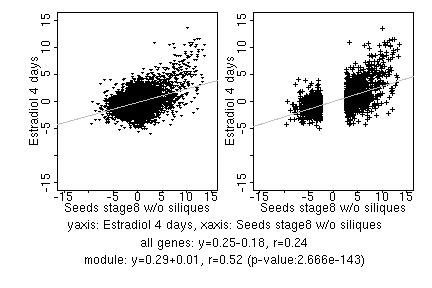
marker gene list
| Seeds stage8 w/o siliques |
| Estradiol4 days |
| probeID | AGICode | Annotation | gene expression |
|
|---|---|---|---|---|
| 1 | 261577_at | AT1G01080 | 33 kDa ribonucleoprotein, chloroplast, putative / RNA-binding protein cp33, putative | -2.74 |
| 2 | 261581_at | AT1G01140 | CIPK9 (CBL-INTERACTING PROTEIN KINASE 9); kinase | -2.95 |
| 3 | 261026_at | AT1G01240 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G46550.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO48635.1) | 2.89 |
| 4 | 261059_at | AT1G01250 | AP2 domain-containing transcription factor, putative | 5.51 |
| 5 | 259439_at | AT1G01480 | ACS2 (1-Amino-cyclopropane-1-carboxylate synthase 2) | 7.59 |
| 6 | 259431_at | AT1G01620 | PIP1C (PLASMA MEMBRANE INTRINSIC PROTEIN 1;3); water channel | -2.87 |
| 7 | 261564_at | AT1G01720 | ATAF1 (Arabidopsis NAC domain containing protein 2); transcription factor | 3.97 |
| 8 | 261535_at | AT1G01725 | similar to unnamed protein product [Vitis vinifera] (GB:CAO39583.1) | 2.94 |
| 9 | 261536_at | AT1G01790 | KEA1 (K EFFLUX ANTIPORTER 1); potassium:hydrogen antiporter | -3.01 |
| 10 | 264146_at | AT1G02205 | CER1 (ECERIFERUM 1) | -3.68 |
| 11 | 264147_at | AT1G02205 | CER1 (ECERIFERUM 1) | -5.99 |
| 12 | 259442_at | AT1G02310 | glycosyl hydrolase family protein 5 / cellulase family protein / (1-4)-beta-mannan endohydrolase, putative | 4.55 |
| 13 | 260917_at | AT1G02700 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G02140.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN70483.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO40074.1) | 8.84 |
| 14 | 262121_at | AT1G02800 | ATCEL2 (Arabidopsis thaliana Cellulase 2); hydrolase, hydrolyzing O-glycosyl compounds | -2.90 |
| 15 | 262116_at | AT1G02816 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G02370.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO40037.1); contains InterPro domain Protein of unknown function DUF538 (InterPro:IPR007493) | 3.08 |
| 16 | 262113_at | AT1G02820 | late embryogenesis abundant 3 family protein / LEA3 family protein | -4.50 |
| 17 | 262118_at | AT1G02850 | glycosyl hydrolase family 1 protein | 5.84 |
| 18 | 262120_at | AT1G02950 | ATGSTF4 (GLUTATHIONE S-TRANSFERASE 31); glutathione transferase | -3.06 |
| 19 | 263114_at | AT1G03130 | PSAD-2 (photosystem I subunit D-2) | -2.58 |
| 20 | 264821_at | AT1G03470 | kinase interacting family protein | 4.21 |
| 21 | 264839_at | AT1G03630 | POR C (PROTOCHLOROPHYLLIDE OXIDOREDUCTASE); NADPH dehydrogenase/ oxidoreductase/ protochlorophyllide reductase | -4.57 |
| 22 | 264832_at | AT1G03660 | similar to ankyrin repeat family protein [Arabidopsis thaliana] (TAIR:AT1G03670.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO23951.1); contains domain ANK REPEAT-CONTAINING (PTHR18958:SF80); contains domain ANKYRIN REPEAT-CONTAINING (PTHR18958) | -3.29 |
| 23 | 265086_at | AT1G03770 | protein binding / zinc ion binding | 3.07 |
| 24 | 265084_at | AT1G03790 | zinc finger (CCCH-type) family protein | 8.81 |
| 25 | 265066_at | AT1G03870 | FLA9 | -3.79 |
| 26 | 265095_at | AT1G03880 | CRU2 (CRUCIFERIN 2); nutrient reservoir | 12.63 |
| 27 | 265094_at | AT1G03890 | cupin family protein | 10.86 |
| 28 | 265099_at | AT1G03990 | alcohol oxidase-related | 4.12 |
| 29 | 265042_at | AT1G04040 | acid phosphatase class B family protein | -6.29 |
| 30 | 264330_at | AT1G04120 | ATMRP5 (Arabidopsis thaliana multidrug resistance-associated protein 5) | 2.81 |
| 31 | 264318_at | AT1G04220 | beta-ketoacyl-CoA synthase, putative | 3.79 |
| 32 | 263656_at | AT1G04240 | SHY2 (SHORT HYPOCOTYL 2); transcription factor | -3.20 |
| 33 | 263653_at | AT1G04310 | ERS2 (ETHYLENE RESPONSE SENSOR 2); receptor | 3.64 |
| 34 | 263652_at | AT1G04330 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G23170.1); similar to At1g04330-like protein [Arabidopsis lyrata] (GB:ABY68375.1) | 5.23 |
| 35 | 263649_at | AT1G04380 | 2-oxoglutarate-dependent dioxygenase, putative | 4.16 |
| 36 | 263662_at | AT1G04430 | dehydration-responsive protein-related | -3.56 |
| 37 | 263677_at | AT1G04520 | 33 kDa secretory protein-related | -2.56 |
| 38 | 264612_at | AT1G04560 | AWPM-19-like membrane family protein | 9.73 |
| 39 | 264610_at | AT1G04645 | self-incompatibility protein-related | 3.78 |
| 40 | 264606_at | AT1G04660 | glycine-rich protein | 7.55 |
| 41 | 264611_at | AT1G04680 | pectate lyase family protein | -3.58 |
| 42 | 261169_at | AT1G04920 | ATSPS3F (sucrose phosphate synthase 3F); sucrose-phosphate synthase/ transferase, transferring glycosyl groups | 4.20 |
| 43 | 265194_at | AT1G05010 | EFE (ETHYLENE FORMING ENZYME) | -5.49 |
| 44 | 265216_at | AT1G05100 | MAPKKK18 (Mitogen-activated protein kinase kinase kinase 18); kinase | 3.26 |
| 45 | 264575_at | AT1G05190 | EMB2394 (EMBRYO DEFECTIVE 2394); structural constituent of ribosome | -4.04 |
| 46 | 264587_at | AT1G05200 | ATGLR3.4 (Arabidopsis thaliana glutamate receptor 3.4) | -5.35 |
| 47 | 264565_at | AT1G05280 | fringe-related protein | 4.47 |
| 48 | 264580_at | AT1G05340 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G32210.1) | 6.20 |
| 49 | 261385_at | AT1G05450 | protease inhibitor/seed storage/lipid transfer protein (LTP)-related | 6.54 |
| 50 | 263175_at | AT1G05510 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G31985.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO45247.1); contains InterPro domain Protein of unknown function DUF1264 (InterPro:IPR010686) | 11.82 |
| 51 | 261256_at | AT1G05760 | RTM1 (RESTRICTED TEV MOVEMENT 1) | -2.89 |
| 52 | 262629_at | AT1G06460 | ACD32.1 (ALPHA-CRYSTALLIN DOMAIN 31.2) | -3.79 |
| 53 | 262630_at | AT1G06520 | ATGPAT1/GPAT1 (GLYCEROL-3-PHOSPHATE ACYLTRANSFERASE 1); 1-acylglycerol-3-phosphate O-acyltransferase/ acyltransferase | 3.48 |
| 54 | 262632_at | AT1G06680 | PSBP-1 (OXYGEN-EVOLVING ENHANCER PROTEIN 2); poly(U) binding | -2.37 |
| 55 | 256045_at | AT1G07150 | MAPKKK13 (Mitogen-activated protein kinase kinase kinase 13); kinase | 2.85 |
| 56 | 256053_at | AT1G07260 | UDP-glucoronosyl/UDP-glucosyl transferase family protein | 2.87 |
| 57 | 261078_at | AT1G07320 | RPL4 (ribosomal protein L4); poly(U) binding / structural constituent of ribosome | -2.82 |
| 58 | 261077_at | AT1G07430 | protein phosphatase 2C, putative / PP2C, putative | 5.30 |
| 59 | 261084_at | AT1G07440 | tropinone reductase, putative / tropine dehydrogenase, putative | -3.40 |
| 60 | 261065_at | AT1G07500 | unknown protein | 7.46 |
| 61 | 261460_at | AT1G07880 | ATMPK13 (ARABIDOPSIS THALIANA MAP KINASE 13); MAP kinase/ kinase | -3.29 |
| 62 | 261412_at | AT1G07890 | APX1 (ASCORBATE PEROXIDASE 1); L-ascorbate peroxidase | -2.62 |
| 63 | 261755_at | AT1G08170 | histone H2B family protein | 5.97 |
| 64 | 261815_at | AT1G08320 | bZIP family transcription factor | 4.72 |
| 65 | 261746_at | AT1G08380 | PSAO (photosystem I subunit O) | -2.54 |
| 66 | 261692_at | AT1G08450 | CRT3 (CALRETICULIN 3); calcium ion binding | -2.90 |
| 67 | 264781_at | AT1G08540 | SIGB (SIGMA FACTOR B); DNA binding / DNA-directed RNA polymerase/ transcription factor | -4.22 |
| 68 | 264782_at | AT1G08810 | MYB60 (myb domain protein 60); DNA binding / transcription factor | 3.63 |
| 69 | 264624_at | AT1G08930 | ERD6 (EARLY RESPONSE TO DEHYDRATION 6); carbohydrate transmembrane transporter/ sugar:hydrogen ion symporter | -3.69 |
| 70 | 264643_at | AT1G08990 | PGSIP5 (PLANT GLYCOGENIN-LIKE STARCH INITIATION PROTEIN 5); transferase, transferring glycosyl groups | 6.25 |
| 71 | 264263_at | AT1G09155 | ATPP2-B15 (Phloem protein 2-B15); carbohydrate binding | 3.92 |
| 72 | 264266_at | AT1G09160 | protein phosphatase 2C-related / PP2C-related | -3.26 |
| 73 | 264262_at | AT1G09200 | histone H3 | -2.35 |
| 74 | 263709_at | AT1G09310 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G56580.1); similar to unknown [Populus trichocarpa] (GB:ABK94207.1); contains InterPro domain Protein of unknown function DUF538 (InterPro:IPR007493) | -6.67 |
| 75 | 263676_at | AT1G09340 | CRB; binding / catalytic/ coenzyme binding | -5.71 |
| 76 | 264500_at | AT1G09370 | enzyme inhibitor/ pectinesterase | 5.37 |
| 77 | 264505_at | AT1G09380 | integral membrane family protein / nodulin MtN21-related | 5.95 |
| 78 | 264514_at | AT1G09500 | cinnamyl-alcohol dehydrogenase family / CAD family | 4.65 |
| 79 | 264669_at | AT1G09630 | ATRAB11C (ARABIDOPSIS RAB GTPASE HOMOLOG A2A); GTP binding | -2.41 |
| 80 | 264668_at | AT1G09780 | 2,3-biphosphoglycerate-independent phosphoglycerate mutase, putative / phosphoglyceromutase, putative | -3.51 |
| 81 | 264710_at | AT1G09790 | COBL6 (COBRA-LIKE PROTEIN 6 PRECURSOR) | 3.26 |
| 82 | 264687_at | AT1G09850 | XBCP3 (XYLEM BARK CYSTEINE PEPTIDASE 3); cysteine-type peptidase | -3.12 |
| 83 | 264661_at | AT1G09950 | transcription factor-related | 3.20 |
| 84 | 264524_at | AT1G10070 | ATBCAT-2; branched-chain-amino-acid transaminase/ catalytic | 3.07 |
| 85 | 264462_at | AT1G10200 | WLIM1; transcription factor | -2.40 |
| 86 | 264434_at | AT1G10340 | ankyrin repeat family protein | -5.25 |
| 87 | 264435_at | AT1G10360 | ATGSTU18 (GLUTATHIONE S-TRANSFERASE 29); glutathione transferase | -2.60 |
| 88 | 263259_at | AT1G10560 | armadillo/beta-catenin repeat family protein / U-box domain-containing protein | -2.84 |
| 89 | 263210_at | AT1G10585 | transcription factor | -2.85 |
| 90 | 262788_at | AT1G10690 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G60783.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN59885.1) | 3.16 |
| 91 | 262784_at | AT1G10760 | SEX1 (STARCH EXCESS 1) | -2.68 |
| 92 | 260481_at | AT1G10960 | ATFD1 (FERREDOXIN 1); 2 iron, 2 sulfur cluster binding / electron carrier/ iron-sulfur cluster binding | -2.43 |
| 93 | 260461_at | AT1G10980 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G61670.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO41520.1); contains InterPro domain Transmembrane receptor, eukaryota; (InterPro:IPR009637) | -2.65 |
| 94 | 260472_at | AT1G10990 | unknown protein | -3.23 |
| 95 | 260483_at | AT1G11000 | MLO4 (MILDEW RESISTANCE LOCUS O 4); calmodulin binding | -3.08 |
| 96 | 262478_at | AT1G11170 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G61240.2); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G61240.3); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G61240.4); similar to unknown [Picea sitchensis] (GB:ABK25584.1); contains InterPro domain Protein of unknown function DUF707 (InterPro:IPR007877) | 6.42 |
| 97 | 262454_at | AT1G11190 | BFN1 (BIFUNCTIONAL NUCLEASE I); nucleic acid binding | 4.20 |
| 98 | 262508_at | AT1G11300 | carbohydrate binding / kinase | -3.00 |
| 99 | 261848_at | AT1G11590 | pectin methylesterase, putative | 5.15 |
| 100 | 264343_at | AT1G11850 | unknown protein | -6.14 |
| 101 | 264346_at | AT1G12010 | 1-aminocyclopropane-1-carboxylate oxidase, putative / ACC oxidase, putative | 7.46 |
| 102 | 257421_at | AT1G12030 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G62420.1); similar to Protein of unknown function DUF506, plant [Medicago truncatula] (GB:ABN05810.1); contains InterPro domain Protein of unknown function DUF506, plant (InterPro:IPR006502) | 4.25 |
| 103 | 264371_at | AT1G12090 | ELP (EXTENSIN-LIKE PROTEIN); lipid binding | -3.46 |
| 104 | 260994_at | AT1G12130 | flavin-containing monooxygenase family protein / FMO family protein | 7.08 |
| 105 | 260968_at | AT1G12250 | thylakoid lumenal protein-related | -2.53 |
| 106 | 259523_at | AT1G12500 | phosphate translocator-related | -2.47 |
| 107 | 259527_at | AT1G12600 | similar to ATUTR2/UTR2 (UDP-GALACTOSE TRANSPORTER 2) [Arabidopsis thaliana] (TAIR:AT4G23010.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO63660.1); contains InterPro domain UAA transporter (InterPro:IPR013657) | 3.10 |
| 108 | 255938_at | AT1G12620 | pentatricopeptide (PPR) repeat-containing protein | -2.87 |
| 109 | 261206_at | AT1G12800 | S1 RNA-binding domain-containing protein | -2.88 |
| 110 | 261197_at | AT1G12900 | GAPA-2; glyceraldehyde-3-phosphate dehydrogenase | -3.26 |
| 111 | 262793_at | AT1G13110 | CYP71B7 (cytochrome P450, family 71, subfamily B, polypeptide 7); oxygen binding | -6.54 |
| 112 | 259364_at | AT1G13260 | RAV1 (Related to ABI3/VP1 1); DNA binding / transcription factor | -3.25 |
| 113 | 259363_at | AT1G13270 | MAP1C (METHIONINE AMINOPEPTIDASE 1B); metalloexopeptidase | -2.60 |
| 114 | 259366_at | AT1G13280 | AOC4 (ALLENE OXIDE CYCLASE 4) | -2.74 |
| 115 | 256158_at | AT1G13590 | ATPSK1 (PHYTOSULFOKINE 1 PRECURSOR); growth factor | 3.29 |
| 116 | 256096_at | AT1G13650 | similar to 18S pre-ribosomal assembly protein gar2-related [Arabidopsis thaliana] (TAIR:AT2G03810.4); similar to 18S pre-ribosomal assembly protein gar2-related [Arabidopsis thaliana] (TAIR:AT2G03810.2); similar to 18S pre-ribosomal assembly protein gar2-related [Arabidopsis thaliana] (TAIR:AT2G03810.3); similar to hypothetical protein Tc00.1047053503999.40 [Trypanosoma cruzi strain CL Brener] (GB:XP_813437.1) | -3.68 |
| 117 | 256069_at | AT1G13740 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G69260.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO41856.1); contains InterPro domain Protein of unknown function DUF1675 (InterPro:IPR012463) | 3.38 |
| 118 | 259448_at | AT1G13790 | XH/XS domain-containing protein / XS zinc finger domain-containing protein | -3.53 |
| 119 | 262609_at | AT1G13930 | similar to nodulin-related [Arabidopsis thaliana] (TAIR:AT2G03440.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN63832.1) | -3.58 |
| 120 | 262615_at | AT1G13950 | EIF-5A (eukaryotic translation initiation factor 5A-1); translation initiation factor | -4.56 |
| 121 | 262648_at | AT1G14030 | ribulose-1,5 bisphosphate carboxylase oxygenase large subunit N-methyltransferase, putative | -4.17 |
| 122 | 262612_at | AT1G14150 | oxygen evolving enhancer 3 (PsbQ) family protein | -4.17 |
| 123 | 262654_at | AT1G14180 | unknown protein | -2.70 |
| 124 | 261480_at | AT1G14280 | PKS2 (PHYTOCHROME KINASE SUBSTRATE 2) | -6.69 |
| 125 | 261488_at | AT1G14345 | oxidoreductase | -3.85 |
| 126 | 261473_at | AT1G14490 | DNA-binding protein-related | 4.75 |
| 127 | 261482_at | AT1G14530 | (TOM THREE HOMOLOG); virion binding | 3.26 |
| 128 | 262888_at | AT1G14790 | RDR1 (RNA-DEPENDENT RNA POLYMERASE 1); RNA-directed RNA polymerase/ nucleic acid binding | -2.40 |
| 129 | 262857_at | AT1G14930 | major latex protein-related / MLP-related | 10.99 |
| 130 | 262858_at | AT1G14940 | major latex protein-related / MLP-related | 9.37 |
| 131 | 262828_at | AT1G14950 | major latex protein-related / MLP-related | 9.07 |
| 132 | 260739_at | AT1G15000 | SCPL50 (serine carboxypeptidase-like 50); serine carboxypeptidase | -2.35 |
| 133 | 260742_at | AT1G15050 | IAA34 (indoleacetic acid-induced protein 34); transcription factor | 3.31 |
| 134 | 262596_at | AT1G15080 | ATPAP2 (PHOSPHATIDIC ACID PHOSPHATASE 2); phosphatidate phosphatase | -2.63 |
| 135 | 262598_at | AT1G15260 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G16070.1) | -2.55 |
| 136 | 262580_at | AT1G15330 | CBS domain-containing protein | 5.16 |
| 137 | 262603_at | AT1G15380 | lactoylglutathione lyase family protein / glyoxalase I family protein | 4.37 |
| 138 | 261768_at | AT1G15550 | GA4 (GA REQUIRING 4); gibberellin 3-beta-dioxygenase | -3.62 |
| 139 | 259491_at | AT1G15820 | LHCB6 (LIGHT HARVESTING COMPLEX PSII); chlorophyll binding | -2.83 |
| 140 | 261845_at | AT1G15960 | NRAMP6 (NRAMP metal ion transporter 6); metal ion transmembrane transporter | 3.64 |
| 141 | 261788_at | AT1G15980 | similar to unnamed protein product [Vitis vinifera] (GB:CAO49411.1); contains domain G3DSA:3.40.50.2000 (G3DSA:3.40.50.2000); contains domain SSF53756 (SSF53756) | -5.14 |
| 142 | 261838_at | AT1G16030 | HSP70B (heat shock protein 70B); ATP binding | 4.70 |
| 143 | 262705_at | AT1G16260 | protein kinase family protein | -2.57 |
| 144 | 255763_at | AT1G16730 | similar to unknown [Picea sitchensis] (GB:ABK21208.1) | 6.33 |
| 145 | 255761_at | AT1G16770 | similar to unnamed protein product [Vitis vinifera] (GB:CAO41707.1) | 6.34 |
| 146 | 256114_at | AT1G16850 | unknown protein | 6.79 |
| 147 | 256115_at | AT1G16880 | uridylyltransferase-related | -4.54 |
| 148 | 262527_at | AT1G17010 | oxidoreductase, 2OG-Fe(II) oxygenase family protein | 7.97 |
| 149 | 262482_at | AT1G17020 | SRG1 (SENESCENCE-RELATED GENE 1); oxidoreductase, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors | 3.02 |
| 150 | 262525_at | AT1G17060 | CYP72C1 (cytochrome P450, family 72, subfamily C, polypeptide 1); oxygen binding | 4.56 |
| 151 | 262533_at | AT1G17090 | unknown protein | 3.59 |
| 152 | 262483_at | AT1G17220 | FUG1 (FU-GAERI1); translation initiation factor | -2.47 |
| 153 | 262531_at | AT1G17230 | protein binding / protein kinase | -2.83 |
| 154 | 260845_at | AT1G17310 | MADS-box protein (AGL100) | 5.38 |
| 155 | 261032_at | AT1G17430 | hydrolase, alpha/beta fold family protein | 3.23 |
| 156 | 259399_at | AT1G17710 | phosphoric monoester hydrolase | 3.13 |
| 157 | 259403_at | AT1G17745 | PGDH (3-PHOSPHOGLYCERATE DEHYDROGENASE); phosphoglycerate dehydrogenase | -2.67 |
| 158 | 255905_at | AT1G17810 | BETA-TIP (BETA-TONOPLAST INTRINSIC PROTEIN); water channel | 11.94 |
| 159 | 255900_at | AT1G17830 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G73210.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO45300.1); contains InterPro domain Protein of unknown function DUF789 (InterPro:IPR008507) | 4.39 |
| 160 | 255903_at | AT1G17950 | MYB52 (myb domain protein 52); DNA binding / transcription factor | 6.11 |
| 161 | 256073_at | AT1G18100 | E12A11; phosphatidylethanolamine binding | 6.29 |
| 162 | 256125_at | AT1G18250 | ATLP-1 (Arabidopsis thaumatin-like protein 1) | -2.71 |
| 163 | 255773_at | AT1G18590 | sulfotransferase family protein | -4.68 |
| 164 | 255774_at | AT1G18620 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G74160.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO49165.1) | -2.47 |
| 165 | 261422_at | AT1G18730 | similar to unknown [Populus trichocarpa] (GB:ABK95263.1) | -2.93 |
| 166 | 261407_at | AT1G18810 | phytochrome kinase substrate-related | -3.55 |
| 167 | 259462_at | AT1G18940 | nodulin family protein | 3.86 |
| 168 | 259466_at | AT1G19050 | ARR7 (RESPONSE REGULATOR 7); transcription regulator/ two-component response regulator | -3.01 |
| 169 | 256015_at | AT1G19150 | LHCA6 (Photosystem I light harvesting complex gene 6); chlorophyll binding | -7.75 |
| 170 | 256039_at | AT1G19190 | hydrolase | 4.31 |
| 171 | 256014_at | AT1G19200 | senescence-associated protein-related | 4.46 |
| 172 | 256009_at | AT1G19210 | AP2 domain-containing transcription factor, putative | 3.23 |
| 173 | 256011_at | AT1G19230 | respiratory burst oxidase protein E (RbohE) / NADPH oxidase | 3.94 |
| 174 | 260661_at | AT1G19500 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G35765.1) | 5.52 |
| 175 | 260668_at | AT1G19530 | similar to unnamed protein product [Vitis vinifera] (GB:CAO61141.1) | 9.88 |
| 176 | 260662_at | AT1G19540 | isoflavone reductase, putative | 6.54 |
| 177 | 255786_at | AT1G19670 | ATCLH1 (CORONATINE-INDUCED PROTEIN 1) | -4.33 |
| 178 | 255815_at | AT1G19890 | ATMGH3/MGH3 (MALE-GAMETE-SPECIFIC HISTONE H3); DNA binding | 3.56 |
| 179 | 255814_at | AT1G19900 | glyoxal oxidase-related | 8.82 |
| 180 | 255785_at | AT1G19920 | APS2 (ATP SULFURYLASE PRECURSOR) | -2.64 |
| 181 | 261221_at | AT1G19960 | similar to transmembrane receptor [Arabidopsis thaliana] (TAIR:AT2G32140.1) | -4.57 |
| 182 | 261230_at | AT1G20010 | TUB5 (tubulin beta-5 chain) | -4.38 |
| 183 | 261218_at | AT1G20020 | ATLFNR2 (LEAF FNR 2); NADPH dehydrogenase/ oxidoreductase/ poly(U) binding | -7.58 |
| 184 | 261224_at | AT1G20160 | ATSBT5.2; subtilase | -2.56 |
| 185 | 261243_at | AT1G20180 | Identical to UPF0496 protein At1g20180 [Arabidopsis Thaliana] (GB:Q6DYE5;GB:Q5BQ01;GB:Q9LNU2); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G49070.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO60936.1); contains InterPro domain Protein of unknown function DUF677 (InterPro:IPR007749) | 4.40 |
| 186 | 261226_at | AT1G20190 | ATEXPA11 (ARABIDOPSIS THALIANA EXPANSIN A11) | -5.54 |
| 187 | 255886_at | AT1G20340 | DRT112 (DNA-damage-repair/toleration protein 112); copper ion binding / electron carrier | -2.88 |
| 188 | 259570_at | AT1G20440 | COR47 (cold regulated 47) | -3.00 |
| 189 | 259518_at | AT1G20510 | OPCL1 (OPC-8:0 COA LIGASE1); 4-coumarate-CoA ligase | -2.33 |
| 190 | 262796_at | AT1G20850 | XCP2 (XYLEM CYSTEINE PEPTIDASE 2); cysteine-type peptidase/ peptidase | -5.34 |
| 191 | 262854_at | AT1G20870 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G54850.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO66281.1); contains InterPro domain HSP20-like chaperone (InterPro:IPR008978) | 2.90 |
| 192 | 262806_at | AT1G20950 | pyrophosphate--fructose-6-phosphate 1-phosphotransferase-related / pyrophosphate-dependent 6-phosphofructose-1-kinase-related | -3.68 |
| 193 | 262803_at | AT1G21000 | zinc-binding family protein | 3.51 |
| 194 | 261449_at | AT1G21120 | O-methyltransferase, putative | -3.54 |
| 195 | 261453_at | AT1G21130 | O-methyltransferase, putative | -5.61 |
| 196 | 259560_at | AT1G21270 | WAK2 (wall-associated kinase 2); protein serine/threonine kinase | -3.38 |
| 197 | 260877_at | AT1G21500 | similar to hypothetical protein OsI_030994 [Oryza sativa (indica cultivar-group)] (GB:EAZ09762.1); similar to Os09g0517000 [Oryza sativa (japonica cultivar-group)] (GB:NP_001063677.1); similar to unknown protein [Oryza sativa (japonica cultivar-group)] (GB:BAD33829.1) | -3.00 |
| 198 | 260854_at | AT1G21970 | LEC1 (LEAFY COTYLEDON 1); transcription factor | 3.47 |
| 199 | 255950_at | AT1G22110 | structural constituent of ribosome | 2.98 |
| 200 | 255923_at | AT1G22180 | SEC14 cytosolic factor family protein / phosphoglyceride transfer family protein | 2.89 |
| 201 | 261936_at | AT1G22600 | similar to late embryogenesis abundant domain-containing protein / LEA domain-containing protein [Arabidopsis thaliana] (TAIR:AT1G72100.1); similar to seed maturation protein PM27 [Glycine max] (GB:AAD30426.1); contains domain PTHR23241:SF1 (PTHR23241:SF1); contains domain PTHR23241 (PTHR23241) | 6.38 |
| 202 | 264201_at | AT1G22630 | heat shock protein binding / unfolded protein binding | -4.33 |
| 203 | 264200_at | AT1G22650 | beta-fructofuranosidase, putative / invertase, putative / saccharase, putative / beta-fructosidase, putative | -3.05 |
| 204 | 264726_at | AT1G22985 | AP2 domain-containing transcription factor, putative | 3.17 |
| 205 | 264729_at | AT1G22990 | heavy-metal-associated domain-containing protein / copper chaperone (CCH)-related | 6.76 |
| 206 | 264752_at | AT1G23010 | LPR1 (LOW PHOSPHATE ROOT1); copper ion binding / oxidoreductase, acting on diphenols and related substances as donors, oxygen as acceptor | 3.89 |
| 207 | 264889_at | AT1G23050 | hydroxyproline-rich glycoprotein family protein | 4.80 |
| 208 | 264888_at | AT1G23070 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G38360.2); similar to unnamed protein product [Vitis vinifera] (GB:CAO65220.1); contains InterPro domain Protein of unknown function DUF300 (InterPro:IPR005178) | 6.61 |
| 209 | 264900_at | AT1G23080 | PIN7 (PIN-FORMED 7); auxin:hydrogen symporter/ transporter | -5.87 |
| 210 | 264887_at | AT1G23120 | major latex protein-related / MLP-related | 3.87 |
| 211 | 264899_at | AT1G23130 | Bet v I allergen family protein | -5.43 |
| 212 | 264891_at | AT1G23200 | pectinesterase family protein | 3.76 |
| 213 | 262988_at | AT1G23310 | GGT1 (ALANINE-2-OXOGLUTARATE AMINOTRANSFERASE 1) | -3.77 |
| 214 | 265175_at | AT1G23480 | ATCSLA03 (Cellulose synthase-like A3); transferase, transferring glycosyl groups | -6.31 |
| 215 | 257401_at | AT1G23550 | SRO2 (SIMILAR TO RCD ONE 2); NAD+ ADP-ribosyltransferase | 3.10 |
| 216 | 265177_at | AT1G23630 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G23660.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO66084.1); contains InterPro domain Protein of unknown function DUF220 (InterPro:IPR003863) | 4.92 |
| 217 | 265182_at | AT1G23740 | oxidoreductase, zinc-binding dehydrogenase family protein | -2.34 |
| 218 | 265171_at | AT1G23790 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G70340.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO40888.1); contains InterPro domain Protein of unknown function DUF936, plant (InterPro:IPR010341) | -2.81 |
| 219 | 263027_at | AT1G24010 | Identical to Uncharacterized protein At1g24010 [Arabidopsis Thaliana] (GB:P0C0B1;GB:Q9LR93); similar to Bet v I allergen family protein [Arabidopsis thaliana] (TAIR:AT1G24000.1); contains InterPro domain Bet v I allergen; (InterPro:IPR000916) | 3.22 |
| 220 | 263034_at | AT1G24020 | MLP423 (MLP-LIKE PROTEIN 423) | -4.08 |
| 221 | 264857_at | AT1G24170 | GATL8/LGT9 (Galacturonosyltransferase-like 8); polygalacturonate 4-alpha-galacturonosyltransferase/ transferase, transferring glycosyl groups / transferase, transferring hexosyl groups | -2.59 |
| 222 | 265014_at | AT1G24430 | transferase family protein | 4.55 |
| 223 | 265020_at | AT1G24540 | CYP86C1 (cytochrome P450, family 86, subfamily C, polypeptide 1); oxygen binding | 4.22 |
| 224 | 265025_at | AT1G24575 | unknown protein | -4.60 |
| 225 | 265024_at | AT1G24600 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G67920.1) | 4.21 |
| 226 | 245650_at | AT1G24735 | caffeoyl-CoA 3-O-methyltransferase, putative | 6.04 |
| 227 | 245637_at | AT1G25230 | purple acid phosphatase family protein | -3.65 |
| 228 | 245634_at | AT1G25270 | similar to nodulin MtN21 family protein [Arabidopsis thaliana] (TAIR:AT1G68170.1); similar to Protein of unknown function DUF6, transmembrane [Medicago truncatula] (GB:ABD28596.1); contains InterPro domain Protein of unknown function DUF6, transmembrane; (InterPro:IPR000620) | 5.37 |
| 229 | 245642_at | AT1G25275 | similar to unnamed protein product [Vitis vinifera] (GB:CAO43403.1) | -4.14 |
| 230 | 255741_at | AT1G25410 | ATIPT6 (Arabidopsis thaliana isopentenyltransferase 6); adenylate dimethylallyltransferase | 6.71 |
| 231 | 255732_at | AT1G25450 | very-long-chain fatty acid condensing enzyme, putative | -2.43 |
| 232 | 255726_at | AT1G25530 | lysine and histidine specific transporter, putative | 3.27 |
| 233 | 255734_at | AT1G25550 | myb family transcription factor | -2.35 |
| 234 | 255742_at | AT1G25560 | AP2 domain-containing transcription factor, putative | -3.77 |
| 235 | 245877_at | AT1G26220 | GCN5-related N-acetyltransferase (GNAT) family protein | -4.16 |
| 236 | 245876_at | AT1G26230 | chaperonin, putative | -2.68 |
| 237 | 261007_at | AT1G26400 | FAD-binding domain-containing protein | 7.18 |
| 238 | 261260_at | AT1G26680 | transcriptional factor B3 family protein | 4.96 |
| 239 | 263689_at | AT1G26820 | RNS3 (RIBONUCLEASE 3); endoribonuclease | 4.13 |
| 240 | 264973_at | AT1G27040 | nitrate transporter, putative | 3.73 |
| 241 | 264447_at | AT1G27300 | similar to unnamed protein product [Vitis vinifera] (GB:CAO66149.1) | 4.02 |
| 242 | 264439_at | AT1G27450 | APT1; adenine phosphoribosyltransferase | -2.56 |
| 243 | 264494_at | AT1G27461 | similar to unnamed protein product [Vitis vinifera] (GB:CAO61483.1) | 11.60 |
| 244 | 261648_at | AT1G27730 | STZ (SALT TOLERANCE ZINC FINGER); nucleic acid binding / transcription factor/ zinc ion binding | 3.90 |
| 245 | 259588_at | AT1G27930 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G67330.1); similar to unknown [Populus trichocarpa] (GB:ABK93495.1); contains InterPro domain Protein of unknown function DUF579, plant (InterPro:IPR006514) | -3.72 |
| 246 | 259607_at | AT1G27940 | PGP13 (P-GLYCOPROTEIN 13); ATPase, coupled to transmembrane movement of substances | -3.11 |
| 247 | 259578_at | AT1G27990 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G52420.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO41629.1) | 8.56 |
