPH3 family protein | -5.10 |
| 1559 | 253069_at | AT4G37840 | hexokinase, putative | 6.13 |
| 1560 | 253014_at | AT4G37940 | AGL21 (AGAMOUS-LIKE 21); transcription factor | 7.74 |
| 1561 | 252983_at | AT4G37980 | ELI3-1 (ELICITOR-ACTIVATED GENE 3); binding / catalytic/ oxidoreductase/ zinc ion binding | -3.98 |
| 1562 | 252984_at | AT4G37990 | ELI3-2 (ELICITOR-ACTIVATED GENE 3) | 5.77 |
| 1563 | 253031_at | AT4G38190 | ATCSLD4 (Cellulose synthase-like D4); cellulose synthase/ transferase, transferring glycosyl groups | 8.08 |
| 1564 | 253033_at | AT4G38220 | aminoacylase, putative / N-acyl-L-amino-acid amidohydrolase, putative | -3.87 |
| 1565 | 252979_at | AT4G38225 | similar to unnamed protein product [Vitis vinifera] (GB:CAO47538.1); contains domain PTHR11014 (PTHR11014); contains domain PTHR11014:SF1 (PTHR11014:SF1) | -5.54 |
| 1566 | 253034_at | AT4G38230 | CPK26 (calcium-dependent protein kinase 26); calmodulin-dependent protein kinase/ kinase | 7.38 |
| 1567 | 252987_at | AT4G38390 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G76270.1); similar to axi 1 [Nicotiana tabacum] (GB:CAA56570.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO47467.1); contains InterPro domain Protein of unknown function DUF246, plant (InterPro:IPR004348) | 4.88 |
| 1568 | 252992_at | AT4G38520 | protein phosphatase 2C family protein / PP2C family protein | -4.83 |
| 1569 | 252976_s_at | AT4G38550 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G20950.1); contains InterPro domain Phospholipase-like, arabidopsis (InterPro:IPR007942) | -5.02 |
| 1570 | 252950_at | AT4G38690 | 1-phosphatidylinositol phosphodiesterase-related | -4.19 |
| 1571 | 252971_at | AT4G38770 | PRP4 (PROLINE-RICH PROTEIN 4) | -6.55 |
| 1572 | 252965_at | AT4G38860 | auxin-responsive protein, putative | -5.10 |
| 1573 | 252929_at | AT4G38970 | fructose-bisphosphate aldolase, putative | -5.34 |
| 1574 | 252933_at | AT4G39110 | protein kinase family protein | 10.45 |
| 1575 | 252937_at | AT4G39180 | SEC14 (secretion 14) | 8.45 |
| 1576 | 252910_at | AT4G39590 | kelch repeat-containing F-box family protein | 4.95 |
| 1577 | 252881_at | AT4G39610 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G21990.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO71771.1); contains InterPro domain Protein of unknown function DUF617, plant (InterPro:IPR006460) | 9.78 |
| 1578 | 252908_at | AT4G39670 | glycolipid binding / glycolipid transporter | 7.66 |
| 1579 | 252865_at | AT4G39753 | kelch repeat-containing F-box family protein | 4.94 |
| 1580 | 252867_at | AT4G39870 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G05590.2); similar to unnamed protein product [Vitis vinifera] (GB:CAO50031.1); contains InterPro domain TLDc (InterPro:IPR006571) | 5.86 |
| 1581 | 252873_at | AT4G40020 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G16730.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN70653.1); contains domain PTHR23160 (PTHR23160) | 7.66 |
| 1582 | 251140_at | AT5G01090 | legume lectin family protein | -5.26 |
| 1583 | 251082_at | AT5G01530 | chlorophyll A-B binding protein CP29 (LHCB4) | -7.18 |
| 1584 | 251083_at | AT5G01590 | similar to unnamed protein product [Vitis vinifera] (GB:CAO63840.1) | -4.99 |
| 1585 | 251107_at | AT5G01610 | Identical to Uncharacterized protein At5g01610 [Arabidopsis Thaliana] (GB:Q9M015;GB:Q0WT00;GB:Q84VV4); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G08890.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G08890.2); similar to hypothetical protein [Vitis vinifera] (GB:CAN63135.1); contains InterPro domain Protein of unknown function DUF538 (InterPro:IPR007493) | 7.35 |
| 1586 | 251102_at | AT5G01690 | ATCHX27 (cation/hydrogen exchanger 27); monovalent cation:proton antiporter | 6.31 |
| 1587 | 251103_at | AT5G01700 | protein phosphatase 2C, putative / PP2C, putative | 7.24 |
| 1588 | 251074_at | AT5G01800 | saposin B domain-containing protein | 4.63 |
| 1589 | 251060_at | AT5G01820 | ATSR1 (SERINE/THREONINE PROTEIN KINASE 1); kinase | 5.00 |
| 1590 | 251039_at | AT5G02020 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G55646.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G59080.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO15731.1) | 4.71 |
| 1591 | 251037_at | AT5G02100 | UNE18 (unfertilized embryo sac 18); oxysterol binding | 4.79 |
| 1592 | 251031_at | AT5G02120 | OHP (ONE HELIX PROTEIN) | -6.93 |
| 1593 | 251024_at | AT5G02180 | amino acid transporter family protein | 5.17 |
| 1594 | 251047_at | AT5G02390 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G07620.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO15645.1); contains domain PTHR21726:SF3 (PTHR21726:SF3); contains domain PTHR21726 (PTHR21726) | 5.39 |
| 1595 | 251049_at | AT5G02430 | WD-40 repeat family protein | 5.41 |
| 1596 | 251015_at | AT5G02480 | similar to SLT1 (SODIUM- AND LITHIUM-TOLERANT 1) [Arabidopsis thaliana] (TAIR:AT2G37570.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO15631.1); contains InterPro domain HSP20-like chaperone (InterPro:IPR008978); contains InterPro domain Heat shock protein Hsp20 (InterPro:IPR002068) | -4.67 |
| 1597 | 250997_at | AT5G02570 | histone H2B, putative | 8.22 |
| 1598 | 251008_at | AT5G02710 | similar to unnamed protein product [Vitis vinifera] (GB:CAO18074.1); contains InterPro domain Protein of unknown function UPF0153 (InterPro:IPR005358) | -6.13 |
| 1599 | 251004_at | AT5G02720 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G53490.1); similar to unknown [Picea sitchensis] (GB:ABK22243.1) | 4.79 |
| 1600 | 250983_at | AT5G02780 | In2-1 protein, putative | 5.45 |
| 1601 | 250968_at | AT5G02890 | transferase family protein | -4.19 |
| 1602 | 250957_at | AT5G03250 | phototropic-responsive NPH3 family protein | 5.03 |
| 1603 | 250942_at | AT5G03350 | legume lectin family protein | -4.94 |
| 1604 | 250897_at | AT5G03430 | phosphoadenosine phosphosulfate (PAPS) reductase family protein | -5.60 |
| 1605 | 250904_at | AT5G03620 | subtilase family protein | 5.95 |
| 1606 | 250917_at | AT5G03690 | fructose-bisphosphate aldolase, putative | 5.96 |
| 1607 | 250892_at | AT5G03760 | ATCSLA09 (RESISTANT TO AGROBACTERIUM TRANSFORMATION 4); transferase, transferring glycosyl groups | 5.67 |
| 1608 | 250884_at | AT5G03940 | FFC (FIFTY-FOUR CHLOROPLAST HOMOLOGUE); 7S RNA binding / GTP binding / mRNA binding | -5.39 |
| 1609 | 250875_at | AT5G04020 | calmodulin-binding protein-related (PICBP) | -3.77 |
| 1610 | 245701_at | AT5G04140 | GLS1/GLU1/GLUS (FERREDOXIN-DEPENDENT GLUTAMATE SYNTHASE 1); glutamate synthase (ferredoxin) | -7.83 |
| 1611 | 245700_at | AT5G04180 | carbonic anhydrase family protein | 10.02 |
| 1612 | 245693_at | AT5G04260 | thioredoxin family protein | -4.82 |
| 1613 | 245705_at | AT5G04390 | zinc finger (C2H2 type) family protein | 6.43 |
| 1614 | 250888_at | AT5G04460 | protein binding / zinc ion binding | 5.13 |
