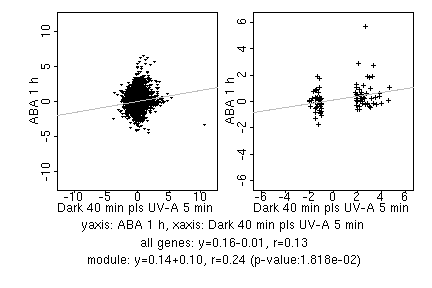
marker gene list
| Dark 40 min pls UV-A 5 min |
| ABA 1 h |
| probeID | AGICode | Annotation | gene expression |
|
|---|---|---|---|---|
| 1 | 260955_at | AT1G06000 | UDP-glucoronosyl/UDP-glucosyl transferase family protein | 2.57 |
| 2 | 260819_at | AT1G06850 | ATBZIP52 (ARABIDOPSIS THALIANA BASIC LEUCINE ZIPPER 52); DNA binding / transcription factor | -1.00 |
| 3 | 256062_at | AT1G07090 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G58500.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN60458.1); contains InterPro domain Protein of unknown function DUF640 (InterPro:IPR006936) | -1.00 |
| 4 | 263207_at | AT1G10550 | XTH33 (xyloglucan:xyloglucosyl transferase 33); hydrolase, acting on glycosyl bonds | -1.30 |
| 5 | 256097_at | AT1G13670 | unknown protein | 2.23 |
| 6 | 262850_at | AT1G14920 | GAI (GA INSENSITIVE); transcription factor | -1.10 |
| 7 | 261663_at | AT1G18330 | EPR1 (EARLY-PHYTOCHROME-RESPONSIVE1); DNA binding / transcription factor | 2.50 |
| 8 | 259787_at | AT1G29460 | auxin-responsive protein, putative | -0.98 |
| 9 | 259773_at | AT1G29500 | auxin-responsive protein, putative | -1.05 |
| 10 | 264415_at | AT1G43160 | RAP2.6 (related to AP2 6); DNA binding / transcription factor | 3.35 |
| 11 | 246371_at | AT1G51940 | protein kinase family protein / peptidoglycan-binding LysM domain-containing protein | -0.89 |
| 12 | 263151_at | AT1G54120 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G14060.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO45609.1) | -0.97 |
| 13 | 263738_at | AT1G60060 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G53900.2); similar to unnamed protein product [Vitis vinifera] (GB:CAO40921.1); contains domain PTHR13902:SF3 (PTHR13902:SF3); contains domain PTHR13902 (PTHR13902) | -1.43 |
| 14 | 261975_at | AT1G64640 | plastocyanin-like domain-containing protein | -0.90 |
| 15 | 260167_at | AT1G71970 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G22680.1); similar to unknown [Populus trichocarpa] (GB:ABK94498.1) | -0.93 |
| 16 | 260070_at | AT1G73830 | BEE3 (BR ENHANCED EXPRESSION 3); DNA binding / transcription factor | -0.94 |
| 17 | 259729_at | AT1G77640 | AP2 domain-containing transcription factor, putative | -1.05 |
| 18 | 260773_at | AT1G78440 | ATGA2OX1 (GIBBERELLIN 2-OXIDASE 1); gibberellin 2-beta-dioxygenase | 3.10 |
| 19 | 266745_at | AT2G02950 | PKS1 (PHYTOCHROME KINASE SUBSTRATE 1) | 2.76 |
| 20 | 265342_at | AT2G18300 | basic helix-loop-helix (bHLH) family protein | -0.92 |
| 21 | 266688_at | AT2G19660 | DC1 domain-containing protein | -1.01 |
| 22 | 263374_at | AT2G20560 | DNAJ heat shock family protein | 2.04 |
| 23 | 264021_at | AT2G21200 | auxin-responsive protein, putative | -1.28 |
| 24 | 263739_at | AT2G21320 | zinc finger (B-box type) family protein | 3.94 |
| 25 | 263796_at | AT2G24540 | AFR (ATTENUATED FAR-RED RESPONSE) | 2.60 |
| 26 | 266841_at | AT2G26150 | ATHSFA2 (Arabidopsis thaliana heat shock transcription factor A2); DNA binding / transcription factor | 4.73 |
| 27 | 267614_at | AT2G26710 | BAS1/CYP734A1 (PHYB ACTIVATION TAGGED SUPPRESSOR 1); oxygen binding / steroid hydroxylase | -1.37 |
| 28 | 266832_at | AT2G30040 | MAPKKK14 (Mitogen-activated protein kinase kinase kinase 14); kinase | 1.98 |
| 29 | 267516_at | AT2G30520 | RPT2 (ROOT PHOTOTROPISM 2); protein binding | 2.97 |
| 30 | 267517_at | AT2G30520 | RPT2 (ROOT PHOTOTROPISM 2); protein binding | 3.40 |
| 31 | 263252_at | AT2G31380 | STH (salt tolerance homologue); transcription factor/ zinc ion binding | 2.35 |
| 32 | 266097_at | AT2G37970 | SOUL-1; binding | 2.10 |
| 33 | 267364_at | AT2G40080 | ELF4 (EARLY FLOWERING 4) | 2.12 |
| 34 | 265245_at | AT2G43060 | transcription factor | -0.95 |