| 248 | 259580_at | AT1G28030 | oxidoreductase, 2OG-Fe(II) oxygenase family protein | 3.67 |
| 249 | 259596_at | AT1G28130 | GH3.17; indole-3-acetic acid amido synthetase | -4.17 |
| 250 | 245662_at | AT1G28190 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G12340.1); similar to unknown [Picea sitchensis] (GB:ABK21062.1) | -4.83 |
| 251 | 245688_at | AT1G28290 | AGP31 (ARABINOGALACTAN-PROTEIN 31); structural constituent of cell wall | -4.15 |
| 252 | 261496_at | AT1G28360 | ATERF12/ERF12 (ERF domain protein 12); DNA binding / transcription factor/ transcription repressor | 4.25 |
| 253 | 261441_at | AT1G28470 | ANAC010 (Arabidopsis NAC domain containing protein 10); transcription factor | 3.42 |
| 254 | 262740_at | AT1G28590 | lipase, putative | 2.90 |
| 255 | 262734_at | AT1G28640 | GDSL-motif lipase, putative | 4.03 |
| 256 | 262739_at | AT1G28650 | lipase, putative | 3.93 |
| 257 | 260898_at | AT1G29070 | ribosomal protein L34 family protein | -2.52 |
| 258 | 260887_at | AT1G29160 | Dof-type zinc finger domain-containing protein | 3.54 |
| 259 | 260897_at | AT1G29330 | ERD2 (ER lumen protein retaining receptor 2); receptor | 3.89 |
| 260 | 259768_at | AT1G29390 | COR314-TM2 (cold regulated 314 thylakoid membrane 2) | -2.79 |
| 261 | 259784_at | AT1G29450 | auxin-responsive protein, putative | -4.03 |
| 262 | 259773_at | AT1G29500 | auxin-responsive protein, putative | -5.14 |
| 263 | 259783_at | AT1G29510 | SAUR68 (SMALL AUXIN UPREGULATED 68) | -3.28 |
| 264 | 259786_at | AT1G29660 | GDSL-motif lipase/hydrolase family protein | -2.95 |
| 265 | 259788_at | AT1G29670 | GDSL-motif lipase/hydrolase family protein | -9.21 |
| 266 | 259782_at | AT1G29680 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G45690.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO47983.1); contains InterPro domain Protein of unknown function DUF1264 (InterPro:IPR010686) | 7.35 |
| 267 | 255994_at | AT1G29760 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G34380.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO48004.1); contains domain BERNARDINELLI-SEIP CONGENITAL LIPODYSTROPHY 2 HOMOLOG (BSCL2 PROTEIN) (PTHR21212) | 2.80 |
| 268 | 256190_at | AT1G30100 | NCED5 (NINE-CIS-EPOXYCAROTENOID DIOXYGENASE 5) | 3.40 |
| 269 | 261803_at | AT1G30500 | CCAAT-binding transcription factor (CBF-B/NF-YA) family protein | 2.81 |
| 270 | 261801_at | AT1G30520 | AAE14 (ACYL-ACTIVATING ENZYME 14); AMP binding | -3.40 |
| 271 | 263228_at | AT1G30700 | FAD-binding domain-containing protein | 7.08 |
| 272 | 263215_at | AT1G30710 | FAD-binding domain-containing protein | 4.13 |
| 273 | 265128_at | AT1G30860 | protein binding / zinc ion binding | 6.61 |
| 274 | 262557_at | AT1G31330 | PSAF (photosystem I subunit F) | -2.33 |
| 275 | 256489_at | AT1G31550 | GDSL-motif lipase, putative | -4.76 |
| 276 | 256497_at | AT1G31580 | ECS1 | -8.22 |
| 277 | 246574_at | AT1G31670 | copper amine oxidase, putative | 6.14 |
| 278 | 246601_at | AT1G31710 | copper amine oxidase, putative | -2.92 |
| 279 | 246602_at | AT1G31710 | copper amine oxidase, putative | -2.76 |
| 280 | 246582_at | AT1G31750 | proline-rich family protein | 6.91 |
| 281 | 246268_at | AT1G31800 | CYP97A3/LUT5 (CYTOCHROME P450-TYPE MONOOXYGENASE 97A3); carotene beta-ring hydroxylase/ oxygen binding | -3.20 |
| 282 | 255720_at | AT1G32060 | PRK (PHOSPHORIBULOKINASE); ATP binding / phosphoribulokinase/ protein binding | -3.22 |
| 283 | 255718_at | AT1G32070 | ATNSI (NUCLEAR SHUTTLE INTERACTING); N-acetyltransferase | -2.68 |
| 284 | 255719_at | AT1G32080 | membrane protein, putative | -4.27 |
| 285 | 260704_at | AT1G32470 | glycine cleavage system H protein, mitochondrial, putative | -6.30 |
| 286 | 256464_at | AT1G32560 | late embryogenesis abundant group 1 domain-containing protein / LEA group 1 domain-containing protein | 10.71 |
| 287 | 261190_at | AT1G32990 | PRPL11 (PLASTID RIBOSOMAL PROTEIN L11); structural constituent of ribosome | -2.69 |
| 288 | 261567_at | AT1G33055 | unknown protein | 4.21 |
| 289 | 256529_at | AT1G33260 | protein kinase family protein | 2.94 |
| 290 | 245768_at | AT1G33590 | disease resistance protein-related / LRR protein-related | -5.38 |
| 291 | 245765_at | AT1G33600 | leucine-rich repeat family protein | -5.86 |
| 292 | 261991_at | AT1G33700 | catalytic | 5.78 |
| 293 | 262001_at | AT1G33790 | jacalin lectin family protein | 4.33 |
| 294 | 260116_at | AT1G33960 | AIG1 (AVRRPT2-INDUCED GENE 1); GTP binding | -3.27 |
| 295 | 262542_at | AT1G34180 | ANAC016 (Arabidopsis NAC domain containing protein 16); transcription factor | 2.96 |
| 296 | 262563_at | AT1G34210 | SERK2 (SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE 2); kinase | -3.04 |
| 297 | 245756_at | AT1G35190 | oxidoreductase, 2OG-Fe(II) oxygenase family protein | 4.53 |
| 298 | 259546_at | AT1G35350 | similar to SHB1 (SHORT HYPOCOTYL UNDER BLUE1) [Arabidopsis thaliana] (TAIR:AT4G25350.1); similar to EXS family protein / ERD1/XPR1/SYG1 family protein [Arabidopsis thaliana] (TAIR:AT1G26730.1); similar to ATP binding / ATPase, coupled to transmembrane movement of substances [Arabidopsis thaliana] (TAIR:AT1G14040.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO68453.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO68461.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN73334.1); contains InterPro domain SPX, N-terminal (InterPro:IPR004331); contains InterPro domain EXS, C-terminal; (InterPro:IPR004342) | -3.87 |
| 299 | 262029_at | AT1G35680 | 50S ribosomal protein L21, chloroplast / CL21 (RPL21) | -2.46 |
| 300 | 263194_at | AT1G36060 | AP2 domain-containing transcription factor, putative | 3.74 |
| 301 | 261979_at | AT1G37130 | NIA2 (NITRATE REDUCTASE 2) | -5.09 |
| 302 | 262032_at | AT1G37150 | HCS2 (HOLOCARBOXYLASE SYNTHETASE 2); catalytic | 3.42 |
| 303 | 259625_at | AT1G42970 | GAPB (GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE B SUBUNIT); glyceraldehyde-3-phosphate dehydrogenase | -4.12 |
| 304 | 264415_at | AT1G43160 | RAP2.6 (related to AP2 6); DNA binding / transcription factor | 8.59 |
| 305 | 262721_at | AT1G43560 | ATY2 (Arabidopsis thioredoxin y2); thiol-disulfide exchange intermediate | -2.71 |
| 306 | 262726_at | AT1G43640 | AtTLP5 (TUBBY LIKE PROTEIN 5); phosphoric diester hydrolase/ transcription factor | 4.12 |
| 307 | 260869_at | AT1G43800 | acyl-(acyl-carrier-protein) desaturase, putative / stearoyl-ACP desaturase, putative | 4.36 |
| 308 | 245243_at | AT1G44414 | similar to unnamed protein product [Vitis vinifera] (GB:CAO61874.1) | 3.53 |
| 309 | 245213_at | AT1G44575 | NPQ4 (NONPHOTOCHEMICAL QUENCHING) | -2.36 |
| 310 | 261330_at | AT1G44900 | ATP binding / DNA binding / DNA-dependent ATPase | -3.44 |
| 311 | 245807_at | AT1G46768 | RAP2.1 (related to AP2 1); DNA binding / transcription factor | 3.74 |
| 312 | 262431_at | AT1G47540 | trypsin inhibitor, putative | 10.13 |
| 313 | 259615_at | AT1G47980 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G62730.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO44946.1) | 6.02 |
| 314 | 260716_at | AT1G48130 | ATPER1 (Arabidopsis thaliana 1-cysteine peroxiredoxin 1); antioxidant | 12.26 |
| 315 | 261305_at | AT1G48470 | GLN1;5 (GLUTAMINE SYNTHETASE 1;5); glutamate-ammonia ligase | 7.17 |
| 316 | 256139_at | AT1G48660 | auxin-responsive GH3 family protein | 7.96 |
| 317 | 245831_at | AT1G48840 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G18350.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO68070.1); similar to unknown protein [Oryza sativa (japonica cultivar-group)] (GB:AAT01350.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN84081.1); contains InterPro domain Protein of unknown function DUF639 (InterPro:IPR006927) | 3.98 |
| 318 | 246624_at | AT1G48910 | YUC10; monooxygenase | 2.83 |
| 319 | 262416_at | AT1G49390 | oxidoreductase, 2OG-Fe(II) oxygenase family protein | 4.46 |
| 320 | 261598_at | AT1G49750 | leucine-rich repeat family protein | -5.68 |
| 321 | 259809_at | AT1G49800 | unknown protein | 3.62 |
| 322 | 261640_at | AT1G49960 | xanthine/uracil permease family protein | 3.50 |
| 323 | 261638_at | AT1G49975 | similar to hypothetical protein [Vitis vinifera] (GB:CAN66219.1) | -2.96 |
| 324 | 262473_at | AT1G50250 | FTSH1 (FtsH protease 1); ATP-dependent peptidase/ ATPase/ metallopeptidase | -2.48 |
| 325 | 245744_at | AT1G51110 | plastid-lipid associated protein PAP / fibrillin family protein | -5.27 |
| 326 | 245745_at | AT1G51110 | plastid-lipid associated protein PAP / fibrillin family protein | -3.99 |
| 327 | 256176_at | AT1G51640 | ATEXO70G2 (exocyst subunit EXO70 family protein G2); protein binding | 2.91 |
| 328 | 256170_at | AT1G51790 | kinase | -2.40 |
| 329 | 256168_at | AT1G51805 | leucine-rich repeat protein kinase, putative | -3.76 |
| 330 | 246371_at | AT1G51940 | protein kinase family protein / peptidoglycan-binding LysM domain-containing protein | -3.06 |
| 331 | 265053_at | AT1G52000 | jacalin lectin family protein | -6.57 |
| 332 | 259839_at | AT1G52190 | proton-dependent oligopeptide transport (POT) family protein | -7.58 |
| 333 | 259841_at | AT1G52200 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G18470.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO45876.1); contains InterPro domain Protein of unknown function Cys-rich (InterPro:IPR006461) | -3.60 |
| 334 | 259838_at | AT1G52220 | similar to PSAP/PSI-P/PTAC8/TMP14 (THYLAKOID MEMBRANE PHOSPHOPROTEIN OF 14 KDA) [Arabidopsis thaliana] (TAIR:AT2G46820.2); similar to PSAP/PSI-P/PTAC8/TMP14 (THYLAKOID MEMBRANE PHOSPHOPROTEIN OF 14 KDA), DNA binding [Arabidopsis thaliana] (TAIR:AT2G46820.1); similar to unknown [Populus trichocarpa] (GB:ABK95860.1) | -3.42 |
| 335 | 259840_at | AT1G52230 | PSAH-2/PSAH2/PSI-H (PHOTOSYSTEM I SUBUNIT H-2) | -3.82 |
| 336 | 259611_at | AT1G52280 | AtRABG3d (Arabidopsis Rab GTPase homolog G3d); GTP binding | -2.96 |
| 337 | 259640_at | AT1G52400 | BGL1 (BETA-GLUCOSIDASE HOMOLOG 1); hydrolase, hydrolyzing O-glycosyl compounds | -5.05 |
| 338 | 259609_at | AT1G52410 | TSA1 (TSK-ASSOCIATING PROTEIN 1); calcium ion binding / protein binding | -3.09 |
| 339 | 262148_at | AT1G52560 | 26.5 kDa class I small heat shock protein-like (HSP26.5-P) | 7.21 |
| 340 | 262128_at | AT1G52690 | late embryogenesis abundant protein, putative / LEA protein, putative | 9.44 |
| 341 | 260146_at | AT1G52770 | phototropic-responsive NPH3 family protein | -3.22 |
| 342 | 260156_at | AT1G52880 | NAM (Arabidopsis NAC domain containing protein 18); transcription factor | 3.33 |
| 343 | 260203_at | AT1G52890 | ANAC019 (Arabidopsis NAC domain containing protein 19); transcription factor | 4.01 |
| 344 | 260145_at | AT1G52920 | GCR2/GPCR (G PROTEIN COUPLED RECEPTOR); abscisic acid binding/ catalytic | 3.13 |
| 345 | 261366_at | AT1G53100 | acetylglucosaminyltransferase | 3.49 |
| 346 | 260643_at | AT1G53270 | ABC transporter family protein | 6.92 |
| 347 | 260975_at | AT1G53430 | leucine-rich repeat family protein / protein kinase family protein | -4.27 |
| 348 | 259962_at | AT1G53690 | DNA-directed RNA polymerases I, II, and III 7 kDa subunit, putative | 4.18 |
| 349 | 263174_at | AT1G54040 | ESP (EPITHIOSPECIFIER PROTEIN) | -5.97 |
| 350 | 263150_at | AT1G54050 | 17.4 kDa class III heat shock protein (HSP17.4-CIII) | 3.07 |
| 351 | 263157_at | AT1G54100 | ALDH7B4 (ALDEHYDE DEHYDROGENASE 7B4); 3-chloroallyl aldehyde dehydrogenase | 2.95 |
| 352 | 263005_at | AT1G54540 | similar to harpin-induced protein-related / HIN1-related / harpin-responsive protein-related [Arabidopsis thaliana] (TAIR:AT1G65690.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO62044.1); contains InterPro domain Harpin-induced 1 (InterPro:IPR010847) | 5.40 |
| 353 | 264185_at | AT1G54780 | thylakoid lumen 18.3 kDa protein | -3.26 |
| 354 | 264240_at | AT1G54820 | protein kinase family protein | -3.15 |
| 355 | 264187_at | AT1G54860 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G19250.1); similar to GPI-anchored protein-like protein II [Cucumis melo] (GB:ABR67421.1) | 12.50 |
| 356 | 256354_at | AT1G54870 | oxidoreductase | 9.87 |
| 357 | 259663_at | AT1G55190 | prenylated rab acceptor (PRA1) family protein | 3.33 |
| 358 | 259660_at | AT1G55260 | lipid binding | -2.91 |
| 359 | 265075_at | AT1G55450 | embryo-abundant protein-related | -4.41 |
| 360 | 265073_at | AT1G55480 | binding / protein binding | -3.03 |
| 361 | 265076_at | AT1G55490 | CPN60B (CHAPERONIN 60 BETA); ATP binding / protein binding / unfolded protein binding | -4.25 |
| 362 | 262083_at | AT1G56100 | pectinesterase inhibitor domain-containing protein | 8.86 |
| 363 | 262098_at | AT1G56170 | HAP5B (Heme activator protein (yeast) homolog 5B); DNA binding / transcription factor | 3.37 |
| 364 | 256228_at | AT1G56190 | phosphoglycerate kinase, putative | -2.49 |
| 365 | 256217_at | AT1G56320 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G49350.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO44880.1) | 4.66 |
| 366 | 259603_at | AT1G56500 | haloacid dehalogenase-like hydrolase family protein | -2.96 |
| 367 | 245627_at | AT1G56600 | ATGOLS2 (ARABIDOPSIS THALIANA GALACTINOL SYNTHASE 2); transferase, transferring glycosyl groups / transferase, transferring hexosyl groups | 8.19 |
| 368 | 245828_at | AT1G57820 | ORTH2/VIM1 (VARIANT IN METHYLATION 1); DNA binding / chromatin binding / double-stranded methylated DNA binding / histone binding / methyl-CpG binding / methyl-CpNpG binding / methyl-CpNpN binding | -2.43 |
| 369 | 262072_at | AT1G59590 | ZCF37 | -3.42 |
| 370 | 262899_at | AT1G59870 | PDR8/PEN3 (PLEIOTROPIC DRUG RESISTANCE8); ATPase, coupled to transmembrane movement of substances / cadmium ion transmembrane transporter | -4.50 |
| 371 | 263679_at | AT1G59990 | DEAD/DEAH box helicase, putative (RH22) | -2.47 |
| 372 | 263737_at | AT1G60010 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G10530.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO40951.1) | -2.68 |
| 373 | 264931_at | AT1G60590 | polygalacturonase, putative / pectinase, putative | -2.72 |
| 374 | 264963_at | AT1G60600 | ABC4 (ABERRANT CHLOROPLAST DEVELOPMENT 4); 1,4-dihydroxy-2-naphthoate octaprenyltransferase/ prenyltransferase | -2.52 |
| 375 | 264941_at | AT1G60680 | AGD2 (ARF-GAP DOMAIN 2); aldo-keto reductase | 6.42 |
| 376 | 259728_at | AT1G60970 | clathrin adaptor complex small chain family protein | 5.31 |
| 377 | 264401_at | AT1G61720 | BAN (BANYULS) | 7.61 |
| 378 | 264433_at | AT1G61810 | BGLU45; hydrolase, hydrolyzing O-glycosyl compounds | 3.98 |
| 379 | 264310_at | AT1G62030 | DC1 domain-containing protein | -4.04 |
| 380 | 264740_at | AT1G62070 | unknown protein | 9.30 |
| 381 | 264737_at | AT1G62210 | unknown protein | 6.73 |
| 382 | 264741_at | AT1G62290 | aspartyl protease family protein | 7.91 |
| 383 | 265117_at | AT1G62500 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | 5.11 |
| 384 | 265111_at | AT1G62510 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | 7.42 |
| 385 | 262644_at | AT1G62710 | BETA-VPE (vacuolar processing enzyme beta); cysteine-type endopeptidase | 6.51 |
| 386 | 262645_at | AT1G62750 | ATSCO1/ATSCO1/CPEF-G/SCO1 (SNOWY COTYLEDON1); translation elongation factor/ translation factor, nucleic acid binding | -3.57 |
| 387 | 262693_at | AT1G62780 | similar to hypothetical protein [Vitis vinifera] (GB:CAN83165.1) | -3.72 |
| 388 | 261106_at | AT1G62990 | KNAT7 (Knotted-like Arabidopsis thaliana); DNA binding / transcription factor | 3.17 |
| 389 | 259694_at | AT1G63180 | UGE3 (UDP-D-GLUCOSE/UDP-D-GALACTOSE 4-EPIMERASE 3); UDP-glucose 4-epimerase/ protein dimerization | 3.20 |
| 390 | 260296_at | AT1G63750 | ATP binding / nucleoside-triphosphatase/ nucleotide binding / protein binding | -3.63 |
| 391 | 262347_at | AT1G64110 | AAA-type ATPase family protein | 5.69 |
| 392 | 259765_at | AT1G64370 | unknown protein | -6.81 |
| 393 | 261958_at | AT1G64500 | glutaredoxin family protein | -3.56 |
| 394 | 261954_at | AT1G64510 | ribosomal protein S6 family protein | -2.97 |
| 395 | 261975_at | AT1G64640 | plastocyanin-like domain-containing protein | -3.29 |
| 396 | 261957_at | AT1G64660 | ATMGL; catalytic/ methionine gamma-lyase | 4.39 |
| 397 | 262872_at | AT1G64690 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G11590.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO62905.1) | -2.89 |
| 398 | 262878_at | AT1G64770 | carbohydrate binding / catalytic | -2.34 |
| 399 | 262883_at | AT1G64780 | ATAMT1;2 (AMMONIUM TRANSPORTER 1;2); ammonium transmembrane transporter | -5.17 |
| 400 | 262879_at | AT1G64860 | SIGA (SIGMA FACTOR A); DNA binding / DNA-directed RNA polymerase/ transcription factor | -2.34 |
| 401 | 263138_at | AT1G65090 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G36100.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G36100.2); similar to hypothetical protein [Vitis vinifera] (GB:CAN81170.1) | 8.95 |
| 402 | 263142_at | AT1G65230 | similar to hypothetical protein [Vitis vinifera] (GB:CAN76393.1) | -3.62 |
| 403 | 264636_at | AT1G65490 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G65486.1) | -3.98 |
| 404 | 264634_at | AT1G65670 | CYP702A1 (CYTOCHROME P450, FAMILY 702, SUBFAMILY A, POLYPEPTIDE 1); oxygen binding | 6.83 |
| 405 | 261915_at | AT1G65880 | BZO1; benzoate-CoA ligase | 6.67 |
| 406 | 261921_at | AT1G65900 | similar to unnamed protein product [Vitis vinifera] (GB:CAO22185.1); contains domain Helical backbone metal receptor (SSF53807) | -2.71 |
| 407 | 261970_at | AT1G65960 | GAD2 (GLUTAMATE DECARBOXYLASE 2); calmodulin binding | -6.25 |
| 408 | 256516_at | AT1G66150 | TMK1 (TRANSMEMBRANE KINASE 1) | -2.99 |
| 409 | 260136_at | AT1G66360 | C2 domain-containing protein | 2.80 |
| 410 | 256359_at | AT1G66460 | protein kinase family protein | 4.87 |
| 411 | 256371_at | AT1G66770 | nodulin MtN3 family protein | 4.63 |
| 412 | 256366_at | AT1G66880 | serine/threonine protein kinase family protein | -5.25 |
| 413 | 255856_at | AT1G66940 | protein kinase-related | -2.63 |
| 414 | 255852_at | AT1G66970 | glycerophosphoryl diester phosphodiesterase family protein | -3.89 |
| 415 | 255913_at | AT1G66980 | protein kinase family protein / glycerophosphoryl diester phosphodiesterase family protein | -3.08 |
| 416 | 255854_at | AT1G67050 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G38320.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G38320.2); similar to At1g67050-like protein [Arabidopsis lyrata subsp. petraea] (GB:ABY59212.1) | -2.69 |
| 417 | 264469_at | AT1G67100 | LBD40 (LOB DOMAIN-CONTAINING PROTEIN 40) | 7.96 |
| 418 | 260201_at | AT1G67600 | similar to catalytic [Arabidopsis thaliana] (TAIR:AT1G24350.1); similar to unknown [Picea sitchensis] (GB:ABK26930.1); contains InterPro domain Acid phosphatase/vanadium-dependent haloperoxidase related (InterPro:IPR003832) | 3.46 |
| 419 | 245189_at | AT1G67670 | unknown protein | 3.32 |
| 420 | 245196_at | AT1G67750 | pectate lyase family protein | -3.62 |
| 421 | 245193_at | AT1G67810 | Fe-S metabolism associated domain-containing protein | 3.22 |
| 422 | 245194_at | AT1G67820 | protein phosphatase 2C, putative / PP2C, putative | 3.06 |
| 423 | 245201_at | AT1G67840 | ATP-binding region, ATPase-like domain-containing protein | -2.64 |
| 424 | 260012_at | AT1G67865 | unknown protein | -4.12 |
| 425 | 260007_at | AT1G67870 | glycine-rich protein | -7.29 |
| 426 | 259996_at | AT1G67910 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G24577.1); similar to unknown [Populus trichocarpa] (GB:ABK93048.1) | -3.55 |
| 427 | 260005_at | AT1G67920 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G24600.1) | 3.34 |
| 428 | 260014_at | AT1G68010 | HPR (HYDROXYPYRUVATE REDUCTASE); glycerate dehydrogenase/ poly(U) binding | -4.14 |
| 429 | 260433_at | AT1G68170 | nodulin MtN21 family protein | 6.74 |
| 430 | 260458_at | AT1G68250 | unknown protein | 3.07 |
| 431 | 260437_at | AT1G68380 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G68390.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO42175.1); contains InterPro domain Protein of unknown function DUF266, plant (InterPro:IPR004949) | 6.85 |
| 432 | 260261_at | AT1G68450 | VQ motif-containing protein | 3.30 |
| 433 | 260266_at | AT1G68520 | zinc finger (B-box type) family protein | -3.48 |
| 434 | 262230_at | AT1G68560 | ATXYL1/XYL1 (ALPHA-XYLOSIDASE 1); alpha-N-arabinofuranosidase/ hydrolase, hydrolyzing O-glycosyl compounds / xylan 1,4-beta-xylosidase | -3.71 |
| 435 | 262283_at | AT1G68590 | plastid-specific 30S ribosomal protein 3, putative / PSRP-3, putative | -3.66 |
| 436 | 262232_at | AT1G68600 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G25480.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G17470.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO42118.1); contains InterPro domain Protein of unknown function UPF0005 (InterPro:IPR006214) | -3.46 |
| 437 | 260035_at | AT1G68850 | peroxidase, putative | 7.28 |
| 438 | 259371_at | AT1G69080 | universal stress protein (USP) family protein | -2.54 |
| 439 | 259373_at | AT1G69160 | unknown protein | -3.71 |
| 440 | 260357_at | AT1G69260 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G13740.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO41856.1); contains InterPro domain Protein of unknown function DUF1675 (InterPro:IPR012463) | 4.06 |
| 441 | 260352_at | AT1G69295 | beta-1,3-glucanase-related | 3.54 |
| 442 | 256293_at | AT1G69440 | AGO7 (ARGONAUTE7); nucleic acid binding | -2.86 |
| 443 | 256303_at | AT1G69550 | disease resistance protein (TIR-NBS class), putative | -3.19 |
| 444 | 259831_at | AT1G69600 | ATHB29/ZFHD1 (ZINC FINGER HOMEODOMAIN 1); DNA binding / transcription activator/ transcription factor | 3.21 |
| 445 | 260371_at | AT1G69690 | TCP family transcription factor, putative | -2.63 |
| 446 | 260419_at | AT1G69730 | protein kinase family protein | -3.76 |
| 447 | 260395_at | AT1G69780 | ATHB13; DNA binding / transcription factor | -3.98 |
| 448 | 260413_at | AT1G69800 | CBS domain-containing protein | 3.83 |
| 449 | 264717_at | AT1G70140 | ATFH8 (FORMIN 8); actin binding / actin filament binding / profilin binding | 3.48 |
| 450 | 264696_at | AT1G70230 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G01430.1); similar to Os09g0375300 [Oryza sativa (japonica cultivar-group)] (GB:NP_001063036.1); similar to Os06g0235200 [Oryza sativa (japonica cultivar-group)] (GB:NP_001057240.1); similar to leaf senescence protein-like [Oryza sativa (japonica cultivar-group)] (GB:BAD26032.1); contains InterPro domain Protein of unknown function DUF231, plant (InterPro:IPR004253) | -4.76 |
| 451 | 264340_at | AT1G70280 | NHL repeat-containing protein | -3.41 |
| 452 | 264315_at | AT1G70370 | BURP domain-containing protein / polygalacturonase, putative | -2.83 |
| 453 | 264313_at | AT1G70410 | carbonic anhydrase, putative / carbonate dehydratase, putative | -4.44 |
| 454 | 260181_at | AT1G70710 | AtGH9B1 (ARABIDOPSIS THALIANA GLYCOSYL HYDROLASE 9B1); hydrolase, hydrolyzing O-glycosyl compounds | -2.39 |
| 455 | 260182_at | AT1G70750 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G16720.1); similar to unknown [Populus trichocarpa] (GB:ABK92464.1); similar to Os03g0259900 [Oryza sativa (japonica cultivar-group)] (GB:NP_001049616.1); contains InterPro domain Protein of unknown function DUF593 (InterPro:IPR007656) | -2.71 |
| 456 | 262288_at | AT1G70760 | inorganic carbon transport protein-related | -3.65 |
| 457 | 262309_at | AT1G70820 | phosphoglucomutase, putative / glucose phosphomutase, putative | -6.43 |
| 458 | 262312_at | AT1G70830 | MLP28 (MLP-LIKE PROTEIN 28) | -3.79 |
| 459 | 262310_at | AT1G70840 | MLP31 (MLP-LIKE PROTEIN 31) | 7.16 |
| 460 | 262304_at | AT1G70890 | MLP43 (MLP-LIKE PROTEIN 43) | -3.81 |
| 461 | 262263_at | AT1G70940 | PIN3 (PIN-FORMED 3); auxin:hydrogen symporter/ transporter | -2.71 |
| 462 | 262290_at | AT1G70985 | hydroxyproline-rich glycoprotein family protein | 7.73 |
| 463 | 259742_at | AT1G71120 | GLIP6 (GDSL-motif lipase/hydrolase 6); carboxylesterase | 4.29 |
| 464 | 259935_at | AT1G71250 | GDSL-motif lipase/hydrolase family protein | 4.84 |
| 465 | 259896_at | AT1G71500 | Rieske (2Fe-2S) domain-containing protein | -2.34 |
| 466 | 261503_at | AT1G71691 | GDSL-motif lipase/hydrolase family protein | 3.18 |
| 467 | 261505_at | AT1G71696 | SOL1 (suppressor of LLP1 1); carboxypeptidase A | -3.18 |
| 468 | 260143_at | AT1G71880 | SUC1 (SUCROSE-PROTON SYMPORTER 1); carbohydrate transmembrane transporter/ sucrose:hydrogen symporter/ sugar:hydrogen ion symporter | -6.83 |
| 469 | 256338_at | AT1G72100 | late embryogenesis abundant domain-containing protein / LEA domain-containing protein | 9.44 |
| 470 | 259848_at | AT1G72180 | leucine-rich repeat transmembrane protein kinase, putative | -2.52 |
| 471 | 259854_at | AT1G72200 | zinc finger (C3HC4-type RING finger) family protein | 4.11 |
| 472 | 259828_at | AT1G72220 | zinc finger (C3HC4-type RING finger) family protein | 5.22 |
| 473 | 259802_at | AT1G72260 | THI2.1 (THIONIN 2.1); toxin receptor binding | 3.50 |
| 474 | 259892_at | AT1G72610 | GLP1 (GERMIN-LIKE PROTEIN 1); manganese ion binding / metal ion binding / nutrient reservoir | -5.93 |
| 475 | 259913_at | AT1G72660 | developmentally regulated GTP-binding protein, putative | 3.07 |
| 476 | 259911_at | AT1G72680 | cinnamyl-alcohol dehydrogenase, putative | 4.43 |
| 477 | 262381_at | AT1G72900 | disease resistance protein (TIR-NBS class), putative | -2.68 |
| 478 | 262356_at | AT1G73000 | similar to Bet v I allergen family protein [Arabidopsis thaliana] (TAIR:AT2G26040.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO43790.1); contains InterPro domain Streptomyces cyclase/dehydrase (InterPro:IPR005031) | 4.17 |
| 479 | 262369_at | AT1G73010 | phosphoric monoester hydrolase | 4.94 |
| 480 | 262357_at | AT1G73040 | jacalin lectin family protein | -2.49 |
| 481 | 262377_at | AT1G73110 | ribulose bisphosphate carboxylase/oxygenase activase, putative / RuBisCO activase, putative | -4.63 |
| 482 | 260088_at | AT1G73190 | ALPHA-TIP/TIP3;1 (ALPHA-TONOPLAST INTRINSIC PROTEIN); water channel | 12.27 |
| 483 | 260097_at | AT1G73220 | ATOCT1 (ARABIDOPSIS THALIANA ORGANIC CATION/CARNITINE TRANSPORTER1); carbohydrate transmembrane transporter/ carnitine transporter/ transporter | 9.63 |
| 484 | 260101_at | AT1G73260 | trypsin and protease inhibitor family protein / Kunitz family protein | 3.21 |
| 485 | 260091_at | AT1G73290 | SCPL5 (serine carboxypeptidase-like 5); serine carboxypeptidase | 7.28 |
| 486 | 260389_at | AT1G74055 | unknown protein | -3.35 |
| 487 | 260388_at | AT1G74070 | peptidyl-prolyl cis-trans isomerase cyclophilin-type family protein | -2.58 |
| 488 | 260387_at | AT1G74100 | sulfotransferase family protein | -2.63 |
| 489 | 260248_at | AT1G74310 | ATHSP101 (HEAT SHOCK PROTEIN 101); ATP binding / ATPase | 6.01 |
| 490 | 260237_at | AT1G74430 | MYB95 (myb domain protein 95); DNA binding / transcription factor | -3.79 |
| 491 | 260234_at | AT1G74460 | GDSL-motif lipase/hydrolase family protein | 4.81 |
| 492 | 260230_at | AT1G74500 | bHLH family protein | 4.20 |
| 493 | 262168_at | AT1G74730 | similar to unknown [Populus trichocarpa x Populus deltoides] (GB:ABK96654.1); contains InterPro domain Protein of unknown function DUF1118 (InterPro:IPR009500) | -2.60 |
| 494 | 262175_at | AT1G74880 | NDH-O (NAD(P)H:PLASTOQUINONE DEHYDROGENASE COMPLEX SUBUNIT O) | -4.50 |
| 495 | 262172_at | AT1G74970 | RPS9 (RIBOSOMAL PROTEIN S9); structural constituent of ribosome | -3.37 |
| 496 | 259954_at | AT1G75130 | CYP721A1 (cytochrome P450, family 721, subfamily A, polypeptide 1); oxygen binding | -4.42 |
| 497 | 262977_at | AT1G75490 | DNA binding / transcription factor | 7.40 |
| 498 | 262951_at | AT1G75500 | nodulin MtN21 family protein | -4.42 |
| 499 | 257460_at | AT1G75580 | auxin-responsive protein, putative | 3.19 |