| 1615 | 250889_at | AT5G04500 | glycosyltransferase family protein 47 | 5.28 |
| 1616 | 250860_at | AT5G04770 | ATCAT6/CAT6 (CATIONIC AMINO ACID TRANSPORTER 6); amino acid transmembrane transporter/ basic amino acid transmembrane transporter/ cationic amino acid transmembrane transporter | -3.90 |
| 1617 | 246955_at | AT5G04870 | CPK1 (calcium-dependent protein kinase isoform AK1); calmodulin-dependent protein kinase/ kinase | -4.79 |
| 1618 | 250806_at | AT5G05070 | zinc ion binding | 10.98 |
| 1619 | 250823_at | AT5G05180 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G10880.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO41532.1) | -5.13 |
| 1620 | 250824_at | AT5G05200 | ABC1 family protein | -4.74 |
| 1621 | 250793_at | AT5G05600 | oxidoreductase, 2OG-Fe(II) oxygenase family protein | -4.30 |
| 1622 | 250752_at | AT5G05690 | CPD (CABBAGE 3); oxygen binding | -4.12 |
| 1623 | 250738_at | AT5G05730 | ASA1 (ANTHRANILATE SYNTHASE ALPHA SUBUNIT 1); anthranilate synthase | -5.34 |
| 1624 | 250750_at | AT5G05870 | UGT76C1 (UDP-GLUCOSYL TRANSFERASE 76C1); UDP-glycosyltransferase/ transferase, transferring glycosyl groups | -4.24 |
| 1625 | 250733_at | AT5G06290 | 2-cys peroxiredoxin, chloroplast, putative | -3.84 |
| 1626 | 250692_at | AT5G06560 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G11850.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G11850.2); similar to unnamed protein product [Vitis vinifera] (GB:CAO39886.1); contains InterPro domain Protein of unknown function DUF593 (InterPro:IPR007656) | 4.87 |
| 1627 | 250645_at | AT5G06700 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G12060.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO23412.1); contains InterPro domain Protein of unknown function DUF231, plant (InterPro:IPR004253) | -4.63 |
| 1628 | 250648_at | AT5G06760 | late embryogenesis abundant group 1 domain-containing protein / LEA group 1 domain-containing protein | 5.79 |
| 1629 | 250669_at | AT5G06870 | PGIP2 (POLYGALACTURONASE INHIBITING PROTEIN 2); protein binding | -3.90 |
| 1630 | 250581_at | AT5G07300 | BON2 (BONZAI 2) | -4.21 |
| 1631 | 250608_at | AT5G07420 | pectinesterase family protein | 12.91 |
| 1632 | 250631_at | AT5G07430 | pectinesterase family protein | 12.98 |
| 1633 | 250580_at | AT5G07440 | GDH2 (GLUTAMATE DEHYDROGENASE 2); oxidoreductase | 4.94 |
| 1634 | 250561_at | AT5G08030 | glycerophosphoryl diester phosphodiesterase family protein | 6.65 |
| 1635 | 250575_at | AT5G08240 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G23160.1) | 5.30 |
| 1636 | 250517_at | AT5G08260 | SCPL35 (serine carboxypeptidase-like 35); serine carboxypeptidase | -4.36 |
| 1637 | 246011_at | AT5G08330 | TCP family transcription factor, putative | -4.59 |
| 1638 | 250524_at | AT5G08520 | myb family transcription factor | -4.97 |
| 1639 | 245880_at | AT5G09430 | hydrolase | 8.52 |
| 1640 | 245883_at | AT5G09500 | 40S ribosomal protein S15 (RPS15C) | 5.80 |
| 1641 | 250514_at | AT5G09550 | RAB GDP-dissociation inhibitor | 8.66 |
| 1642 | 250515_at | AT5G09570 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G64400.1); similar to unknown [Populus trichocarpa] (GB:ABK94558.1); contains InterPro domain CHCH (InterPro:IPR010625) | 6.06 |
| 1643 | 250498_at | AT5G09660 | PMDH2 (PEROXISOMAL NAD-MALATE DEHYDROGENASE 2); malate dehydrogenase | -4.75 |
| 1644 | 250493_at | AT5G09800 | U-box domain-containing protein | 5.09 |
| 1645 | 250485_at | AT5G09990 | PROPEP5 (Elicitor peptide 5 precursor) | -3.81 |
| 1646 | 250466_at | AT5G10090 | tetratricopeptide repeat (TPR)-containing protein | 9.93 |
| 1647 | 250475_at | AT5G10180 | AST68 (Sulfate transporter 2.1) | -4.47 |
| 1648 | 250479_at | AT5G10260 | AtRABH1e (Arabidopsis Rab GTPase homolog H1e); GTP binding | 6.97 |
| 1649 | 250480_at | AT5G10290 | leucine-rich repeat family protein / protein kinase family protein | -5.35 |
| 1650 | 250433_at | AT5G10400 | histone H3 | -4.49 |
| 1651 | 250439_at | AT5G10450 | GRF6 (G-BOX REGULATING FACTOR 6); protein phosphorylated amino acid binding | -5.78 |
| 1652 | 250429_at | AT5G10470 | kinesin motor protein-related | -6.12 |
| 1653 | 250427_at | AT5G10500 | kinase interacting family protein | 9.33 |
| 1654 | 246012_at | AT5G10650 | zinc finger (C3HC4-type RING finger) family protein | 5.12 |
| 1655 | 246013_at | AT5G10660 | calmodulin-binding protein-related | 7.86 |
| 1656 | 250400_at | AT5G10740 | protein phosphatase 2C-related / PP2C-related | 5.44 |
| 1657 | 250409_at | AT5G10860 | CBS domain-containing protein | -4.07 |
| 1658 | 250395_at | AT5G10950 | cylicin-related | 5.30 |
| 1659 | 250366_at | AT5G11420 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G25460.1); similar to unknown [Ricinus communis] (GB:CAB02653.1); contains InterPro domain Protein of unknown function DUF642 (InterPro:IPR006946); contains InterPro domain Galactose-binding like (InterPro:IPR008979) | -5.73 |
| 1660 | 250336_at | AT5G11720 | alpha-glucosidase 1 (AGLU1) | -4.42 |
| 1661 | 250300_at | AT5G11890 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G17620.1); similar to unknown [Populus trichocarpa] (GB:ABK93997.1); contains InterPro domain Harpin-induced 1 (InterPro:IPR010847) | -5.25 |
| 1662 | 250349_at | AT5G12000 | kinase | 9.83 |
| 1663 | 250350_at | AT5G12010 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G29780.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO43835.1); contains domain PTHR22930 (PTHR22930) | -4.00 |
| 1664 | 250296_at | AT5G12020 | HSP17.6II (17.6 KDA CLASS II HEAT SHOCK PROTEIN) | 4.58 |
| 1665 | 250351_at | AT5G12030 | AT-HSP17.6A (Arabidopsis thaliana heat shock protein 17.6A) | 4.64 |
| 1666 | 250308_at | AT5G12180 | CPK17 (calcium-dependent protein kinase 17); calmodulin-dependent protein kinase/ kinase | 11.04 |
| 1667 | 250313_at | AT5G12210 | geranylgeranyl transferase type II beta subunit, putative / RAB geranylgeranyltransferase beta subunit, putative | -4.56 |
| 1668 | 245178_at | AT5G12390 | binding | 5.27 |
| 1669 | 245980_at | AT5G13140 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G26960.1); similar to unknown [Euphorbia pulcherrima] (GB:ABO43960.1); contains InterPro domain Pollen Ole e 1 allergen and extensin (InterPro:IPR006041) | -5.38 |
| 1670 | 245979_at | AT5G13150 | ATEXO70C1 (exocyst subunit EXO70 family protein C1); protein binding | 9.48 |
| 1671 | 245987_at | AT5G13180 | ANAC083 (Arabidopsis NAC domain containing protein 83); transcription factor | -3.88 |
| 1672 | 250262_at | AT5G13410 | immunophilin / FKBP-type peptidyl-prolyl cis-trans isomerase family protein | -4.59 |
| 1673 | 245852_at | AT5G13510 | ribosomal protein L10 family protein | -4.18 |
| 1674 | 250243_at | AT5G13630 | GUN5 (GENOMES UNCOUPLED 5) | -5.08 |
| 1675 | 250244_at | AT5G13680 | ABO1/ELO2 (ABA-OVERLY SENSITIVE 1); transcription elongation regulator | -4.38 |
| 1676 | 250259_at | AT5G13800 | hydrolase, alpha/beta fold family protein | -3.74 |