| 35 | 266125_at | AT2G45050 | zinc finger (GATA type) family protein | -1.04 |
| 36 | 263779_at | AT2G46340 | SPA1 (SUPPRESSOR OF PHYA-105 1); signal transducer | 2.92 |
| 37 | 258497_at | AT3G02380 | COL2 (CONSTANS-LIKE 2); transcription factor/ zinc ion binding | 2.41 |
| 38 | 258609_at | AT3G02910 | Identical to UPF0131 protein At3g02910 [Arabidopsis Thaliana] (GB:Q9M8T3); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G46720.1); similar to Os03g0854000 [Oryza sativa (japonica cultivar-group)] (GB:NP_001051934.1); similar to unknown [Picea sitchensis] (GB:ABK22133.1); contains InterPro domain Butirosin biosynthesis, BtrG-like (InterPro:IPR013024); contains InterPro domain Protein of unknown function UPF0131 (InterPro:IPR005347) | 3.17 |
| 39 | 258742_at | AT3G05800 | transcription factor | 2.29 |
| 40 | 256266_at | AT3G12320 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G06980.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN75675.1) | 2.33 |
| 41 | 256245_at | AT3G12580 | HSP70 (heat shock protein 70); ATP binding | 2.87 |
| 42 | 257858_at | AT3G12920 | protein binding / zinc ion binding | -0.98 |
| 43 | 256999_at | AT3G14200 | DNAJ heat shock N-terminal domain-containing protein | 2.11 |
| 44 | 258399_at | AT3G15540 | IAA19 (indoleacetic acid-induced protein 19); transcription factor | -0.98 |
| 45 | 257932_at | AT3G17040 | HCF107 (HIGH CHLOROPHYLL FLUORESCENT 107); binding | 2.12 |
| 46 | 258349_at | AT3G17609 | HYH (HY5-HOMOLOG); DNA binding / transcription factor | 4.12 |
| 47 | 258167_at | AT3G21560 | UGT84A2; UDP-glycosyltransferase/ sinapate 1-glucosyltransferase | 2.06 |
| 48 | 257262_at | AT3G21890 | zinc finger (B-box type) family protein | 3.43 |
| 49 | 258325_at | AT3G22830 | AT-HSFA6B (Arabidopsis thaliana heat shock transcription factor A6B); DNA binding / transcription factor | 2.74 |
| 50 | 258321_at | AT3G22840 | ELIP1 (EARLY LIGHT-INDUCABLE PROTEIN); chlorophyll binding | 3.31 |
| 51 | 258133_at | AT3G24500 | ATMBF1C/MBF1C (MULTIPROTEIN BRIDGING FACTOR 1C); DNA binding / transcription coactivator/ transcription factor | 2.43 |
| 52 | 252558_at | AT3G45990 | actin-depolymerizing factor, putative | -1.20 |
| 53 | 252429_at | AT3G47500 | CDF3 (CYCLING DOF FACTOR 3); DNA binding / protein binding / transcription factor | 2.06 |
| 54 | 252123_at | AT3G51240 | F3H (TRANSPARENT TESTA 6); naringenin 3-dioxygenase | 3.61 |
| 55 | 251977_at | AT3G53250 | auxin-responsive family protein | -1.70 |
| 56 | 251811_at | AT3G54990 | SMZ (SCHLAFMUTZE); DNA binding / transcription factor | 2.09 |
| 57 | 251289_at | AT3G61830 | ARF18 (AUXIN RESPONSE FACTOR 18); transcription factor | -0.91 |
| 58 | 255064_at | AT4G08950 | phosphate-responsive protein, putative (EXO) | -1.19 |
| 59 | 254876_at | AT4G11610 | C2 domain-containing protein | -0.89 |
| 60 | 254839_at | AT4G12400 | stress-inducible protein, putative | 2.69 |
| 61 | 254809_at | AT4G12410 | auxin-responsive family protein | 2.05 |
| 62 | 254685_at | AT4G13790 | auxin-responsive protein, putative | -1.83 |
| 63 | 245328_at | AT4G14465 | DNA-binding protein-related | -1.23 |
| 64 | 245439_at | AT4G16670 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G17350.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO39969.1); contains InterPro domain Pleckstrin-like, plant (InterPro:IPR013666); contains InterPro domain Protein of unknown function DUF828, plant (InterPro:IPR008546); contains InterPro domain Pleckstrin-like (InterPro:IPR001849) | -1.32 |
| 65 | 254356_at | AT4G22190 | similar to unknown [Populus trichocarpa] (GB:ABK94177.1) | -1.10 |
| 66 | 254110_at | AT4G25260 | invertase/pectin methylesterase inhibitor family protein | -0.89 |