| 500 | 262970_at | AT1G75690 | chaperone protein dnaJ-related | -3.44 |
| 501 | 262978_at | AT1G75780 | TUB1 (tubulin beta-1 chain); structural molecule | -2.88 |
| 502 | 262728_at | AT1G75820 | CLV1 (CLAVATA 1); ATP binding / kinase/ protein serine/threonine kinase | -4.86 |
| 503 | 262679_at | AT1G75830 | LCR67/PDF1.1 (Low-molecular-weight cysteine-rich 67) | 12.07 |
| 504 | 262680_at | AT1G75880 | family II extracellular lipase 1 (EXL1) | -2.33 |
| 505 | 262681_at | AT1G75890 | family II extracellular lipase 2 (EXL2) | -2.93 |
| 506 | 261751_at | AT1G76080 | ATCDSP32/CDSP32 (CHLOROPLASTIC DROUGHT-INDUCED STRESS PROTEIN OF 32 KD); thiol-disulfide exchange intermediate | -4.16 |
| 507 | 261769_at | AT1G76100 | plastocyanin | -2.69 |
| 508 | 261777_at | AT1G76210 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G20520.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN69930.1); contains InterPro domain Protein of unknown function DUF241, plant (InterPro:IPR004320) | 2.81 |
| 509 | 261779_at | AT1G76230 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G20100.1) | 5.69 |
| 510 | 261752_at | AT1G76290 | AMP-dependent synthetase and ligase family protein | 4.12 |
| 511 | 259981_at | AT1G76450 | oxygen-evolving complex-related | -2.45 |
| 512 | 259980_at | AT1G76520 | auxin efflux carrier family protein | -2.43 |
| 513 | 259977_at | AT1G76590 | zinc-binding family protein | 3.85 |
| 514 | 259880_at | AT1G76730 | 5-formyltetrahydrofolate cyclo-ligase family protein | -2.95 |
| 515 | 259868_at | AT1G76760 | ATY1 (Arabidopsis thioredoxin y1); thiol-disulfide exchange intermediate | 3.02 |
| 516 | 264953_at | AT1G77120 | ADH1 (ALCOHOL DEHYDROGENASE 1) | 6.06 |
| 517 | 264477_at | AT1G77240 | AMP-binding protein, putative | 7.70 |
| 518 | 259705_at | AT1G77450 | ANAC032 (Arabidopsis NAC domain containing protein 32); transcription factor | 5.58 |
| 519 | 259680_at | AT1G77690 | amino acid permease, putative | -3.29 |
| 520 | 259676_at | AT1G77730 | pleckstrin homology (PH) domain-containing protein | 2.94 |
| 521 | 259681_at | AT1G77760 | NIA1 (NITRATE REDUCTASE 1) | -2.62 |
| 522 | 262185_at | AT1G77950 | AGL67; transcription factor | 10.42 |
| 523 | 262162_at | AT1G78020 | senescence-associated protein-related | -3.15 |
| 524 | 262181_at | AT1G78060 | glycosyl hydrolase family 3 protein | -3.74 |
| 525 | 260797_at | AT1G78390 | NCED9 (NINE-CIS-EPOXYCAROTENOID DIOXYGENASE 9) | 7.63 |
| 526 | 260804_at | AT1G78410 | VQ motif-containing protein | -3.03 |
| 527 | 260801_at | AT1G78430 | Identical to Uncharacterized protein At1g78430 [Arabidopsis Thaliana] (GB:Q9M9F9); similar to tropomyosin-related [Arabidopsis thaliana] (TAIR:AT1G17140.1); similar to tropomyosin-related [Arabidopsis thaliana] (TAIR:AT1G17140.2); similar to hypothetical protein [Vitis vinifera] (GB:CAN73256.1); contains InterPro domain Prefoldin (InterPro:IPR009053) | -3.70 |
| 528 | 263133_at | AT1G78450 | SOUL heme-binding family protein | -4.16 |
| 529 | 263126_at | AT1G78460 | SOUL heme-binding family protein | -2.75 |
| 530 | 263120_at | AT1G78490 | CYP708A3 (cytochrome P450, family 708, subfamily A, polypeptide 3); oxygen binding | -2.62 |
| 531 | 263123_at | AT1G78500 | pentacyclic triterpene synthase, putative | 6.63 |
| 532 | 264301_at | AT1G78780 | pathogenesis-related family protein | 3.36 |
| 533 | 264100_at | AT1G78970 | LUP1 (LUPEOL SYNTHASE 1); lupeol synthase | -2.82 |
| 534 | 264096_at | AT1G78995 | similar to unnamed protein product [Vitis vinifera] (GB:CAO41778.1) | -4.72 |
| 535 | 264143_at | AT1G79330 | AMC6/ATMC5/ATMCP2B (TYPE-II METACASPASES); caspase/ cysteine-type endopeptidase | 3.86 |
| 536 | 264124_at | AT1G79360 | ATOCT2 (ARABIDOPSIS THALIANA ORGANIC CATION/CARNITINE TRANSPORTER2); carbohydrate transmembrane transporter/ sugar:hydrogen ion symporter | 2.99 |
| 537 | 261399_at | AT1G79620 | leucine-rich repeat transmembrane protein kinase, putative | -3.73 |
| 538 | 260166_at | AT1G79840 | GL2 (GLABRA 2); DNA binding / transcription factor | 3.05 |
| 539 | 262069_at | AT1G80090 | CBS domain-containing protein | 7.12 |
| 540 | 262047_at | AT1G80160 | lactoylglutathione lyase family protein / glyoxalase I family protein | 6.62 |
| 541 | 262060_at | AT1G80170 | polygalacturonase, putative / pectinase, putative | -4.33 |
| 542 | 260299_at | AT1G80330 | ATGA3OX4 (GIBBERELLIN 3-OXIDASE 4); gibberellin 3-beta-dioxygenase | 3.51 |
| 543 | 260272_at | AT1G80570 | F-box family protein (FBL14) | 3.40 |
| 544 | 261899_at | AT1G80820 | CCR2 (CINNAMOYL COA REDUCTASE) | 3.13 |
| 545 | 266346_at | AT2G01430 | homeobox-leucine zipper protein 17 (HB-17) / HD-ZIP transcription factor 17 | 3.69 |
| 546 | 266344_at | AT2G01580 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G07510.2); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G07510.1); similar to unknown [Populus trichocarpa] (GB:ABK93765.1); contains InterPro domain Ankyrin (InterPro:IPR002110) | 4.61 |
| 547 | 265872_at | AT2G01670 | ATNUDT17 (Arabidopsis thaliana Nudix hydrolase homolog 17); hydrolase | -2.66 |
| 548 | 265869_at | AT2G01760 | ARR14 (ARABIDOPSIS RESPONSE REGULATOR 14); transcription factor/ two-component response regulator | -2.74 |
| 549 | 265863_at | AT2G01770 | VIT1 (VACUOLAR IRON TRANSPORTER 1); iron ion transmembrane transporter | 3.75 |
| 550 | 266141_at | AT2G02120 | LCR70/PDF2.1 (Low-molecular-weight cysteine-rich 70); protease inhibitor | 10.18 |
| 551 | 267218_at | AT2G02515 | unknown protein | 3.95 |
| 552 | 267267_at | AT2G02580 | CYP71B9 (cytochrome P450, family 71, subfamily B, polypeptide 9); oxygen binding | 4.16 |
| 553 | 267472_at | AT2G02850 | ARPN (PLANTACYANIN); copper ion binding | 4.39 |
| 554 | 266743_at | AT2G02990 | RNS1 (RIBONUCLEASE 1); endoribonuclease | 6.32 |
| 555 | 266708_at | AT2G03200 | aspartyl protease family protein | 3.43 |
| 556 | 266707_at | AT2G03310 | unknown protein | -4.49 |
| 557 | 265704_at | AT2G03420 | similar to expressed protein [Oryza sativa (japonica cultivar-group)] (GB:ABF94594.1); similar to hypothetical protein OsI_010237 [Oryza sativa (indica cultivar-group)] (GB:EAY89004.1) | -2.72 |
| 558 | 265714_at | AT2G03520 | ATUPS4 (ARABIDOPSIS THALIANA UREIDE PERMEASE 4) | 7.26 |
| 559 | 263410_at | AT2G04039 | similar to unnamed protein product [Vitis vinifera] (GB:CAO41070.1) | -2.48 |
| 560 | 263403_at | AT2G04040 | ATDTX1; antiporter/ multidrug efflux pump/ multidrug transporter/ transporter | 4.21 |
| 561 | 263409_at | AT2G04063 | glycine-rich protein | 3.65 |
| 562 | 263809_at | AT2G04570 | GDSL-motif lipase/hydrolase family protein | -4.63 |
| 563 | 263046_at | AT2G05380 | GRP3S (GLYCINE-RICH PROTEIN 3 SHORT ISOFORM) | -7.49 |
| 564 | 265560_at | AT2G05520 | GRP-3 (GLYCINE-RICH PROTEIN 3) | -5.96 |
| 565 | 265511_at | AT2G05540 | glycine-rich protein | 3.55 |
| 566 | 265567_at | AT2G05580 | pseudogene, glycine-rich protein | 8.94 |
| 567 | 266021_at | AT2G05910 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G20640.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN71378.1); contains InterPro domain Protein of unknown function DUF567 (InterPro:IPR007612) | 6.09 |
| 568 | 266034_at | AT2G06005 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G20580.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G20580.2); similar to unknown [Populus trichocarpa] (GB:ABK93352.1) | 3.49 |
| 569 | 265530_at | AT2G06050 | OPR3 (OPDA-REDUCTASE 3); 12-oxophytodienoate reductase | -3.23 |
| 570 | 263769_at | AT2G06390 | transposable element gene | 2.91 |
| 571 | 265374_at | AT2G06520 | PSBX (photosystem II subunit X) | -2.37 |
| 572 | 266215_at | AT2G06850 | EXGT-A1 (ENDOXYLOGLUCAN TRANSFERASE); hydrolase, acting on glycosyl bonds | -5.55 |
| 573 | 266428_at | AT2G07180 | protein kinase, putative | -3.18 |
| 574 | 265400_at | AT2G10940 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | -4.48 |
| 575 | 265362_at | AT2G13230 | transposable element gene | 4.20 |
| 576 | 265366_at | AT2G13290 | glycosyl transferase family 17 protein | 2.89 |
| 577 | 263350_at | AT2G13360 | AGT (ALANINE:GLYOXYLATE AMINOTRANSFERASE) | -3.06 |
| 578 | 265656_at | AT2G13820 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | -3.23 |
| 579 | 263282_at | AT2G14095 | unknown protein | 4.03 |
| 580 | 265828_at | AT2G14520 | CBS domain-containing protein | 4.75 |
| 581 | 265891_at | AT2G15010 | thionin, putative | 9.28 |
| 582 | 265894_at | AT2G15050 | LTP; lipid binding | -3.04 |
| 583 | 263297_at | AT2G15310 | ATARFB1A (ADP-ribosylation factor B1A); GTP binding | -3.72 |
| 584 | 265472_at | AT2G15580 | zinc finger (C3HC4-type RING finger) family protein | -4.73 |
| 585 | 265475_at | AT2G15620 | NIR1 (NITRITE REDUCTASE); ferredoxin-nitrate reductase | -2.84 |
| 586 | 265480_at | AT2G15970 | COR413-PM1 (cold regulated 413 plasma membrane 1) | -2.88 |
| 587 | 263096_at | AT2G16060 | AHB1 (ARABIDOPSIS HEMOGLOBIN 1) | 3.09 |
| 588 | 263553_at | AT2G16430 | ATPAP10/PAP10; protein serine/threonine phosphatase | -3.67 |
| 589 | 263612_at | AT2G16440 | DNA replication licensing factor, putative | -2.73 |
| 590 | 265355_at | AT2G16760 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G47370.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO40115.1); contains InterPro domain Six-bladed beta-propeller, TolB-like (InterPro:IPR011042) | 3.22 |
| 591 | 266532_at | AT2G16890 | UDP-glucoronosyl/UDP-glucosyl transferase family protein | 3.35 |
| 592 | 264850_at | AT2G17340 | pantothenate kinase-related | -2.61 |
| 593 | 264590_at | AT2G17710 | similar to unnamed protein product [Vitis vinifera] (GB:CAO42932.1) | 3.23 |
| 594 | 264846_at | AT2G17850 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G66170.2); similar to unnamed protein product [Vitis vinifera] (GB:CAO48196.1); contains InterPro domain Rhodanese-like (InterPro:IPR001763) | 3.73 |
| 595 | 265342_at | AT2G18300 | basic helix-loop-helix (bHLH) family protein | -4.39 |
| 596 | 265379_at | AT2G18340 | late embryogenesis abundant domain-containing protein / LEA domain-containing protein | 5.62 |
| 597 | 265341_at | AT2G18360 | hydrolase, alpha/beta fold family protein | 6.29 |
| 598 | 265334_at | AT2G18370 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | 7.35 |
| 599 | 265922_at | AT2G18480 | mannitol transporter, putative | 4.00 |
| 600 | 265949_at | AT2G18540 | cupin family protein | 5.48 |
| 601 | 267321_at | AT2G19320 | unknown protein | 10.24 |
| 602 | 265943_at | AT2G19570 | CDA1 (CYTIDINE DEAMINASE 1) | 3.48 |
| 603 | 266687_at | AT2G19670 | protein arginine N-methyltransferase, putative | -2.55 |
| 604 | 266690_at | AT2G19900 | ATNADP-ME1 (NADP-MALIC ENZYME 1); malate dehydrogenase (oxaloacetate-decarboxylating) (NADP+)/ malic enzyme/ oxidoreductase, acting on NADH or NADPH, NAD or NADP as acceptor | 7.09 |
| 605 | 266689_at | AT2G19930 | RNA-dependent RNA polymerase family protein | 4.46 |
| 606 | 265257_at | AT2G20420 | succinyl-CoA ligase (GDP-forming) beta-chain, mitochondrial, putative / succinyl-CoA synthetase, beta chain, putative / SCS-beta, putative | -2.33 |
| 607 | 263369_at | AT2G20480 | similar to unknown [Populus trichocarpa] (GB:ABK93010.1); similar to hypothetical protein OsI_030526 [Oryza sativa (indica cultivar-group)] (GB:EAZ09294.1); similar to Os09g0446000 [Oryza sativa (japonica cultivar-group)] (GB:NP_001063306.1) | -3.13 |
| 608 | 265425_at | AT2G20770 | GCL2 (GCR2-LIKE 2); catalytic | 4.14 |
| 609 | 265440_at | AT2G20960 | pEARLI4 | -2.36 |
| 610 | 264010_at | AT2G21100 | disease resistance-responsive protein-related / dirigent protein-related | 4.29 |
| 611 | 264007_at | AT2G21140 | ATPRP2 (PROLINE-RICH PROTEIN 2) | -3.66 |
| 612 | 264024_at | AT2G21180 | similar to Os03g0249700 [Oryza sativa (japonica cultivar-group)] (GB:NP_001049562.1) | 3.42 |
| 613 | 264014_at | AT2G21210 | auxin-responsive protein, putative | -4.82 |
| 614 | 263760_at | AT2G21280 | GC1 (GIANT CHLOROPLAST 1); binding / catalytic/ coenzyme binding | -4.43 |
| 615 | 263761_at | AT2G21330 | fructose-bisphosphate aldolase, putative | -3.12 |
| 616 | 263773_at | AT2G21370 | xylulose kinase, putative | -4.57 |
| 617 | 263753_at | AT2G21490 | LEA (DEHYDRIN LEA) | 10.25 |
| 618 | 263754_at | AT2G21510 | DNAJ heat shock N-terminal domain-containing protein | 2.81 |
| 619 | 263765_at | AT2G21540 | SEC14 cytosolic factor, putative / phosphoglyceride transfer protein, putative | -2.83 |
| 620 | 263549_at | AT2G21650 | MEE3 (maternal effect embryo arrest 3); DNA binding / transcription factor | -4.02 |
| 621 | 263548_at | AT2G21660 | ATGRP7 (COLD, CIRCADIAN RHYTHM, AND RNA BINDING 2); RNA binding / double-stranded DNA binding / single-stranded DNA binding | -3.51 |
| 622 | 263877_at | AT2G21780 | unknown protein | 2.96 |
| 623 | 263881_at | AT2G21820 | similar to hypothetical protein MtrDRAFT_AC155884g16v2 [Medicago truncatula] (GB:ABN08202.1) | 10.23 |
| 624 | 257432_at | AT2G21850 | protein binding / zinc ion binding | -2.90 |
| 625 | 263433_at | AT2G22240 | inositol-3-phosphate synthase isozyme 2 / myo-inositol-1-phosphate synthase 2 / MI-1-P synthase 2 / IPS 2 | 3.74 |
| 626 | 264044_at | AT2G22480 | phosphofructokinase family protein | -3.83 |
| 627 | 263998_at | AT2G22510 | hydroxyproline-rich glycoprotein family protein | 6.85 |
| 628 | 264054_at | AT2G22540 | SVP (SHORT VEGETATIVE PHASE); transcription factor | -3.87 |
| 629 | 265350_at | AT2G22620 | lyase | 3.24 |
| 630 | 266797_at | AT2G22840 | AtGRF1 (GROWTH-REGULATING FACTOR 1) | 2.91 |
| 631 | 267265_at | AT2G22980 | SCPL13; serine carboxypeptidase | -4.48 |
| 632 | 267263_at | AT2G23110 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G23120.1); similar to seed maturation protein PM35 [Glycine max] (GB:AAD51623.1) | 9.30 |
| 633 | 245074_at | AT2G23200 | protein kinase family protein | -3.06 |
| 634 | 245070_at | AT2G23240 | plant EC metallothionein-like family 15 protein | 9.26 |
| 635 | 245068_at | AT2G23260 | UGT84B1 (UDP-GLUCOSYL TRANSFERASE 84B1); UDP-glycosyltransferase/ abscisic acid glucosyltransferase/ indole-3-acetate beta-glucosyltransferase/ transferase, transferring glycosyl groups | 3.81 |
| 636 | 267137_at | AT2G23410 | ACPT (ARABIDOPSIS CIS-PRENYLTRANSFERASE); dehydrodolichyl diphosphate synthase | 4.56 |
| 637 | 267150_at | AT2G23510 | transferase family protein | 7.84 |
| 638 | 267121_at | AT2G23540 | GDSL-motif lipase/hydrolase family protein | 8.20 |
| 639 | 267125_at | AT2G23580 | hydrolase, alpha/beta fold family protein | 4.85 |
| 640 | 267286_at | AT2G23640 | reticulon family protein (RTNLB13) | 8.03 |
| 641 | 267288_at | AT2G23680 | stress-responsive protein, putative | -2.83 |
| 642 | 267295_at | AT2G23800 | GGPS2 (GERANYLGERANYL PYROPHOSPHATE SYNTHASE 2); farnesyltranstransferase | 3.07 |
| 643 | 266572_at | AT2G23840 | HNH endonuclease domain-containing protein | -3.10 |
| 644 | 265682_at | AT2G24390 | similar to avirulence-responsive protein-related / avirulence induced gene (AIG) protein-related [Arabidopsis thaliana] (TAIR:AT4G31310.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO43968.1); contains InterPro domain Butirosin biosynthesis, BtrG-like (InterPro:IPR013024); contains InterPro domain AIG2-like (InterPro:IPR009288) | -3.46 |
| 645 | 265685_at | AT2G24430 | ANAC038/ANAC039; transcription factor | 4.74 |
| 646 | 263537_at | AT2G24790 | COL3 (CONSTANS-LIKE 3); protein binding / transcription factor/ zinc ion binding | -2.82 |
| 647 | 263587_at | AT2G25340 | ATVAMP712 (Arabidopsis thaliana vesicle-associated membrane protein 712) | 5.32 |
| 648 | 265611_at | AT2G25510 | unknown protein | -5.88 |
| 649 | 265605_at | AT2G25540 | CESA10 (CELLULOSE SYNTHASE 10); transferase, transferring glycosyl groups | 3.02 |
| 650 | 265886_at | AT2G25620 | protein phosphatase 2C, putative / PP2C, putative | 2.94 |
| 651 | 265913_at | AT2G25625 | similar to unnamed protein product [Vitis vinifera] (GB:CAO44157.1) | 3.54 |
| 652 | 266658_at | AT2G25735 | unknown protein | -3.64 |
| 653 | 266662_at | AT2G25830 | YebC-related | -2.38 |
| 654 | 266648_at | AT2G25840 | OVA4 (OVULE ABORTION 4); ATP binding / aminoacyl-tRNA ligase | -2.33 |
| 655 | 266654_at | AT2G25890 | glycine-rich protein / oleosin | 11.48 |
| 656 | 266849_at | AT2G25940 | ALPHA-VPE (ALPHA-VACUOLAR PROCESSING ENZYME); cysteine-type endopeptidase | 7.08 |
| 657 | 257365_x_at | AT2G26020 | PDF1.2b (plant defensin 1.2b) | 3.22 |
| 658 | 266892_at | AT2G26080 | ATGLDP2 (ARABIDOPSIS THALIANA GLYCINE DECARBOXYLASE P-PROTEIN 2); glycine dehydrogenase (decarboxylating) | -2.86 |
| 659 | 266841_at | AT2G26150 | ATHSFA2 (Arabidopsis thaliana heat shock transcription factor A2); DNA binding / transcription factor | 5.00 |
| 660 | 245044_at | AT2G26500 | cytochrome b6f complex subunit (petM), putative | -2.53 |
| 661 | 245038_at | AT2G26560 | PLP2 (PHOSPHOLIPASE A 2A); nutrient reservoir | -3.48 |
| 662 | 267610_at | AT2G26650 | AKT1 (ARABIDOPSIS K TRANSPORTER 1); cyclic nucleotide binding / inward rectifier potassium channel | 4.66 |
| 663 | 267612_at | AT2G26690 | nitrate transporter (NTP2) | -3.11 |
| 664 | 267614_at | AT2G26710 | BAS1/CYP734A1 (PHYB ACTIVATION TAGGED SUPPRESSOR 1); oxygen binding / steroid hydroxylase | -3.46 |
| 665 | 267618_at | AT2G26760 | CYCB1;4; cyclin-dependent protein kinase regulator | -3.79 |
| 666 | 266857_at | AT2G26900 | bile acid:sodium symporter family protein | -3.05 |
| 667 | 263083_at | AT2G27190 | PAP1 (PURPLE ACID PHOSPHATASE 1); acid phosphatase/ protein serine/threonine phosphatase | -3.83 |
| 668 | 265625_at | AT2G27240 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G08440.1); similar to putative aluminum activated malate transporter [Brassica napus] (GB:BAE97280.1); contains InterPro domain Protein of unknown function UPF0005 (InterPro:IPR006214) | 3.08 |
| 669 | 265645_at | AT2G27370 | integral membrane family protein | 4.90 |
| 670 | 265644_at | AT2G27380 | ATEPR1 (Arabidopsis thaliana extensin proline-rich 1) | 9.80 |
| 671 | 266246_at | AT2G27690 | CYP94C1 (cytochrome P450, family 94, subfamily C, polypeptide 1); oxygen binding | 2.99 |
| 672 | 266244_at | AT2G27740 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G36410.2); similar to unnamed protein product [Vitis vinifera] (GB:CAO16676.1); contains InterPro domain Protein of unknown function DUF662 (InterPro:IPR007033) | -3.65 |
| 673 | 264066_at | AT2G27880 | argonaute protein, putative / AGO, putative | 3.17 |
| 674 | 257429_at | AT2G27940 | zinc finger (C3HC4-type RING finger) family protein | 4.34 |
| 675 | 265570_at | AT2G28310 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G08040.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G08040.2); similar to unnamed protein product [Vitis vinifera] (GB:CAO21860.1); contains InterPro domain Protein of unknown function DUF707 (InterPro:IPR007877) | -2.60 |
| 676 | 265276_at | AT2G28400 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G60680.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO21845.1); contains InterPro domain Protein of unknown function DUF584 (InterPro:IPR007608) | 3.66 |
| 677 | 265255_at | AT2G28420 | lactoylglutathione lyase family protein / glyoxalase I family protein | 7.78 |
| 678 | 264079_at | AT2G28490 | cupin family protein | 11.22 |
| 679 | 264057_at | AT2G28550 | RAP2.7/TOE1 (TARGET OF EAT1 1); DNA binding / transcription factor | -4.14 |
| 680 | 263442_at | AT2G28605 | Identical to PsbP-related thylakoid lumenal protein 3, chloroplast precursor [Arabidopsis Thaliana] (GB:Q8VY52;GB:P83050;GB:Q8S8B1); similar to unnamed protein product [Vitis vinifera] (GB:CAO71682.1); contains InterPro domain Mog1/PsbP/DUF1795, alpha/beta/alpha sandwich (InterPro:IPR016124); contains InterPro domain Mog1/PsbP, alpha/beta/alpha sandwich (InterPro:IPR016123) | -2.56 |
| 681 | 263439_at | AT2G28650 | ATEXO70H8 (exocyst subunit EXO70 family protein H8); protein binding | 4.16 |
| 682 | 266222_at | AT2G28780 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G09450.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO21693.1) | 5.97 |
| 683 | 266229_at | AT2G28840 | ankyrin repeat family protein | -2.52 |
| 684 | 266790_at | AT2G28950 | ATEXPA6 (ARABIDOPSIS THALIANA EXPANSIN A6) | -4.90 |
| 685 | 266778_at | AT2G29090 | CYP707A2 (cytochrome P450, family 707, subfamily A, polypeptide 2); oxygen binding | 3.07 |
| 686 | 266285_at | AT2G29180 | similar to 80C09_19 [Brassica rapa subsp. pekinensis] (GB:AAZ41830.1) | -2.95 |
| 687 | 266280_at | AT2G29260 | tropinone reductase, putative / tropine dehydrogenase, putative | -2.99 |
| 688 | 266279_at | AT2G29290 | tropinone reductase, putative / tropine dehydrogenase, putative | -3.08 |
| 689 | 266278_at | AT2G29300 | tropinone reductase, putative / tropine dehydrogenase, putative | 5.09 |
| 690 | 266277_at | AT2G29310 | tropinone reductase, putative / tropine dehydrogenase, putative | -2.71 |
| 691 | 266291_at | AT2G29320 | tropinone reductase, putative / tropine dehydrogenase, putative | -2.56 |
| 692 | 266293_at | AT2G29360 | tropinone reductase, putative / tropine dehydrogenase, putative | -3.47 |
| 693 | 266274_at | AT2G29380 | protein phosphatase 2C, putative / PP2C, putative | 4.18 |
| 694 | 266267_at | AT2G29460 | ATGSTU4 (GLUTATHIONE S-TRANSFERASE 22); glutathione transferase | 4.40 |
| 695 | 266270_at | AT2G29470 | ATGSTU3 (GLUTATHIONE S-TRANSFERASE 21); glutathione transferase | 2.94 |
| 696 | 266297_at | AT2G29570 | PCNA2 (PROLIFERATING CELL NUCLEAR ANTIGEN 2); DNA binding / DNA polymerase processivity factor | -2.48 |
| 697 | 266673_at | AT2G29630 | thiamine biosynthesis family protein / thiC family protein | -3.47 |
| 698 | 266866_at | AT2G29940 | ATPDR3/PDR3 (PLEIOTROPIC DRUG RESISTANCE 3); ATPase, coupled to transmembrane movement of substances | 4.76 |
| 699 | 266805_at | AT2G30010 | similar to PMR5 (POWDERY MILDEW RESISTANT 5) [Arabidopsis thaliana] (TAIR:AT5G58600.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO21434.1); contains InterPro domain Protein of unknown function DUF231, plant (InterPro:IPR004253) | -2.53 |
| 700 | 267276_at | AT2G30130 | ASL5 (phosphoenolpyruvate carboxykinase1) | 5.08 |
| 701 | 267299_at | AT2G30150 | UDP-glucoronosyl/UDP-glucosyl transferase family protein | -4.46 |
| 702 | 255862_at | AT2G30300 | nodulin-related | 3.09 |
| 703 | 255873_at | AT2G30340 | LBD13 (LOB DOMAIN-CONTAINING PROTEIN 13) | 4.53 |
| 704 | 267516_at | AT2G30520 | RPT2 (ROOT PHOTOTROPISM 2); protein binding | -4.03 |
| 705 | 267517_at | AT2G30520 | RPT2 (ROOT PHOTOTROPISM 2); protein binding | -5.11 |
| 706 | 267497_at | AT2G30540 | glutaredoxin family protein | -4.08 |
| 707 | 267198_at | AT2G30810 | gibberellin-regulated family protein | 4.27 |
| 708 | 267154_at | AT2G30870 | ATGSTF10 (EARLY DEHYDRATION-INDUCED 13); glutathione transferase | -5.18 |
| 709 | 267152_at | AT2G31040 | ATP synthase protein I -related | -2.43 |
| 710 | 266469_at | AT2G31180 | AtMYB14/Myb14at (myb domain protein 14); DNA binding / transcription factor | 3.20 |
| 711 | 263473_at | AT2G31750 | UGT74D1 (UDP-GLUCOSYL TRANSFERASE 74D1); UDP-glycosyltransferase/ abscisic acid glucosyltransferase/ transferase, transferring glycosyl groups / transferase, transferring hexosyl groups | -2.87 |
| 712 | 263477_at | AT2G31790 | UDP-glucoronosyl/UDP-glucosyl transferase family protein | -3.65 |
| 713 | 263478_at | AT2G31880 | leucine-rich repeat transmembrane protein kinase, putative | -2.69 |
| 714 | 263475_at | AT2G31945 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G05575.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO22015.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN61524.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN71356.1) | 5.50 |
| 715 | 265672_at | AT2G31980 | cysteine proteinase inhibitor-related | 6.27 |
| 716 | 265728_at | AT2G31990 | exostosin family protein | 3.65 |
| 717 | 265675_at | AT2G32120 | HSP70T-2; ATP binding | 4.47 |
| 718 | 267548_at | AT2G32660 | disease resistance family protein / LRR family protein | 3.11 |
| 719 | 267545_at | AT2G32690 | pseudogene, glycine-rich protein | -4.82 |
| 720 | 245161_at | AT2G33070 | jacalin lectin family protein | 7.54 |
| 721 | 245164_at | AT2G33210 | chaperonin, putative | -2.70 |
| 722 | 255793_at | AT2G33250 | similar to unnamed protein product [Vitis vinifera] (GB:CAO71510.1) | -3.58 |
| 723 | 255840_at | AT2G33520 | similar to proline-rich family protein [Arabidopsis thaliana] (TAIR:AT1G12810.1) | 8.18 |
| 724 | 255842_at | AT2G33530 | SCPL46 (serine carboxypeptidase-like 46); serine carboxypeptidase | -3.89 |
| 725 | 255787_at | AT2G33590 | cinnamoyl-CoA reductase family | 5.07 |
| 726 | 267460_at | AT2G33810 | SPL3 (SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 3); transcription factor | -4.81 |
| 727 | 267041_at | AT2G34315 | disease resistance protein-related | 6.47 |
| 728 | 266899_at | AT2G34620 | mitochondrial transcription termination factor-related / mTERF-related | -3.88 |
| 729 | 267317_at | AT2G34700 | pollen Ole e 1 allergen and extensin family protein | 8.81 |
| 730 | 267313_at | AT2G34740 | protein phosphatase 2C, putative / PP2C, putative | 6.37 |
| 731 | 267414_at | AT2G34790 | EDA28/MEE23 (MATERNAL EFFECT EMBRYO ARREST 23); electron carrier | 4.21 |
| 732 | 267425_at | AT2G34810 | FAD-binding domain-containing protein | -3.52 |
| 733 | 267430_at | AT2G34860 | EDA3 (embryo sac development arrest 3); heat shock protein binding / unfolded protein binding | -2.64 |
| 734 | 267431_at | AT2G34870 | MEE26 (maternal effect embryo arrest 26) | 7.39 |
| 735 | 266489_at | AT2G35190 | NPSN11 (NOVEL PLANT SNARE 11); protein transporter | -5.65 |
| 736 | 266551_at | AT2G35260 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G17840.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN74782.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO63413.1) | -3.30 |
| 737 | 266544_at | AT2G35300 | late embryogenesis abundant group 1 domain-containing protein / LEA group 1 domain-containing protein | 12.30 |
| 738 | 266636_at | AT2G35370 | GDCH (Glycine decarboxylase complex H) | -8.55 |
| 739 | 266608_at | AT2G35500 | shikimate kinase-related | -3.66 |
| 740 | 265824_at | AT2G35650 | ATCSLA07 (CELLULOSE SYNTHASE LIKE); transferase, transferring glycosyl groups | -3.40 |
| 741 | 265796_at | AT2G35730 | heavy-metal-associated domain-containing protein | 3.11 |
| 742 | 263935_at | AT2G35930 | U-box domain-containing protein | -2.69 |
| 743 | 263951_at | AT2G35960 | NHL12 (NDR1/HIN1-like 12) | -3.78 |
| 744 | 263285_at | AT2G36120 | pseudogene, glycine-rich protein | -3.00 |
| 745 | 263959_at | AT2G36210 | auxin-responsive family protein | 5.00 |
| 746 | 263931_at | AT2G36220 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G52710.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO15901.1) | 3.00 |
| 747 | 263902_at | AT2G36230 | APG10 (ALBINO AND PALE GREEN 10); 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino]imidazole-4-carboxamide isomerase | -2.49 |
| 748 | 263907_at | AT2G36270 | ABI5 (ABA INSENSITIVE 5); DNA binding / transcription activator/ transcription factor | 5.95 |
| 749 | 263962_at | AT2G36350 | protein kinase, putative | 2.81 |
| 750 | 263915_at | AT2G36430 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G22540.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO15956.1); contains InterPro domain Protein of unknown function DUF247, plant (InterPro:IPR004158) | -3.81 |
| 751 | 265211_at | AT2G36640 | ATECP63 (EMBRYONIC CELL PROTEIN 63) | 11.96 |
| 752 | 265198_at | AT2G36760 | UGT73C2 (UDP-GLUCOSYL TRANSFERASE 73C2); UDP-glycosyltransferase/ transferase, transferring glycosyl groups / transferase, transferring hexosyl groups | 2.82 |
| 753 | 263867_at | AT2G36830 | GAMMA-TIP (Tonoplast intrinsic protein (TIP) gamma); water channel | -5.78 |
| 754 | 263846_at | AT2G36990 | SIGF (RNA POLYMERASE SIGMA-70 FACTOR); DNA binding / DNA-directed RNA polymerase/ transcription factor | -3.52 |
