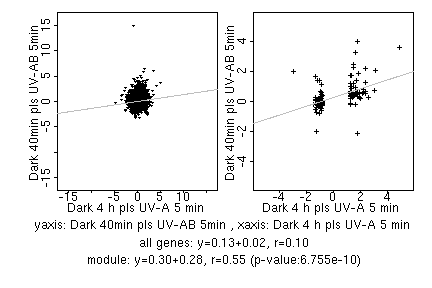
marker gene list
| Dark 4 h pls UV-A 5 min |
| Dark 40min pls UV-AB 5min |
| probeID | AGICode | Annotation | gene expression |
|
|---|---|---|---|---|
| 1 | 261581_at | AT1G01140 | CIPK9 (CBL-INTERACTING PROTEIN KINASE 9); kinase | -0.74 |
| 2 | 261558_at | AT1G01770 | similar to hypothetical protein [Vitis vinifera] (GB:CAN74802.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO39542.1); similar to hypothetical protein OsI_024980 [Oryza sativa (indica cultivar-group)] (GB:EAZ03748.1); contains InterPro domain Protein of unknown function DUF1446 (InterPro:IPR010839) | -0.87 |
| 3 | 263114_at | AT1G03130 | PSAD-2 (photosystem I subunit D-2) | 1.34 |
| 4 | 262635_at | AT1G06570 | PDS1 (PHYTOENE DESATURATION 1) | -0.91 |
| 5 | 261746_at | AT1G08380 | PSAO (photosystem I subunit O) | 2.47 |
| 6 | 264524_at | AT1G10070 | ATBCAT-2; branched-chain-amino-acid transaminase/ catalytic | -1.15 |
| 7 | 261197_at | AT1G12900 | GAPA-2; glyceraldehyde-3-phosphate dehydrogenase | 1.34 |
| 8 | 262609_at | AT1G13930 | similar to nodulin-related [Arabidopsis thaliana] (TAIR:AT2G03440.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN63832.1) | 1.90 |
| 9 | 259491_at | AT1G15820 | LHCB6 (LIGHT HARVESTING COMPLEX PSII); chlorophyll binding | 2.33 |
| 10 | 255905_at | AT1G17810 | BETA-TIP (BETA-TONOPLAST INTRINSIC PROTEIN); water channel | -1.23 |
| 11 | 261428_at | AT1G18870 | ICS2; isochorismate synthase | 1.35 |
| 12 | 260668_at | AT1G19530 | similar to unnamed protein product [Vitis vinifera] (GB:CAO61141.1) | -1.04 |
| 13 | 261136_at | AT1G19600 | pfkB-type carbohydrate kinase family protein | -0.75 |
| 14 | 255886_at | AT1G20340 | DRT112 (DNA-damage-repair/toleration protein 112); copper ion binding / electron carrier | 1.84 |
| 15 | 261272_at | AT1G26665 | similar to RNA polymerase II mediator complex protein-related [Arabidopsis thaliana] (TAIR:AT5G41910.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO62915.1); contains domain PTHR13345:SF2 (PTHR13345:SF2); contains domain PD331788 (PD331788); contains domain PTHR13345 (PTHR13345) | -0.74 |
| 16 | 256309_at | AT1G30380 | PSAK (PHOTOSYSTEM I SUBUNIT K) | 2.32 |
| 17 | 262557_at | AT1G31330 | PSAF (photosystem I subunit F) | 1.48 |
| 18 | 262264_at | AT1G42470 | patched family protein | -0.84 |
| 19 | 259460_at | AT1G44000 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G11911.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN81476.1) | 1.33 |
| 20 | 260506_at | AT1G47210 | CYCA3;2; cyclin-dependent protein kinase | -0.73 |
| 21 | 261603_at | AT1G49600 | ATRBP47A (ARABIDOPSIS THALIANA RNA-BINDING PROTEIN 47A); RNA binding | -0.77 |
| 22 | 259840_at | AT1G52230 | PSAH-2/PSAH2/PSI-H (PHOTOSYSTEM I SUBUNIT H-2) | 2.44 |
| 23 | 259962_at | AT1G53690 | DNA-directed RNA polymerases I, II, and III 7 kDa subunit, putative | -0.77 |
| 24 | 262198_at | AT1G53830 | ATPME2 (Arabidopsis thaliana pectin methylesterase 2) | -0.91 |
| 25 | 264545_at | AT1G55670 | PSAG | 1.57 |
| 26 | 256359_at | AT1G66460 | protein kinase family protein | -1.24 |
| 27 | 260460_at | AT1G68230 | reticulon family protein (RTNLB14) | -0.87 |
| 28 | 260181_at | AT1G70710 | AtGH9B1 (ARABIDOPSIS THALIANA GLYCOSYL HYDROLASE 9B1); hydrolase, hydrolyzing O-glycosyl compounds | -1.03 |
| 29 | 259892_at | AT1G72610 | GLP1 (GERMIN-LIKE PROTEIN 1); manganese ion binding / metal ion binding / nutrient reservoir | 1.82 |