| 1677 | 250207_at | AT5G13930 | ATCHS/CHS/TT4 (CHALCONE SYNTHASE); naringenin-chalcone synthase | -5.34 |
| 1678 | 250204_at | AT5G13990 | ATEXO70C2 (exocyst subunit EXO70 family protein C2); protein binding | 8.69 |
| 1679 | 250190_at | AT5G14320 | 30S ribosomal protein S13, chloroplast (CS13) | -5.13 |
| 1680 | 250174_at | AT5G14380 | AGP6 (ARABINOGALACTAN PROTEINS 6) | 10.02 |
| 1681 | 250143_at | AT5G14670 | ATARFA1B (ADP-RIBOSYLATION FACTOR A1B); GTP binding / phospholipase activator/ protein binding | 6.08 |
| 1682 | 246596_at | AT5G14740 | CA2 (BETA CARBONIC ANHYDRASE 2); carbonate dehydratase/ zinc ion binding | -4.77 |
| 1683 | 246590_at | AT5G14870 | ATCNGC18/CNGC18 (CYCLIC NUCLEOTIDE-GATED CHANNEL 18); calmodulin binding / cyclic nucleotide binding / ion channel | 6.38 |
| 1684 | 246592_at | AT5G14890 | NHL repeat-containing protein | 10.65 |
| 1685 | 246550_at | AT5G14920 | gibberellin-regulated family protein | -7.09 |
| 1686 | 246544_at | AT5G15100 | PIN8 (PIN-FORMED 8); auxin:hydrogen symporter/ transporter | 5.63 |
| 1687 | 246545_at | AT5G15110 | pectate lyase family protein | 11.24 |
| 1688 | 250154_at | AT5G15140 | aldose 1-epimerase family protein | 10.46 |
| 1689 | 250109_at | AT5G15230 | GASA4 (GAST1 PROTEIN HOMOLOG 4) | -6.89 |
| 1690 | 250110_at | AT5G15350 | plastocyanin-like domain-containing protein | -3.77 |
| 1691 | 246555_at | AT5G15470 | GAUT14 (Galacturonosyltransferase 14); polygalacturonate 4-alpha-galacturonosyltransferase/ transferase, transferring glycosyl groups / transferase, transferring hexosyl groups | 4.80 |
| 1692 | 246540_at | AT5G15600 | SP1L4 (SPIRAL1-LIKE4) | 8.36 |
| 1693 | 246491_at | AT5G16100 | similar to hypothetical protein [Cleome spinosa] (GB:ABD96916.1) | 7.55 |
| 1694 | 246506_at | AT5G16110 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G02555.1); similar to hypothetical protein [Cleome spinosa] (GB:ABD96917.1) | 4.91 |
| 1695 | 246500_at | AT5G16270 | ATRAD21.3/SYN4 (ARABIDOPSIS HOMOLOG OF RAD21 3) | -3.96 |
| 1696 | 250124_at | AT5G16480 | tyrosine specific protein phosphatase family protein | 5.79 |
| 1697 | 250121_at | AT5G16500 | protein kinase family protein | 12.56 |
| 1698 | 250128_at | AT5G16540 | ZFN3 (ZINC FINGER PROTEIN 3); nucleic acid binding | 6.77 |
| 1699 | 246509_at | AT5G16715 | EMB2247 (EMBRYO DEFECTIVE 2247); ATP binding / aminoacyl-tRNA ligase | -4.85 |
| 1700 | 246476_at | AT5G16730 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G02930.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO62148.1); contains InterPro domain Spectrin repeat (InterPro:IPR002017) | -5.20 |
| 1701 | 250095_at | AT5G17230 | PSY (PHYTOENE SYNTHASE); geranylgeranyl-diphosphate geranylgeranyltransferase | -3.73 |
| 1702 | 250091_at | AT5G17340 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G03272.1); similar to M3.4 protein [Brassica napus] (GB:AAD24197.1); contains domain PROKAR_LIPOPROTEIN (PS51257) | 6.67 |
| 1703 | 246427_at | AT5G17400 | ADP, ATP carrier protein, mitochondrial, putative / ADP/ATP translocase, putative / adenine nucleotide translocator, putative | 6.98 |
| 1704 | 246431_at | AT5G17480 | polcalcin, putative / calcium-binding pollen allergen, putative | 11.19 |
| 1705 | 250062_at | AT5G17760 | AAA-type ATPase family protein | -4.21 |
| 1706 | 250058_at | AT5G17870 | PSRP6 (PLASTID-SPECIFIC 50S RIBOSOMAL PROTEIN 6) | -3.91 |
| 1707 | 250014_at | AT5G17990 | TRP1 (TRYPTOPHAN BIOSYNTHESIS 1); anthranilate phosphoribosyltransferase | -4.02 |
| 1708 | 250018_at | AT5G18150 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G14602.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO23480.1); contains domain PTHR12133 (PTHR12133) | 4.73 |
| 1709 | 249986_at | AT5G18460 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G23340.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G23340.2); similar to unnamed protein product [Vitis vinifera] (GB:CAO38766.1); contains InterPro domain Protein of unknown function DUF239, plant (InterPro:IPR004314) | -4.00 |
| 1710 | 249981_at | AT5G18510 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G50830.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO38989.1); contains InterPro domain Protein of unknown function DUF1723 (InterPro:IPR013541) | 8.23 |
| 1711 | 249996_at | AT5G18600 | glutaredoxin family protein | -6.07 |
| 1712 | 250006_at | AT5G18660 | DVR (PALE-GREEN AND CHLOROPHYLL B REDUCED 2); 3,8-divinyl protochlorophyllide a 8-vinyl reductase | -6.31 |
| 1713 | 249950_at | AT5G18910 | protein kinase family protein | 12.02 |
| 1714 | 249923_at | AT5G19120 | aspartic-type endopeptidase/ pepsin A | -3.94 |
| 1715 | 249922_at | AT5G19140 | auxin/aluminum-responsive protein, putative | -3.82 |
| 1716 | 249919_at | AT5G19250 | Identical to Uncharacterized GPI-anchored protein At5g19250 precursor [Arabidopsis Thaliana] (GB:P59833;GB:Q67ZQ2;GB:Q6ID89); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G06035.1); similar to unknown [Populus trichocarpa] (GB:ABK94712.1) | -6.15 |
| 1717 | 246065_at | AT5G19360 | CPK34 (calcium-dependent protein kinase 34); calmodulin-dependent protein kinase/ kinase | 9.29 |
| 1718 | 245947_at | AT5G19530 | ACL5 (ACAULIS 5) | -5.54 |
| 1719 | 245946_at | AT5G19580 | glyoxal oxidase-related | 9.53 |
| 1720 | 245958_at | AT5G19610 | sec7 domain-containing protein | 7.25 |
| 1721 | 245959_at | AT5G19640 | proton-dependent oligopeptide transport (POT) family protein | 4.60 |
| 1722 | 245937_at | AT5G19750 | peroxisomal membrane 22 kDa family protein | -3.90 |
| 1723 | 246154_at | AT5G19940 | plastid-lipid associated protein PAP-related / fibrillin-related | -5.01 |
| 1724 | 246152_at | AT5G20040 | ATIPT9 (Arabidopsis thaliana isopentenyltransferase 9); ATP binding / tRNA isopentenyltransferase | -3.99 |
| 1725 | 246099_at | AT5G20230 | ATBCB (ARABIDOPSIS BLUE-COPPER-BINDING PROTEIN); copper ion binding | 5.02 |
| 1726 | 246076_at | AT5G20280 | ATSPS1F (SUCROSE PHOSPHATE SYNTHASE 1F); sucrose-phosphate synthase/ transferase, transferring glycosyl groups | -3.82 |
| 1727 | 246096_at | AT5G20330 | BETAG4 (BETA-1,3-GLUCANASE 4); hydrolase, hydrolyzing O-glycosyl compounds | 7.11 |
| 1728 | 246118_at | AT5G20340 | BG5 (BETA-1,3-GLUCANASE 5); hydrolase, hydrolyzing O-glycosyl compounds | 7.35 |
| 1729 | 246123_at | AT5G20390 | beta-1,3-glucanase, putative | 10.83 |
| 1730 | 246075_at | AT5G20410 | MGD2 (monogalactosyldiacylglycerol synthase 2); 1,2-diacylglycerol 3-beta-galactosyltransferase/ transferase, transferring glycosyl groups | 8.86 |
| 1731 | 246080_at | AT5G20460 | unknown protein | 8.29 |
| 1732 | 246004_at | AT5G20630 | GLP3 (GERMIN-LIKE PROTEIN 3); manganese ion binding / metal ion binding / nutrient reservoir | -6.15 |
| 1733 | 245992_at | AT5G20690 | leucine-rich repeat transmembrane protein kinase, putative | 7.42 |
| 1734 | 245993_at | AT5G20700 | senescence-associated protein-related | -4.29 |