| 755 | 266006_at | AT2G37360 | ABC transporter family protein | 3.79 |
| 756 | 265967_at | AT2G37450 | nodulin MtN21 family protein | -4.14 |
| 757 | 265962_at | AT2G37460 | nodulin MtN21 family protein | 3.34 |
| 758 | 265965_at | AT2G37500 | arginine biosynthesis protein ArgJ family | -2.34 |
| 759 | 267158_at | AT2G37640 | ATEXPA3 (ARABIDOPSIS THALIANA EXPANSIN A3) | -5.11 |
| 760 | 267172_at | AT2G37660 | binding / catalytic/ coenzyme binding | -4.00 |
| 761 | 267160_at | AT2G37670 | WD-40 repeat family protein | 3.34 |
| 762 | 267178_at | AT2G37750 | unknown protein | 2.92 |
| 763 | 266097_at | AT2G37970 | SOUL-1; binding | 3.64 |
| 764 | 267094_at | AT2G38080 | IRX12/LAC4 (laccase 4); copper ion binding / oxidoreductase | -3.21 |
| 765 | 267093_at | AT2G38170 | CAX1 (CATION EXCHANGER 1); calcium ion transmembrane transporter/ calcium:hydrogen antiporter | -3.09 |
| 766 | 267147_at | AT2G38240 | oxidoreductase, 2OG-Fe(II) oxygenase family protein | 3.18 |
| 767 | 267142_at | AT2G38290 | ATAMT2 (AMMONIUM TRANSPORTER 2); ammonium transmembrane transporter | -3.18 |
| 768 | 267026_at | AT2G38340 | AP2 domain-containing transcription factor, putative (DRE2B) | 6.99 |
| 769 | 267035_at | AT2G38400 | AGT3 (ALANINE:GLYOXYLATE AMINOTRANSFERASE 3); alanine-glyoxylate transaminase | 3.56 |
| 770 | 267036_at | AT2G38465 | similar to unnamed protein product [Vitis vinifera] (GB:CAO61577.1) | 5.31 |
| 771 | 267032_at | AT2G38490 | CIPK22 (CBL-INTERACTING PROTEIN KINASE 22); kinase | 3.25 |
| 772 | 266415_at | AT2G38530 | LTP2 (LIPID TRANSFER PROTEIN 2); lipid binding | 4.97 |
| 773 | 266421_at | AT2G38540 | LP1 (nonspecific lipid transfer protein 1) | -3.89 |
| 774 | 266418_at | AT2G38750 | ANNAT4 (ANNEXIN ARABIDOPSIS 4); calcium ion binding / calcium-dependent phospholipid binding | -2.38 |
| 775 | 263265_at | AT2G38820 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G22970.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO22614.1); contains InterPro domain Protein of unknown function DUF506, plant (InterPro:IPR006502) | 3.12 |
| 776 | 266168_at | AT2G38870 | protease inhibitor, putative | 3.99 |
| 777 | 266169_at | AT2G38900 | serine protease inhibitor, potato inhibitor I-type family protein | 11.05 |
| 778 | 266143_at | AT2G38905 | hydrophobic protein, putative / low temperature and salt responsive protein, putative | 8.68 |
| 779 | 266200_at | AT2G38920 | SPX (SYG1/Pho81/XPR1) domain-containing protein / zinc finger (C3HC4-type RING finger) protein-related | 4.61 |
| 780 | 266172_at | AT2G39010 | PIP2;6/PIP2E (plasma membrane intrinsic protein 2;6); water channel | -4.25 |
| 781 | 266192_at | AT2G39080 | binding / catalytic | -2.65 |
| 782 | 266993_at | AT2G39210 | nodulin family protein | -2.71 |
| 783 | 266987_at | AT2G39280 | RAB GTPase activator | -2.53 |
| 784 | 266979_at | AT2G39470 | PPL2 (PSBP-LIKE PROTEIN 2); calcium ion binding | -3.44 |
| 785 | 245061_at | AT2G39730 | RCA (RUBISCO ACTIVASE) | -4.58 |
| 786 | 245088_at | AT2G39850 | subtilase | -4.27 |
| 787 | 267355_at | AT2G39900 | LIM domain-containing protein | -2.70 |
| 788 | 263385_at | AT2G40170 | ATEM6/GEA6 (ARABIDOPSIS EARLY METHIONINE-LABELLED 6) | 12.82 |
| 789 | 263377_at | AT2G40220 | ABI4 (ABA INSENSITIVE 4); DNA binding / transcription factor | 2.96 |
| 790 | 263825_at | AT2G40370 | LAC5 (laccase 5); copper ion binding / oxidoreductase | 4.40 |
| 791 | 255877_at | AT2G40460 | proton-dependent oligopeptide transport (POT) family protein | 2.88 |
| 792 | 255823_at | AT2G40470 | LBD15 (LOB DOMAIN-CONTAINING PROTEIN 15) | 3.19 |
| 793 | 255824_at | AT2G40530 | unknown protein | -3.09 |
| 794 | 255822_at | AT2G40610 | ATEXPA8 (ARABIDOPSIS THALIANA EXPANSIN A8) | -3.37 |
| 795 | 257382_at | AT2G40750 | WRKY54 (WRKY DNA-binding protein 54); transcription factor | -5.36 |
| 796 | 267075_at | AT2G41070 | EEL (ENHANCED EM LEVEL); DNA binding / transcription factor | 6.76 |
| 797 | 267076_at | AT2G41090 | calmodulin-like calcium-binding protein, 22 kDa (CaBP-22) | -5.67 |
| 798 | 267083_at | AT2G41100 | TCH3 (TOUCH 3) | -5.04 |
| 799 | 266364_at | AT2G41230 | unknown protein | 6.67 |
| 800 | 266393_at | AT2G41260 | M17 | 8.62 |
| 801 | 266392_at | AT2G41280 | M10 | 12.07 |
| 802 | 266391_at | AT2G41290 | strictosidine synthase family protein | 3.92 |
| 803 | 245110_at | AT2G41550 | transcription termination factor | 3.44 |
| 804 | 245107_at | AT2G41690 | AT-HSFB3 (Arabidopsis thaliana heat shock transcription factor B3); DNA binding / transcription factor | 3.01 |
| 805 | 260494_at | AT2G41820 | leucine-rich repeat transmembrane protein kinase, putative | -2.86 |
| 806 | 267579_at | AT2G42000 | plant EC metallothionein-like family 15 protein | 8.25 |
| 807 | 267636_at | AT2G42110 | similar to unknown [Populus trichocarpa] (GB:ABK94541.1); similar to unknown [Populus trichocarpa] (GB:ABK92893.1); contains domain PTHR10270:SF18 (PTHR10270:SF18); contains domain PTHR10270 (PTHR10270) | -3.53 |
| 808 | 267631_at | AT2G42150 | DNA-binding bromodomain-containing protein | 3.20 |
| 809 | 267639_at | AT2G42200 | squamosa promoter-binding protein-like 9 (SPL9) | -3.21 |
| 810 | 267635_at | AT2G42220 | rhodanese-like domain-containing protein | -3.27 |
| 811 | 265852_at | AT2G42350 | zinc finger (C3HC4-type RING finger) family protein | 4.12 |
| 812 | 265877_at | AT2G42380 | bZIP transcription factor family protein | -2.52 |
| 813 | 265856_at | AT2G42430 | LBD16 (ASYMMETRIC LEAVES2-LIKE18) | 3.14 |
| 814 | 263495_at | AT2G42530 | COR15B | -3.18 |
| 815 | 263497_at | AT2G42540 | COR15A (COLD-REGULATED 15A) | -3.76 |
| 816 | 263492_at | AT2G42560 | late embryogenesis abundant domain-containing protein / LEA domain-containing protein | 11.74 |
| 817 | 263498_at | AT2G42610 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G07090.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN66524.1); contains InterPro domain Protein of unknown function DUF640 (InterPro:IPR006936) | -4.71 |
| 818 | 263987_at | AT2G42690 | lipase, putative | -3.95 |
| 819 | 263980_at | AT2G42770 | peroxisomal membrane 22 kDa family protein | -3.55 |
| 820 | 263986_at | AT2G42790 | CSY3 (CITRATE SYNTHASE 3); citrate (SI)-synthase | 3.03 |
| 821 | 263982_at | AT2G42860 | unknown protein | 3.42 |
| 822 | 265269_at | AT2G42950 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G29820.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G29820.2); similar to unnamed protein product [Vitis vinifera] (GB:CAO66903.1) | 3.58 |
| 823 | 265268_at | AT2G42960 | protein kinase family protein | -3.71 |
| 824 | 260535_at | AT2G43390 | unknown protein | 7.14 |
| 825 | 260540_at | AT2G43500 | RWP-RK domain-containing protein | 3.42 |
| 826 | 260546_at | AT2G43520 | ATTI2 (ARABIDOPSIS THALIANA TRYPSIN INHIBITOR PROTEIN 2); trypsin inhibitor | 5.27 |
| 827 | 260541_at | AT2G43530 | trypsin inhibitor, putative | -2.38 |
| 828 | 260547_at | AT2G43550 | trypsin inhibitor, putative | -5.43 |
| 829 | 260542_at | AT2G43560 | immunophilin / FKBP-type peptidyl-prolyl cis-trans isomerase family protein | -3.00 |
| 830 | 260556_at | AT2G43620 | chitinase, putative | -3.90 |
| 831 | 260611_at | AT2G43670 | glycosyl hydrolase family protein 17 | 4.41 |
| 832 | 267226_at | AT2G44010 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G59880.1) | 5.74 |
| 833 | 267191_at | AT2G44110 | MLO15 (MILDEW RESISTANCE LOCUS O 15); calmodulin binding | 3.21 |
| 834 | 267367_at | AT2G44210 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G55360.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO67164.1); contains InterPro domain Protein of unknown function DUF239, plant (InterPro:IPR004314) | -2.81 |
| 835 | 267343_at | AT2G44260 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G44230.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G01880.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G01870.1); similar to Protein of unknown function DUF946, plant [Medicago truncatula] (GB:ABN08110.1); similar to Protein of unknown function DUF946, plant [Medicago truncatula] (GB:ABN08108.1); contains InterPro domain Protein of unknown function DUF946, plant (InterPro:IPR009291) | 5.69 |
| 836 | 267388_at | AT2G44450 | glycosyl hydrolase family 1 protein | 4.76 |
| 837 | 267390_at | AT2G44470 | glycosyl hydrolase family 1 protein | 4.96 |
| 838 | 267392_at | AT2G44490 | PEN2 (PENETRATION 2); hydrolase, hydrolyzing O-glycosyl compounds | -4.97 |
| 839 | 266887_at | AT2G44650 | CHL-CPN10 (chloroplast chaperonin 10) | -2.34 |
| 840 | 266813_at | AT2G44920 | thylakoid lumenal 15 kDa protein, chloroplast | -2.41 |
| 841 | 266126_at | AT2G45040 | matrix metalloproteinase | 2.98 |
| 842 | 266123_at | AT2G45180 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | -6.65 |
| 843 | 245136_at | AT2G45210 | auxin-responsive protein-related | 8.56 |
| 844 | 245148_at | AT2G45220 | pectinesterase family protein | 3.42 |
| 845 | 245089_at | AT2G45290 | transketolase, putative | 4.31 |
| 846 | 245140_at | AT2G45420 | LBD18 (LOB DOMAIN-CONTAINING PROTEIN 18) | 3.50 |
| 847 | 245139_at | AT2G45430 | DNA-binding protein-related | 3.04 |
| 848 | 267559_at | AT2G45570 | CYP76C2 (cytochrome P450, family 76, subfamily C, polypeptide 2); oxygen binding | 4.49 |
| 849 | 267514_at | AT2G45630 | oxidoreductase family protein | -2.92 |
| 850 | 267509_at | AT2G45660 | AGL20 (AGAMOUS-LIKE 20); transcription factor | -3.65 |
| 851 | 266928_at | AT2G45790 | ATPMM; phosphomannomutase | -2.74 |
| 852 | 266597_at | AT2G46130 | WRKY43 (WRKY DNA-binding protein 43); transcription factor | 4.51 |
| 853 | 266592_at | AT2G46210 | delta-8 sphingolipid desaturase, putative | 3.21 |
| 854 | 266552_at | AT2G46330 | AGP16 (ARABINOGALACTAN PROTEIN 16) | -2.43 |
| 855 | 265460_at | AT2G46600 | calcium-binding protein, putative | -3.75 |
| 856 | 266326_at | AT2G46650 | B5 #1 (cytochrome b5 family protein #1); heme binding / transition metal ion binding | -2.75 |
| 857 | 266712_at | AT2G46750 | FAD-binding domain-containing protein | 5.61 |
| 858 | 266716_at | AT2G46820 | PSAP/PSI-P/PTAC8/TMP14 (THYLAKOID MEMBRANE PHOSPHOPROTEIN OF 14 KDA); DNA binding | -2.92 |
| 859 | 266756_at | AT2G46950 | CYP709B2 (cytochrome P450, family 709, subfamily B, polypeptide 2); oxygen binding | 4.01 |
| 860 | 266736_at | AT2G46960 | CYP709B1 (cytochrome P450, family 709, subfamily B, polypeptide 1); oxygen binding | 5.62 |
| 861 | 266762_at | AT2G47120 | short-chain dehydrogenase/reductase (SDR) family protein | 9.26 |
| 862 | 263320_at | AT2G47180 | ATGOLS1 (ARABIDOPSIS THALIANA GALACTINOL SYNTHASE 1); transferase, transferring hexosyl groups | 4.61 |
| 863 | 260582_at | AT2G47200 | unknown protein | 6.24 |
| 864 | 245123_at | AT2G47450 | CAO (CHAOS); chromatin binding | -4.06 |
| 865 | 245174_at | AT2G47500 | microtubule motor | -4.02 |
| 866 | 245173_at | AT2G47520 | AP2 domain-containing transcription factor, putative | 3.29 |
| 867 | 245150_at | AT2G47590 | PHR2 (PHOTOLYASE/BLUE-LIGHT RECEPTOR 2) | -2.91 |
| 868 | 266488_at | AT2G47670 | invertase/pectin methylesterase inhibitor family protein | 3.73 |
| 869 | 266465_at | AT2G47750 | GH3.9 (PUTATIVE INDOLE-3-ACETIC ACID-AMIDO SYNTHETASE GH3.9) | 3.21 |
| 870 | 266462_at | AT2G47770 | benzodiazepine receptor-related | 8.61 |
| 871 | 266503_at | AT2G47780 | rubber elongation factor (REF) protein-related | 3.46 |
| 872 | 266509_at | AT2G47940 | DEGP2 (DEGP PROTEASE 2); serine-type peptidase/ trypsin | -3.15 |
| 873 | 265772_at | AT2G48010 | RKF3 (RECEPTOR-LIKE KINASE IN IN FLOWERS 3); kinase | 3.08 |
| 874 | 262349_at | AT2G48130 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | 4.90 |
| 875 | 262317_at | AT2G48140 | EDA4 (embryo sac development arrest 4); lipid binding | 7.20 |
| 876 | 259277_at | AT3G01180 | ATSS2 (STARCH SYNTHASE 2); transferase, transferring glycosyl groups | -3.81 |
| 877 | 259272_at | AT3G01290 | band 7 family protein | -4.74 |
| 878 | 258957_at | AT3G01420 | ALPHA-DOX1 (ALPHA-DIOXYGENASE 1) | 6.12 |
| 879 | 258956_at | AT3G01440 | oxygen evolving enhancer 3 (PsbQ) family protein | -2.71 |
| 880 | 259193_at | AT3G01480 | CYP38 (CYCLOPHILIN 38); peptidyl-prolyl cis-trans isomerase | -3.06 |
| 881 | 259161_at | AT3G01500 | CA1 (CARBONIC ANHYDRASE 1); carbonate dehydratase/ zinc ion binding | -9.39 |
| 882 | 259188_at | AT3G01510 | 5'-AMP-activated protein kinase beta-1 subunit-related | -2.34 |
| 883 | 259185_at | AT3G01550 | triose phosphate/phosphate translocator, putative | -2.70 |
| 884 | 259167_at | AT3G01570 | glycine-rich protein / oleosin | 9.84 |
| 885 | 259178_at | AT3G01650 | RGLG1 (RING DOMAIN LIGASE1); protein binding / zinc ion binding | 3.03 |
| 886 | 259166_at | AT3G01670 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G01680.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN83922.1) | -2.82 |
| 887 | 259180_at | AT3G01680 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G01670.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO14653.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO14657.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO14661.1); contains domain PTHR13871:SF2 (PTHR13871:SF2); contains domain PTHR13871 (PTHR13871) | -3.18 |
| 888 | 259189_at | AT3G01700 | AGP11 (ARABINOGALACTAN PROTEIN 11) | 4.30 |
| 889 | 259000_at | AT3G01860 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G27210.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO14583.1) | -4.57 |
| 890 | 258971_at | AT3G01990 | ACR6 (ACT Domain Repeat 6) | -2.96 |
| 891 | 258856_at | AT3G02040 | SRG3 (SENESCENCE-RELATED GENE 3); glycerophosphodiester phosphodiesterase | 3.23 |
| 892 | 259124_at | AT3G02310 | SEP2 (SEPALLATA2); DNA binding / transcription factor | 5.47 |
| 893 | 258498_at | AT3G02480 | ABA-responsive protein-related | 10.36 |
| 894 | 258487_at | AT3G02550 | LBD41 (LOB DOMAIN-CONTAINING PROTEIN 41) | 3.22 |
| 895 | 258480_at | AT3G02640 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G16250.1); similar to unknown [Populus trichocarpa] (GB:ABK93711.1) | -2.42 |
| 896 | 258607_at | AT3G02730 | ATF1/TRXF1 (THIOREDOXIN F-TYPE 1); thiol-disulfide exchange intermediate | -3.22 |
| 897 | 258621_at | AT3G02830 | ZFN1 (ZINC FINGER PROTEIN 1); nucleic acid binding | -2.42 |
| 898 | 258613_at | AT3G02870 | VTC4; 3'(2'),5'-bisphosphate nucleotidase/ L-galactose-1-phosphate phosphatase/ inositol or phosphatidylinositol phosphatase | -3.82 |
| 899 | 258620_at | AT3G02940 | MYB107 (myb domain protein 107); DNA binding / transcription factor | 5.64 |
| 900 | 258603_at | AT3G02990 | ATHSFA1E (Arabidopsis thaliana heat shock transcription factor A1E); DNA binding / transcription factor | 3.48 |
| 901 | 258617_at | AT3G03000 | calmodulin, putative | -4.66 |
| 902 | 258878_at | AT3G03170 | similar to hypothetical protein [Vitis vinifera] (GB:CAN71065.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO22226.1) | 2.90 |
| 903 | 258874_at | AT3G03230 | esterase/lipase/thioesterase family protein | 3.60 |
| 904 | 258873_at | AT3G03240 | esterase/lipase/thioesterase family protein | 7.56 |
| 905 | 259057_at | AT3G03310 | lecithin:cholesterol acyltransferase family protein / LACT family protein | 3.20 |
| 906 | 259053_at | AT3G03320 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G43470.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO46227.1); contains InterPro domain ProFAR isomerase-like (InterPro:IPR010759) | 3.00 |
| 907 | 259169_at | AT3G03520 | phosphoesterase family protein | 3.71 |
| 908 | 259171_at | AT3G03590 | SWIB complex BAF60b domain-containing protein | 3.44 |
| 909 | 259217_at | AT3G03620 | MATE efflux family protein | 4.21 |
| 910 | 259344_at | AT3G03710 | RIF10 (RESISTANT TO INHIBITION WITH FSM 10); 3'-5'-exoribonuclease/ RNA binding / nucleic acid binding | -2.40 |
| 911 | 258807_at | AT3G04030 | myb family transcription factor | -4.40 |
| 912 | 258582_at | AT3G04150 | germin-like protein, putative | 3.91 |
| 913 | 258580_at | AT3G04170 | germin-like protein, putative | 7.01 |
| 914 | 258578_at | AT3G04200 | germin-like protein, putative | 3.74 |
| 915 | 258537_at | AT3G04210 | disease resistance protein (TIR-NBS class), putative | -4.01 |
| 916 | 258590_at | AT3G04280 | ARR22 (ARABIDOPSIS RESPONSE REGULATOR 22); transcription regulator/ two-component response regulator | 7.52 |
| 917 | 258589_at | AT3G04290 | ATLTL1/LTL1 (LI-TOLERANT LIPASE 1); carboxylesterase | -7.74 |
| 918 | 258625_at | AT3G04370 | Identical to Cysteine-rich repeat secretory protein 39 precursor (CRRSP39) [Arabidopsis Thaliana] (GB:Q6E263;GB:Q6E264;GB:Q9M849;GB:Q9M8Z3); similar to 33 kDa secretory protein-related [Arabidopsis thaliana] (TAIR:AT2G33330.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN81929.1); contains InterPro domain Protein of unknown function DUF26 (InterPro:IPR002902) | 5.22 |
| 919 | 258795_at | AT3G04570 | DNA-binding protein-related | 4.30 |
| 920 | 258797_at | AT3G04730 | IAA16 (indoleacetic acid-induced protein 16); transcription factor | -2.82 |
| 921 | 259098_at | AT3G04790 | ribose 5-phosphate isomerase-related | -2.78 |
| 922 | 259350_at | AT3G05140 | RBK2 (ROP BINDING PROTEIN KINASES 2); kinase | 3.03 |
| 923 | 259314_at | AT3G05260 | short-chain dehydrogenase/reductase (SDR) family protein | 9.26 |
| 924 | 259106_at | AT3G05490 | RALFL22 (RALF-LIKE 22) | -2.46 |
| 925 | 259105_at | AT3G05500 | rubber elongation factor (REF) family protein | 3.19 |
| 926 | 259113_at | AT3G05510 | phospholipid/glycerol acyltransferase family protein | 3.44 |
| 927 | 258887_at | AT3G05630 | PLDP2 (PHOSPHOLIPASE D ZETA 2); phospholipase D | 4.06 |
| 928 | 258890_at | AT3G05690 | ATHAP2B/HAP2B/UNE8 (HEME ACTIVATOR PROTEIN (YEAST) HOMOLOG 2B); transcription factor | -2.88 |
| 929 | 258736_at | AT3G05900 | neurofilament protein-related | -4.18 |
| 930 | 256402_at | AT3G06130 | heavy-metal-associated domain-containing protein | -4.92 |
| 931 | 256418_at | AT3G06160 | transcriptional factor B3 family protein | -3.55 |
| 932 | 258914_at | AT3G06360 | AGP27 (ARABINOGALACTAN PROTEIN 27) | 4.03 |
| 933 | 258905_at | AT3G06390 | integral membrane family protein | 8.04 |
| 934 | 258880_at | AT3G06420 | ATG8H (AUTOPHAGY 8H); microtubule binding | 4.75 |
| 935 | 259010_at | AT3G07340 | basic helix-loop-helix (bHLH) family protein | -3.13 |
| 936 | 258639_at | AT3G07820 | polygalacturonase 3 (PGA3) / pectinase | 3.19 |
| 937 | 258631_at | AT3G07970 | polygalacturonase, putative / pectinase, putative | 3.42 |
| 938 | 258641_at | AT3G08030 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G41800.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO40802.1); contains InterPro domain Protein of unknown function DUF642 (InterPro:IPR006946) | -2.34 |
| 939 | 258682_at | AT3G08720 | ATPK19 (ARABIDOPSIS THALIANA PROTEIN KINASE 19); kinase | 3.20 |
| 940 | 258675_at | AT3G08770 | LTP6 (Lipid transfer protein 6); lipid binding | 3.28 |
| 941 | 258983_at | AT3G08860 | alanine--glyoxylate aminotransferase, putative / beta-alanine-pyruvate aminotransferase, putative / AGT, putative | 6.47 |
| 942 | 258993_at | AT3G08940 | LHCB4.2 (LIGHT HARVESTING COMPLEX PSII); chlorophyll binding | -2.62 |
| 943 | 259211_at | AT3G09020 | alpha 1,4-glycosyltransferase family protein / glycosyltransferase sugar-binding DXD motif-containing protein | -2.82 |
| 944 | 258658_at | AT3G09820 | ADK1 (ADENOSINE KINASE 1) | -2.50 |
| 945 | 258652_at | AT3G09910 | AtRABC2b/AtRab18C (Arabidopsis Rab GTPase homolog C2b); GTP binding | 2.86 |
| 946 | 258930_at | AT3G10040 | transcription factor | 3.12 |
| 947 | 258929_at | AT3G10060 | immunophilin, putative / FKBP-type peptidyl-prolyl cis-trans isomerase, putative | -3.12 |
| 948 | 258938_at | AT3G10080 | germin-like protein, putative | -2.81 |
| 949 | 258935_at | AT3G10120 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G03890.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN62766.1) | -3.73 |
| 950 | 259142_at | AT3G10200 | dehydration-responsive protein-related | 3.77 |
| 951 | 259150_at | AT3G10320 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G41640.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO23616.1); contains InterPro domain Protein of unknown function DUF563 (InterPro:IPR007657) | 4.40 |
| 952 | 259149_at | AT3G10340 | phenylalanine ammonia-lyase, putative | 5.09 |
| 953 | 258923_at | AT3G10450 | SCPL7; serine carboxypeptidase | 4.21 |
| 954 | 258920_at | AT3G10520 | AHB2 (NON-SYMBIOTIC HAEMOGLOBIN 2) | -2.98 |
| 955 | 258969_at | AT3G10680 | heat shock protein-related | -2.50 |
| 956 | 258764_at | AT3G10720 | pectinesterase, putative | -5.11 |
| 957 | 258760_at | AT3G10780 | emp24/gp25L/p24 family protein | 2.86 |
| 958 | 256416_at | AT3G11050 | ATFER2 (FERRITIN 2); ferric iron binding | 8.00 |
| 959 | 256435_at | AT3G11180 | oxidoreductase, 2OG-Fe(II) oxygenase family protein | 5.13 |
| 960 | 256255_at | AT3G11280 | myb family transcription factor | -3.63 |
| 961 | 259282_at | AT3G11430 | ATGPAT5/GPAT5 (GLYCEROL-3-PHOSPHATE ACYLTRANSFERASE 5); 1-acylglycerol-3-phosphate O-acyltransferase/ acyltransferase/ organic anion transmembrane transporter | 4.13 |
| 962 | 259237_at | AT3G11630 | 2-cys peroxiredoxin, chloroplast (BAS1) | -2.39 |
| 963 | 258727_at | AT3G11930 | universal stress protein (USP) family protein | 2.98 |
| 964 | 258755_at | AT3G11945 | ATHST; prenyltransferase | -2.52 |
| 965 | 256290_at | AT3G12203 | SCPL17 (serine carboxypeptidase-like 17); serine carboxypeptidase | 7.54 |
| 966 | 256285_at | AT3G12510 | similar to At3g12510-like protein [Boechera stricta] (GB:ABB89768.1); contains domain PTHR11945 (PTHR11945); contains domain PTHR11945:SF13 (PTHR11945:SF13) | 3.31 |
| 967 | 256245_at | AT3G12580 | HSP70 (heat shock protein 70); ATP binding | 6.40 |
| 968 | 256237_at | AT3G12610 | DRT100 (DNA-DAMAGE REPAIR/TOLERATION 100); protein binding | -4.58 |
| 969 | 257704_at | AT3G12720 | AtMYB67/AtY53 (myb domain protein 67); DNA binding / transcription factor | 6.86 |
| 970 | 257858_at | AT3G12920 | protein binding / zinc ion binding | 3.04 |
| 971 | 257856_at | AT3G12930 | similar to unknown [Populus trichocarpa] (GB:ABK94112.1); contains InterPro domain Iojap-related protein (InterPro:IPR004394) | -2.54 |
| 972 | 257859_at | AT3G12955 | auxin-responsive protein-related | 5.97 |
| 973 | 257853_at | AT3G12960 | similar to seed maturation protein [Glycine tomentella] (GB:ABB72392.1) | 13.02 |
| 974 | 257194_at | AT3G13110 | AtSerat2;2 (SERINE ACETYLTRANSFERASE 1); serine O-acetyltransferase | -2.47 |
| 975 | 257180_at | AT3G13180 | NOL1/NOP2/sun family protein / antitermination NusB domain-containing protein | -4.42 |
| 976 | 256985_at | AT3G13540 | ATMYB5 (MYB DOMAIN PROTEIN 5); DNA binding / transcription factor | 3.00 |
| 977 | 256647_at | AT3G13610 | oxidoreductase, 2OG-Fe(II) oxygenase family protein | -2.45 |
| 978 | 256780_at | AT3G13640 | ATRLI1 (Arabidopsis thaliana RNase L inhibitor protein 1) | 2.89 |
| 979 | 256781_at | AT3G13650 | disease resistance response | -3.05 |
| 980 | 256789_at | AT3G13672 | seven in absentia (SINA) family protein | 3.54 |
| 981 | 256772_at | AT3G13750 | BGAL1 (BETA GALACTOSIDASE 1); beta-galactosidase | -3.00 |
| 982 | 257011_at | AT3G14070 | CAX9 (CATION EXCHANGER 9); cation:cation antiporter | 3.36 |
| 983 | 257008_at | AT3G14210 | ESM1 (EPITHIOSPECIFIER MODIFIER 1); carboxylesterase | -5.98 |
| 984 | 258369_at | AT3G14310 | ATPME3 (Arabidopsis thaliana pectin methylesterase 3) | -3.43 |
| 985 | 258374_at | AT3G14360 | lipase class 3 family protein | 4.70 |
| 986 | 258092_at | AT3G14595 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G17080.1); similar to unknown [Populus trichocarpa] (GB:ABK95219.1); contains domain PTHR10052 (PTHR10052); contains domain PTHR10052:SF2 (PTHR10052:SF2) | 2.85 |
| 987 | 258111_at | AT3G14630 | CYP72A9 (cytochrome P450, family 72, subfamily A, polypeptide 9); oxygen binding | 3.96 |
| 988 | 258113_at | AT3G14650 | CYP72A11 (cytochrome P450, family 72, subfamily A, polypeptide 11); oxygen binding | -2.58 |
| 989 | 258064_at | AT3G14680 | CYP72A14 (cytochrome P450, family 72, subfamily A, polypeptide 14); oxygen binding | 7.82 |
| 990 | 256574_at | AT3G14780 | similar to ATGSL04 (GLUCAN SYNTHASE-LIKE 4), 1,3-beta-glucan synthase/ transferase, transferring glycosyl groups [Arabidopsis thaliana] (TAIR:AT3G14570.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN81673.1) | 2.81 |
| 991 | 256547_at | AT3G14840 | leucine-rich repeat family protein / protein kinase family protein | -2.69 |
| 992 | 257234_at | AT3G14880 | similar to DOG1 (DELAY OF GERMINATION 1) [Arabidopsis thaliana] (TAIR:AT5G45830.1); similar to tumor-related protein-like, putative [Medicago truncatula] (GB:ABD32966.1) | 3.27 |
| 993 | 257210_at | AT3G14950 | TTL2 (TETRATRICOPETIDE-REPEAT THIOREDOXIN-LIKE 2); binding | 3.10 |
| 994 | 257050_at | AT3G15260 | protein phosphatase 2C, putative / PP2C, putative | 4.61 |
| 995 | 257059_at | AT3G15280 | similar to unnamed protein product [Vitis vinifera] (GB:CAO66421.1) | 7.81 |
| 996 | 257058_at | AT3G15352 | ATCOX17 (Arabidopsis thaliana cytochrome c oxidase 17) | 3.16 |
| 997 | 258398_at | AT3G15360 | ATHM4 (Arabidopsis thioredoxin M-type 4); thiol-disulfide exchange intermediate | -3.44 |
| 998 | 258395_at | AT3G15500 | ATNAC3 (ARABIDOPSIS NAC DOMAIN CONTAINING PROTEIN 55); transcription factor | 5.08 |
| 999 | 257294_at | AT3G15570 | phototropic-responsive NPH3 family protein | -4.50 |
| 1000 | 258224_at | AT3G15670 | late embryogenesis abundant protein, putative / LEA protein, putative | 13.19 |
| 1001 | 258223_at | AT3G15840 | PIFI (POST-ILLUMINATION CHLOROPHYLL FLUORESCENCE INCREASE) | -4.35 |
| 1002 | 258333_at | AT3G16000 | MFP1 (MAR BINDING FILAMENT-LIKE PROTEIN 1) | -3.26 |
| 1003 | 258052_at | AT3G16190 | isochorismatase hydrolase family protein | 3.75 |
| 1004 | 258049_at | AT3G16220 | similar to RNA binding / catalytic [Arabidopsis thaliana] (TAIR:AT3G16230.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO45862.1); contains InterPro domain Predicted eukaryotic LigT; (InterPro:IPR009210) | -3.21 |
| 1005 | 258054_at | AT3G16240 | DELTA-TIP (delta tonoplast integral protein); water channel | -6.32 |
| 1006 | 258055_at | AT3G16250 | ferredoxin-related | -7.16 |
| 1007 | 259375_at | AT3G16370 | GDSL-motif lipase/hydrolase family protein | -3.48 |
| 1008 | 259328_at | AT3G16440 | ATMLP-300B (MYROSINASE-BINDING PROTEIN-LIKE PROTEIN-300B) | 3.25 |
| 1009 | 259383_at | AT3G16470 | JR1 (Jacalin lectin family protein) | -6.43 |
| 1010 | 257206_at | AT3G16530 | legume lectin family protein | -4.81 |
| 1011 | 258434_at | AT3G16770 | ATEBP/ERF72/RAP2.3 (RELATED TO AP2 3); DNA binding / protein binding / transcription activator/ transcription factor | 3.88 |
| 1012 | 257896_at | AT3G16920 | chitinase | -4.93 |
| 1013 | 257876_at | AT3G17130 | invertase/pectin methylesterase inhibitor family protein | 3.82 |
| 1014 | 257891_at | AT3G17170 | RFC3 (REGULATOR OF FATTY-ACID COMPOSITION 3); structural constituent of ribosome | -3.30 |
| 1015 | 258347_at | AT3G17520 | late embryogenesis abundant domain-containing protein / LEA domain-containing protein | 11.45 |
| 1016 | 258352_at | AT3G17600 | IAA31 (indoleacetic acid-induced protein 31); transcription factor | 2.84 |
| 1017 | 258377_at | AT3G17690 | ATCNGC19 (cyclic nucleotide gated channel 19); calmodulin binding / cyclic nucleotide binding / ion channel | -2.69 |
| 1018 | 258156_at | AT3G18050 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G28100.1); similar to unknown [Populus trichocarpa] (GB:ABK95654.1) | -3.67 |
| 1019 | 257066_at | AT3G18280 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | -3.71 |
| 1020 | 257717_at | AT3G18390 | EMB1865 (EMBRYO DEFECTIVE 1865) | -2.50 |
| 1021 | 257718_at | AT3G18400 | ANAC058 (Arabidopsis NAC domain containing protein 58); transcription factor | 5.27 |
| 1022 | 256827_at | AT3G18570 | glycine-rich protein / oleosin | 9.61 |
| 1023 | 257749_at | AT3G18780 | ACT2 (ACTIN 2); structural constituent of cytoskeleton | -3.61 |
