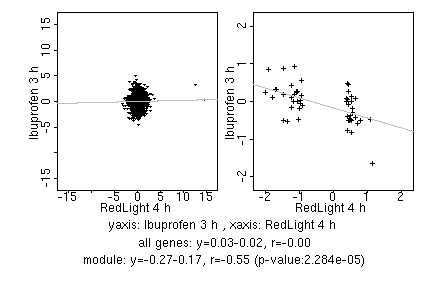
marker gene list
| Red Light 4 h |
| Ibuprofen 3 h |
| probeID | AGICode | Annotation | gene expression |
|
|---|---|---|---|---|
| 1 | 262525_at | AT1G17060 | CYP72C1 (cytochrome P450, family 72, subfamily C, polypeptide 1); oxygen binding | 0.56 |
| 2 | 261927_at | AT1G22500 | zinc finger (C3HC4-type RING finger) family protein | 0.73 |
| 3 | 260516_at | AT1G51450 | SPla/RYanodine receptor (SPRY) domain-containing protein | 0.43 |
| 4 | 262643_at | AT1G62770 | invertase/pectin methylesterase inhibitor family protein | 0.50 |
| 5 | 260072_at | AT1G73650 | oxidoreductase, acting on the CH-CH group of donors | -0.88 |
| 6 | 263128_at | AT1G78600 | zinc finger (B-box type) family protein | -0.91 |
| 7 | 262045_at | AT1G80240 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G25460.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO44183.1); contains InterPro domain Protein of unknown function DUF642 (InterPro:IPR006946) | 1.19 |
| 8 | 265443_at | AT2G20750 | ATEXPB1 (ARABIDOPSIS THALIANA EXPANSIN B1) | 0.43 |
| 9 | 266656_at | AT2G25900 | ATCTH (Arabidopsis thaliana Cys3His zinc finger protein); transcription factor | 0.52 |
| 10 | 266097_at | AT2G37970 | SOUL-1; binding | -0.95 |
| 11 | 263787_at | AT2G46420 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G61700.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO39750.1); contains InterPro domain Conserved hypothetical protein CHP01589, plant (InterPro:IPR006476) | -1.12 |
| 12 | 258609_at | AT3G02910 | Identical to UPF0131 protein At3g02910 [Arabidopsis Thaliana] (GB:Q9M8T3); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G46720.1); similar to Os03g0854000 [Oryza sativa (japonica cultivar-group)] (GB:NP_001051934.1); similar to unknown [Picea sitchensis] (GB:ABK22133.1); contains InterPro domain Butirosin biosynthesis, BtrG-like (InterPro:IPR013024); contains InterPro domain Protein of unknown function UPF0131 (InterPro:IPR005347) | -1.20 |
| 13 | 257701_at | AT3G12710 | methyladenine glycosylase family protein | 0.60 |
| 14 | 256887_at | AT3G15150 | similar to unnamed protein product [Vitis vinifera] (GB:CAO39331.1); contains domain PTHR21330 (PTHR21330); contains domain SSF57850 (SSF57850); contains domain PTHR21330:SF1 (PTHR21330:SF1) | 0.60 |
| 15 | 258349_at | AT3G17609 | HYH (HY5-HOMOLOG); DNA binding / transcription factor | -1.19 |
| 16 | 257033_at | AT3G19170 | ATPREP1/ATZNMP (PRESEQUENCE PROTEASE 1); metalloendopeptidase | -1.02 |
| 17 | 258167_at | AT3G21560 | UGT84A2; UDP-glycosyltransferase/ sinapate 1-glucosyltransferase | -1.04 |
| 18 | 258321_at | AT3G22840 | ELIP1 (EARLY LIGHT-INDUCABLE PROTEIN); chlorophyll binding | -1.43 |
| 19 | 257868_at | AT3G25070 | RIN4 (RPM1 INTERACTING PROTEIN 4); protein binding | 0.41 |
| 20 | 252607_at | AT3G44990 | XTR8 (xyloglucan:xyloglucosyl transferase 8); hydrolase, acting on glycosyl bonds | -1.05 |
| 21 | 252415_at | AT3G47340 | ASN1 (DARK INDUCIBLE 6) | 0.84 |
| 22 | 252250_at | AT3G49790 | ATP binding | 0.49 |