| 67 | 254066_at | AT4G25480 | DREB1A (DEHYDRATION RESPONSE ELEMENT B1A); DNA binding / transcription activator/ transcription factor | 2.96 |
| 68 | 253423_at | AT4G32280 | IAA29 (indoleacetic acid-induced protein 29); transcription factor | -1.90 |
| 69 | 253150_at | AT4G35660 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G35680.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO23714.1); contains InterPro domain Protein of unknown function DUF241, plant (InterPro:IPR004320) | 2.01 |
| 70 | 253155_at | AT4G35720 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G35690.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO23717.1); contains InterPro domain Protein of unknown function DUF241, plant (InterPro:IPR004320) | -1.08 |
| 71 | 253104_at | AT4G36010 | pathogenesis-related thaumatin family protein | -1.35 |
| 72 | 253103_at | AT4G36110 | auxin-responsive protein, putative | -1.38 |
| 73 | 253065_at | AT4G37740 | AtGRF2 (GROWTHREGULATING FACTOR 2) | -1.04 |
| 74 | 252997_at | AT4G38400 | ATEXLA2 (ARABIDOPSIS THALIANA EXPANSIN-LIKE A2) | -1.11 |
| 75 | 252965_at | AT4G38860 | auxin-responsive protein, putative | -1.04 |
| 76 | 251058_at | AT5G01790 | unknown protein | -1.72 |
| 77 | 251020_at | AT5G02270 | ATNAP9 (Non-intrinsic ABC protein 9) | 3.72 |
| 78 | 251012_at | AT5G02580 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G55240.1); similar to hypothetical protein OsI_019565 [Oryza sativa (indica cultivar-group)] (GB:EAY98332.1); similar to Os05g0462000 [Oryza sativa (japonica cultivar-group)] (GB:NP_001055765.1); contains InterPro domain Conserved hypothetical protein CHP01589, plant (InterPro:IPR006476) | -1.93 |
| 79 | 245904_at | AT5G11110 | ATSPS2F/SPS1 (SUCROSE PHOSPHATE SYNTHASE 1); sucrose-phosphate synthase | 2.20 |
| 80 | 250420_at | AT5G11260 | HY5 (ELONGATED HYPOCOTYL 5); DNA binding / transcription factor | 3.12 |
| 81 | 246468_at | AT5G17050 | UDP-glucoronosyl/UDP-glucosyl transferase family protein | 2.33 |
| 82 | 250099_at | AT5G17300 | myb family transcription factor | 2.04 |
| 83 | 250012_x_at | AT5G18060 | auxin-responsive protein, putative | -1.41 |
| 84 | 249862_at | AT5G22920 | zinc finger (C3HC4-type RING finger) family protein | -0.94 |
| 85 | 249769_at | AT5G24120 | SIGE (RNA polymerase sigma subunit E); DNA binding / DNA-directed RNA polymerase/ sigma factor/ transcription factor | 3.21 |
| 86 | 249741_at | AT5G24470 | APRR5 (PSEUDO-RESPONSE REGULATOR 5); transcription regulator | 2.61 |
| 87 | 246966_at | AT5G24850 | CRY3 (CRYPTOCHROME 3); DNA binding / DNA photolyase/ FMN binding | 2.48 |
| 88 | 246932_at | AT5G25190 | ethylene-responsive element-binding protein, putative | -1.44 |
| 89 | 249383_at | AT5G39860 | PRE1 (PACLOBUTRAZOL RESISTANCE1); DNA binding / transcription factor | -1.12 |
| 90 | 249063_at | AT5G44110 | POP1 | 3.92 |
| 91 | 249035_at | AT5G44190 | GLK2 (GOLDEN2-LIKE 2); DNA binding / transcription factor | 2.28 |
| 92 | 249065_at | AT5G44260 | zinc finger (CCCH-type) family protein | -1.56 |
| 93 | 248759_at | AT5G47610 | zinc finger (C3HC4-type RING finger) family protein | 1.98 |
| 94 | 248347_at | AT5G52250 | transducin family protein / WD-40 repeat family protein | 2.54 |
| 95 | 248213_at | AT5G53660 | AtGRF7 (GROWTH-REGULATING FACTOR 7) | -1.26 |
| 96 | 248208_at | AT5G53980 | ATHB52 (ARABIDOPSIS THALIANA HOMEOBOX PROTEIN 52); transcription factor | -1.38 |
| 97 | 247947_at | AT5G57090 | EIR1 (ETHYLENE INSENSITIVE ROOT 1); auxin:hydrogen symporter/ transporter | -1.14 |
| 98 | 247352_at | AT5G63650 | SNRK2-5/SNRK2.5/SRK2H (SNF1-RELATED PROTEIN KINASE 2.5); kinase | -0.96 |
| 99 | 247323_at | AT5G64170 | dentin sialophosphoprotein-related | 4.81 |
| 100 | 247087_at | AT5G66330 | leucine-rich repeat family protein | -0.93 |