| 1024 | 256655_at | AT3G18890 | binding / catalytic/ coenzyme binding | -3.22 |
| 1025 | 256922_at | AT3G19010 | oxidoreductase, 2OG-Fe(II) oxygenase family protein | -2.98 |
| 1026 | 258006_at | AT3G19400 | cysteine proteinase, putative | -3.37 |
| 1027 | 258025_at | AT3G19480 | D-3-phosphoglycerate dehydrogenase, putative / 3-PGDH, putative | -4.65 |
| 1028 | 257022_at | AT3G19580 | AZF2 (ARABIDOPSIS ZINC-FINGER PROTEIN 2); nucleic acid binding / transcription factor/ zinc ion binding | 3.74 |
| 1029 | 257017_at | AT3G19620 | glycosyl hydrolase family 3 protein | -4.27 |
| 1030 | 257994_at | AT3G19920 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G64230.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO44392.1) | 11.65 |
| 1031 | 257130_at | AT3G20210 | DELTA-VPE (delta vacuolar processing enzyme); cysteine-type endopeptidase | 6.20 |
| 1032 | 257670_at | AT3G20340 | Expression of the gene is downregulated in the presence of paraquat, an inducer of photoxidative stress. | 4.37 |
| 1033 | 257671_at | AT3G20450 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G11905.1); similar to unknown [Populus trichocarpa] (GB:ABK95443.1); similar to Os05g0272900 [Oryza sativa (japonica cultivar-group)] (GB:NP_001055067.1); contains InterPro domain B-cell receptor-associated 31-like; (InterPro:IPR008417) | -2.96 |
| 1034 | 257089_at | AT3G20520 | glycerophosphoryl diester phosphodiesterase family protein | 3.26 |
| 1035 | 256697_at | AT3G20660 | ATOCT4 (ARABIDOPSIS THALIANA ORGANIC CATION/CARNITINE TRANSPORTER4); carbohydrate transmembrane transporter/ sugar:hydrogen ion symporter | 3.91 |
| 1036 | 257983_at | AT3G20790 | oxidoreductase family protein | -2.86 |
| 1037 | 256806_at | AT3G20910 | CCAAT-binding transcription factor (CBF-B/NF-YA) family protein | 3.10 |
| 1038 | 256814_at | AT3G21370 | glycosyl hydrolase family 1 protein | 10.81 |
| 1039 | 256815_at | AT3G21380 | similar to MBP1 (MYROSINASE-BINDING PROTEIN 1) [Arabidopsis thaliana] (TAIR:AT1G52040.1); similar to jasmonate inducible protein [Brassica napus] (GB:CAA72270.1); contains InterPro domain Mannose-binding lectin (InterPro:IPR001229) | 7.88 |
| 1040 | 257540_at | AT3G21520 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G21550.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO71058.1); contains InterPro domain Protein of unknown function DUF679 (InterPro:IPR007770) | 4.22 |
| 1041 | 258167_at | AT3G21560 | UGT84A2; UDP-glycosyltransferase/ sinapate 1-glucosyltransferase | 3.60 |
| 1042 | 257946_at | AT3G21710 | unknown protein | 3.49 |
| 1043 | 257947_at | AT3G21720 | isocitrate lyase, putative | 12.24 |
| 1044 | 257943_at | AT3G21840 | ASK7 (ARABIDOPSIS SKP1-LIKE 7); ubiquitin-protein ligase | 2.92 |
| 1045 | 257944_at | AT3G21850 | ASK9 (ARABIDOPSIS SKP1-LIKE 9); ubiquitin-protein ligase | 7.26 |
| 1046 | 257945_at | AT3G21860 | ASK10 (ARABIDOPSIS SKP1-LIKE 10); ubiquitin-protein ligase | 3.28 |
| 1047 | 256822_at | AT3G22190 | IQD5 (IQ-domain 5); calmodulin binding | -2.33 |
| 1048 | 256796_at | AT3G22210 | unknown protein | -4.42 |
| 1049 | 258445_at | AT3G22400 | LOX5; lipoxygenase | 4.55 |
| 1050 | 256931_at | AT3G22490 | late embryogenesis abundant protein, putative / LEA protein, putative | 10.21 |
| 1051 | 256938_at | AT3G22500 | ATECP31 (late embryogenesis abundant protein ECP31) | 10.43 |
| 1052 | 256937_at | AT3G22620 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | 4.20 |
| 1053 | 258327_at | AT3G22640 | cupin family protein | 10.15 |
| 1054 | 258322_at | AT3G22740 | HMT3 (Homocysteine S-methyltransferase 3); homocysteine S-methyltransferase | 3.16 |
| 1055 | 256832_at | AT3G22880 | ATDMC1 (RECA-LIKE GENE); ATP binding / DNA-dependent ATPase/ damaged DNA binding | -3.16 |
| 1056 | 257761_at | AT3G23090 | similar to WDL1 (WVD2-LIKE 1) [Arabidopsis thaliana] (TAIR:AT3G04630.2); similar to WDL1 (WVD2-LIKE 1) [Arabidopsis thaliana] (TAIR:AT3G04630.3); similar to seed specific protein Bn15D14A [Brassica napus] (GB:AAP37969.1); contains InterPro domain Targeting for Xklp2 (InterPro:IPR009675) | 4.84 |
| 1057 | 258300_at | AT3G23340 | CKL10 (Casein Kinase I-like 10); casein kinase I/ kinase | 3.66 |
| 1058 | 258289_at | AT3G23450 | pseudogene, similar to unnamed protein product, blastp match of 19% identity and 2.8e-15 P-value to GP|28802461|emb|CAD70027.1||AX647825 unnamed protein product {Homo sapiens} | -2.68 |
| 1059 | 257172_at | AT3G23700 | S1 RNA-binding domain-containing protein | -3.12 |
| 1060 | 257203_at | AT3G23730 | xyloglucan:xyloglucosyl transferase, putative / xyloglucan endotransglycosylase, putative / endo-xyloglucan transferase, putative | -2.65 |
| 1061 | 256865_at | AT3G23820 | GAE6 (UDP-D-GLUCURONATE 4-EPIMERASE 6); catalytic | -3.70 |
| 1062 | 257242_at | AT3G24220 | NCED6 (NINE-CIS-EPOXYCAROTENOID DIOXYGENASE 6) | 7.90 |
| 1063 | 257168_at | AT3G24430 | HCF101 (HIGH-CHLOROPHYLL-FLUORESCENCE 101); ATP binding | -2.42 |
| 1064 | 258130_at | AT3G24510 | Encodes a defensin-like (DEFL) family protein. | 4.78 |
| 1065 | 256898_at | AT3G24650 | ABI3 (ABA INSENSITIVE 3); DNA binding / transcription activator/ transcription factor | 6.79 |
| 1066 | 257869_at | AT3G25160 | ER lumen protein retaining receptor family protein | 4.29 |
| 1067 | 257841_at | AT3G25260 | proton-dependent oligopeptide transport (POT) family protein | 4.67 |
| 1068 | 257824_at | AT3G25290 | auxin-responsive family protein | 3.14 |
| 1069 | 257911_at | AT3G25530 | ATGHBDH/GHBDH; 3-hydroxybutyrate dehydrogenase/ phosphogluconate dehydrogenase (decarboxylating) | -3.03 |
| 1070 | 256756_at | AT3G25610 | haloacid dehalogenase-like hydrolase family protein | -2.85 |
| 1071 | 256754_at | AT3G25690 | CHUP1 (CHLOROPLAST UNUSUAL POSITIONING 1) | -2.46 |
| 1072 | 258078_at | AT3G25870 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G13360.1); similar to unknown [Populus trichocarpa] (GB:ABK92948.1) | 4.00 |
| 1073 | 258080_at | AT3G25930 | universal stress protein (USP) family protein | 3.35 |
| 1074 | 257552_at | AT3G26130 | glycosyl hydrolase family 5 protein / cellulase family protein | 3.12 |
| 1075 | 256875_at | AT3G26330 | CYP71B37 (cytochrome P450, family 71, subfamily B, polypeptide 37); oxygen binding | 5.65 |
| 1076 | 256883_at | AT3G26440 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G13000.2); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G13000.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO42517.1); contains InterPro domain Protein of unknown function DUF707 (InterPro:IPR007877) | 4.48 |
| 1077 | 256880_at | AT3G26450 | major latex protein-related / MLP-related | -5.42 |
| 1078 | 256878_at | AT3G26460 | major latex protein-related / MLP-related | -2.75 |
| 1079 | 257313_at | AT3G26520 | TIP2 (TONOPLAST INTRINSIC PROTEIN 2); water channel | -5.87 |
| 1080 | 257311_at | AT3G26570 | PHT2;1 (phosphate transporter 2;1) | -2.67 |
| 1081 | 257832_at | AT3G26740 | CCL (CCR-LIKE) | -3.29 |
| 1082 | 258257_at | AT3G26770 | short-chain dehydrogenase/reductase (SDR) family protein | 5.73 |
| 1083 | 258258_at | AT3G26790 | FUS3 (FUSCA 3); DNA binding / transcription factor | 5.98 |
| 1084 | 257793_at | AT3G26960 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G41050.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO49052.1) | -3.10 |
| 1085 | 257794_at | AT3G27050 | similar to unnamed protein product [Vitis vinifera] (GB:CAO49017.1) | -2.74 |
| 1086 | 257809_at | AT3G27060 | TSO2 (TSO MEANING 'UGLY' IN CHINESE); ribonucleoside-diphosphate reductase | -2.50 |
| 1087 | 256753_at | AT3G27160 | GHS1 (GLUCOSE HYPERSENSITIVE 1); structural constituent of ribosome | -2.61 |
| 1088 | 257151_at | AT3G27200 | plastocyanin-like domain-containing protein | 3.26 |
| 1089 | 257735_at | AT3G27400 | pectate lyase family protein | -3.79 |
| 1090 | 257737_at | AT3G27440 | uracil phosphoribosyltransferase, putative / UMP pyrophosphorylase, putative / UPRTase, putative | 3.78 |
| 1091 | 258029_at | AT3G27580 | ATPK7 (Arabidopsis thaliana serine/threonine-protein kinase 7); kinase | 2.91 |
| 1092 | 258240_at | AT3G27660 | OLEO4 (OLEOSIN4) | 10.68 |
| 1093 | 258239_at | AT3G27690 | LHCB2:4 (Photosystem II light harvesting complex gene 2.3); chlorophyll binding | -3.56 |
| 1094 | 258232_at | AT3G27750 | similar to vacuolar sorting protein 9 domain-containing protein / VPS9 domain-containing protein [Arabidopsis thaliana] (TAIR:AT5G09320.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO45867.1) | -2.69 |
| 1095 | 258233_at | AT3G27785 | MYB118 (myb domain protein 118); DNA binding / transcription factor | 2.84 |
| 1096 | 257220_at | AT3G27810 | ATMYB21 (MYB DOMAIN PROTEIN 21); DNA binding / transcription factor | 3.09 |
| 1097 | 257224_at | AT3G27870 | haloacid dehalogenase-like hydrolase family protein | 3.95 |
| 1098 | 257298_at | AT3G28155 | similar to MOR1 (MICROTUBULE ORGANIZATION 1) [Arabidopsis thaliana] (TAIR:AT2G35630.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO67627.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO70817.1); similar to microtubule bundling polypeptide TMBP200 [Nicotiana tabacum] (GB:BAB88648.1) | 3.21 |
| 1099 | 256577_at | AT3G28220 | meprin and TRAF homology domain-containing protein / MATH domain-containing protein | -2.42 |
| 1100 | 257872_at | AT3G28360 | PGP16 (P-GLYCOPROTEIN 16); ATPase, coupled to transmembrane movement of substances | 3.60 |
| 1101 | 258002_at | AT3G28930 | AIG2 (AVRRPT2-INDUCED GENE 2) | -4.11 |
| 1102 | 257773_at | AT3G29185 | similar to hypothetical protein MtrDRAFT_AC147482g2v2 [Medicago truncatula] (GB:ABD32485.1) | -2.55 |
| 1103 | 257776_at | AT3G29190 | lyase/ magnesium ion binding | 4.73 |
| 1104 | 257775_at | AT3G29260 | short-chain dehydrogenase/reductase (SDR) family protein | 4.02 |
| 1105 | 256746_at | AT3G29320 | glucan phosphorylase, putative | -2.66 |
| 1106 | 245228_at | AT3G29810 | COBL2 (COBRA-LIKE PROTEIN 2 PRECURSOR) | 4.31 |
| 1107 | 257081_at | AT3G30460 | zinc finger (C3HC4-type RING finger) family protein | 3.90 |
| 1108 | 256940_at | AT3G30720 | unknown protein | -2.50 |
| 1109 | 252770_at | AT3G42860 | zinc knuckle (CCHC-type) family protein | 4.77 |
| 1110 | 252698_at | AT3G43670 | copper amine oxidase, putative | -2.68 |
| 1111 | 252712_at | AT3G43800 | ATGSTU27 (Arabidopsis thaliana Glutathione S-transferase (class tau) 27); glutathione transferase | -2.68 |
| 1112 | 252713_at | AT3G43810 | CAM7 (CALMODULIN 7); calcium ion binding | -2.48 |
| 1113 | 252691_at | AT3G44050 | kinesin motor protein-related | -3.26 |
| 1114 | 252684_at | AT3G44400 | disease resistance protein (TIR-NBS-LRR class), putative | -4.67 |
| 1115 | 252661_at | AT3G44450 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G52740.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO70571.1) | -2.96 |
| 1116 | 252645_at | AT3G44460 | DPBF2 (BASIC LEUCINE ZIPPER TRANSCRIPTION FACTOR 67); DNA binding / transcription factor | 6.22 |
| 1117 | 252638_at | AT3G44540 | oxidoreductase, acting on the CH-CH group of donors | 6.34 |
| 1118 | 252648_at | AT3G44630 | disease resistance protein RPP1-WsB-like (TIR-NBS-LRR class), putative | -2.66 |
| 1119 | 246332_at | AT3G44830 | lecithin:cholesterol acyltransferase family protein / LACT family protein | 4.69 |
| 1120 | 246339_at | AT3G44890 | RPL9 (ribosomal protein L9); structural constituent of ribosome | -2.41 |
| 1121 | 252618_at | AT3G45140 | LOX2 (LIPOXYGENASE 2) | -6.29 |
| 1122 | 252612_at | AT3G45160 | contains domain PROKAR_LIPOPROTEIN (PS51257) | -4.74 |
| 1123 | 252622_at | AT3G45310 | cysteine proteinase, putative | -5.00 |
| 1124 | 252534_at | AT3G46130 | MYB111 (myb domain protein 111); DNA binding / transcription factor | -3.28 |
| 1125 | 252515_at | AT3G46230 | ATHSP17.4 (Arabidopsis thaliana heat shock protein 17.4) | 7.87 |
| 1126 | 252485_at | AT3G46530 | RPP13 (RECOGNITION OF PERONOSPORA PARASITICA 13); ATP binding | -4.76 |
| 1127 | 252478_at | AT3G46540 | epsin N-terminal homology (ENTH) domain-containing protein / clathrin assembly protein-related | -2.45 |
| 1128 | 252487_at | AT3G46660 | UDP-glucoronosyl/UDP-glucosyl transferase family protein | 5.78 |
| 1129 | 252441_at | AT3G46780 | PTAC16 (PLASTID TRANSCRIPTIONALLY ACTIVE18); binding / catalytic | -4.54 |
| 1130 | 252468_at | AT3G46970 | ATPHS2/PHS2 (ALPHA-GLUCAN PHOSPHORYLASE 2); phosphorylase/ transferase, transferring glycosyl groups | -2.57 |
| 1131 | 252462_at | AT3G47250 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G47200.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G47200.2); similar to unnamed protein product [Vitis vinifera] (GB:CAO15956.1); contains InterPro domain Protein of unknown function DUF247, plant (InterPro:IPR004158) | -2.90 |
| 1132 | 252415_at | AT3G47340 | ASN1 (DARK INDUCIBLE 6) | 3.02 |
| 1133 | 252439_at | AT3G47400 | pectinesterase family protein | -3.00 |
| 1134 | 252433_at | AT3G47560 | esterase/lipase/thioesterase family protein | -3.11 |
| 1135 | 252427_at | AT3G47640 | basic helix-loop-helix (bHLH) family protein | 3.00 |
| 1136 | 252409_at | AT3G47650 | bundle-sheath defective protein 2 family / bsd2 family | -3.50 |
| 1137 | 252391_at | AT3G47860 | apolipoprotein D-related | -3.93 |
| 1138 | 252395_at | AT3G47950 | AHA4 (Arabidopsis H(+)-ATPase 4); ATPase | 4.16 |
| 1139 | 252373_at | AT3G48090 | EDS1 (ENHANCED DISEASE SUSCEPTIBILITY 1); signal transducer/ triacylglycerol lipase | -4.29 |
| 1140 | 252367_at | AT3G48360 | BT2 (BTB AND TAZ DOMAIN PROTEIN 2); protein binding / transcription factor/ transcription regulator | -2.85 |
| 1141 | 252366_at | AT3G48420 | haloacid dehalogenase-like hydrolase family protein | -3.98 |
| 1142 | 252321_at | AT3G48510 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G63350.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO68256.1) | 2.99 |
| 1143 | 252320_at | AT3G48580 | xyloglucan:xyloglucosyl transferase, putative / xyloglucan endotransglycosylase, putative / endo-xyloglucan transferase, putative | 6.06 |
| 1144 | 252346_at | AT3G48650 | pseudogene, At14a-related protein, similar to At14a (GI:11994571 and GI:11994573) (Arabidopsis thaliana) | -3.82 |
| 1145 | 252317_at | AT3G48720 | transferase family protein | -3.70 |
| 1146 | 252318_at | AT3G48730 | GSA2 (GLUTAMATE-1-SEMIALDEHYDE 2,1-AMINOMUTASE 2); glutamate-1-semialdehyde 2,1-aminomutase | -2.84 |
| 1147 | 252255_at | AT3G49220 | pectinesterase family protein | -3.65 |
| 1148 | 252306_at | AT3G49270 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G49305.1) | 4.07 |
| 1149 | 252253_at | AT3G49300 | proline-rich family protein | 4.04 |
| 1150 | 252234_at | AT3G49780 | ATPSK4 (PHYTOSULFOKINE 4 PRECURSOR); growth factor | 3.86 |
| 1151 | 252227_at | AT3G49900 | BTB/POZ domain-containing protein | -2.52 |
| 1152 | 252237_at | AT3G49950 | scarecrow transcription factor family protein | 3.06 |
| 1153 | 252199_at | AT3G50270 | transferase family protein | -4.37 |
| 1154 | 252173_at | AT3G50650 | scarecrow-like transcription factor 7 (SCL7) | 3.08 |
| 1155 | 252178_at | AT3G50750 | brassinosteroid signalling positive regulator-related | -3.60 |
| 1156 | 252130_at | AT3G50820 | PSBO-2/PSBO2 (PHOTOSYSTEM II SUBUNIT O-2); oxygen evolving/ poly(U) binding | -3.63 |
| 1157 | 252133_at | AT3G50900 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G66490.1); similar to hypothetical protein [Thellungiella halophila] (GB:ABB45855.1) | -4.94 |
| 1158 | 252102_at | AT3G50970 | LTI30 (LOW TEMPERATURE-INDUCED 30) | 5.37 |
| 1159 | 252137_at | AT3G50980 | XERO1 (DEHYDRIN XERO 1) | 9.33 |
| 1160 | 252115_at | AT3G51600 | LTP5 (LIPID TRANSFER PROTEIN 5); lipid transporter | -4.16 |
| 1161 | 252073_at | AT3G51750 | unknown protein | 4.75 |
| 1162 | 246299_at | AT3G51810 | ATEM1 (Early methionine labelled) | 12.32 |
| 1163 | 246308_at | AT3G51820 | ATG4/CHLG/G4 (CHLOROPHYLL SYNTHASE); chlorophyll synthetase | -2.96 |
| 1164 | 252082_at | AT3G51940 | oxidoreductase/ transition metal ion binding | 2.94 |
| 1165 | 256676_at | AT3G52180 | ATPTPKIS1/DSP4/SEX4 (STARCH-EXCESS 4); polysaccharide binding / protein tyrosine/serine/threonine phosphatase | -2.35 |
| 1166 | 252048_at | AT3G52500 | aspartyl protease family protein | -2.67 |
| 1167 | 252061_at | AT3G52620 | unknown protein | 3.12 |
| 1168 | 252011_at | AT3G52720 | carbonic anhydrase family protein | -6.25 |
| 1169 | 252001_at | AT3G52750 | FTSZ2-2 (FtsZ2-2); structural molecule | -2.79 |
| 1170 | 252003_at | AT3G52770 | ZPR3 (LITTLE ZIPPER 3); protein binding | 3.32 |
| 1171 | 252004_at | AT3G52780 | ATPAP20/PAP20; acid phosphatase/ protein serine/threonine phosphatase | 6.72 |
| 1172 | 252014_at | AT3G52870 | calmodulin-binding family protein | -3.08 |
| 1173 | 252025_at | AT3G52900 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G36355.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN75481.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO46538.1); contains InterPro domain Protein of unknown function DUF662 (InterPro:IPR007033) | -4.26 |
| 1174 | 252019_at | AT3G53040 | late embryogenesis abundant protein, putative / LEA protein, putative | 9.68 |
| 1175 | 251982_at | AT3G53190 | pectate lyase family protein | -2.38 |
| 1176 | 251975_at | AT3G53230 | cell division cycle protein 48, putative / CDC48, putative | 4.10 |
| 1177 | 251990_at | AT3G53320 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G37070.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO18110.1) | -2.46 |
| 1178 | 251964_at | AT3G53370 | DNA-binding S1FA family protein | 3.34 |
| 1179 | 251962_at | AT3G53420 | PIP2A (PLASMA MEMBRANE INTRINSIC PROTEIN 2A); water channel | -3.89 |
| 1180 | 251942_at | AT3G53480 | ATPDR9/PDR9 (PLEIOTROPIC DRUG RESISTANCE 9); ATPase, coupled to transmembrane movement of substances | 4.47 |
| 1181 | 251944_at | AT3G53510 | ABC transporter family protein | 6.33 |
| 1182 | 251952_at | AT3G53650 | histone H2B, putative | -2.74 |
| 1183 | 251929_at | AT3G53920 | SIGC (RNA polymerase sigma subunit C); DNA binding / DNA-directed RNA polymerase/ transcription factor | -2.91 |
| 1184 | 251928_at | AT3G53980 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | -2.85 |
| 1185 | 251885_at | AT3G54050 | fructose-1,6-bisphosphatase, putative / D-fructose-1,6-bisphosphate 1-phosphohydrolase, putative / FBPase, putative | -4.74 |
| 1186 | 251892_at | AT3G54320 | WRI1 (WRINKLED 1); DNA binding / transcription factor | 2.85 |
| 1187 | 251898_at | AT3G54340 | AP3 (APETALA 3); DNA binding / transcription factor | 4.78 |
| 1188 | 251899_at | AT3G54400 | aspartyl protease family protein | -2.88 |
| 1189 | 251870_at | AT3G54510 | early-responsive to dehydration protein-related / ERD protein-related | 4.20 |
| 1190 | 251846_at | AT3G54560 | HTA11; DNA binding | -2.46 |
| 1191 | 251847_at | AT3G54640 | TSA1 (TRYPTOPHAN SYNTHASE ALPHA CHAIN); tryptophan synthase | -3.96 |
| 1192 | 251814_at | AT3G54890 | LHCA1; chlorophyll binding | -2.77 |
| 1193 | 251838_at | AT3G54940 | cysteine proteinase, putative | 9.96 |
| 1194 | 251811_at | AT3G54990 | SMZ (SCHLAFMUTZE); DNA binding / transcription factor | 3.01 |
| 1195 | 251830_at | AT3G55010 | ATPURM/PUR5; phosphoribosylformylglycinamidine cyclo-ligase | -3.95 |
| 1196 | 251824_at | AT3G55090 | ATPase, coupled to transmembrane movement of substances | 3.30 |
| 1197 | 251785_at | AT3G55130 | ATWBC19 (WHITE-BROWN COMPLEX HOMOLOG 19); ATPase, coupled to transmembrane movement of substances | -2.55 |
| 1198 | 251784_at | AT3G55330 | PPL1 (PSBP-LIKE PROTEIN 1); calcium ion binding | -4.22 |
| 1199 | 251807_at | AT3G55400 | OVA1 (OVULE ABORTION 1); ATP binding / aminoacyl-tRNA ligase | -3.14 |
| 1200 | 251759_at | AT3G55630 | ATDFD (A. THALIANA DHFS-FPGS HOMOLOG D); tetrahydrofolylpolyglutamate synthase | -3.28 |
| 1201 | 251753_at | AT3G55760 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G42430.1); similar to hypothetical protein [Trifolium pratense] (GB:BAE71279.1) | -2.69 |
| 1202 | 251762_at | AT3G55800 | SBPASE (SEDOHEPTULOSE-BISPHOSPHATASE); phosphoric ester hydrolase/ sedoheptulose-bisphosphatase | -2.43 |
| 1203 | 251746_at | AT3G56060 | glucose-methanol-choline (GMC) oxidoreductase family protein | -3.89 |
| 1204 | 251735_at | AT3G56090 | ATFER3 (FERRITIN 3); ferric iron binding | -2.65 |
| 1205 | 251733_at | AT3G56240 | CCH (COPPER CHAPERONE) | -3.61 |
| 1206 | 251731_at | AT3G56350 | superoxide dismutase (Mn), putative / manganese superoxide dismutase, putative | 9.60 |
| 1207 | 251705_at | AT3G56400 | WRKY70 (WRKY DNA-binding protein 70); transcription factor | -4.38 |
| 1208 | 246289_at | AT3G56880 | VQ motif-containing protein | 2.83 |
| 1209 | 251664_at | AT3G56940 | AT103 (DICARBOXYLATE DIIRON 1) | -2.44 |
| 1210 | 251668_at | AT3G57010 | strictosidine synthase family protein | -2.70 |
| 1211 | 251658_at | AT3G57020 | strictosidine synthase family protein | 4.33 |
| 1212 | 251665_at | AT3G57040 | ARR9 (RESPONSE REACTOR 4); transcription regulator | -4.30 |
| 1213 | 251626_at | AT3G57220 | UDP-GlcNAc:dolichol phosphate N-acetylglucosamine-1-phosphate transferase, putative | -2.98 |
| 1214 | 251673_at | AT3G57240 | BG3 (BETA-1,3-GLUCANASE 3); hydrolase, hydrolyzing O-glycosyl compounds | 4.32 |
| 1215 | 251649_at | AT3G57330 | ACA11 (AUTOINHIBITED CA2+-ATPASE 11); calcium-transporting ATPase/ calmodulin binding | -2.51 |
| 1216 | 251623_at | AT3G57390 | AGL18 (AGAMOUS-LIKE 18); transcription factor | 3.17 |
| 1217 | 251620_at | AT3G58060 | cation efflux family protein / metal tolerance protein, putative (MTPc3) | 4.91 |
| 1218 | 251575_at | AT3G58120 | bZIP transcription factor family protein | -5.47 |
| 1219 | 251576_at | AT3G58200 | meprin and TRAF homology domain-containing protein / MATH domain-containing protein | 4.53 |
| 1220 | 251580_at | AT3G58450 | universal stress protein (USP) family protein | 11.26 |
| 1221 | 251531_at | AT3G58550 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | 7.32 |
| 1222 | 251540_at | AT3G58740 | CSY1 (CITRATE SYNTHASE 1); citrate (SI)-synthase | 6.82 |
| 1223 | 251555_at | AT3G58780 | SHP1 (SHATTERPROOF 1); DNA binding / transcription factor | 4.45 |
| 1224 | 251497_at | AT3G59060 | PIL6 (PHYTOCHROME-INTERACTING FACTOR 5); DNA binding / transcription factor | -3.90 |
| 1225 | 251519_at | AT3G59400 | GUN4 (Genomes uncoupled 4) | -2.45 |
| 1226 | 251490_at | AT3G59490 | similar to unnamed protein product [Vitis vinifera] (GB:CAO64819.1) | -2.84 |
| 1227 | 251472_at | AT3G59580 | RWP-RK domain-containing protein | 3.88 |
| 1228 | 251479_at | AT3G59700 | ATHLECRK (ARABIDOPSIS THALIANA LECTIN-RECEPTOR KINASE); kinase | -3.18 |
| 1229 | 251481_at | AT3G59730 | receptor lectin kinase, putative | -2.33 |
| 1230 | 251487_at | AT3G59760 | OASC (O-ACETYLSERINE (THIOL) LYASE ISOFORM C); cysteine synthase | -2.61 |
| 1231 | 251436_at | AT3G59900 | (ARGOS); unknown protein | 4.18 |
| 1232 | 251428_at | AT3G60140 | DIN2 (DARK INDUCIBLE 2); hydrolase, hydrolyzing O-glycosyl compounds | 3.03 |
| 1233 | 251412_at | AT3G60220 | ATL4 (Arabidopsis T?xicos en Levadura 4); protein binding / zinc ion binding | 3.15 |
| 1234 | 251402_at | AT3G60290 | oxidoreductase, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors | 3.21 |
| 1235 | 251370_at | AT3G60450 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G60440.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO70569.1); contains InterPro domain Phosphoglycerate mutase (InterPro:IPR013078); contains InterPro domain PRIB5 (InterPro:IPR012398) | -2.66 |
| 1236 | 251377_at | AT3G60650 | unknown protein | 3.60 |
| 1237 | 251342_at | AT3G60690 | auxin-responsive family protein | 3.22 |
| 1238 | 251382_at | AT3G60730 | pectinesterase family protein | 9.53 |
| 1239 | 251339_at | AT3G60780 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G45360.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO24498.1); contains InterPro domain Protein of unknown function DUF1442 (InterPro:IPR009902) | 6.07 |
| 1240 | 251350_at | AT3G61040 | CYP76C7 (cytochrome P450, family 76, subfamily C, polypeptide 7); oxygen binding | 7.46 |
| 1241 | 251324_at | AT3G61430 | PIP1A (PLASMA MEMBRANE INTRINSIC PROTEIN 1A); water channel | -2.82 |
| 1242 | 251274_at | AT3G61700 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G46420.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO39750.1); contains InterPro domain Conserved hypothetical protein CHP01589, plant (InterPro:IPR006476) | -2.90 |
| 1243 | 251277_at | AT3G61760 | dynamin-like protein B (DL1B) | -2.64 |
| 1244 | 251287_at | AT3G61820 | aspartyl protease family protein | -3.51 |
| 1245 | 251301_at | AT3G61880 | CYP78A9 (CYTOCHROME P450 78A9); oxygen binding | 5.82 |
| 1246 | 251291_at | AT3G61900 | auxin-responsive family protein | 3.62 |
| 1247 | 251293_at | AT3G61930 | unknown protein | 5.16 |
| 1248 | 251299_at | AT3G61950 | basic helix-loop-helix (bHLH) family protein | -2.90 |
| 1249 | 251245_at | AT3G62090 | PIL2 (PHYTOCHROME INTERACTING FACTOR 3-LIKE 2); transcription factor | 4.13 |
| 1250 | 251246_at | AT3G62100 | IAA30 (indoleacetic acid-induced protein 30); transcription factor | 3.92 |
| 1251 | 251248_at | AT3G62150 | PGP21 (P-GLYCOPROTEIN 21); ATPase, coupled to transmembrane movement of substances | -2.44 |
| 1252 | 251259_at | AT3G62260 | protein phosphatase 2C, putative / PP2C, putative | 3.69 |
| 1253 | 251218_at | AT3G62410 | CP12-2 | -6.08 |
| 1254 | 251221_at | AT3G62550 | universal stress protein (USP) family protein | -4.42 |
| 1255 | 251188_at | AT3G62730 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G47980.1); similar to hypothetical protein OsI_011212 [Oryza sativa (indica cultivar-group)] (GB:EAY89979.1) | 8.94 |
| 1256 | 251193_at | AT3G62910 | APG3 (ALBINO AND PALE GREEN); translation release factor | -7.10 |
| 1257 | 251200_at | AT3G63010 | ATGID1B/GID1B (GA INSENSITIVE DWARF1B); hydrolase | -3.85 |
| 1258 | 251202_at | AT3G63040 | unknown protein | 9.62 |
| 1259 | 251157_at | AT3G63140 | mRNA-binding protein, putative | -8.64 |
| 1260 | 251155_at | AT3G63160 | unknown protein | -2.40 |
| 1261 | 251160_at | AT3G63240 | endonuclease/exonuclease/phosphatase family protein | -2.81 |
| 1262 | 251166_at | AT3G63350 | AT-HSFA7B (Arabidopsis thaliana heat shock transcription factor A7B); DNA binding / transcription factor | 3.00 |
| 1263 | 251178_at | AT3G63440 | ATCKX6/ATCKX7/CKX6 (CYTOKININ OXIDASE/DEHYDROGENASE 6); cytokinin dehydrogenase | -3.23 |
| 1264 | 255695_at | AT4G00080 | UNE11 (unfertilized embryo sac 11); pectinesterase inhibitor | -2.48 |
| 1265 | 255701_at | AT4G00220 | JLO/LBD30 (JAGGED LATERAL ORGANS) | 6.43 |
| 1266 | 255692_at | AT4G00400 | GPAT8 (GLYCEROL-3-PHOSPHATE ACYLTRANSFERASE 8); acyltransferase/ glycerol-3-phosphate O-acyltransferase | -3.10 |
| 1267 | 255676_at | AT4G00490 | BAM2/BMY9 (BETA-AMYLASE 2); beta-amylase | -2.38 |
| 1268 | 255626_at | AT4G00780 | meprin and TRAF homology domain-containing protein / MATH domain-containing protein | -4.21 |
| 1269 | 255645_at | AT4G00880 | auxin-responsive family protein | -5.38 |
| 1270 | 255627_at | AT4G00955 | similar to protein kinase family protein [Arabidopsis thaliana] (TAIR:AT2G23450.2); similar to protein kinase family protein [Arabidopsis thaliana] (TAIR:AT2G23450.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO41616.1); contains InterPro domain EGF (InterPro:IPR006210) | -3.69 |
| 1271 | 255656_at | AT4G00990 | transcription factor jumonji (jmjC) domain-containing protein | -3.32 |
| 1272 | 255567_at | AT4G01150 | Identical to Uncharacterized protein At4g01150, chloroplast precursor [Arabidopsis Thaliana] (GB:O04616;GB:Q38835); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G38100.1); similar to unknown [Populus trichocarpa] (GB:ABK95729.1) | -2.43 |
| 1273 | 255577_at | AT4G01410 | harpin-induced family protein / HIN1 family protein / harpin-responsive family protein | 4.00 |
| 1274 | 255575_at | AT4G01430 | nodulin MtN21 family protein | 3.67 |