| 1735 | 245970_at | AT5G20710 | BGAL7 (beta-galactosidase 7); beta-galactosidase | 7.61 |
| 1736 | 245971_at | AT5G20730 | NPH4 (NON-PHOTOTROPHIC HYPOCOTYL); transcription factor | -3.73 |
| 1737 | 245997_at | AT5G20810 | auxin-responsive protein, putative / small auxin up RNA (SAUR_B) | 8.01 |
| 1738 | 246184_at | AT5G20950 | glycosyl hydrolase family 3 protein | -5.60 |
| 1739 | 246188_at | AT5G21050 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G64090.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO67384.1) | 6.37 |
| 1740 | 245684_at | AT5G22000 | CIC7E11; protein binding / zinc ion binding | 4.81 |
| 1741 | 249886_at | AT5G22320 | leucine-rich repeat family protein | -4.18 |
| 1742 | 249931_at | AT5G22370 | EMB1705/QQT1 (QUATRE-QUART1); ATP binding | -4.17 |
| 1743 | 249894_at | AT5G22580 | Identical to Uncharacterized protein At5g22580 [Arabidopsis Thaliana] (GB:Q9FK81); similar to stable protein 1-related [Arabidopsis thaliana] (TAIR:AT3G17210.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN68720.1); contains InterPro domain Dimeric alpha-beta barrel (InterPro:IPR011008); contains InterPro domain Stress responsive alpha-beta barrel (InterPro:IPR013097) | -5.09 |
| 1744 | 249910_at | AT5G22630 | ADT5 (AROGENATE DEHYDRATASE 5); arogenate dehydratase/ prephenate dehydratase | -3.79 |
| 1745 | 249864_at | AT5G22830 | GMN10 (Arabidopsis thaliana Mg transporter 10) | -4.42 |
| 1746 | 249866_at | AT5G23010 | MAM1 (2-isopropylmalate synthase 3); 2-isopropylmalate synthase | -5.47 |
| 1747 | 249876_at | AT5G23060 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G59780.1); similar to extracellular Ca2+ sensing receptor [Glycine max] (GB:ABY57763.1); contains InterPro domain Rhodanese-like (InterPro:IPR001763) | -5.66 |
| 1748 | 249847_at | AT5G23210 | SCPL34; serine carboxypeptidase | -5.26 |
| 1749 | 249852_at | AT5G23270 | STP11 (SUGAR TRANSPORTER 11); carbohydrate transmembrane transporter/ sugar:hydrogen ion symporter | 9.62 |
| 1750 | 249834_at | AT5G23440 | FTRA1 (ferredoxin/thioredoxin reductase subunit A (variable subunit) 1); ferredoxin:thioredoxin reductase | -3.74 |
| 1751 | 249837_at | AT5G23480 | similar to SWIB complex BAF60b domain-containing protein / plus-3 domain-containing protein / GYF domain-containing protein [Arabidopsis thaliana] (TAIR:AT5G08430.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN80413.1); contains InterPro domain GYF (InterPro:IPR003169) | 4.78 |
| 1752 | 249799_at | AT5G23670 | LCB2 (Serine palmitoyltransferase LCB2 (long chain base) subunit gene); protein binding / serine C-palmitoyltransferase | 4.57 |
| 1753 | 249798_at | AT5G23730 | nucleotide binding | -3.85 |
| 1754 | 249817_at | AT5G23820 | MD-2-related lipid recognition domain-containing protein / ML domain-containing protein | -4.61 |
| 1755 | 249808_at | AT5G23890 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G52410.2); similar to unnamed protein product [Vitis vinifera] (GB:CAO49433.1) | -5.56 |
| 1756 | 249780_at | AT5G24240 | phosphatidylinositol 3- and 4-kinase family protein / ubiquitin family protein | 10.13 |
| 1757 | 249742_at | AT5G24490 | 30S ribosomal protein, putative | -3.96 |
| 1758 | 249754_at | AT5G24530 | oxidoreductase, 2OG-Fe(II) oxygenase family protein | -5.80 |
| 1759 | 249745_at | AT5G24560 | ATPP2-B12 (Phloem protein 2-B12); carbohydrate binding | 4.60 |
| 1760 | 246969_at | AT5G24880 | similar to calmodulin-binding protein-related [Arabidopsis thaliana] (TAIR:AT5G10660.1) | 7.77 |
| 1761 | 246977_at | AT5G24930 | zinc finger (B-box type) family protein | -7.95 |
| 1762 | 246971_at | AT5G24940 | protein phosphatase 2C, putative / PP2C, putative | 8.19 |
| 1763 | 246917_at | AT5G25280 | serine-rich protein-related | -4.27 |
| 1764 | 246918_at | AT5G25340 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G32270.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO44248.1); contains domain G3DSA:3.10.20.90 (G3DSA:3.10.20.90); contains domain SSF54236 (SSF54236) | 9.38 |
| 1765 | 246940_at | AT5G25400 | phosphate translocator-related | 4.76 |
| 1766 | 246942_at | AT5G25430 | anion exchange protein family | 7.99 |
| 1767 | 246919_at | AT5G25460 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G11420.1); similar to unknown [Ricinus communis] (GB:CAB02653.1); contains InterPro domain Protein of unknown function DUF642 (InterPro:IPR006946) | -7.26 |
| 1768 | 246894_at | AT5G25530 | DNAJ heat shock protein, putative | 6.89 |
| 1769 | 246896_at | AT5G25550 | leucine-rich repeat family protein / extensin family protein | 5.11 |
| 1770 | 246908_at | AT5G25610 | RD22 (RESPONSIVE TO DESSICATION 22) | -4.06 |
| 1771 | 246915_at | AT5G25880 | ATNADP-ME3 (NADP-MALIC ENZYME 3); malate dehydrogenase (oxaloacetate-decarboxylating) (NADP+)/ malic enzyme/ oxidoreductase, acting on NADH or NADPH, NAD or NADP as acceptor | 6.76 |
| 1772 | 246878_at | AT5G26060 | S1 self-incompatibility protein-related | 12.69 |
| 1773 | 246877_at | AT5G26150 | protein kinase family protein | 10.42 |
| 1774 | 246841_at | AT5G26700 | germin-like protein, putative | 11.37 |
| 1775 | 246802_at | AT5G27000 | ATK4 (ARABIDOPSIS THALIANA KINESIN 4); microtubule motor | 6.24 |
| 1776 | 246791_at | AT5G27280 | zinc finger (DNL type) family protein | -4.97 |
| 1777 | 246781_at | AT5G27350 | SFP1; carbohydrate transmembrane transporter/ sugar:hydrogen ion symporter | -4.53 |
| 1778 | 246736_at | AT5G27560 | similar to unnamed protein product [Vitis vinifera] (GB:CAO17622.1) | -5.15 |
| 1779 | 246790_at | AT5G27610 | DNA binding / transcription factor | 5.76 |
| 1780 | 246749_at | AT5G27830 | similar to hypothetical protein [Vitis vinifera] (GB:CAN74239.1); contains InterPro domain Folate receptor (InterPro:IPR004269) | -3.85 |
| 1781 | 246751_at | AT5G27870 | pectinesterase family protein | 11.19 |
| 1782 | 246761_at | AT5G27980 | seed maturation family protein | 10.40 |
| 1783 | 246727_at | AT5G28010 | Bet v I allergen family protein | 5.10 |
| 1784 | 246701_at | AT5G28020 | ATCYSD2 (Arabidopsis thaliana cysteine synthase D2); cysteine synthase | -4.61 |
| 1785 | 246136_at | AT5G28470 | transporter | 6.38 |
| 1786 | 246106_at | AT5G28680 | protein kinase family protein | 12.29 |
| 1787 | 245924_at | AT5G28750 | thylakoid assembly protein, putative | -3.85 |
| 1788 | 246673_at | AT5G30510 | RPS1 (ribosomal protein S1); RNA binding | -5.02 |
| 1789 | 246170_at | AT5G32450 | RNA recognition motif (RRM)-containing protein | -3.83 |
| 1790 | 246646_at | AT5G35090 | unknown protein | 11.67 |
| 1791 | 246608_at | AT5G35380 | protein kinase family protein | 7.34 |
| 1792 | 246609_at | AT5G35390 | leucine-rich repeat transmembrane protein kinase, putative | 7.24 |
| 1793 | 246614_at | AT5G35410 | SOS2 (SALT OVERLY SENSITIVE 2); kinase | 5.28 |
| 1794 | 249710_at | AT5G35630 | GS2 (GLUTAMINE SYNTHETASE 2); glutamate-ammonia ligase | -5.58 |
| 1795 | 246616_at | AT5G36260 | aspartyl protease family protein | 10.25 |