| 23 | 252123_at | AT3G51240 | F3H (TRANSPARENT TESTA 6); naringenin 3-dioxygenase | -1.10 |
| 24 | 251727_at | AT3G56290 | similar to hypothetical protein [Vitis vinifera] (GB:CAN75527.1) | -1.44 |
| 25 | 251658_at | AT3G57020 | strictosidine synthase family protein | -1.98 |
| 26 | 251670_at | AT3G57190 | peptide chain release factor, putative | -0.93 |
| 27 | 251148_at | AT3G63180 | ATTKL/TKL (TICKLE) | 0.48 |
| 28 | 255549_at | AT4G01950 | ATGPAT3/GPAT3 (GLYCEROL-3-PHOSPHATE ACYLTRANSFERASE 3); acyltransferase | 0.54 |
| 29 | 255298_at | AT4G04840 | methionine sulfoxide reductase domain-containing protein / SeIR domain-containing protein | -1.43 |
| 30 | 253943_at | AT4G27030 | small conjugating protein ligase | -1.13 |
| 31 | 253908_at | AT4G27260 | GH3.5/WES1; indole-3-acetic acid amido synthetase | 0.69 |
| 32 | 253667_at | AT4G30170 | peroxidase, putative | 1.12 |
| 33 | 253406_at | AT4G32890 | zinc finger (GATA type) family protein | 0.57 |
| 34 | 253039_at | AT4G37760 | SQE3 (SQUALENE EPOXIDASE 3); oxidoreductase | -1.02 |
| 35 | 252991_at | AT4G38470 | protein kinase family protein | 0.44 |
| 36 | 251020_at | AT5G02270 | ATNAP9 (Non-intrinsic ABC protein 9) | -1.66 |
| 37 | 250810_at | AT5G05090 | myb family transcription factor | 0.68 |
| 38 | 250207_at | AT5G13930 | ATCHS/CHS/TT4 (CHALCONE SYNTHASE); naringenin-chalcone synthase | -1.64 |
| 39 | 246506_at | AT5G16110 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G02555.1); similar to hypothetical protein [Cleome spinosa] (GB:ABD96917.1) | 0.53 |
| 40 | 246151_at | AT5G19950 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G63540.2); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G63540.1); similar to unknown protein [Oryza sativa (japonica cultivar-group)] (GB:BAD67825.1); contains InterPro domain Region of unknown function DUF1767 (InterPro:IPR013894) | 0.57 |
| 41 | 249915_at | AT5G22870 | harpin-induced protein-related / HIN1-related / harpin-responsive protein-related | 0.46 |
| 42 | 249798_at | AT5G23730 | nucleotide binding | -1.30 |
| 43 | 249769_at | AT5G24120 | SIGE (RNA polymerase sigma subunit E); DNA binding / DNA-directed RNA polymerase/ sigma factor/ transcription factor | -1.21 |
| 44 | 246966_at | AT5G24850 | CRY3 (CRYPTOCHROME 3); DNA binding / DNA photolyase/ FMN binding | -0.96 |
| 45 | 246911_at | AT5G25810 | TNY (TINY); DNA binding / transcription factor | -0.99 |
| 46 | 249415_at | AT5G39660 | CDF2 (CYCLING DOF FACTOR 2); DNA binding / protein binding / transcription factor | 0.43 |
| 47 | 249063_at | AT5G44110 | POP1 | -1.75 |
| 48 | 248683_at | AT5G48490 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | 0.56 |
| 49 | 248049_at | AT5G56090 | COX15 (CYTOCHROME C OXIDASE 15) | -0.91 |
| 50 | 247830_at | AT5G58530 | glutaredoxin family protein | 0.45 |
| 51 | 247780_at | AT5G58770 | dehydrodolichyl diphosphate synthase, putative / DEDOL-PP synthase, putative | -1.86 |
| 52 | 247586_at | AT5G60660 | PIP2;4/PIP2F (plasma membrane intrinsic protein 2;4); water channel | 0.48 |