| 1275 | 255550_at | AT4G01970 | ATSTS (ARABIDOPSIS THALIANA STACHYOSE SYNTHASE); galactinol-raffinose galactosyltransferase/ hydrolase, hydrolyzing O-glycosyl compounds | 5.73 |
| 1276 | 255529_at | AT4G02120 | CTP synthase, putative / UTP--ammonia ligase, putative | -2.52 |
| 1277 | 255506_at | AT4G02130 | GATL6/LGT10; polygalacturonate 4-alpha-galacturonosyltransferase/ transferase, transferring glycosyl groups / transferase, transferring hexosyl groups | -4.66 |
| 1278 | 255521_at | AT4G02280 | SUS3; UDP-glycosyltransferase/ sucrose synthase/ transferase, transferring glycosyl groups | 7.41 |
| 1279 | 255524_at | AT4G02330 | ATPMEPCRB; pectinesterase | -4.68 |
| 1280 | 255527_at | AT4G02360 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G02813.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO40037.1); contains InterPro domain Protein of unknown function DUF538 (InterPro:IPR007493) | 4.66 |
| 1281 | 255479_at | AT4G02380 | SAG21 (SENESCENCE-ASSOCIATED GENE 21) | -2.92 |
| 1282 | 255503_at | AT4G02420 | lectin protein kinase, putative | -3.58 |
| 1283 | 255440_at | AT4G02530 | chloroplast thylakoid lumen protein | -3.52 |
| 1284 | 255493_at | AT4G02690 | glutamate binding / | 5.83 |
| 1285 | 255457_at | AT4G02770 | PSAD-1 (photosystem I subunit D-1) | -2.87 |
| 1286 | 255460_at | AT4G02800 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G30050.1); similar to Os04g0228100 [Oryza sativa (japonica cultivar-group)] (GB:NP_001052288.1); similar to H0209A05.2 [Oryza sativa (indica cultivar-group)] (GB:CAH66085.1) | -3.04 |
| 1287 | 255471_at | AT4G03050 | AOP3 (2-oxoglutarate?dependent dioxygenase 3); oxidoreductase, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors | 6.61 |
| 1288 | 255417_at | AT4G03190 | GRH1 (GRR1-LIKE PROTEIN 1); ubiquitin-protein ligase | -2.40 |
| 1289 | 255418_at | AT4G03200 | catalytic | 3.05 |
| 1290 | 255433_at | AT4G03210 | XTH9 (XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 9); hydrolase, acting on glycosyl bonds | -5.13 |
| 1291 | 255430_at | AT4G03320 | TIC20-IV (TRANSLOCON AT THE INNER ENVELOPE MEMBRANE OF CHLOROPLASTS 20-IV); P-P-bond-hydrolysis-driven protein transmembrane transporter | 4.07 |
| 1292 | 255357_at | AT4G03930 | pectinesterase | 4.12 |
| 1293 | 255331_at | AT4G04330 | similar to unnamed protein product [Vitis vinifera] (GB:CAO69665.1) | -2.72 |
| 1294 | 255345_at | AT4G04460 | aspartyl protease family protein | 5.80 |
| 1295 | 255290_at | AT4G04640 | ATPC1 (ATP synthase gamma chain 1) | -3.74 |
| 1296 | 255295_at | AT4G04760 | sugar transporter family protein | 3.34 |
| 1297 | 255298_at | AT4G04840 | methionine sulfoxide reductase domain-containing protein / SeIR domain-containing protein | 3.85 |
| 1298 | 255300_at | AT4G04870 | CLS (CARDIOLIPIN SYNTHASE); cardiolipin synthase/ phosphatidyltransferase | 3.22 |
| 1299 | 255250_at | AT4G05100 | AtMYB74 (myb domain protein 74); DNA binding / transcription factor | -3.19 |
| 1300 | 255104_at | AT4G08685 | SAH7 | -3.04 |
| 1301 | 255116_at | AT4G08850 | leucine-rich repeat family protein / protein kinase family protein | -4.80 |
| 1302 | 255078_at | AT4G09010 | APX4 (ASCORBATE PEROXIDASE 4); peroxidase | -3.37 |
| 1303 | 255080_at | AT4G09030 | AGP10 (Arabinogalactan protein 10) | 3.98 |
| 1304 | 255048_at | AT4G09600 | GASA3 (GAST1 PROTEIN HOMOLOG 3) | 12.26 |
| 1305 | 255049_at | AT4G09610 | GASA2 (GAST1 PROTEIN HOMOLOG 2) | 11.48 |
| 1306 | 255046_at | AT4G09650 | ATP synthase delta chain, chloroplast, putative / H(+)-transporting two-sector ATPase, delta (OSCP) subunit, putative | -3.82 |
| 1307 | 255056_at | AT4G09820 | TT8 (TRANSPARENT TESTA 8); DNA binding / transcription factor | 4.91 |
| 1308 | 255014_at | AT4G09960 | STK (SEEDSTICK); transcription factor | 5.32 |
| 1309 | 255007_at | AT4G10020 | ATHSD5 (HYDROXYSTEROID DEHYDROGENASE 5); oxidoreductase | 11.53 |
| 1310 | 255008_at | AT4G10060 | catalytic | -3.50 |
| 1311 | 255807_at | AT4G10270 | wound-responsive family protein | 6.22 |
| 1312 | 254974_at | AT4G10490 | oxidoreductase, 2OG-Fe(II) oxygenase family protein | 3.81 |
| 1313 | 254956_at | AT4G10850 | nodulin MtN3 family protein | 5.90 |
| 1314 | 254911_at | AT4G11100 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G03060.1) | -3.15 |
| 1315 | 254934_at | AT4G11140 | CRF1 (CYTOKININ RESPONSE FACTOR 1); DNA binding / transcription factor | 3.71 |
| 1316 | 254924_at | AT4G11330 | ATMPK5 (MAP KINASE 5); MAP kinase/ kinase | -3.39 |
| 1317 | 254853_at | AT4G12080 | DNA-binding family protein | 2.81 |
| 1318 | 254854_at | AT4G12130 | aminomethyltransferase | 4.10 |
| 1319 | 254815_at | AT4G12420 | SKU5 (skewed 5); copper ion binding | -3.51 |
| 1320 | 254818_at | AT4G12470 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | -3.50 |
| 1321 | 254790_at | AT4G12800 | PSAL (photosystem I subunit L) | -2.43 |
| 1322 | 254789_at | AT4G12880 | plastocyanin-like domain-containing protein | -2.62 |
| 1323 | 254786_at | AT4G12890 | gamma interferon responsive lysosomal thiol reductase family protein / GILT family protein | 3.88 |
| 1324 | 254777_at | AT4G12960 | gamma interferon responsive lysosomal thiol reductase family protein / GILT family protein | 8.77 |
| 1325 | 254797_at | AT4G13030 | similar to unnamed protein product [Vitis vinifera] (GB:CAO18001.1); contains domain SSF52540 (SSF52540) | 4.67 |
| 1326 | 254727_at | AT4G13670 | PTAC5 (PLASTID TRANSCRIPTIONALLY ACTIVE5); heat shock protein binding / unfolded protein binding | -4.22 |
| 1327 | 254687_at | AT4G13770 | CYP83A1 (CYTOCHROME P450 83A1); oxygen binding | -2.35 |
| 1328 | 245624_at | AT4G14090 | UDP-glucoronosyl/UDP-glucosyl transferase family protein | -2.68 |
| 1329 | 245594_at | AT4G14570 | acylaminoacyl-peptidase-related | -2.43 |
| 1330 | 245574_at | AT4G14750 | IQD19 (IQ-DOMAIN 19); calmodulin binding | -2.84 |
| 1331 | 245577_at | AT4G14780 | protein kinase, putative | 3.07 |
| 1332 | 245270_at | AT4G14960 | TUA6 (tubulin alpha-6 chiain) | -2.51 |
| 1333 | 245586_at | AT4G14980 | DC1 domain-containing protein | 6.84 |
| 1334 | 245309_at | AT4G15140 | similar to unknown [Populus trichocarpa] (GB:ABK96081.1) | -3.00 |
| 1335 | 245555_at | AT4G15390 | transferase family protein | 8.16 |
| 1336 | 245556_at | AT4G15400 | transferase family protein | 5.35 |
| 1337 | 245253_at | AT4G15440 | HPL1 (HYDROPEROXIDE LYASE 1); heme binding / iron ion binding / monooxygenase | -3.26 |
| 1338 | 245528_at | AT4G15530 | PPDK (PYRUVATE ORTHOPHOSPHATE DIKINASE); kinase/ pyruvate, phosphate dikinase | 3.51 |
| 1339 | 245505_at | AT4G15690 | glutaredoxin family protein | -3.35 |
| 1340 | 245371_at | AT4G15750 | invertase/pectin methylesterase inhibitor family protein | 6.16 |
| 1341 | 245516_at | AT4G15820 | wound-responsive protein-related | -2.94 |
| 1342 | 245343_at | AT4G15830 | binding | -2.47 |
| 1343 | 245524_at | AT4G15920 | nodulin MtN3 family protein | -4.87 |
| 1344 | 245335_at | AT4G16160 | ATOEP16-2/ATOEP16-S; P-P-bond-hydrolysis-driven protein transmembrane transporter | 11.96 |
| 1345 | 245388_at | AT4G16410 | similar to hypothetical protein [Vitis vinifera] (GB:CAN73304.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO39941.1); contains InterPro domain Protein of unknown function DUF751 (InterPro:IPR008470) | -2.85 |
| 1346 | 245436_at | AT4G16620 | integral membrane family protein / nodulin MtN21-related | 5.52 |
| 1347 | 245345_at | AT4G16640 | matrix metalloproteinase, putative | 3.86 |
| 1348 | 245448_at | AT4G16860 | RPP4 (RECOGNITION OF PERONOSPORA PARASITICA 4) | -6.05 |
| 1349 | 245456_at | AT4G16950 | RPP5 (RECOGNITION OF PERONOSPORA PARASITICA 5) | -2.82 |
| 1350 | 245458_at | AT4G16970 | kinase | -2.98 |
| 1351 | 245318_at | AT4G16980 | arabinogalactan-protein family | -3.94 |
| 1352 | 245460_at | AT4G16990 | disease resistance protein (TIR-NBS class), putative | -3.64 |
| 1353 | 245346_at | AT4G17090 | CT-BMY (BETA-AMYLASE 3, BETA-AMYLASE 8); beta-amylase | -3.80 |
| 1354 | 245432_at | AT4G17100 | serine protease protein -related, expressed pseudogene, blastp match of 64% identity and 1.9e-114 P-value to GP|29367523|gb|AAO72617.1||AY224498 putative serine protease-like protein {Oryza sativa (japonica cultivar-group)} | -2.68 |
| 1355 | 245305_at | AT4G17215 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G47635.1); similar to hypothetical protein 40.t00006 [Brassica oleracea] (GB:ABD65131.1) | 5.18 |
| 1356 | 245264_at | AT4G17245 | zinc finger (C3HC4-type RING finger) family protein | -2.68 |
| 1357 | 245412_at | AT4G17280 | auxin-responsive family protein | 3.86 |
| 1358 | 245399_at | AT4G17340 | DELTA-TIP2/TIP2;2 (tonoplast intrinsic protein 2;2); water channel | -2.47 |
| 1359 | 245362_at | AT4G17460 | HAT1 (homeobox-leucine zipper protein 1); DNA binding / transcription factor | -6.59 |
| 1360 | 245357_at | AT4G17560 | ribosomal protein L19 family protein | -2.73 |
| 1361 | 245354_at | AT4G17600 | LIL3:1; transcription factor | -2.73 |
| 1362 | 254691_at | AT4G17840 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G35260.1); similar to hypothetical protein 40.t00061 [Brassica oleracea] (GB:ABD65174.1) | 3.07 |
| 1363 | 254693_at | AT4G17880 | basic helix-loop-helix (bHLH) family protein | -3.55 |
| 1364 | 254701_at | AT4G18030 | dehydration-responsive family protein | -2.64 |
| 1365 | 254668_at | AT4G18350 | NCED2 (NINE-CIS-EPOXYCAROTENOID DIOXYGENASE 2) | 3.06 |
| 1366 | 254669_at | AT4G18370 | DEG5/DEGP5/HHOA (DEGP PROTEASE 5); serine-type peptidase/ trypsin | -2.39 |
| 1367 | 254629_at | AT4G18425 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G46090.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO64139.1); contains InterPro domain Protein of unknown function DUF679 (InterPro:IPR007770) | 6.35 |
| 1368 | 254623_at | AT4G18480 | CHLI1 (CHLORINA 42); magnesium chelatase | -3.01 |
| 1369 | 254646_at | AT4G18530 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G11170.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO46884.1); contains InterPro domain Protein of unknown function DUF707 (InterPro:IPR007877) | 3.75 |
| 1370 | 254634_at | AT4G18650 | transcription factor-related | 8.14 |
| 1371 | 254618_at | AT4G18780 | CESA8 (CELLULOSE SYNTHASE 8); cellulose synthase/ transferase, transferring glycosyl groups | -4.15 |
| 1372 | 254608_at | AT4G18910 | NIP1;2/NLM2 (NOD26-like intrinsic protein 1;2); water channel | 3.45 |
| 1373 | 254607_at | AT4G18920 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G45690.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO47983.1); contains InterPro domain Protein of unknown function DUF1264 (InterPro:IPR010686) | 4.93 |
| 1374 | 254609_at | AT4G18970 | GDSL-motif lipase/hydrolase family protein | -3.73 |
| 1375 | 254612_at | AT4G19100 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G52780.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN79943.1) | -3.88 |
| 1376 | 254563_at | AT4G19120 | ERD3 (EARLY-RESPONSIVE TO DEHYDRATION 3) | -2.59 |
| 1377 | 254572_at | AT4G19380 | alcohol oxidase-related | 4.25 |
| 1378 | 254580_at | AT4G19390 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G13720.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO63752.1); contains InterPro domain Uncharacterised conserved protein UCP022348 (InterPro:IPR016804); contains InterPro domain Protein of unknown function UPF0114 (InterPro:IPR005134) | 3.13 |
| 1379 | 254551_at | AT4G19840 | ATPP2-A1 (Arabidopsis thaliana phloem protein 2-A1) | -3.83 |
| 1380 | 254498_at | AT4G20090 | EMB1025 (EMBRYO DEFECTIVE 1025) | -2.39 |
| 1381 | 254474_at | AT4G20390 | integral membrane family protein | 5.09 |
| 1382 | 254485_at | AT4G20760 | short-chain dehydrogenase/reductase (SDR) family protein | -5.09 |
| 1383 | 254448_at | AT4G20870 | fatty acid hydroxylase, putative | -2.68 |
| 1384 | 254440_at | AT4G21020 | late embryogenesis abundant domain-containing protein / LEA domain-containing protein | 13.32 |
| 1385 | 254398_at | AT4G21280 | PSBQ/PSBQ-1/PSBQA; calcium ion binding | -4.25 |
| 1386 | 254377_at | AT4G21650 | subtilase family protein | -5.39 |
| 1387 | 254396_at | AT4G21680 | proton-dependent oligopeptide transport (POT) family protein | 6.77 |
| 1388 | 254369_at | AT4G21720 | similar to unknown [Picea sitchensis] (GB:ABK23300.1) | -3.49 |
| 1389 | 254384_at | AT4G21870 | 26.5 kDa class P-related heat shock protein (HSP26.5-P) | -2.98 |
| 1390 | 254339_at | AT4G22100 | glycosyl hydrolase family 1 protein | 5.80 |
| 1391 | 254362_at | AT4G22160 | unknown protein | -5.57 |
| 1392 | 254305_at | AT4G22200 | AKT2 (Arabidopsis K+ transporter 2); cyclic nucleotide binding / inward rectifier potassium channel | -3.56 |
| 1393 | 254328_at | AT4G22570 | APT3 (ADENINE PHOSPHORIBOSYL TRANSFERASE 3); adenine phosphoribosyltransferase | -4.31 |
| 1394 | 254277_at | AT4G22680 | MYB85 (myb domain protein 85); DNA binding / transcription factor | 4.07 |
| 1395 | 254298_at | AT4G22890 | PGR5-LIKE A | -2.65 |
| 1396 | 254299_at | AT4G22920 | ATNYE1/NYE1 (NON-YELLOWING 1) | 4.28 |
| 1397 | 254266_at | AT4G23130 | CRK5 (CYSTEINE-RICH RLK5); kinase | -3.17 |
| 1398 | 254256_at | AT4G23180 | CRK10 (CYSTEINE-RICH RLK10); kinase | -2.76 |
| 1399 | 254255_at | AT4G23220 | protein kinase family protein | -3.49 |
| 1400 | 254248_at | AT4G23270 | protein kinase family protein | -3.14 |
| 1401 | 254250_at | AT4G23290 | protein kinase family protein | -2.42 |
| 1402 | 254224_at | AT4G23650 | CDPK6 (CALCIUM-DEPENDENT PROTEIN KINASE 6); calmodulin-dependent protein kinase/ kinase | -2.78 |
| 1403 | 254225_at | AT4G23670 | major latex protein-related / MLP-related | -7.32 |
| 1404 | 254234_at | AT4G23680 | major latex protein-related / MLP-related | -2.67 |
| 1405 | 254221_at | AT4G23820 | glycoside hydrolase family 28 protein / polygalacturonase (pectinase) family protein | -3.46 |
| 1406 | 254187_at | AT4G23890 | similar to unnamed protein product [Vitis vinifera] (GB:CAO69542.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN73381.1) | -2.45 |
| 1407 | 254197_at | AT4G24040 | ATTRE1/TRE1 (TREHALASE 1); alpha,alpha-trehalase/ trehalase | 3.50 |
| 1408 | 254200_at | AT4G24110 | similar to unnamed protein product [Vitis vinifera] (GB:CAO62924.1) | 5.25 |
| 1409 | 254174_at | AT4G24120 | YSL1 (YELLOW STRIPE LIKE 1); oligopeptide transporter | -4.24 |
| 1410 | 254201_at | AT4G24130 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G56580.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO62919.1); contains InterPro domain Protein of unknown function DUF538 (InterPro:IPR007493) | 3.97 |
| 1411 | 254202_at | AT4G24140 | hydrolase, alpha/beta fold family protein | 3.59 |
| 1412 | 254203_at | AT4G24150 | AtGRF8 (GROWTH-REGULATING FACTOR 8) | 5.86 |
| 1413 | 254132_at | AT4G24660 | ATHB22/MEE68 (ARABIDOPSIS THALIANA HOMEOBOX PROTEIN 22); DNA binding / transcription factor | 4.03 |
| 1414 | 254119_at | AT4G24780 | pectate lyase family protein | -5.28 |
| 1415 | 254104_at | AT4G25040 | integral membrane family protein | 5.31 |
| 1416 | 254105_at | AT4G25080 | CHLM (MAGNESIUM-PROTOPORPHYRIN IX METHYLTRANSFERASE); magnesium protoporphyrin IX methyltransferase | -2.52 |
| 1417 | 254095_at | AT4G25140 | OLEO1 (OLEOSIN1) | 11.11 |
| 1418 | 254057_at | AT4G25170 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G61490.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO60860.1); contains InterPro domain Uncharacterised conserved protein UCP012943 (InterPro:IPR016606) | 3.09 |
| 1419 | 254072_at | AT4G25370 | Clp amino terminal domain-containing protein | -2.36 |
| 1420 | 245233_at | AT4G25580 | stress-responsive protein-related | 7.41 |
| 1421 | 254029_at | AT4G25870 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G57270.3); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G57270.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G57270.2); similar to unnamed protein product [Vitis vinifera] (GB:CAO64039.1); contains InterPro domain Protein of unknown function DUF266, plant (InterPro:IPR004949) | -2.80 |
| 1422 | 254040_at | AT4G25900 | aldose 1-epimerase family protein | -2.85 |
| 1423 | 254036_at | AT4G25980 | cationic peroxidase, putative | 5.17 |
| 1424 | 254013_at | AT4G26050 | leucine-rich repeat family protein | 6.76 |
| 1425 | 253989_at | AT4G26130 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G56980.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN72576.1) | -2.38 |
| 1426 | 253999_at | AT4G26200 | ACS7 (1-Amino-cyclopropane-1-carboxylate synthase 7); 1-aminocyclopropane-1-carboxylate synthase | 4.20 |
| 1427 | 253985_at | AT4G26220 | caffeoyl-CoA 3-O-methyltransferase, putative | 6.03 |
| 1428 | 253971_at | AT4G26530 | fructose-bisphosphate aldolase, putative | -6.01 |
| 1429 | 253973_at | AT4G26555 | immunophilin / FKBP-type peptidyl-prolyl cis-trans isomerase family protein | -2.98 |
| 1430 | 253984_at | AT4G26590 | ATOPT5 (oligopeptide transporter 5); oligopeptide transporter | 4.75 |
| 1431 | 253930_at | AT4G26740 | ATS1 (ARABIDOPSIS THALIANA SEED GENE 1); calcium ion binding | 10.92 |
| 1432 | 253931_at | AT4G26770 | phosphatidate cytidylyltransferase, putative / CDP-diglyceride synthetase, putative | 3.43 |
| 1433 | 253904_at | AT4G27140 | 2S seed storage protein 1 / 2S albumin storage protein / NWMU1-2S albumin 1 | 10.98 |
| 1434 | 253894_at | AT4G27150 | 2S seed storage protein 2 / 2S albumin storage protein / NWMU2-2S albumin 2 | 13.73 |
| 1435 | 253895_at | AT4G27160 | AT2S3; lipid binding / nutrient reservoir | 12.51 |
| 1436 | 253902_at | AT4G27170 | 2S seed storage protein 4 / 2S albumin storage protein / NWMU2-2S albumin 4 | 13.24 |
| 1437 | 253907_at | AT4G27250 | dihydroflavonol 4-reductase family / dihydrokaempferol 4-reductase family | 4.56 |
| 1438 | 253872_at | AT4G27410 | RD26 (RESPONSIVE TO DESSICATION 26); transcription factor | 3.05 |
| 1439 | 253863_at | AT4G27420 | ABC transporter family protein | 3.17 |
| 1440 | 253871_at | AT4G27440 | PORB (PROTOCHLOROPHYLLIDE OXIDOREDUCTASE B); oxidoreductase/ protochlorophyllide reductase | -2.99 |
| 1441 | 253874_at | AT4G27450 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G15450.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO39242.1); contains domain G3DSA:3.60.20.10 (G3DSA:3.60.20.10); contains domain SSF56235 (SSF56235) | 4.36 |
| 1442 | 253864_at | AT4G27460 | CBS domain-containing protein | 4.90 |
| 1443 | 253870_at | AT4G27530 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G53895.1) | 9.19 |
| 1444 | 253830_at | AT4G27652 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G27657.1) | 2.82 |
| 1445 | 253887_at | AT4G27730 | ATOPT6 (oligopeptide transporter 6); oligopeptide transporter | -3.69 |
| 1446 | 253840_at | AT4G27780 | ACBP2 (ACYL-COA BINDING PROTEIN ACBP 2) | 3.72 |
| 1447 | 253842_at | AT4G27860 | integral membrane family protein | -2.93 |
| 1448 | 253823_at | AT4G28030 | GCN5-related N-acetyltransferase (GNAT) family protein | -2.40 |
| 1449 | 253815_at | AT4G28250 | ATEXPB3 (ARABIDOPSIS THALIANA EXPANSIN B3) | -2.58 |
| 1450 | 253818_at | AT4G28330 | ATP binding / ATPase, coupled to transmembrane movement of substances | 4.04 |
| 1451 | 253780_at | AT4G28400 | protein phosphatase 2C, putative / PP2C, putative | -2.83 |
| 1452 | 253767_at | AT4G28520 | CRU3 (CRUCIFERIN 3); nutrient reservoir | 10.13 |
| 1453 | 253740_at | AT4G28706 | pfkB-type carbohydrate kinase family protein | -2.48 |
| 1454 | 253738_at | AT4G28750 | PSAE-1 (PSA E1 KNOCKOUT); catalytic | -2.83 |
| 1455 | 253736_at | AT4G28780 | GDSL-motif lipase/hydrolase family protein | -4.45 |
| 1456 | 253753_at | AT4G29030 | glycine-rich protein | -3.56 |
| 1457 | 253758_at | AT4G29060 | EMB2726 (EMBRYO DEFECTIVE 2726); translation elongation factor | -3.33 |
| 1458 | 253749_at | AT4G29080 | PAP2 (PHYTOCHROME-ASSOCIATED PROTEIN 2); transcription factor | -2.63 |
| 1459 | 253709_at | AT4G29220 | phosphofructokinase family protein | -3.65 |
| 1460 | 253710_at | AT4G29230 | ANAC075 (Arabidopsis NAC domain containing protein 75); transcription factor | 4.07 |
| 1461 | 253676_at | AT4G29570 | cytidine deaminase, putative / cytidine aminohydrolase, putative | 3.56 |
| 1462 | 253699_at | AT4G29800 | PLA IVD/PLP8 (Patatin-like protein 8) | 3.81 |
| 1463 | 253650_at | AT4G30020 | subtilase family protein | -3.41 |
| 1464 | 253660_at | AT4G30140 | GDSL-motif lipase/hydrolase family protein | -3.23 |
| 1465 | 253617_at | AT4G30410 | transcription factor | -4.09 |
| 1466 | 253627_at | AT4G30650 | hydrophobic protein, putative / low temperature and salt responsive protein, putative | -3.97 |
| 1467 | 253585_at | AT4G30720 | oxidoreductase | -2.83 |
| 1468 | 253575_at | AT4G31060 | AP2 domain-containing transcription factor, putative | 3.87 |
| 1469 | 253534_at | AT4G31500 | CYP83B1 (CYTOCHROME P450 MONOOXYGENASE 83B1); oxygen binding | -7.86 |
| 1470 | 253533_at | AT4G31590 | ATCSLC05 (Cellulose synthase-like C5); transferase, transferring glycosyl groups | 3.19 |
| 1471 | 253494_at | AT4G31830 | similar to unnamed protein product [Vitis vinifera] (GB:CAO44350.1) | 11.90 |
| 1472 | 253461_at | AT4G32170 | CYP96A2 (cytochrome P450, family 96, subfamily A, polypeptide 2); oxygen binding | 2.85 |
| 1473 | 253475_at | AT4G32290 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G25330.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO44242.1); contains InterPro domain Protein of unknown function DUF266, plant (InterPro:IPR004949) | 4.40 |
| 1474 | 253421_at | AT4G32340 | binding | -4.47 |
| 1475 | 253440_at | AT4G32570 | TIFY8 | -3.48 |
| 1476 | 253394_at | AT4G32770 | VTE1 (VITAMIN E DEFICIENT 1) | 3.34 |
| 1477 | 253407_at | AT4G32920 | glycine-rich protein | 3.12 |
| 1478 | 253412_at | AT4G33000 | CBL10 (CALCINEURIN B-LIKE10); calcium ion binding | -3.21 |
| 1479 | 253387_at | AT4G33010 | ATGLDP1 (ARABIDOPSIS THALIANA GLYCINE DECARBOXYLASE P-PROTEIN 1); glycine dehydrogenase (decarboxylating) | -4.05 |
| 1480 | 253414_at | AT4G33050 | EDA39 (embryo sac development arrest 39); calmodulin binding | -5.67 |
| 1481 | 253416_at | AT4G33070 | pyruvate decarboxylase, putative | 2.97 |
| 1482 | 253373_at | AT4G33150 | LKR (SACCHAROPINE DEHYDROGENASE) | 2.97 |
| 1483 | 253372_at | AT4G33220 | pectinesterase family protein | -2.69 |
| 1484 | 253375_at | AT4G33280 | DNA binding / transcription factor | 3.48 |
| 1485 | 253331_at | AT4G33490 | aspartic-type endopeptidase/ pepsin A | -2.44 |
| 1486 | 253330_at | AT4G33530 | KUP5 (K+ uptake permease 5); potassium ion transmembrane transporter | -3.76 |
| 1487 | 253298_at | AT4G33560 | unknown protein | 5.06 |
| 1488 | 253346_at | AT4G33600 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G33590.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN80154.1) | 5.38 |
| 1489 | 253322_at | AT4G33980 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G42900.2); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G42900.3); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G42900.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN64989.1) | 3.05 |
| 1490 | 253282_at | AT4G34120 | LEJ1 (LOSS OF THE TIMING OF ET AND JA BIOSYNTHESIS 1) | -3.12 |
| 1491 | 253272_at | AT4G34190 | SEP1 (STRESS ENHANCED PROTEIN 1) | -2.72 |
| 1492 | 253278_at | AT4G34220 | leucine-rich repeat transmembrane protein kinase, putative | -2.71 |
| 1493 | 253237_at | AT4G34240 | ALDH3I1 (Aldehyde dehydrogenase 3I1); 3-chloroallyl aldehyde dehydrogenase/ aldehyde dehydrogenase (NAD) | -2.55 |
| 1494 | 253285_at | AT4G34250 | fatty acid elongase, putative | 5.79 |
| 1495 | 253289_at | AT4G34320 | Identical to UPF0496 protein At4g34320 [Arabidopsis Thaliana] (GB:Q9SYZ7); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G34330.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO47666.1); contains InterPro domain Protein of unknown function DUF677 (InterPro:IPR007749) | 4.68 |
| 1496 | 253240_at | AT4G34510 | KCS2 (3-ketoacyl-CoA synthase 2); acyltransferase | 5.80 |
| 1497 | 253241_at | AT4G34520 | FAE1 (FATTY ACID ELONGATION1); acyltransferase | 7.89 |
| 1498 | 253209_at | AT4G34830 | binding | -5.40 |
| 1499 | 253218_at | AT4G34980 | SLP2 (subtilisin-like serine protease 2); subtilase | -3.05 |
| 1500 | 253197_at | AT4G35250 | vestitone reductase-related | -4.13 |
| 1501 | 253191_at | AT4G35350 | XCP1 (XYLEM CYSTEINE PEPTIDASE 1); cysteine-type peptidase | -5.22 |
| 1502 | 253164_at | AT4G35725 | unknown protein | 5.69 |
| 1503 | 253083_at | AT4G36250 | ALDH3F1 (ALDEHYDE DEHYDROGENASE 3F1); 3-chloroallyl aldehyde dehydrogenase/ aldehyde dehydrogenase (NAD) | -2.75 |
| 1504 | 253090_at | AT4G36360 | BGAL3 (beta-galactosidase 3); beta-galactosidase | 2.90 |
| 1505 | 246216_at | AT4G36380 | ROT3 (ROTUNDIFOLIA 3); oxygen binding / steroid hydroxylase | 4.68 |
| 1506 | 246270_at | AT4G36500 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G18210.1); similar to hypothetical protein [Thellungiella halophila] (GB:ABB45855.1) | -3.66 |
| 1507 | 246275_at | AT4G36540 | BEE2 (BR ENHANCED EXPRESSION 2); DNA binding / transcription factor | -4.85 |
| 1508 | 246242_at | AT4G36600 | late embryogenesis abundant domain-containing protein / LEA domain-containing protein | 6.62 |
| 1509 | 246203_at | AT4G36610 | hydrolase, alpha/beta fold family protein | 4.04 |
| 1510 | 246274_at | AT4G36620 | zinc finger (GATA type) family protein | 3.65 |
| 1511 | 246273_at | AT4G36700 | cupin family protein | 11.65 |
| 1512 | 246222_at | AT4G36900 | RAP2.10 (related to AP2 10); DNA binding / transcription factor | 3.14 |
| 1513 | 246241_at | AT4G37050 | PLA V/PLP4 (Patatin-like protein 4); nutrient reservoir | 6.76 |
| 1514 | 253091_at | AT4G37360 | CYP81D2 (cytochrome P450, family 81, subfamily D, polypeptide 2); oxygen binding | 3.14 |
| 1515 | 253051_at | AT4G37490 | CYC1 (CYCLIN 1); cyclin-dependent protein kinase regulator | -3.79 |
| 1516 | 253095_at | AT4G37510 | ribonuclease III family protein | -2.42 |
| 1517 | 253040_at | AT4G37800 | xyloglucan:xyloglucosyl transferase, putative / xyloglucan endotransglycosylase, putative / endo-xyloglucan transferase, putative | -4.39 |
| 1518 | 253012_at | AT4G37900 | glycine-rich protein | -3.36 |
| 1519 | 253009_at | AT4G37930 | SHM1 (SERINE HYDROXYMETHYLTRANSFERASE 1); glycine hydroxymethyltransferase/ poly(U) binding | -5.10 |
| 1520 | 253024_at | AT4G38080 | hydroxyproline-rich glycoprotein family protein | 9.97 |
| 1521 | 253033_at | AT4G38220 | aminoacylase, putative / N-acyl-L-amino-acid amidohydrolase, putative | -2.56 |
| 1522 | 252997_at | AT4G38400 | ATEXLA2 (ARABIDOPSIS THALIANA EXPANSIN-LIKE A2) | -3.14 |
| 1523 | 252989_at | AT4G38420 | SKS9 (SKU5 Similar 9); copper ion binding / oxidoreductase | -4.08 |
| 1524 | 252976_s_at | AT4G38550 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G20950.1); contains InterPro domain Phospholipase-like, arabidopsis (InterPro:IPR007942) | -3.25 |
| 1525 | 252954_at | AT4G38660 | thaumatin, putative | -2.43 |
| 1526 | 252950_at | AT4G38690 | 1-phosphatidylinositol phosphodiesterase-related | -6.43 |
| 1527 | 252971_at | AT4G38770 | PRP4 (PROLINE-RICH PROTEIN 4) | -4.70 |
| 1528 | 252972_at | AT4G38840 | auxin-responsive protein, putative | -2.62 |
| 1529 | 252965_at | AT4G38860 | auxin-responsive protein, putative | -6.11 |
| 1530 | 252929_at | AT4G38970 | fructose-bisphosphate aldolase, putative | -3.86 |
| 1531 | 252934_at | AT4G39120 | inositol monophosphatase family protein | -3.27 |
| 1532 | 252914_at | AT4G39130 | dehydrin family protein | 6.72 |
| 1533 | 252943_at | AT4G39330 | mannitol dehydrogenase, putative | -3.26 |
| 1534 | 252898_at | AT4G39500 | CYP96A11 (cytochrome P450, family 96, subfamily A, polypeptide 11); oxygen binding | 2.98 |
| 1535 | 252908_at | AT4G39670 | glycolipid binding / glycolipid transporter | -4.06 |
| 1536 | 252882_at | AT4G39675 | unknown protein | 6.32 |
| 1537 | 252853_at | AT4G39710 | immunophilin, putative / FKBP-type peptidyl-prolyl cis-trans isomerase, putative | -4.55 |
| 1538 | 252827_at | AT4G39950 | CYP79B2 (cytochrome P450, family 79, subfamily B, polypeptide 2); oxygen binding | -3.98 |