| 1796 | 249649_at | AT5G37060 | ATCHX24 (cation/hydrogen exchanger 24); monovalent cation:proton antiporter | 7.13 |
| 1797 | 249604_x_at | AT5G37230 | zinc finger (C3HC4-type RING finger) family protein | 5.18 |
| 1798 | 249612_at | AT5G37290 | armadillo/beta-catenin repeat family protein | -4.09 |
| 1799 | 249581_at | AT5G37600 | ATGSR1 (Arabidopsis thaliana glutamine synthase clone R1); glutamate-ammonia ligase | -6.38 |
| 1800 | 249547_at | AT5G38160 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | 4.78 |
| 1801 | 249548_at | AT5G38170 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | 5.64 |
| 1802 | 249526_at | AT5G38560 | protein kinase family protein | -3.96 |
| 1803 | 249536_at | AT5G38760 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G53820.1); similar to ABA-inducible protein-like protein [Brassica rapa] (GB:ABV89610.1); contains domain PTHR23241:SF11 (PTHR23241:SF11); contains domain PTHR23241 (PTHR23241) | 14.49 |
| 1804 | 249483_at | AT5G38895 | zinc finger (C3HC4-type RING finger) family protein | 4.78 |
| 1805 | 249482_at | AT5G38980 | unknown protein | -4.92 |
| 1806 | 249503_at | AT5G39310 | ATEXPA24 (ARABIDOPSIS THALIANA EXPANSIN A24) | 9.09 |
| 1807 | 249447_at | AT5G39400 | pollen specific phosphatase, putative / phosphatase and tensin, putative (PTEN1) | 10.36 |
| 1808 | 249448_at | AT5G39420 | CDC2CAT (ARABIDOPSIS THALIANA CDC2C); kinase | 11.30 |
| 1809 | 249468_at | AT5G39650 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G09157.1); similar to 52O08_3 [Brassica rapa subsp. pekinensis] (GB:AAZ67548.1); contains InterPro domain Protein of unknown function DUF679 (InterPro:IPR007770) | 8.16 |
| 1810 | 249421_at | AT5G39830 | DEG8/DEGP8 (DEGP PROTEASE 8); peptidase/ serine-type peptidase/ trypsin | -4.17 |
| 1811 | 249428_at | AT5G39870 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G28980.1); contains InterPro domain Protein of unknown function DUF1216 (InterPro:IPR009605) | 10.37 |
| 1812 | 249429_at | AT5G39880 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G28750.1) | 12.44 |
| 1813 | 249381_at | AT5G40040 | 60S acidic ribosomal protein P2 (RPP2E) | 7.87 |
| 1814 | 249402_at | AT5G40155 | Encodes a defensin-like (DEFL) family protein. | 13.71 |
| 1815 | 249401_at | AT5G40260 | nodulin MtN3 family protein | 6.32 |
| 1816 | 249405_at | AT5G40280 | ERA1 (ENHANCED RESPONSE TO ABA 1); protein farnesyltransferase | -4.08 |
| 1817 | 249400_at | AT5G40300 | integral membrane protein, putative | -4.63 |
| 1818 | 249376_at | AT5G40645 | nitrate-responsive NOI protein, putative | 7.77 |
| 1819 | 249375_at | AT5G40730 | AGP24 (ARABINOGALACTAN PROTEIN 24) | 6.94 |
| 1820 | 249325_at | AT5G40850 | UPM1 (UROPHORPHYRIN METHYLASE 1); uroporphyrin-III C-methyltransferase | -4.62 |
| 1821 | 249327_at | AT5G40890 | ATCLC-A (CHLORIDE CHANNEL A); anion channel/ voltage-gated chloride channel | -3.73 |
| 1822 | 249296_at | AT5G41310 | kinesin motor protein-related | 7.04 |
| 1823 | 249306_at | AT5G41400 | zinc finger (C3HC4-type RING finger) family protein | -4.07 |
| 1824 | 249270_at | AT5G41780 | myosin heavy chain-related | 11.15 |
| 1825 | 249252_at | AT5G42010 | WD-40 repeat family protein | 4.63 |
| 1826 | 249237_at | AT5G42050 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G27090.1); similar to unknown [Populus trichocarpa] (GB:ABK95892.1); contains InterPro domain Kelch related (InterPro:IPR013089); contains InterPro domain Development and cell death (InterPro:IPR013989) | -3.88 |
| 1827 | 249226_at | AT5G42170 | carboxylesterase | 8.78 |
| 1828 | 249242_at | AT5G42250 | alcohol dehydrogenase, putative | -3.96 |
| 1829 | 249244_at | AT5G42270 | VAR1 (VARIEGATED 1); ATP-dependent peptidase/ ATPase/ metallopeptidase | -4.79 |
| 1830 | 249250_at | AT5G42340 | binding / ubiquitin-protein ligase | 8.60 |
| 1831 | 249193_at | AT5G42480 | ARC6 (ACCUMULATION AND REPLICATION OF CHLOROPLASTS 6) | -4.92 |
| 1832 | 249194_at | AT5G42490 | kinesin motor family protein | 12.20 |
| 1833 | 249199_at | AT5G42520 | ATBPC6/BBR/BPC6/BPC6 (BASIC PENTACYSTEINE 6); DNA binding / transcription factor | -5.60 |
| 1834 | 249208_at | AT5G42650 | AOS (ALLENE OXIDE SYNTHASE); hydro-lyase/ oxygen binding | -4.50 |
| 1835 | 249167_at | AT5G42860 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G45688.1); similar to H0814G11.12 [Oryza sativa (indica cultivar-group)] (GB:CAJ86345.1); similar to CAA30379.1 protein [Oryza sativa] (GB:CAB53482.1) | -4.21 |
| 1836 | 249169_at | AT5G42880 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G45545.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO61337.1); contains InterPro domain Protein of unknown function DUF827, plant (InterPro:IPR008545) | 6.24 |
| 1837 | 249185_at | AT5G43030 | DC1 domain-containing protein | 7.47 |
| 1838 | 249187_at | AT5G43060 | cysteine proteinase, putative / thiol protease, putative | 4.77 |
| 1839 | 249128_at | AT5G43440 | 2-oxoglutarate-dependent dioxygenase, putative | -4.97 |
| 1840 | 249125_at | AT5G43450 | 2-oxoglutarate-dependent dioxygenase, putative | -4.11 |
| 1841 | 249098_at | AT5G43530 | SNF2 domain-containing protein / helicase domain-containing protein / RING finger domain-containing protein | 5.70 |
| 1842 | 249109_at | AT5G43700 | ATAUX2-11 (indoleacetic acid-induced protein 4); transcription factor | -5.09 |
| 1843 | 249120_at | AT5G43750 | similar to unnamed protein product [Vitis vinifera] (GB:CAO71280.1) | -6.63 |
| 1844 | 249123_at | AT5G43760 | beta-ketoacyl-CoA synthase, putative | -3.83 |
| 1845 | 249117_at | AT5G43840 | AT-HSFA6A (Arabidopsis thaliana heat shock transcription factor A6A); DNA binding / transcription factor | 4.57 |
| 1846 | 249048_at | AT5G44300 | dormancy/auxin associated family protein | 9.55 |
| 1847 | 249009_at | AT5G44610 | MAP18 (MICROTUBULE-ASSOCIATED PROTEIN 18); microtubule binding | 7.16 |
| 1848 | 249008_at | AT5G44680 | methyladenine glycosylase family protein | -4.73 |
| 1849 | 249025_at | AT5G44720 | molybdenum cofactor sulfurase family protein | -4.30 |
| 1850 | 249027_at | AT5G44790 | RAN1 (RESPONSIVE-TO-ANTAGONIST1); ATPase, coupled to transmembrane movement of ions, phosphorylative mechanism | 5.22 |
| 1851 | 248986_at | AT5G45170 | CbbY protein-related | -7.00 |
| 1852 | 248942_at | AT5G45480 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G45470.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G45530.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G45540.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO39193.1); contains InterPro domain Protein of unknown function DUF594 (InterPro:IPR007658) | -3.84 |
| 1853 | 248945_at | AT5G45510 | leucine-rich repeat family protein | -4.59 |
| 1854 | 248912_at | AT5G45670 | GDSL-motif lipase/hydrolase family protein | -4.07 |
| 1855 | 248963_at | AT5G45700 | NLI interacting factor (NIF) family protein | 6.19 |
| 1856 | 248909_at | AT5G45810 | CIPK19 (SNF1-RELATED PROTEIN KINASE 3.5); kinase | 9.31 |