| 1539 | 252876_at | AT4G39970 | haloacid dehalogenase-like hydrolase family protein | -2.64 |
| 1540 | 251142_at | AT5G01015 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G65295.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN82885.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO45347.1) | -2.43 |
| 1541 | 251131_at | AT5G01190 | LAC10 (laccase 10); copper ion binding / oxidoreductase | -2.85 |
| 1542 | 251133_at | AT5G01240 | amino acid permease, putative | -3.77 |
| 1543 | 251137_at | AT5G01300 | phosphatidylethanolamine-binding family protein | 8.28 |
| 1544 | 251106_at | AT5G01500 | mitochondrial substrate carrier family protein | 2.84 |
| 1545 | 251084_at | AT5G01520 | zinc finger (C3HC4-type RING finger) family protein | 4.29 |
| 1546 | 251083_at | AT5G01590 | similar to unnamed protein product [Vitis vinifera] (GB:CAO63840.1) | -5.48 |
| 1547 | 251100_at | AT5G01670 | aldose reductase, putative | 7.24 |
| 1548 | 251073_at | AT5G01750 | Identical to Uncharacterized protein At5g01750 [Arabidopsis Thaliana] (GB:Q9LZX1;GB:Q8LEK9); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G11740.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO48825.1); contains InterPro domain Protein of unknown function DUF567 (InterPro:IPR007612) | -3.30 |
| 1549 | 251039_at | AT5G02020 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G55646.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G59080.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO15731.1) | 2.90 |
| 1550 | 251031_at | AT5G02120 | OHP (ONE HELIX PROTEIN) | -2.48 |
| 1551 | 251036_at | AT5G02160 | unknown protein | -3.03 |
| 1552 | 251035_at | AT5G02220 | similar to unknown [Picea sitchensis] (GB:ABK23883.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN70860.1) | -2.83 |
| 1553 | 251019_at | AT5G02420 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G08180.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN68753.1) | 2.87 |
| 1554 | 251050_at | AT5G02440 | similar to unnamed protein product [Vitis vinifera] (GB:CAO15638.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN65558.1) | -2.65 |
| 1555 | 251012_at | AT5G02580 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G55240.1); similar to hypothetical protein OsI_019565 [Oryza sativa (indica cultivar-group)] (GB:EAY98332.1); similar to Os05g0462000 [Oryza sativa (japonica cultivar-group)] (GB:NP_001055765.1); contains InterPro domain Conserved hypothetical protein CHP01589, plant (InterPro:IPR006476) | 6.94 |
| 1556 | 250968_at | AT5G02890 | transferase family protein | -4.01 |
| 1557 | 250960_at | AT5G02940 | similar to phosphotransferase-related [Arabidopsis thaliana] (TAIR:AT5G43745.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO71282.1); similar to hypothetical protein OsI_009890 [Oryza sativa (indica cultivar-group)] (GB:EAY88657.1); similar to Unknown protein [Oryza sativa (japonica cultivar-group)] (GB:AAN06856.1); contains InterPro domain Protein of unknown function DUF1012 (InterPro:IPR010420) | -5.26 |
| 1558 | 250933_at | AT5G03170 | FLA11 (fasciclin-like arabinogalactan-protein 11) | -2.99 |
| 1559 | 250938_at | AT5G03180 | zinc finger (C3HC4-type RING finger) family protein | 4.06 |
| 1560 | 250898_at | AT5G03300 | ADK2 (ADENOSINE KINASE 2); kinase | -2.58 |
| 1561 | 250942_at | AT5G03350 | legume lectin family protein | -6.62 |
| 1562 | 250920_at | AT5G03390 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G68960.1); contains InterPro domain Protein of unknown function DUF295 (InterPro:IPR005174) | -3.88 |
| 1563 | 250905_at | AT5G03640 | protein kinase family protein | 4.21 |
| 1564 | 250906_at | AT5G03650 | SBE2.2 (STARCH BRANCHING ENZYME 2.2); 1,4-alpha-glucan branching enzyme | -2.94 |
| 1565 | 250893_at | AT5G03795 | oxidoreductase | 4.17 |
| 1566 | 250868_at | AT5G03860 | MLS (MALATE SYNTHASE); malate synthase | 9.51 |
| 1567 | 250884_at | AT5G03940 | FFC (FIFTY-FOUR CHLOROPLAST HOMOLOGUE); 7S RNA binding / GTP binding / mRNA binding | -3.46 |
| 1568 | 250882_at | AT5G04000 | similar to hypothetical protein [Vitis vinifera] (GB:CAN76250.1) | 7.05 |
| 1569 | 250874_at | AT5G04010 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G03920.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO23344.1); contains domain SSF81383 (SSF81383) | 6.80 |
| 1570 | 245701_at | AT5G04140 | GLS1/GLU1/GLUS (FERREDOXIN-DEPENDENT GLUTAMATE SYNTHASE 1); glutamate synthase (ferredoxin) | -5.36 |
| 1571 | 245697_at | AT5G04200 | ATMC9 (METACASPASE 9); caspase/ cysteine-type peptidase | 4.28 |
| 1572 | 245711_at | AT5G04340 | C2H2 (ZINC FINGER OF ARABIDOPSIS THALIANA 6); nucleic acid binding / transcription factor/ zinc ion binding | 3.27 |
| 1573 | 245713_at | AT5G04370 | NAMT1; S-adenosylmethionine-dependent methyltransferase | 6.18 |
| 1574 | 245703_at | AT5G04380 | S-adenosyl-L-methionine:carboxyl methyltransferase family protein | 5.75 |
| 1575 | 250889_at | AT5G04500 | glycosyltransferase family protein 47 | 7.00 |
| 1576 | 250820_at | AT5G05160 | leucine-rich repeat transmembrane protein kinase, putative | -3.94 |
| 1577 | 250826_at | AT5G05220 | similar to hypothetical protein [Vitis vinifera] (GB:CAN82940.1) | 5.77 |
| 1578 | 250776_at | AT5G05320 | monooxygenase, putative (MO3) | 4.52 |
| 1579 | 250781_at | AT5G05410 | DREB2A (DRE-BINDING PROTEIN 2A); DNA binding / transcription activator/ transcription factor | 3.17 |
| 1580 | 250739_at | AT5G05740 | ATEGY2; metalloendopeptidase | -2.56 |
| 1581 | 250746_at | AT5G05880 | UDP-glucoronosyl/UDP-glucosyl transferase family protein | 5.09 |
| 1582 | 250764_at | AT5G05960 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | -2.50 |
| 1583 | 250733_at | AT5G06290 | 2-cys peroxiredoxin, chloroplast, putative | -3.83 |
| 1584 | 250694_at | AT5G06710 | HAT14 (homeobox-leucine zipper protein 14); DNA binding / transcription factor | -4.23 |
| 1585 | 250648_at | AT5G06760 | late embryogenesis abundant group 1 domain-containing protein / LEA group 1 domain-containing protein | 9.70 |
| 1586 | 250669_at | AT5G06870 | PGIP2 (POLYGALACTURONASE INHIBITING PROTEIN 2); protein binding | -3.32 |
| 1587 | 250662_at | AT5G07010 | sulfotransferase family protein | 4.68 |
| 1588 | 250620_at | AT5G07190 | ATS3 (ARABIDOPSIS THALIANA SEED GENE 3) | 9.35 |
| 1589 | 250611_at | AT5G07200 | YAP169 (GIBBERELLIN 20 OXIDASE 3); gibberellin 20-oxidase | 7.37 |
| 1590 | 250613_at | AT5G07240 | IQD24 (IQ-domain 24); calmodulin binding | -2.48 |
| 1591 | 250614_at | AT5G07260 | homeobox protein-related | 7.05 |
| 1592 | 250624_at | AT5G07330 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G63060.1); similar to unknown [Xerophyta humilis] (GB:AAT45004.1) | 10.04 |
| 1593 | 250627_at | AT5G07360 | amidase family protein | 3.38 |
| 1594 | 250629_at | AT5G07390 | ATRBOHA (RESPIRATORY BURST OXIDASE HOMOLOG A); FAD binding / calcium ion binding / iron ion binding / oxidoreductase | 3.69 |
| 1595 | 250608_at | AT5G07420 | pectinesterase family protein | 3.62 |
| 1596 | 250583_at | AT5G07500 | PEI1; nucleic acid binding / transcription factor | 5.82 |
| 1597 | 250582_at | AT5G07580 | DNA binding / transcription factor | -3.10 |
| 1598 | 250597_at | AT5G07680 | ANAC079/ANAC080/ATNAC4 (Arabidopsis NAC domain containing protein 79, Arabidopsis NAC domain containing protein 80); transcription factor | 5.38 |
| 1599 | 250604_at | AT5G07830 | ATGUS2 (ARABIDOPSIS THALIANA GLUCURONIDASE 2); beta-glucuronidase | 3.32 |
| 1600 | 250550_at | AT5G07870 | transferase family protein | -2.86 |
| 1601 | 250563_at | AT5G08050 | similar to unnamed protein product [Vitis vinifera] (GB:CAO62462.1); similar to unknown [Populus trichocarpa] (GB:ABK95457.1); contains InterPro domain Protein of unknown function DUF1118 (InterPro:IPR009500); contains InterPro domain Uncharacterised conserved protein UCP022207 (InterPro:IPR016801) | -3.35 |
| 1602 | 250575_at | AT5G08240 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G23160.1) | 3.54 |
| 1603 | 250576_at | AT5G08250 | cytochrome P450 family protein | 3.03 |
| 1604 | 246054_at | AT5G08360 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G23380.1); similar to Os04g0528000 [Oryza sativa (japonica cultivar-group)] (GB:NP_001053372.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO68101.1); similar to Os10g0494000 [Oryza sativa (japonica cultivar-group)] (GB:NP_001064942.1); contains InterPro domain Protein of unknown function DUF789 (InterPro:IPR008507) | 3.15 |
| 1605 | 246036_at | AT5G08370 | ATAGAL2 (ARABIDOPSIS THALIANA ALPHA-GALACTOSIDASE 2); alpha-galactosidase | -3.52 |
| 1606 | 250519_at | AT5G08460 | GDSL-motif lipase/hydrolase family protein | 4.16 |
| 1607 | 250535_at | AT5G08480 | VQ motif-containing protein | 4.51 |
| 1608 | 245892_at | AT5G09370 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | 3.54 |
| 1609 | 245889_at | AT5G09480 | hydroxyproline-rich glycoprotein family protein | 8.98 |
| 1610 | 250541_at | AT5G09520 | hydroxyproline-rich glycoprotein family protein | 4.43 |
| 1611 | 250500_at | AT5G09530 | hydroxyproline-rich glycoprotein family protein | 9.89 |
| 1612 | 250501_at | AT5G09640 | SNG2 (SINAPOYLGLUCOSE ACCUMULATOR 2); serine carboxypeptidase | 7.48 |
| 1613 | 250496_at | AT5G09650 | ATPPA6 (ARABIDOPSIS THALIANA PYROPHOSPHORYLASE 6); inorganic diphosphatase/ pyrophosphatase | -3.84 |
| 1614 | 250498_at | AT5G09660 | PMDH2 (PEROXISOMAL NAD-MALATE DEHYDROGENASE 2); malate dehydrogenase | -4.77 |
| 1615 | 250503_at | AT5G09820 | plastid-lipid associated protein PAP / fibrillin family protein | -2.60 |
| 1616 | 250509_at | AT5G09970 | CYP78A7 (cytochrome P450, family 78, subfamily A, polypeptide 7); oxygen binding | 6.30 |
| 1617 | 250485_at | AT5G09990 | PROPEP5 (Elicitor peptide 5 precursor) | 3.38 |
| 1618 | 250468_at | AT5G10120 | ethylene insensitive 3 family protein | 4.47 |
| 1619 | 250476_at | AT5G10140 | FLC (FLOWERING LOCUS C); transcription factor | 5.61 |
| 1620 | 250471_at | AT5G10170 | inositol-3-phosphate synthase, putative / myo-inositol-1-phosphate synthase, putative / MI-1-P synthase, putative | -2.34 |
| 1621 | 250473_at | AT5G10220 | ANN6 (ANNEXIN ARABIDOPSIS 6); calcium ion binding / calcium-dependent phospholipid binding | 3.62 |
| 1622 | 250433_at | AT5G10400 | histone H3 | -2.48 |
| 1623 | 250426_at | AT5G10510 | AIL6 (AINTEGUMENTA-LIKE 6); DNA binding / transcription factor | 4.08 |
| 1624 | 250438_at | AT5G10580 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G31330.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN79714.1); contains InterPro domain Protein of unknown function DUF599 (InterPro:IPR006747) | 3.80 |
| 1625 | 246012_at | AT5G10650 | zinc finger (C3HC4-type RING finger) family protein | 3.09 |
| 1626 | 246019_at | AT5G10690 | pentatricopeptide (PPR) repeat-containing protein / CBS domain-containing protein | -2.49 |
| 1627 | 250366_at | AT5G11420 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G25460.1); similar to unknown [Ricinus communis] (GB:CAB02653.1); contains InterPro domain Protein of unknown function DUF642 (InterPro:IPR006946); contains InterPro domain Galactose-binding like (InterPro:IPR008979) | -4.80 |
| 1628 | 250339_at | AT5G11670 | ATNADP-ME2 (NADP-MALIC ENZYME 2); malate dehydrogenase (oxaloacetate-decarboxylating) (NADP+)/ malic enzyme/ oxidoreductase, acting on NADH or NADPH, NAD or NADP as acceptor | -2.43 |
| 1629 | 250336_at | AT5G11720 | alpha-glucosidase 1 (AGLU1) | -4.12 |
| 1630 | 250351_at | AT5G12030 | AT-HSP17.6A (Arabidopsis thaliana heat shock protein 17.6A) | 8.00 |
| 1631 | 245181_at | AT5G12420 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G16350.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO48523.1); contains InterPro domain Protein of unknown function UPF0089 (InterPro:IPR004255); contains InterPro domain Protein of unknown function DUF1298 (InterPro:IPR009721) | 3.73 |
| 1632 | 250264_at | AT5G12890 | UDP-glucoronosyl/UDP-glucosyl transferase family protein | -2.94 |
| 1633 | 250277_at | AT5G12940 | leucine-rich repeat family protein | -4.06 |
| 1634 | 250269_at | AT5G12970 | C2 domain-containing protein | 3.43 |
| 1635 | 245981_at | AT5G13100 | oxidoreductase | -3.03 |
| 1636 | 245982_at | AT5G13170 | nodulin MtN3 family protein | 6.62 |
| 1637 | 250287_at | AT5G13330 | RAP2.6L (related to AP2 6L); DNA binding / transcription factor | 5.38 |
| 1638 | 250239_at | AT5G13580 | ABC transporter family protein | 4.02 |
| 1639 | 250243_at | AT5G13630 | GUN5 (GENOMES UNCOUPLED 5) | -3.28 |
| 1640 | 250255_at | AT5G13730 | SIG4 (SIGMA FACTOR 4); DNA binding / DNA-directed RNA polymerase/ transcription factor | -3.43 |
| 1641 | 250258_at | AT5G13790 | AGL15 (AGAMOUS-LIKE 15); DNA binding / transcription factor | 6.33 |
| 1642 | 250230_at | AT5G13900 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | 2.88 |
| 1643 | 250208_at | AT5G14000 | ANAC084 (Arabidopsis NAC domain containing protein 84); transcription factor | 3.04 |
| 1644 | 250200_at | AT5G14130 | peroxidase, putative | 5.87 |
| 1645 | 250199_at | AT5G14180 | MPL1 (MYZUS PERSICAE-INDUCED LIPASE 1); catalytic | 4.32 |
| 1646 | 250183_at | AT5G14510 | armadillo/beta-catenin repeat family protein | -3.46 |
| 1647 | 250146_at | AT5G14660 | PDF1B (PEPTIDE DEFORMYLASE 1B); peptide deformylase | -2.98 |
| 1648 | 246596_at | AT5G14740 | CA2 (BETA CARBONIC ANHYDRASE 2); carbonate dehydratase/ zinc ion binding | -6.11 |
| 1649 | 246597_at | AT5G14760 | AO (L-ASPARTATE OXIDASE); L-aspartate oxidase | -2.42 |
| 1650 | 246595_at | AT5G14780 | FDH (FORMATE DEHYDROGENASE); NAD binding / binding / catalytic/ cofactor binding / oxidoreductase, acting on the CH-OH group of donors, NAD or NADP as acceptor | 2.98 |
| 1651 | 246550_at | AT5G14920 | gibberellin-regulated family protein | -3.05 |
| 1652 | 250152_at | AT5G15120 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G39890.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO14912.1); contains InterPro domain Protein of unknown function DUF1637 (InterPro:IPR012864) | 5.21 |
| 1653 | 250109_at | AT5G15230 | GASA4 (GAST1 PROTEIN HOMOLOG 4) | -5.80 |
| 1654 | 250161_at | AT5G15240 | amino acid transporter family protein | 3.41 |
| 1655 | 250167_at | AT5G15310 | AtMIXTA/AtMYB16 (myb domain protein 16); DNA binding / transcription factor | -2.35 |
| 1656 | 250110_at | AT5G15350 | plastocyanin-like domain-containing protein | -3.07 |
| 1657 | 246557_at | AT5G15510 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G01015.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO15061.1); contains InterPro domain Targeting for Xklp2 (InterPro:IPR009675) | -2.67 |
| 1658 | 246562_at | AT5G15580 | LNG1 (LONGIFOLIA1) | -2.40 |
| 1659 | 246540_at | AT5G15600 | SP1L4 (SPIRAL1-LIKE4) | -3.00 |
| 1660 | 246531_at | AT5G15800 | SEP1 (SEPALLATA1); DNA binding / transcription factor | 3.13 |
| 1661 | 246487_at | AT5G16030 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G02500.1); similar to hypothetical protein [Cleome spinosa] (GB:ABD96956.1) | -5.36 |
| 1662 | 246498_at | AT5G16230 | acyl-(acyl-carrier-protein) desaturase, putative / stearoyl-ACP desaturase, putative | 3.72 |
| 1663 | 250118_at | AT5G16460 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G29760.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN83562.1); contains domain PTHR21212 (PTHR21212) | 8.31 |
| 1664 | 250100_at | AT5G16570 | GLN1;4 (Glutamine synthetase 1;4); glutamate-ammonia ligase | -2.85 |
| 1665 | 246509_at | AT5G16715 | EMB2247 (EMBRYO DEFECTIVE 2247); ATP binding / aminoacyl-tRNA ligase | -2.80 |
| 1666 | 246477_at | AT5G16770 | AtMYB9 (myb domain protein 9); DNA binding / transcription factor | 3.52 |
| 1667 | 246420_at | AT5G16870 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G03010.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G03010.2); similar to hypothetical protein [Vitis vinifera] (GB:CAN83813.1); contains InterPro domain Peptidyl-tRNA hydrolase, PTH2 (InterPro:IPR002833) | -3.47 |
| 1668 | 246463_at | AT5G16970 | AT-AER (ALKENAL REDUCTASE); 2-alkenal reductase | 2.97 |
| 1669 | 250095_at | AT5G17230 | PSY (PHYTOENE SYNTHASE); geranylgeranyl-diphosphate geranylgeranyltransferase | -2.81 |
| 1670 | 250074_at | AT5G17310 | UTP--glucose-1-phosphate uridylyltransferase, putative / UDP-glucose pyrophosphorylase, putative / UGPase, putative | 3.33 |
| 1671 | 250098_at | AT5G17350 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G03280.1); similar to unknown [Populus trichocarpa] (GB:ABK95625.1) | 3.02 |
| 1672 | 246429_at | AT5G17450 | heavy-metal-associated domain-containing protein / copper chaperone (CCH)-related | -3.97 |
| 1673 | 246432_at | AT5G17490 | RGL3 (RGA-LIKE 3); transcription factor | 4.36 |
| 1674 | 250045_at | AT5G17700 | MATE efflux family protein | 3.17 |
| 1675 | 250053_at | AT5G17850 | cation exchanger, putative (CAX8) | 3.21 |
| 1676 | 250014_at | AT5G17990 | TRP1 (TRYPTOPHAN BIOSYNTHESIS 1); anthranilate phosphoribosyltransferase | -2.82 |
| 1677 | 250024_at | AT5G18270 | ANAC087; transcription factor | 2.90 |
| 1678 | 250010_at | AT5G18450 | AP2 domain-containing transcription factor, putative | 5.08 |
| 1679 | 250006_at | AT5G18660 | DVR (PALE-GREEN AND CHLOROPHYLL B REDUCED 2); 3,8-divinyl protochlorophyllide a 8-vinyl reductase | -3.81 |
| 1680 | 249955_at | AT5G18840 | sugar transporter, putative | 3.59 |
| 1681 | 249972_at | AT5G19040 | ATIPT5 (Arabidopsis thaliana isopentenyltransferase 5); transferase, transferring alkyl or aryl (other than methyl) groups | 3.20 |
| 1682 | 249927_at | AT5G19220 | ADG2 (ADPG PYROPHOSPHORYLASE 2); glucose-1-phosphate adenylyltransferase | -3.46 |
| 1683 | 249918_at | AT5G19240 | Identical to Uncharacterized GPI-anchored protein At5g19240 precursor [Arabidopsis Thaliana] (GB:Q84VZ5;GB:Q8H7A4); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G19230.1); similar to unknown [Populus trichocarpa] (GB:ABK94712.1) | -3.49 |
| 1684 | 249919_at | AT5G19250 | Identical to Uncharacterized GPI-anchored protein At5g19250 precursor [Arabidopsis Thaliana] (GB:P59833;GB:Q67ZQ2;GB:Q6ID89); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G06035.1); similar to unknown [Populus trichocarpa] (GB:ABK94712.1) | -7.42 |
| 1685 | 249920_at | AT5G19260 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G06020.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN75990.1) | -3.20 |
| 1686 | 246067_at | AT5G19410 | ABC transporter family protein | 6.21 |
| 1687 | 245939_at | AT5G19760 | dicarboxylate/tricarboxylate carrier (DTC) | -2.71 |
| 1688 | 246125_at | AT5G19875 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G31940.1); similar to hypothetical protein MtrDRAFT_AC155880g29v2 [Medicago truncatula] (GB:ABN08129.1) | -2.51 |
| 1689 | 246154_at | AT5G19940 | plastid-lipid associated protein PAP-related / fibrillin-related | -3.62 |
| 1690 | 246004_at | AT5G20630 | GLP3 (GERMIN-LIKE PROTEIN 3); manganese ion binding / metal ion binding / nutrient reservoir | -6.38 |
| 1691 | 246002_at | AT5G20740 | invertase/pectin methylesterase inhibitor family protein | -5.08 |
| 1692 | 246001_at | AT5G20790 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G43110.1) | 4.86 |
| 1693 | 245997_at | AT5G20810 | auxin-responsive protein, putative / small auxin up RNA (SAUR_B) | 3.46 |
| 1694 | 246181_at | AT5G20860 | pectinesterase family protein | 4.62 |
| 1695 | 246183_at | AT5G20940 | glycosyl hydrolase family 3 protein | 4.27 |
| 1696 | 246133_at | AT5G20960 | AAO1 (ALDEHYDE OXIDASE 1) | 4.16 |
| 1697 | 246027_at | AT5G21060 | homoserine dehydrogenase family protein | -2.34 |
| 1698 | 246021_at | AT5G21100 | L-ascorbate oxidase, putative | -3.92 |
| 1699 | 249937_at | AT5G22470 | NAD+ ADP-ribosyltransferase | 8.77 |
| 1700 | 249895_at | AT5G22500 | acyl CoA reductase, putative / male-sterility protein, putative | 3.53 |
| 1701 | 249894_at | AT5G22580 | Identical to Uncharacterized protein At5g22580 [Arabidopsis Thaliana] (GB:Q9FK81); similar to stable protein 1-related [Arabidopsis thaliana] (TAIR:AT3G17210.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN68720.1); contains InterPro domain Dimeric alpha-beta barrel (InterPro:IPR011008); contains InterPro domain Stress responsive alpha-beta barrel (InterPro:IPR013097) | -4.20 |
| 1702 | 249900_at | AT5G22640 | EMB1211 (EMBRYO DEFECTIVE 1211) | -2.37 |
| 1703 | 249913_at | AT5G22810 | GDSL-motif lipase, putative | 2.81 |
| 1704 | 249866_at | AT5G23010 | MAM1 (2-isopropylmalate synthase 3); 2-isopropylmalate synthase | -2.44 |
| 1705 | 249876_at | AT5G23060 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G59780.1); similar to extracellular Ca2+ sensing receptor [Glycine max] (GB:ABY57763.1); contains InterPro domain Rhodanese-like (InterPro:IPR001763) | -4.13 |
| 1706 | 249875_at | AT5G23120 | HCF136 (High chlorophyll fluorescence 136) | -3.22 |
| 1707 | 249881_at | AT5G23190 | CYP86B1 (cytochrome P450, family 86, subfamily B, polypeptide 1); oxygen binding | 8.08 |
| 1708 | 249847_at | AT5G23210 | SCPL34; serine carboxypeptidase | -4.68 |
| 1709 | 249849_at | AT5G23230 | NIC2 (NICOTINAMIDASE 2); catalytic/ nicotinamidase | 3.55 |
| 1710 | 249838_at | AT5G23460 | unknown protein | 3.56 |
| 1711 | 249817_at | AT5G23820 | MD-2-related lipid recognition domain-containing protein / ML domain-containing protein | -2.73 |
| 1712 | 249809_at | AT5G23910 | microtubule motor | -3.85 |
| 1713 | 249772_at | AT5G24130 | similar to unnamed protein product [Vitis vinifera] (GB:CAO69379.1) | 4.71 |
| 1714 | 249757_at | AT5G24316 | proline-rich family protein | 5.49 |
| 1715 | 249729_at | AT5G24410 | glucosamine/galactosamine-6-phosphate isomerase-related | 4.82 |
| 1716 | 249742_at | AT5G24490 | 30S ribosomal protein, putative | -2.64 |
| 1717 | 249747_at | AT5G24600 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G18215.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO65983.1); contains InterPro domain Protein of unknown function DUF599 (InterPro:IPR006747) | 3.70 |
| 1718 | 246959_at | AT5G24690 | similar to RER1 (RETICULATA-RELATED 1) [Arabidopsis thaliana] (TAIR:AT5G22790.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO24736.1) | -2.69 |
| 1719 | 246978_at | AT5G24910 | CYP714A1 (cytochrome P450, family 714, subfamily A, polypeptide 1); oxygen binding | 4.85 |
| 1720 | 246973_at | AT5G24970 | ABC1 family protein | 4.21 |
| 1721 | 246948_at | AT5G25130 | CYP71B12 (cytochrome P450, family 71, subfamily B, polypeptide 12); oxygen binding | -2.46 |
| 1722 | 246925_at | AT5G25180 | CYP71B14 (cytochrome P450, family 71, subfamily B, polypeptide 14); oxygen binding | 4.72 |
| 1723 | 246935_at | AT5G25350 | EBF2 (EIN3-BINDING F BOX PROTEIN 2) | 3.40 |
| 1724 | 246943_at | AT5G25440 | protein kinase family protein | -3.00 |
| 1725 | 246919_at | AT5G25460 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G11420.1); similar to unknown [Ricinus communis] (GB:CAB02653.1); contains InterPro domain Protein of unknown function DUF642 (InterPro:IPR006946) | -5.75 |
| 1726 | 246896_at | AT5G25550 | leucine-rich repeat family protein / extensin family protein | -2.39 |
| 1727 | 246905_at | AT5G25570 | similar to unnamed protein product [Vitis vinifera] (GB:CAO44135.1) | -2.84 |
| 1728 | 246908_at | AT5G25610 | RD22 (RESPONSIVE TO DESSICATION 22) | -3.42 |
| 1729 | 246912_at | AT5G25820 | exostosin family protein | 4.91 |
| 1730 | 246915_at | AT5G25880 | ATNADP-ME3 (NADP-MALIC ENZYME 3); malate dehydrogenase (oxaloacetate-decarboxylating) (NADP+)/ malic enzyme/ oxidoreductase, acting on NADH or NADPH, NAD or NADP as acceptor | -3.56 |
| 1731 | 246874_at | AT5G26120 | glycosyl hydrolase family protein 51 | 5.78 |
| 1732 | 246854_at | AT5G26200 | mitochondrial substrate carrier family protein | -3.15 |
| 1733 | 246821_at | AT5G26920 | calmodulin binding | -4.44 |
| 1734 | 246814_at | AT5G27200 | ACP5 (ACYL CARRIER PROTEIN 5); acyl carrier | 2.90 |
| 1735 | 246816_at | AT5G27230 | similar to hydroxyproline-rich glycoprotein family protein [Arabidopsis thaliana] (TAIR:AT3G22440.1); similar to unknown [Picea sitchensis] (GB:ABK24390.1); contains InterPro domain Frigida-like (InterPro:IPR012474) | 3.15 |
| 1736 | 246792_at | AT5G27290 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G54680.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G54680.2); similar to unnamed protein product [Vitis vinifera] (GB:CAO68604.1) | -2.33 |
| 1737 | 246137_at | AT5G28490 | LSH1 (LIGHT-DEPENDENT SHORT HYPOCOTYLS 1) | -2.74 |
| 1738 | 245952_at | AT5G28500 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G04550.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO65032.1) | -3.60 |
| 1739 | 246103_at | AT5G28640 | AN3 (ANGUSITFOLIA3) | 2.91 |
| 1740 | 246651_at | AT5G35170 | adenylate kinase family protein | -2.78 |
| 1741 | 246612_at | AT5G35320 | similar to hypothetical protein [Vitis vinifera] (GB:CAN60849.1) | 2.90 |
| 1742 | 249710_at | AT5G35630 | GS2 (GLUTAMINE SYNTHETASE 2); glutamate-ammonia ligase | -5.65 |
| 1743 | 249708_at | AT5G35660 | pseudogene similar to protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | 6.19 |
| 1744 | 249709_at | AT5G35670 | IQD33 (IQ-domain 33); calmodulin binding | -4.28 |
| 1745 | 249719_at | AT5G35735 | auxin-responsive family protein | -3.27 |
| 1746 | 249676_at | AT5G35960 | protein kinase, putative | 3.14 |
| 1747 | 249677_at | AT5G35970 | DNA-binding protein, putative | -3.65 |
| 1748 | 249645_at | AT5G36910 | THI2.2 (THIONIN 2.2); toxin receptor binding | -2.34 |
| 1749 | 249610_at | AT5G37360 | similar to unnamed protein product [Vitis vinifera] (GB:CAO38751.1) | -2.75 |
| 1750 | 249619_at | AT5G37500 | GORK (GATED OUTWARDLY-RECTIFYING K+ CHANNEL); cyclic nucleotide binding / inward rectifier potassium channel/ outward rectifier potassium channel | -3.94 |
| 1751 | 249575_at | AT5G37670 | 15.7 kDa class I-related small heat shock protein-like (HSP15.7-CI) | 3.53 |
| 1752 | 249576_at | AT5G37690 | GDSL-motif lipase/hydrolase family protein | 4.18 |
| 1753 | 249547_at | AT5G38160 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | 8.65 |
| 1754 | 249548_at | AT5G38170 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | 9.54 |
| 1755 | 249549_at | AT5G38180 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | 6.85 |
| 1756 | 249556_at | AT5G38195 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | 7.80 |
| 1757 | 249515_at | AT5G38530 | tryptophan synthase-related | 3.22 |
| 1758 | 249482_at | AT5G38980 | unknown protein | -6.05 |
| 1759 | 249491_at | AT5G39130 | germin-like protein, putative | 7.24 |
| 1760 | 249472_at | AT5G39210 | CRR7 (CHLORORESPIRATORY REDUCTION 7) | -5.91 |
| 1761 | 249500_at | AT5G39260 | ATEXPA21 (ARABIDOPSIS THALIANA EXPANSIN A21) | 3.31 |
| 1762 | 249503_at | AT5G39310 | ATEXPA24 (ARABIDOPSIS THALIANA EXPANSIN A24) | 3.06 |
| 1763 | 249469_at | AT5G39320 | UDP-glucose 6-dehydrogenase, putative | -2.87 |
| 1764 | 249454_at | AT5G39520 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G39530.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO15021.1) | 5.92 |
| 1765 | 249467_at | AT5G39610 | ANAC092/ATNAC2/ATNAC6 (Arabidopsis NAC domain containing protein 92); protein heterodimerization/ protein homodimerization/ transcription factor | 4.13 |
| 1766 | 249465_at | AT5G39720 | AIG2L (AVIRULENCE INDUCED GENE 2 LIKE PROTEIN) | 9.17 |
| 1767 | 249384_at | AT5G39890 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G15120.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO14912.1); contains InterPro domain Protein of unknown function DUF1637 (InterPro:IPR012864) | 5.42 |
| 1768 | 249439_at | AT5G40020 | pathogenesis-related thaumatin family protein | -2.62 |
| 1769 | 249386_at | AT5G40060 | pseudogene of disease resistance protein (TIR-NBS-LRR class) | -3.39 |
| 1770 | 249391_at | AT5G40140 | armadillo/beta-catenin repeat family protein / U-box domain-containing protein | -3.60 |
| 1771 | 249402_at | AT5G40155 | Encodes a defensin-like (DEFL) family protein. | 4.52 |