| 1857 | 248910_at | AT5G45820 | CIPK20 (CBL-INTERACTING PROTEIN KINASE 20); kinase | 5.80 |
| 1858 | 248916_at | AT5G45840 | leucine-rich repeat transmembrane protein kinase, putative | 8.24 |
| 1859 | 248926_at | AT5G45880 | pollen Ole e 1 allergen and extensin family protein | 11.51 |
| 1860 | 248920_at | AT5G45930 | CHLI2; magnesium chelatase | -4.98 |
| 1861 | 248921_at | AT5G45950 | GDSL-motif lipase/hydrolase family protein | -4.36 |
| 1862 | 248886_at | AT5G46110 | APE2 (ACCLIMATION OF PHOTOSYNTHESIS TO ENVIRONMENT); antiporter/ triose-phosphate transmembrane transporter | -4.40 |
| 1863 | 248880_at | AT5G46200 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G17860.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO14518.1); contains InterPro domain Protein of unknown function DUF239, plant (InterPro:IPR004314) | 7.12 |
| 1864 | 248895_at | AT5G46330 | FLS2 (FLAGELLIN-SENSITIVE 2); ATP binding / kinase/ protein binding / protein serine/threonine kinase/ transmembrane receptor protein serine/threonine kinase | -4.28 |
| 1865 | 248840_at | AT5G46770 | unknown protein | 11.24 |
| 1866 | 248865_at | AT5G46790 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G17870.1); similar to Streptomyces cyclase/dehydrase family protein [Brassica oleracea] (GB:ABD65631.1); contains InterPro domain Streptomyces cyclase/dehydrase (InterPro:IPR005031) | -4.10 |
| 1867 | 248872_at | AT5G46795 | MSP2 (MICROSPORE-SPECIFIC PROMOTER 2) | 5.10 |
| 1868 | 248869_at | AT5G46840 | RNA recognition motif (RRM)-containing protein | 5.24 |
| 1869 | 248814_at | AT5G46910 | transcription factor jumonji (jmj) family protein | 5.05 |
| 1870 | 248824_at | AT5G46940 | invertase/pectin methylesterase inhibitor family protein | 6.78 |
| 1871 | 248822_at | AT5G47000 | peroxidase, putative | 11.48 |
| 1872 | 248828_at | AT5G47110 | lil3 protein, putative | -4.62 |
| 1873 | 248794_at | AT5G47220 | ATERF-2/ATERF2/ERF2 (ETHYLENE RESPONSE FACTOR 2); DNA binding / transcription activator/ transcription factor | -3.92 |
| 1874 | 248804_at | AT5G47470 | nodulin MtN21 family protein | 7.23 |
| 1875 | 248751_at | AT5G47540 | Mo25 family protein | 6.89 |
| 1876 | 248758_at | AT5G47620 | heterogeneous nuclear ribonucleoprotein, putative / hnRNP, putative | 5.14 |
| 1877 | 248764_at | AT5G47640 | CCAAT-box binding transcription factor subunit B (NF-YB) (HAP3 ) (AHAP3) family (Hap3b) | -3.86 |
| 1878 | 248781_at | AT5G47870 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G71310.2); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G71310.1); similar to unknown [Populus trichocarpa] (GB:ABK95546.1) | -4.36 |
| 1879 | 248735_at | AT5G48100 | TT10 (TRANSPARENT TESTA 10); laccase | 5.76 |
| 1880 | 248714_at | AT5G48140 | polygalacturonase, putative / pectinase, putative | 12.02 |
| 1881 | 248711_at | AT5G48270 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G04220.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO41124.1); contains InterPro domain Protein of unknown function DUF868, plant (InterPro:IPR008586) | 8.57 |
| 1882 | 248687_at | AT5G48300 | ADG1 (ADP GLUCOSE PYROPHOSPHORYLASE SMALL SUBUNIT 1); glucose-1-phosphate adenylyltransferase | -5.43 |
| 1883 | 248696_at | AT5G48360 | formin homology 2 domain-containing protein / FH2 domain-containing protein | -4.09 |
| 1884 | 248656_at | AT5G48460 | fimbrin-like protein, putative | -4.67 |
| 1885 | 248684_at | AT5G48485 | DIR1 (DEFECTIVE IN INDUCED RESISTANCE 1); lipid binding | -4.50 |
| 1886 | 248673_at | AT5G48780 | disease resistance protein (TIR-NBS class), putative | 5.20 |
| 1887 | 248645_at | AT5G49150 | ATGEX2/GEX2 (GAMETE EXPRESSED2) | 4.69 |
| 1888 | 248613_at | AT5G49555 | amine oxidase-related | -5.28 |
| 1889 | 248582_at | AT5G49910 | cpHSC70-2 (HEAT SHOCK PROTEIN 70-7); ATP binding / unfolded protein binding | -4.49 |
| 1890 | 248534_at | AT5G50030 | invertase/pectin methylesterase inhibitor family protein | 9.60 |
| 1891 | 248541_at | AT5G50180 | protein kinase, putative | -4.68 |
| 1892 | 248555_at | AT5G50340 | ATP binding / ATP-dependent peptidase/ damaged DNA binding / nucleoside-triphosphatase/ nucleotide binding / serine-type endopeptidase | -3.74 |
| 1893 | 248470_at | AT5G50830 | unknown protein | 11.04 |
| 1894 | 248480_at | AT5G50920 | CLPC (HEAT SHOCK PROTEIN 93-V); ATP binding / ATPase | -4.82 |
| 1895 | 248481_at | AT5G50930 | DNA binding | 5.40 |
| 1896 | 248491_at | AT5G51010 | rubredoxin family protein | -5.75 |
| 1897 | 248484_at | AT5G51030 | short-chain dehydrogenase/reductase (SDR) family protein | 7.88 |
| 1898 | 248449_at | AT5G51110 | similar to dehydratase family [Arabidopsis thaliana] (TAIR:AT1G29810.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO62291.1); contains InterPro domain Transcriptional coactivator/pterin dehydratase (InterPro:IPR001533) | -4.84 |
| 1899 | 248446_at | AT5G51140 | pseudouridine synthase family protein | -4.37 |
| 1900 | 248404_at | AT5G51460 | ATTPPA (Arabidopsis thaliana trehalose-6-phosphate phosphatase); trehalose-phosphatase | 5.14 |
| 1901 | 248377_at | AT5G51720 | similar to Os07g0467200 [Oryza sativa (japonica cultivar-group)] (GB:NP_001059590.1); similar to hypothetical protein OsI_025030 [Oryza sativa (indica cultivar-group)] (GB:EAZ03798.1); contains domain PTHR13680 (PTHR13680); contains domain PTHR13680:SF1 (PTHR13680:SF1) | -5.82 |
| 1902 | 248393_at | AT5G52060 | ATBAG1 (ARABIDOPSIS THALIANA BCL-2-ASSOCIATED ATHANOGENE 1); protein binding | -5.00 |
| 1903 | 248355_at | AT5G52340 | ATEXO70A2 (exocyst subunit EXO70 family protein A2); protein binding | 8.16 |
| 1904 | 248367_at | AT5G52360 | actin-depolymerizing factor, putative | 12.15 |
| 1905 | 248338_at | AT5G52440 | HCF106 (High chlorophyll fluorescence 106) | -5.12 |
| 1906 | 248287_at | AT5G52970 | thylakoid lumen 15.0 kDa protein | -5.47 |
| 1907 | 248252_at | AT5G53250 | AGP22/ATAGP22 (ARABINOGALACTAN PROTEINS 22) | 7.26 |
| 1908 | 248256_at | AT5G53360 | seven in absentia (SINA) family protein | 5.57 |
| 1909 | 248275_at | AT5G53520 | ATOPT8 (oligopeptide transporter 8); oligopeptide transporter | 6.96 |
| 1910 | 248242_at | AT5G53580 | aldo/keto reductase family protein | -5.34 |
| 1911 | 248227_at | AT5G53820 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G38760.1); similar to pollen coat protein [Brassica oleracea] (GB:CAA63531.1); contains domain PTHR23241:SF11 (PTHR23241:SF11); contains domain PTHR23241 (PTHR23241) | 10.77 |
| 1912 | 248207_at | AT5G53970 | aminotransferase, putative | -3.92 |
| 1913 | 248211_at | AT5G54010 | glycosyltransferase family protein | 7.52 |
| 1914 | 248194_at | AT5G54095 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G27580.1) | 14.99 |
| 1915 | 248190_at | AT5G54130 | calcium-binding EF hand family protein | 6.49 |
| 1916 | 248191_at | AT5G54130 | calcium-binding EF hand family protein | 5.45 |