| 1772 | 249393_at | AT5G40170 | disease resistance family protein | -3.29 |
| 1773 | 249410_at | AT5G40380 | protein kinase family protein | -4.83 |
| 1774 | 249353_at | AT5G40420 | OLEO2 (OLEOSIN 2) | 12.34 |
| 1775 | 249378_at | AT5G40450 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G28770.1); similar to hypothetical protein Kpol_1058p4 [Vanderwaltozyma polyspora DSM 70294] (GB:XP_001645325.1) | -3.06 |
| 1776 | 249376_at | AT5G40645 | nitrate-responsive NOI protein, putative | 4.38 |
| 1777 | 249377_at | AT5G40690 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G41730.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO14635.1) | 3.08 |
| 1778 | 249289_at | AT5G41040 | transferase family protein | 6.15 |
| 1779 | 249306_at | AT5G41400 | zinc finger (C3HC4-type RING finger) family protein | -3.88 |
| 1780 | 249253_at | AT5G42060 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G64490.1); contains InterPro domain DEK C terminal (InterPro:IPR014876) | 3.63 |
| 1781 | 249227_at | AT5G42180 | peroxidase 64 (PER64) (P64) (PRXR4) | 4.25 |
| 1782 | 249216_at | AT5G42240 | SCPL42 (serine carboxypeptidase-like 42); serine carboxypeptidase | -2.58 |
| 1783 | 249246_at | AT5G42290 | transcription activator-related | 9.90 |
| 1784 | 249221_at | AT5G42440 | protein kinase family protein | -3.00 |
| 1785 | 249212_at | AT5G42690 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G37080.2); similar to unnamed protein product [Vitis vinifera] (GB:CAO61290.1); contains InterPro domain Protein of unknown function DUF547 (InterPro:IPR006869) | 4.24 |
| 1786 | 249215_at | AT5G42800 | DFR (DIHYDROFLAVONOL 4-REDUCTASE); dihydrokaempferol 4-reductase | 3.00 |
| 1787 | 249167_at | AT5G42860 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G45688.1); similar to H0814G11.12 [Oryza sativa (indica cultivar-group)] (GB:CAJ86345.1); similar to CAA30379.1 protein [Oryza sativa] (GB:CAB53482.1) | -5.07 |
| 1788 | 249135_at | AT5G43160 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G19570.2); similar to unnamed protein product [Vitis vinifera] (GB:CAO63959.1); contains InterPro domain Protein of unknown function DUF566 (InterPro:IPR007573) | -3.25 |
| 1789 | 249144_at | AT5G43270 | SPL2 (SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 2); DNA binding / transcription factor | -3.31 |
| 1790 | 249128_at | AT5G43440 | 2-oxoglutarate-dependent dioxygenase, putative | -4.69 |
| 1791 | 249101_at | AT5G43580 | serine-type endopeptidase inhibitor | 6.10 |
| 1792 | 249108_at | AT5G43690 | sulfotransferase family protein | 2.86 |
| 1793 | 249109_at | AT5G43700 | ATAUX2-11 (indoleacetic acid-induced protein 4); transcription factor | -3.04 |
| 1794 | 249120_at | AT5G43750 | similar to unnamed protein product [Vitis vinifera] (GB:CAO71280.1) | -5.73 |
| 1795 | 249111_at | AT5G43770 | proline-rich family protein | 7.53 |
| 1796 | 249091_at | AT5G43860 | ATCLH2 (Chlorophyll-chlorophyllido hydrolase 2) | 2.82 |
| 1797 | 249073_at | AT5G44020 | acid phosphatase class B family protein | -4.66 |
| 1798 | 249078_at | AT5G44070 | CAD1 (CADMIUM SENSITIVE 1) | -4.00 |
| 1799 | 249082_at | AT5G44120 | CRA1 (CRUCIFERINA); nutrient reservoir | 13.59 |
| 1800 | 249037_at | AT5G44130 | FLA13 (FASCICLIN-LIKE ARABINOGALACTAN PROTEIN 13 PRECURSOR) | -3.91 |
| 1801 | 249065_at | AT5G44260 | zinc finger (CCCH-type) family protein | 4.68 |
| 1802 | 249039_at | AT5G44310 | late embryogenesis abundant domain-containing protein / LEA domain-containing protein | 11.99 |
| 1803 | 249043_at | AT5G44360 | FAD-binding domain-containing protein | 7.01 |
| 1804 | 249045_at | AT5G44380 | FAD-binding domain-containing protein | 5.79 |
| 1805 | 249046_at | AT5G44400 | FAD-binding domain-containing protein | -4.47 |
| 1806 | 249061_at | AT5G44550 | integral membrane family protein | 3.89 |
| 1807 | 249009_at | AT5G44610 | MAP18 (MICROTUBULE-ASSOCIATED PROTEIN 18); microtubule binding | 3.79 |
| 1808 | 249007_at | AT5G44650 | similar to unnamed protein product [Vitis vinifera] (GB:CAO44475.1) | -2.52 |
| 1809 | 249008_at | AT5G44680 | methyladenine glycosylase family protein | -6.34 |
| 1810 | 249013_at | AT5G44700 | EDA23 (embryo sac development arrest 23); ATP binding / protein serine/threonine kinase | 3.26 |
| 1811 | 254521_at | AT5G44820 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G19970.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO46707.1); contains domain PTHR10483:SF6 (PTHR10483:SF6); contains domain PTHR10483 (PTHR10483) | -3.80 |
| 1812 | 248969_at | AT5G45310 | similar to unnamed protein product [Vitis vinifera] (GB:CAO63757.1) | 7.28 |
| 1813 | 248944_at | AT5G45500 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G45520.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO43141.1); similar to Os01g0799000 [Oryza sativa (japonica cultivar-group)] (GB:NP_001044526.1); contains domain SSF52047 (SSF52047); contains domain G3DSA:3.80.10.10 (G3DSA:3.80.10.10) | -2.77 |
| 1814 | 248959_at | AT5G45630 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G18980.1); similar to unknown [Cucumis sativus] (GB:ABY56081.1); similar to unknown [Cucumis melo] (GB:ABQ53634.1); contains InterPro domain Protein of unknown function DUF584 (InterPro:IPR007608) | 4.40 |
| 1815 | 248915_at | AT5G45690 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G18920.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO47983.1); contains InterPro domain Protein of unknown function DUF1264 (InterPro:IPR010686) | 10.58 |
| 1816 | 248911_at | AT5G45830 | DOG1 (DELAY OF GERMINATION 1) | 8.62 |
| 1817 | 248926_at | AT5G45880 | pollen Ole e 1 allergen and extensin family protein | 4.78 |
| 1818 | 248920_at | AT5G45930 | CHLI2; magnesium chelatase | -5.12 |
| 1819 | 248923_at | AT5G45940 | ATNUDT11 (Arabidopsis thaliana Nudix hydrolase homolog 11); hydrolase | -2.40 |
| 1820 | 248921_at | AT5G45950 | GDSL-motif lipase/hydrolase family protein | -5.32 |
| 1821 | 248886_at | AT5G46110 | APE2 (ACCLIMATION OF PHOTOSYNTHESIS TO ENVIRONMENT); antiporter/ triose-phosphate transmembrane transporter | -2.56 |
| 1822 | 248889_at | AT5G46230 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G09310.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO14438.1); contains InterPro domain Protein of unknown function DUF538 (InterPro:IPR007493) | 4.32 |
| 1823 | 248895_at | AT5G46330 | FLS2 (FLAGELLIN-SENSITIVE 2); ATP binding / kinase/ protein binding / protein serine/threonine kinase/ transmembrane receptor protein serine/threonine kinase | -2.73 |
| 1824 | 248865_at | AT5G46790 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G17870.1); similar to Streptomyces cyclase/dehydrase family protein [Brassica oleracea] (GB:ABD65631.1); contains InterPro domain Streptomyces cyclase/dehydrase (InterPro:IPR005031) | -4.98 |
| 1825 | 248838_at | AT5G46800 | BOU (A BOUT DE SOUFFLE); binding | -3.53 |
| 1826 | 248820_at | AT5G47060 | senescence-associated protein-related | 2.94 |
| 1827 | 248828_at | AT5G47110 | lil3 protein, putative | -2.70 |
| 1828 | 248830_at | AT5G47150 | YDG/SRA domain-containing protein | 4.57 |
| 1829 | 248831_at | AT5G47160 | YDG/SRA domain-containing protein | 4.20 |
| 1830 | 248798_at | AT5G47190 | ribosomal protein L19 family protein | -2.70 |
| 1831 | 248791_at | AT5G47350 | palmitoyl protein thioesterase family protein | 5.35 |
| 1832 | 248807_at | AT5G47500 | pectinesterase family protein | -3.51 |
| 1833 | 248763_at | AT5G47550 | cysteine protease inhibitor, putative / cystatin, putative | 3.61 |
| 1834 | 248761_at | AT5G47635 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G40113.1); similar to hypothetical protein 31.t00008 [Brassica oleracea] (GB:ABD65108.1) | 4.06 |
| 1835 | 248754_at | AT5G47670 | CCAAT-box binding transcription factor family protein / leafy cotyledon 1-related (L1L) | 5.27 |
| 1836 | 248780_at | AT5G47760 | ATPGLP2/ATPK5/PGLP2 (ARABIDOPSIS THALIANA SERINE/THREONINE PROTEIN KINASE 5); phosphoglycolate phosphatase/ protein serine/threonine kinase | -4.62 |
| 1837 | 248718_at | AT5G47770 | FPS1 (FARNESYL DIPHOSPHATE SYNTHASE 1); dimethylallyltranstransferase/ geranyltranstransferase | -4.92 |
| 1838 | 248722_at | AT5G47810 | phosphofructokinase family protein | 5.74 |
| 1839 | 248735_at | AT5G48100 | TT10 (TRANSPARENT TESTA 10); laccase | 8.39 |
| 1840 | 248736_at | AT5G48110 | terpene synthase/cyclase family protein | 4.24 |
| 1841 | 248713_at | AT5G48180 | kelch repeat-containing protein | 3.02 |
| 1842 | 248687_at | AT5G48300 | ADG1 (ADP GLUCOSE PYROPHOSPHORYLASE SMALL SUBUNIT 1); glucose-1-phosphate adenylyltransferase | -2.86 |
| 1843 | 248691_at | AT5G48310 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G24610.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G65440.1); similar to predicted protein [Physcomitrella patens subsp. patens] (GB:EDQ82245.1); similar to predicted protein [Physcomitrella patens subsp. patens] (GB:EDQ70121.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO41110.1); contains InterPro domain C2 calcium/lipid-binding region, CaLB (InterPro:IPR008973) | -2.68 |
| 1844 | 248656_at | AT5G48460 | fimbrin-like protein, putative | -2.34 |
| 1845 | 248684_at | AT5G48485 | DIR1 (DEFECTIVE IN INDUCED RESISTANCE 1); lipid binding | -2.35 |
| 1846 | 248674_at | AT5G48800 | phototropic-responsive NPH3 family protein | -3.74 |
| 1847 | 248676_at | AT5G48850 | male sterility MS5 family protein | 4.30 |
| 1848 | 248623_at | AT5G49170 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G06840.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO44815.1) | -2.58 |
| 1849 | 248593_at | AT5G49180 | pectinesterase family protein | 4.71 |
| 1850 | 248647_at | AT5G49190 | SUS2 (SUCROSE SYNTHASE 2); UDP-glycosyltransferase/ sucrose synthase/ transferase, transferring glycosyl groups | 8.28 |
| 1851 | 248621_at | AT5G49350 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G56320.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO44880.1); contains domain PTHR10499 (PTHR10499) | 6.68 |
| 1852 | 248563_at | AT5G49690 | UDP-glucoronosyl/UDP-glucosyl transferase family protein | 3.69 |
| 1853 | 248564_at | AT5G49700 | DNA-binding protein-related | 6.39 |
| 1854 | 248568_at | AT5G49760 | leucine-rich repeat family protein / protein kinase family protein | -4.77 |
| 1855 | 248572_at | AT5G49800 | similar to unnamed protein product [Vitis vinifera] (GB:CAO46940.1); contains InterPro domain Lipid-binding START (InterPro:IPR002913) | -3.15 |
| 1856 | 248576_at | AT5G49810 | MMT (methionine S-methyltransferase); S-adenosylmethionine-dependent methyltransferase | -2.52 |
| 1857 | 248584_at | AT5G49960 | binding / catalytic | -2.56 |
| 1858 | 248557_at | AT5G49980 | AFB5 (AUXIN F-BOX PROTEIN 5); ubiquitin-protein ligase | -2.44 |
| 1859 | 248558_at | AT5G49990 | xanthine/uracil permease family protein | 4.35 |
| 1860 | 248529_at | AT5G50000 | protein kinase, putative | -2.33 |
| 1861 | 248540_at | AT5G50160 | ATFRO8/FRO8 (FERRIC REDUCTION OXIDASE 8); ferric-chelate reductase/ oxidoreductase | -2.52 |
| 1862 | 248553_at | AT5G50170 | C2 domain-containing protein / GRAM domain-containing protein | 4.81 |
| 1863 | 248545_at | AT5G50260 | cysteine proteinase, putative | 7.12 |
| 1864 | 248555_at | AT5G50340 | ATP binding / ATP-dependent peptidase/ damaged DNA binding / nucleoside-triphosphatase/ nucleotide binding / serine-type endopeptidase | -3.71 |
| 1865 | 248505_at | AT5G50360 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G63350.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO68256.1) | 9.13 |
| 1866 | 248468_at | AT5G50750 | RGP4; alpha-1,4-glucan-protein synthase (UDP-forming) | 5.96 |
| 1867 | 248494_at | AT5G50770 | ATHSD6 (HYDROXYSTEROID DEHYDROGENASE 6); oxidoreductase | 5.74 |
| 1868 | 248496_at | AT5G50790 | nodulin MtN3 family protein | 5.01 |
| 1869 | 248476_at | AT5G50890 | similar to lipase class 3 family protein [Arabidopsis thaliana] (TAIR:AT2G05260.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO69997.1); contains domain SSF53474 (SSF53474) | -2.39 |
| 1870 | 248449_at | AT5G51110 | similar to dehydratase family [Arabidopsis thaliana] (TAIR:AT1G29810.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO62291.1); contains InterPro domain Transcriptional coactivator/pterin dehydratase (InterPro:IPR001533) | -2.42 |
| 1871 | 248435_at | AT5G51210 | OLEO3 (OLEOSIN3) | 7.90 |
| 1872 | 248406_at | AT5G51490 | pectinesterase family protein | 7.54 |
| 1873 | 248377_at | AT5G51720 | similar to Os07g0467200 [Oryza sativa (japonica cultivar-group)] (GB:NP_001059590.1); similar to hypothetical protein OsI_025030 [Oryza sativa (indica cultivar-group)] (GB:EAZ03798.1); contains domain PTHR13680 (PTHR13680); contains domain PTHR13680:SF1 (PTHR13680:SF1) | -4.50 |
| 1874 | 248428_at | AT5G51760 | AHG1 (ABA-HYPERSENSITIVE GERMINATION 1); protein serine/threonine phosphatase | 8.40 |
| 1875 | 248381_at | AT5G51830 | pfkB-type carbohydrate kinase family protein | 3.57 |
| 1876 | 248382_at | AT5G51890 | peroxidase | -3.50 |
| 1877 | 248368_at | AT5G51950 | glucose-methanol-choline (GMC) oxidoreductase family protein | 3.77 |
| 1878 | 248395_at | AT5G52120 | ATPP2-A14 (Phloem protein 2-A14); carbohydrate binding | -2.86 |
| 1879 | 248352_at | AT5G52300 | LTI65/RD29B (RESPONSIVE TO DESSICATION 29B) | 9.74 |
| 1880 | 248333_at | AT5G52390 | photoassimilate-responsive protein, putative | -4.13 |
| 1881 | 248336_at | AT5G52420 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G23920.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO49441.1) | 3.98 |
| 1882 | 248338_at | AT5G52440 | HCF106 (High chlorophyll fluorescence 106) | -4.11 |
| 1883 | 248335_at | AT5G52450 | MATE efflux protein-related | -2.46 |
| 1884 | 248311_at | AT5G52570 | BETA-OHASE 2 (BETA-CAROTENE HYDROXYLASE 2); beta-carotene hydroxylase | 3.26 |
| 1885 | 248332_at | AT5G52640 | HSP81-1 (HEAT SHOCK PROTEIN 81-1); ATP binding / unfolded protein binding | 3.54 |
| 1886 | 248322_at | AT5G52760 | heavy-metal-associated domain-containing protein | -6.22 |
| 1887 | 248287_at | AT5G52970 | thylakoid lumen 15.0 kDa protein | -3.17 |
| 1888 | 248302_at | AT5G53160 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G27920.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO69376.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN64668.1); contains InterPro domain Bet v I allergen; (InterPro:IPR000916) | -2.49 |
| 1889 | 248263_at | AT5G53370 | ATPMEPCRF; pectinesterase | -3.29 |
| 1890 | 248224_at | AT5G53490 | thylakoid lumenal 17.4 kDa protein, chloroplast | -3.20 |
| 1891 | 248230_at | AT5G53830 | VQ motif-containing protein | -3.78 |
| 1892 | 248186_at | AT5G53880 | similar to unknown [Populus trichocarpa] (GB:ABK92577.1) | -4.08 |
| 1893 | 248207_at | AT5G53970 | aminotransferase, putative | 3.29 |
| 1894 | 248210_at | AT5G54000 | oxidoreductase, 2OG-Fe(II) oxygenase family protein | 10.35 |
| 1895 | 248188_at | AT5G54070 | AT-HSFA9 (ARABIDOPSIS THALIANA HEAT SHOCK TRANSCRIPTION FACTOR A9); DNA binding / transcription factor | 6.14 |
| 1896 | 248194_at | AT5G54095 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G27580.1) | 3.11 |
| 1897 | 248202_at | AT5G54220 | Encodes a defensin-like (DEFL) family protein. | 3.61 |
| 1898 | 248205_at | AT5G54300 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G61260.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO39207.1); contains InterPro domain Protein of unknown function DUF761, plant (InterPro:IPR008480) | 3.90 |
| 1899 | 248163_at | AT5G54510 | DFL1/GH3.6 (DWARF IN LIGHT 1); indole-3-acetic acid amido synthetase | -3.56 |
| 1900 | 248175_at | AT5G54640 | HTA1/RAT5 (RESISTANT TO AGROBACTERIUM TRANSFORMATION 5); DNA binding | 3.34 |
| 1901 | 248125_at | AT5G54740 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | 10.53 |
| 1902 | 248139_at | AT5G54970 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G26960.1) | -2.91 |
| 1903 | 248094_at | AT5G55220 | trigger factor type chaperone family protein | -4.23 |
| 1904 | 248096_at | AT5G55240 | caleosin-related family protein / embryo-specific protein, putative | 8.13 |
| 1905 | 248062_at | AT5G55450 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | -4.64 |
| 1906 | 248084_at | AT5G55470 | ATNHX3 (Arabidopsis thaliana Na+/H+ exchanger 3); sodium:hydrogen antiporter | 3.33 |
| 1907 | 248086_at | AT5G55490 | ATGEX1/GEX1 (GAMETE EXPRESSED PROTEIN1) | 3.94 |
| 1908 | 248065_at | AT5G55580 | mitochondrial transcription termination factor family protein / mTERF family protein | -3.61 |
| 1909 | 248028_at | AT5G55620 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G09950.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO39940.1) | 3.66 |
| 1910 | 248076_at | AT5G55750 | hydroxyproline-rich glycoprotein family protein | 9.01 |
| 1911 | 248050_at | AT5G56100 | glycine-rich protein / oleosin | 4.21 |
| 1912 | 248011_at | AT5G56300 | GAMT2; S-adenosylmethionine-dependent methyltransferase/ gibberellin carboxyl-O-methyltransferase | 5.37 |
| 1913 | 248020_at | AT5G56490 | FAD-binding domain-containing protein | -3.02 |
| 1914 | 247965_at | AT5G56540 | AGP14 (ARABINOGALACTAN PROTEIN 14) | 6.77 |
| 1915 | 247964_at | AT5G56600 | PFN3/PRF3 (PROFILIN 3); actin binding | -2.71 |
| 1916 | 247977_at | AT5G56850 | similar to unknown [Populus trichocarpa x Populus deltoides] (GB:ABK96358.1) | -6.07 |
| 1917 | 247936_at | AT5G57030 | LUT2 (LUTEIN DEFICIENT 2); lycopene epsilon cyclase | -2.55 |
| 1918 | 247957_at | AT5G57050 | ABI2 (ABA INSENSITIVE 2); protein serine/threonine phosphatase | 2.96 |
| 1919 | 247946_at | AT5G57180 | CIA2 (CHLOROPLAST IMPORT APPARATUS 2) | -3.21 |
| 1920 | 247951_at | AT5G57240 | oxysterol-binding family protein | 4.02 |
| 1921 | 247953_at | AT5G57260 | CYP71B10 (cytochrome P450, family 71, subfamily B, polypeptide 10); oxygen binding | 6.05 |
| 1922 | 247899_at | AT5G57345 | similar to unknown [Populus trichocarpa x Populus deltoides] (GB:ABK96592.1) | -2.70 |
| 1923 | 247904_at | AT5G57390 | AIL5 (AINTEGUMENTA-LIKE 5); DNA binding / transcription factor | 4.59 |
| 1924 | 247905_at | AT5G57400 | unknown protein | 4.30 |
| 1925 | 247866_at | AT5G57550 | XTR3 (XYLOGLUCAN ENDOTRANSGLYCOSYLASE 3); hydrolase, acting on glycosyl bonds | 9.16 |
| 1926 | 247880_at | AT5G57780 | transcription regulator | -3.12 |
| 1927 | 247882_at | AT5G57785 | unknown protein | -3.06 |
| 1928 | 247883_at | AT5G57790 | unknown protein | 6.88 |
| 1929 | 247897_at | AT5G57810 | TET15 (TETRASPANIN15) | 4.73 |
| 1930 | 247851_at | AT5G58070 | lipocalin, putative | 3.84 |
| 1931 | 247853_at | AT5G58140 | PHOT2 (NON PHOTOTROPIC HYPOCOTYL 1-LIKE); kinase | -3.46 |
| 1932 | 247861_at | AT5G58160 | actin binding | 3.20 |
| 1933 | 247855_at | AT5G58210 | hydroxyproline-rich glycoprotein family protein | 3.29 |
| 1934 | 247816_at | AT5G58260 | Encodes subunit NDH-N of NAD(P)H:plastoquinone dehydrogenase complex (Ndh complex) present in the thylakoid membrane of chloroplasts. This subunit is thought to be required for Ndh complex assembly. | -2.71 |
| 1935 | 247856_at | AT5G58300 | leucine-rich repeat transmembrane protein kinase, putative | -3.81 |
| 1936 | 247793_at | AT5G58650 | PSY1 (PLANT PEPTIDE CONTAINING SULFATED TYROSINE 1) | 3.28 |
| 1937 | 247774_at | AT5G58660 | oxidoreductase, 2OG-Fe(II) oxygenase family protein | 4.21 |
| 1938 | 247788_at | AT5G58730 | pfkB-type carbohydrate kinase family protein | 2.89 |
| 1939 | 247765_at | AT5G58860 | CYP86A1 (cytochrome P450, family 86, subfamily A, polypeptide 1); oxygen binding | 4.89 |
| 1940 | 247741_at | AT5G58960 | GIL1 (GRAVITROPIC IN THE LIGHT) | -4.10 |
| 1941 | 247743_at | AT5G59010 | protein kinase-related | -4.63 |
| 1942 | 247755_at | AT5G59090 | ATSBT4.12; subtilase | -3.10 |
| 1943 | 247762_at | AT5G59170 | proline-rich family protein | 8.03 |
| 1944 | 247723_at | AT5G59220 | protein phosphatase 2C, putative / PP2C, putative | 4.13 |
| 1945 | 247709_at | AT5G59250 | sugar transporter family protein | -5.94 |
| 1946 | 247731_at | AT5G59590 | UDP-glucoronosyl/UDP-glucosyl transferase family protein | 3.27 |
| 1947 | 247657_at | AT5G59845 | gibberellin-regulated family protein | 5.26 |
| 1948 | 247651_at | AT5G59870 | HTA6; DNA binding | -2.61 |
| 1949 | 247648_at | AT5G60020 | LAC17 (laccase 17); copper ion binding / oxidoreductase | -4.36 |
| 1950 | 247666_at | AT5G60140 | transcriptional factor B3 family protein | 3.97 |
| 1951 | 247672_at | AT5G60220 | TET4 (TETRASPANIN4) | 6.18 |
| 1952 | 247632_at | AT5G60460 | sec61beta family protein | 4.75 |
| 1953 | 247634_at | AT5G60520 | late embryogenesis abundant protein-related / LEA protein-related | 2.95 |
| 1954 | 247586_at | AT5G60660 | PIP2;4/PIP2F (plasma membrane intrinsic protein 2;4); water channel | -2.96 |
| 1955 | 247582_at | AT5G60760 | 2-phosphoglycerate kinase-related | 8.70 |
| 1956 | 247563_at | AT5G61130 | glycosyl hydrolase family protein 17 | -2.76 |
| 1957 | 247549_at | AT5G61420 | MYB28 (MYB DOMAIN PROTEIN 28); DNA binding / transcription factor | -2.97 |
| 1958 | 247531_at | AT5G61550 | protein kinase family protein | 2.92 |
| 1959 | 247541_at | AT5G61660 | glycine-rich protein | -4.31 |
| 1960 | 247474_at | AT5G62280 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G45360.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO49726.1); contains InterPro domain Protein of unknown function DUF1442 (InterPro:IPR009902) | -3.98 |
| 1961 | 247450_at | AT5G62350 | invertase/pectin methylesterase inhibitor family protein / DC 1.2 homolog (FL5-2I22) | -3.40 |
| 1962 | 247478_at | AT5G62360 | invertase/pectin methylesterase inhibitor family protein | -2.41 |
| 1963 | 247435_at | AT5G62480 | ATGSTU9 (GLUTATHIONE S-TRANSFERASE TAU 9); glutathione transferase | 4.49 |
| 1964 | 247437_at | AT5G62490 | ATHVA22B (Arabidopsis thaliana HVA22 homologue B) | 10.51 |
| 1965 | 247425_at | AT5G62550 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G28770.1); similar to unknown [Populus trichocarpa] (GB:ABK92769.1) | -4.28 |
| 1966 | 247429_at | AT5G62620 | galactosyltransferase family protein | 2.85 |
| 1967 | 247402_at | AT5G62750 | unknown protein | 4.43 |
| 1968 | 247421_at | AT5G62800 | seven in absentia (SINA) family protein | 7.30 |
| 1969 | 247424_at | AT5G62850 | ATVEX1 (VEGETATIVE CELL EXPRESSED1) | 3.06 |
| 1970 | 247406_at | AT5G62920 | ARR6 (RESPONSE REGULATOR 6); transcription regulator/ two-component response regulator | -2.96 |
| 1971 | 247418_at | AT5G63030 | glutaredoxin, putative | 4.40 |
| 1972 | 247374_at | AT5G63190 | MA3 domain-containing protein | 2.99 |
| 1973 | 247376_at | AT5G63310 | NDPK2 (NUCLEOSIDE DIPHOSPHATE KINASE 2); ATP binding / nucleoside diphosphate kinase | -3.50 |
| 1974 | 247386_at | AT5G63420 | EMB2746 (EMBRYO DEFECTIVE 2746); catalytic | -2.77 |
| 1975 | 247391_at | AT5G63540 | similar to hypothetical protein OsJ_014026 [Oryza sativa (japonica cultivar-group)] (GB:EAZ30543.1); similar to OSJNBa0055C08.4 [Oryza sativa (japonica cultivar-group)] (GB:CAE02280.2); contains InterPro domain Region of unknown function DUF1767 (InterPro:IPR013894) | -2.65 |
| 1976 | 247359_at | AT5G63560 | transferase family protein | 6.61 |
| 1977 | 247354_at | AT5G63590 | FLS (Flavonol synthase); flavonol synthase | 6.44 |
| 1978 | 247333_at | AT5G63600 | flavonol synthase, putative | 4.33 |
| 1979 | 247357_at | AT5G63710 | leucine-rich repeat transmembrane protein kinase, putative | -3.38 |
| 1980 | 247348_at | AT5G63810 | BGAL10 (beta-galactosidase 10); beta-galactosidase | -3.02 |
| 1981 | 247312_at | AT5G63970 | copine-related | 3.01 |
| 1982 | 247320_at | AT5G64040 | PSAN (photosystem I reaction center subunit PSI-N); calmodulin binding | -3.44 |
| 1983 | 247268_at | AT5G64080 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | 4.36 |
| 1984 | 247327_at | AT5G64120 | peroxidase, putative | -3.68 |
| 1985 | 247324_at | AT5G64190 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G40390.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO41445.1) | 6.78 |
| 1986 | 247269_at | AT5G64210 | AOX2 (alternative oxidase 2); alternative oxidase | 3.62 |
| 1987 | 247287_at | AT5G64230 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G19920.1); similar to hypothetical protein [Trifolium pratense] (GB:BAE71289.1) | 4.27 |
| 1988 | 247261_at | AT5G64460 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G58280.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO67833.1); contains InterPro domain Phosphoglycerate mutase (InterPro:IPR013078) | -2.84 |
| 1989 | 247243_at | AT5G64700 | nodulin MtN21 family protein | -2.77 |
| 1990 | 247252_at | AT5G64770 | similar to 80C09_10 [Brassica rapa subsp. pekinensis] (GB:AAZ41821.1) | -4.86 |
| 1991 | 247255_at | AT5G64780 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G09830.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO71731.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN64859.1); contains InterPro domain Uncharacterised conserved protein UCP009193 (InterPro:IPR016549) | 3.02 |
| 1992 | 247216_at | AT5G64860 | DPE1 (DISPROPORTIONATING ENZYME); 4-alpha-glucanotransferase | -2.97 |
| 1993 | 247232_at | AT5G64940 | ATATH13 (ABC2 homolog 13) | -2.88 |
| 1994 | 247218_at | AT5G65010 | ASN2 (ASPARAGINE SYNTHETASE 2); asparagine synthase (glutamine-hydrolyzing) | -2.48 |
| 1995 | 247223_at | AT5G65070 | MAF4 (MADS AFFECTING FLOWERING 4) | 3.53 |
| 1996 | 247226_at | AT5G65100 | ethylene insensitive 3 family protein | 3.71 |
| 1997 | 247228_at | AT5G65140 | trehalose-6-phosphate phosphatase, putative | 3.50 |
| 1998 | 247192_at | AT5G65360 | histone H3 | -2.37 |
| 1999 | 247170_at | AT5G65530 | protein kinase, putative | 5.31 |
| 2000 | 247172_at | AT5G65550 | UDP-glucoronosyl/UDP-glucosyl transferase family protein | 5.76 |
| 2001 | 247162_at | AT5G65730 | xyloglucan:xyloglucosyl transferase, putative / xyloglucan endotransglycosylase, putative / endo-xyloglucan transferase, putative | -6.53 |
| 2002 | 247161_at | AT5G65810 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G49720.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G49720.2); similar to hypothetical protein [Vitis vinifera] (GB:CAN74732.1); contains domain S-adenosyl-L-methionine-dependent methyltransferases (SSF53335) | -2.36 |
| 2003 | 247118_at | AT5G65890 | ACR1 (ACT DOMAIN REPEAT 1) | 3.82 |
| 2004 | 247120_at | AT5G65990 | amino acid transporter family protein | -3.15 |
| 2005 | 247128_at | AT5G66110 | metal ion binding | 7.08 |
| 2006 | 247136_at | AT5G66170 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G17850.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO48196.1); contains InterPro domain Rhodanese-like (InterPro:IPR001763) | 3.24 |
| 2007 | 247095_at | AT5G66400 | RAB18 (RESPONSIVE TO ABA 18) | 8.23 |
| 2008 | 247096_at | AT5G66430 | S-adenosyl-L-methionine:carboxyl methyltransferase family protein | 4.16 |
| 2009 | 247098_at | AT5G66470 | GTP binding / RNA binding | -3.04 |
| 2010 | 247061_at | AT5G66780 | similar to unknown [Ammopiptanthus mongolicus] (GB:AAW33981.1) | 10.33 |
| 2011 | 247024_at | AT5G66985 | unknown protein | 7.40 |
| 2012 | 247026_at | AT5G67080 | MAPKKK19 (Mitogen-activated protein kinase kinase kinase 19); kinase | 3.11 |
| 2013 | 247028_at | AT5G67100 | ICU2 (INCURVATA2); DNA-directed DNA polymerase | -2.59 |
| 2014 | 246997_at | AT5G67390 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G69360.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO71983.1) | -4.42 |
| 2015 | 246996_at | AT5G67420 | LBD37 (LOB DOMAIN-CONTAINING PROTEIN 37) | -3.06 |
| 2016 | 247013_at | AT5G67480 | BT4 (BTB AND TAZ DOMAIN PROTEIN 4); protein binding / transcription regulator | 3.16 |
| 2017 | 247009_at | AT5G67600 | unknown protein | 3.00 |
| 2018 | 245007_at | ATCG00350 | Encodes psaA protein comprising the reaction center for photosystem I along with psaB protein; hydrophobic protein encoded by the chloroplast genome. | -2.45 |