| 1917 | 248153_at | AT5G54250 | ATCNGC4 (DEFENSE, NO DEATH 2); calmodulin binding / cation channel/ cyclic nucleotide binding | -5.16 |
| 1918 | 248151_at | AT5G54270 | LHCB3 (LIGHT-HARVESTING CHLOROPHYLL BINDING PROTEIN 3) | -9.38 |
| 1919 | 248174_at | AT5G54600 | 50S ribosomal protein L24, chloroplast (CL24) | -4.53 |
| 1920 | 248129_at | AT5G54780 | RAB GTPase activator | 7.96 |
| 1921 | 248138_at | AT5G54960 | PDC2 (PYRUVATE DECARBOXYLASE-2); pyruvate decarboxylase | -4.74 |
| 1922 | 248095_at | AT5G55230 | ATMAP65-1 (MICROTUBULE-ASSOCIATED PROTEINS 65-1); microtubule binding | -3.81 |
| 1923 | 248062_at | AT5G55450 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | -4.59 |
| 1924 | 248086_at | AT5G55490 | ATGEX1/GEX1 (GAMETE EXPRESSED PROTEIN1) | 7.07 |
| 1925 | 248037_at | AT5G55930 | ATOPT1 (oligopeptide transporter 1); oligopeptide transporter | 8.00 |
| 1926 | 248038_at | AT5G55980 | serine-rich protein-related | 8.61 |
| 1927 | 247995_at | AT5G56160 | transporter | 6.73 |
| 1928 | 247964_at | AT5G56600 | PFN3/PRF3 (PROFILIN 3); actin binding | -5.80 |
| 1929 | 247981_at | AT5G56640 | MIOX5 (MYO-INOSITOL OXYGENASE 5) | 9.94 |
| 1930 | 247979_at | AT5G56750 | Ndr family protein | 7.26 |
| 1931 | 247974_at | AT5G56780 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G56770.1); similar to effector of transcription [Brassica napus] (GB:AAT00536.1) | 5.46 |
| 1932 | 247941_at | AT5G57200 | epsin N-terminal homology (ENTH) domain-containing protein / clathrin assembly protein-related | 6.32 |
| 1933 | 247951_at | AT5G57240 | oxysterol-binding family protein | 5.86 |
| 1934 | 247928_at | AT5G57320 | villin, putative | 6.31 |
| 1935 | 247899_at | AT5G57345 | similar to unknown [Populus trichocarpa x Populus deltoides] (GB:ABK96592.1) | -4.04 |
| 1936 | 247873_at | AT5G57690 | diacylglycerol kinase | 8.87 |
| 1937 | 247874_at | AT5G57710 | heat shock protein-related | -4.65 |
| 1938 | 247897_at | AT5G57810 | TET15 (TETRASPANIN15) | 9.24 |
| 1939 | 247885_at | AT5G57830 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G30830.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO66637.1); contains InterPro domain Protein of unknown function DUF593 (InterPro:IPR007656) | 6.63 |
| 1940 | 247892_at | AT5G57970 | methyladenine glycosylase family protein | -3.95 |
| 1941 | 247843_at | AT5G58050 | glycerophosphoryl diester phosphodiesterase family protein | 12.86 |
| 1942 | 247804_at | AT5G58170 | glycerophosphoryl diester phosphodiesterase family protein | 10.69 |
| 1943 | 247808_at | AT5G58190 | ECT10 | 4.70 |
| 1944 | 247813_at | AT5G58330 | malate dehydrogenase (NADP), chloroplast, putative | -5.77 |
| 1945 | 247820_at | AT5G58380 | CIPK10 (CBL-INTERACTING PROTEIN KINASE 10); kinase | 5.77 |
| 1946 | 247824_at | AT5G58460 | ATCHX25 (cation/hydrogen exchanger 25); monovalent cation:proton antiporter | 5.78 |
| 1947 | 247830_at | AT5G58530 | glutaredoxin family protein | 5.03 |
| 1948 | 247801_at | AT5G58560 | phosphatidate cytidylyltransferase family protein | -4.94 |
| 1949 | 247776_at | AT5G58700 | phosphoinositide-specific phospholipase C family protein | 5.18 |
| 1950 | 247778_at | AT5G58750 | wound-responsive protein-related | 5.76 |
| 1951 | 247780_at | AT5G58770 | dehydrodolichyl diphosphate synthase, putative / DEDOL-PP synthase, putative | -4.32 |
| 1952 | 247783_at | AT5G58800 | quinone reductase family protein | -4.29 |
| 1953 | 247750_at | AT5G58950 | protein kinase family protein | 4.88 |
| 1954 | 247741_at | AT5G58960 | GIL1 (GRAVITROPIC IN THE LIGHT) | -4.03 |
| 1955 | 247759_at | AT5G59040 | COPT3 (Copper transporter 3); copper ion transmembrane transporter | 5.63 |
| 1956 | 247709_at | AT5G59250 | sugar transporter family protein | -5.27 |
| 1957 | 247736_at | AT5G59370 | ACT4 (ACTIN 4) | 9.18 |
| 1958 | 247693_at | AT5G59730 | ATEXO70H7 (EXOCYST SUBUNIT EXO70 FAMILY PROTEIN H7); protein binding | -4.11 |
| 1959 | 247658_at | AT5G59880 | ADF3 (ACTIN DEPOLYMERIZING FACTOR 3); actin binding | -4.27 |
| 1960 | 247647_at | AT5G60010 | ferric reductase-like transmembrane component family protein | 6.40 |
| 1961 | 247660_at | AT5G60070 | ankyrin repeat family protein | 7.23 |
| 1962 | 247615_at | AT5G60250 | zinc finger (C3HC4-type RING finger) family protein | 5.81 |
| 1963 | 247616_at | AT5G60260 | unknown protein | 5.51 |
| 1964 | 247627_at | AT5G60360 | AALP (ARABIDOPSIS ALEURAIN-LIKE PROTEASE); cysteine-type peptidase | -5.93 |
| 1965 | 247589_at | AT5G60690 | REV (REVOLUTA); DNA binding / lipid binding / transcription factor | -5.27 |
| 1966 | 247613_at | AT5G60740 | ABC transporter family protein | 7.04 |
| 1967 | 247582_at | AT5G60760 | 2-phosphoglycerate kinase-related | 5.35 |
| 1968 | 247563_at | AT5G61130 | glycosyl hydrolase family protein 17 | -5.11 |
| 1969 | 247581_at | AT5G61350 | protein kinase family protein | 5.57 |
| 1970 | 247547_at | AT5G61360 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G07730.1) | 7.16 |
| 1971 | 247525_at | AT5G61380 | TOC1 (TIMING OF CAB EXPRESSION 1); transcription regulator | -4.94 |
| 1972 | 247549_at | AT5G61420 | MYB28 (MYB DOMAIN PROTEIN 28); DNA binding / transcription factor | -4.61 |
| 1973 | 247540_at | AT5G61590 | AP2 domain-containing transcription factor family protein | -3.95 |
| 1974 | 247535_at | AT5G61620 | myb family transcription factor | 5.38 |
| 1975 | 247541_at | AT5G61660 | glycine-rich protein | -4.44 |
| 1976 | 247538_at | AT5G61700 | ATATH16 (ABC2 homolog 16); ATPase, coupled to transmembrane movement of substances | 5.65 |
| 1977 | 247539_at | AT5G61710 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G02160.1) | 12.36 |
| 1978 | 247512_at | AT5G61720 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G28830.1); contains InterPro domain Protein of unknown function DUF1216 (InterPro:IPR009605) | 14.43 |
| 1979 | 247506_at | AT5G61960 | AML1 (ARABIDOPSIS MEI2-LIKE PROTEIN 1); RNA binding | 5.36 |
| 1980 | 247503_at | AT5G61980 | AGD1 (ARF-GAP DOMAIN 1); ARF GTPase activator/ protein binding / zinc ion binding | 6.59 |
| 1981 | 247468_at | AT5G62000 | ARF2 (AUXIN RESPONSE FACTOR 2); transcription factor | -5.03 |
| 1982 | 247508_at | AT5G62000 | ARF2 (AUXIN RESPONSE FACTOR 2); transcription factor | -5.42 |
| 1983 | 247472_at | AT5G62250 | microtubule associated protein (MAP65/ASE1) family protein | 7.79 |
| 1984 | 247475_at | AT5G62310 | IRE (INCOMPLETE ROOT HAIR ELONGATION); kinase | 6.10 |
| 1985 | 247450_at | AT5G62350 | invertase/pectin methylesterase inhibitor family protein / DC 1.2 homolog (FL5-2I22) | -6.53 |
| 1986 | 247478_at | AT5G62360 | invertase/pectin methylesterase inhibitor family protein | -3.96 |
| 1987 | 247402_at | AT5G62750 | unknown protein | 13.47 |
| 1988 | 247401_at | AT5G62790 | DXR (1-DEOXY-D-XYLULOSE 5-PHOSPHATE REDUCTOISOMERASE) | -6.20 |
| 1989 | 247424_at | AT5G62850 | ATVEX1 (VEGETATIVE CELL EXPRESSED1) | 7.25 |
| 1990 | 247377_at | AT5G63180 | pectate lyase family protein | -4.51 |
| 1991 | 247376_at | AT5G63310 | NDPK2 (NUCLEOSIDE DIPHOSPHATE KINASE 2); ATP binding / nucleoside diphosphate kinase | -3.90 |
| 1992 | 247341_at | AT5G63720 | unknown protein | 4.95 |
| 1993 | 247342_at | AT5G63730 | IBR domain-containing protein | 5.53 |
| 1994 | 247320_at | AT5G64040 | PSAN (photosystem I reaction center subunit PSI-N); calmodulin binding | -7.46 |
| 1995 | 247322_at | AT5G64090 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G21050.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO43471.1) | -4.21 |
| 1996 | 247293_at | AT5G64510 | similar to unnamed protein product [Vitis vinifera] (GB:CAO49799.1) | 8.67 |
| 1997 | 247266_at | AT5G64570 | XYL4 (beta-xylosidase 4); hydrolase, hydrolyzing O-glycosyl compounds | -4.53 |
| 1998 | 247235_at | AT5G64580 | AAA-type ATPase family protein | -3.80 |
| 1999 | 247242_at | AT5G64690 | neurofilament triplet H protein-related | 6.99 |
| 2000 | 247253_at | AT5G64790 | glycosyl hydrolase family 17 protein | 10.23 |
| 2001 | 247222_at | AT5G64840 | ATGCN5 (Arabidopsis thaliana general control non-repressible 5) | -4.13 |
| 2002 | 247225_at | AT5G65090 | BST1/DER4/MRH3 (BRISTLED1); hydrolase | 5.30 |
| 2003 | 247229_at | AT5G65160 | tetratricopeptide repeat (TPR)-containing protein | 7.28 |
| 2004 | 247201_at | AT5G65220 | ribosomal protein L29 family protein | -4.10 |
| 2005 | 247189_at | AT5G65390 | AGP7 (Arabinogalactan protein 7) | -4.21 |
| 2006 | 247188_at | AT5G65430 | GRF8 (GENERAL REGULATORY FACTOR 8); protein phosphorylated amino acid binding | -5.59 |
| 2007 | 247170_at | AT5G65530 | protein kinase, putative | 8.94 |
| 2008 | 247149_at | AT5G65660 | hydroxyproline-rich glycoprotein family protein | -6.33 |
| 2009 | 247148_at | AT5G65670 | IAA9 (indoleacetic acid-induced protein 9); transcription factor | -5.19 |
| 2010 | 247162_at | AT5G65730 | xyloglucan:xyloglucosyl transferase, putative / xyloglucan endotransglycosylase, putative / endo-xyloglucan transferase, putative | -4.29 |
| 2011 | 247156_at | AT5G65760 | serine carboxypeptidase S28 family protein | -4.22 |
| 2012 | 247168_at | AT5G65860 | ankyrin repeat family protein | -3.83 |
| 2013 | 247122_at | AT5G66020 | ATSAC1B/IBS2 (IMPAIRED IN BABA-INDUCED STERILITY 2); phosphoinositide 5-phosphatase | 7.11 |
| 2014 | 247107_at | AT5G66040 | STR16 (SULFURTRANSFERASE PROTEIN 16) | -6.29 |
| 2015 | 247131_at | AT5G66190 | ATLFNR1 (LEAF FNR 1); NADPH dehydrogenase/ electron transporter, transferring electrons within the cyclic electron transport pathway of photosynthesis/ electron transporter, transferring electrons within the noncyclic electron transport pathway of photosy | -4.69 |
| 2016 | 247073_at | AT5G66570 | OE33/OEE1/OEE33/PSBO-1/PSBO1 (OXYGEN-EVOLVING ENHANCER 33); oxygen evolving/ poly(U) binding | -7.17 |
| 2017 | 247055_at | AT5G66740 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G75160.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN74294.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO22871.1); contains InterPro domain Protein of unknown function DUF620 (InterPro:IPR006873) | 7.78 |
| 2018 | 247025_at | AT5G67030 | ABA1 (ABA DEFICIENT 1); zeaxanthin epoxidase | -3.78 |
| 2019 | 247026_at | AT5G67080 | MAPKKK19 (Mitogen-activated protein kinase kinase kinase 19); kinase | 5.30 |
| 2020 | 247040_at | AT5G67150 | transferase family protein | -4.17 |
| 2021 | 247042_at | AT5G67220 | nitrogen regulation family protein | -5.88 |
| 2022 | 246987_at | AT5G67300 | ATMYB44/ATMYBR1/MYBR1 (MYB DOMAIN PROTEIN 44); DNA binding / transcription factor | -6.67 |
| 2023 | 246998_at | AT5G67370 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G11840.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN75014.1); contains InterPro domain Protein of unknown function DUF1230 (InterPro:IPR009631) | -4.01 |
| 2024 | 246995_at | AT5G67470 | formin homology 2 domain-containing protein / FH2 domain-containing protein | -5.35 |
| 2025 | 247013_at | AT5G67480 | BT4 (BTB AND TAZ DOMAIN PROTEIN 4); protein binding / transcription regulator | 4.61 |
| 2026 | 247005_at | AT5G67520 | adenylylsulfate kinase, putative | 5.83 |
| 2027 | 247003_at | AT5G67550 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G71110.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN65326.1) | 5.22 |
| 2028 | 245047_at | ATCG00020 | Encodes chlorophyll binding protein D1, a part of the photosystem II reaction center core | -4.03 |
| 2029 | 245024_at | ATCG00120 | Encodes the ATPase alpha subunit, which is a subunit of ATP synthase and part of the CF1 portion which catalyzes the conversion of ADP to ATP using the proton motive force. This complex is located in the thylakoid membrane of the chloroplast. | -4.30 |
| 2030 | 245025_at | ATCG00130 | ATPase F subunit. | -4.01 |
| 2031 | 245026_at | ATCG00140 | ATPase III subunit | -4.54 |
| 2032 | 244996_at | ATCG00160 | Chloroplast ribosomal protein S2 | -3.93 |
| 2033 | 245001_at | ATCG00220 | PSII low MW protein | -4.23 |
| 2034 | 245003_at | ATCG00280 | chloroplast gene encoding a CP43 subunit of the photosystem II reaction center. promoter contains a blue-light responsive element. | -4.41 |
| 2035 | 245004_at | ATCG00300 | encodes PsbZ, which is a subunit of photosystem II. In Chlamydomonas, this protein has been shown to be essential in the interaction between PS II and the light harvesting complex II. | -3.88 |
| 2036 | 245006_at | ATCG00340 | Encodes the D1 subunit of photosystem I and II reaction centers. | -4.03 |
| 2037 | 245007_at | ATCG00350 | Encodes psaA protein comprising the reaction center for photosystem I along with psaB protein; hydrophobic protein encoded by the chloroplast genome. | -5.43 |
| 2038 | 245009_at | ATCG00380 | Chloroplast encoded ribosomal protein S4 | -4.05 |
| 2039 | 245010_at | ATCG00420 | Encodes NADH dehydrogenase subunit J. Its transcription is increased upon sulfur depletion. | -6.34 |
| 2040 | 244963_at | ATCG00570 | PSII cytochrome b559 | -3.82 |
| 2041 | 244964_at | ATCG00580 | PSII cytochrome b559. There have been many speculations about the function of Cyt b559, but the most favored at present is that it plays a protective role by acting as an electron acceptor or electron donor under conditions when electron flow through PSII is not optimized. | -4.65 |
| 2042 | 244967_at | ATCG00630 | Encodes subunit J of photosystem I. | -3.98 |
| 2043 | 244972_at | ATCG00680 | encodes for CP47, subunit of the photosystem II reaction center. | -4.06 |
| 2044 | 244974_at | ATCG00700 | PSII low MW protein | -4.24 |
| 2045 | 244961_at | ATCG01040 | hypothetical protein | -3.81 |
| 2046 | 244932_at | ATCG01060 | Encodes the PsaC subunit of photosystem I. | -3.91 |
| 2047 | 244935_at | ATCG01090 | Encodes subunit of the chloroplast NAD(P)H dehydrogenase complex | -3.76 |
| 2048 | 244937_at | ATCG01110 | Encodes the 49KDa plastid NAD(P)H dehydrogenase subunit H protein. Its transcription is regulated by an ndhF-specific plastid sigma factor, SIG4. | -3.77 |