| 30 | 260072_at | AT1G73650 | oxidoreductase, acting on the CH-CH group of donors | 1.52 |
| 31 | 262168_at | AT1G74730 | similar to unknown [Populus trichocarpa x Populus deltoides] (GB:ABK96654.1); contains InterPro domain Protein of unknown function DUF1118 (InterPro:IPR009500) | 1.69 |
| 32 | 262970_at | AT1G75690 | chaperone protein dnaJ-related | 1.80 |
| 33 | 261751_at | AT1G76080 | ATCDSP32/CDSP32 (CHLOROPLASTIC DROUGHT-INDUCED STRESS PROTEIN OF 32 KD); thiol-disulfide exchange intermediate | 1.43 |
| 34 | 261769_at | AT1G76100 | plastocyanin | 1.34 |
| 35 | 246390_at | AT1G77330 | 1-aminocyclopropane-1-carboxylate oxidase, putative / ACC oxidase, putative | -1.16 |
| 36 | 262047_at | AT1G80160 | lactoylglutathione lyase family protein / glyoxalase I family protein | -1.18 |
| 37 | 265374_at | AT2G06520 | PSBX (photosystem II subunit X) | 1.44 |
| 38 | 263894_at | AT2G21910 | CYP96A5 (cytochrome P450, family 96, subfamily A, polypeptide 5); oxygen binding | -0.77 |
| 39 | 264045_at | AT2G22450 | riboflavin biosynthesis protein, putative | -0.77 |
| 40 | 267517_at | AT2G30520 | RPT2 (ROOT PHOTOTROPISM 2); protein binding | 1.68 |
| 41 | 267526_at | AT2G30570 | PSBW (PHOTOSYSTEM II REACTION CENTER W) | 1.35 |
| 42 | 256712_at | AT2G34020 | calcium ion binding | -0.92 |
| 43 | 266901_at | AT2G34600 | JAZ7/TIFY5B (JASMONATE-ZIM-DOMAIN PROTEIN 7) | -0.80 |
| 44 | 263841_at | AT2G36870 | xyloglucan:xyloglucosyl transferase, putative / xyloglucan endotransglycosylase, putative / endo-xyloglucan transferase, putative | 1.36 |
| 45 | 266097_at | AT2G37970 | SOUL-1; binding | 1.55 |
| 46 | 245061_at | AT2G39730 | RCA (RUBISCO ACTIVASE) | 1.33 |
| 47 | 260557_at | AT2G43610 | glycoside hydrolase family 19 protein | -1.03 |
| 48 | 258589_at | AT3G04290 | ATLTL1/LTL1 (LI-TOLERANT LIPASE 1); carboxylesterase | 1.69 |
| 49 | 259143_at | AT3G10190 | calmodulin, putative | -0.71 |
| 50 | 256433_at | AT3G10985 | SAG20 (WOUND-INDUCED PROTEIN 12) | -0.71 |
| 51 | 256266_at | AT3G12320 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G06980.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN75675.1) | -2.86 |
| 52 | 258395_at | AT3G15500 | ATNAC3 (ARABIDOPSIS NAC DOMAIN CONTAINING PROTEIN 55); transcription factor | -0.82 |
| 53 | 257932_at | AT3G17040 | HCF107 (HIGH CHLOROPHYLL FLUORESCENT 107); binding | 1.59 |
| 54 | 256979_at | AT3G21055 | PSBTN (photosystem II subunit T) | 2.67 |
| 55 | 257197_at | AT3G23800 | selenium-binding family protein | -1.12 |
| 56 | 257311_at | AT3G26570 | PHT2;1 (phosphate transporter 2;1) | 1.32 |
| 57 | 252430_at | AT3G47470 | LHCA4 (Photosystem I light harvesting complex gene 4); chlorophyll binding | 1.81 |
| 58 | 252170_at | AT3G50480 | HR4 (HOMOLOG OF RPW8 4) | -0.79 |
| 59 | 252130_at | AT3G50820 | PSBO-2/PSBO2 (PHOTOSYSTEM II SUBUNIT O-2); oxygen evolving/ poly(U) binding | 1.86 |
| 60 | 252123_at | AT3G51240 | F3H (TRANSPARENT TESTA 6); naringenin 3-dioxygenase | 1.85 |
| 61 | 252054_at | AT3G52540 | ATOFP18/OFP18 (Arabidopsis thaliana ovate family protein 18) | -0.83 |
| 62 | 251977_at | AT3G53250 | auxin-responsive family protein | 1.85 |
| 63 | 251814_at | AT3G54890 | LHCA1; chlorophyll binding | 3.18 |
| 64 | 251658_at | AT3G57020 | strictosidine synthase family protein | 1.67 |
| 65 | 251272_at | AT3G61890 | ATHB-12 (ARABIDOPSIS THALIANA HOMEOBOX PROTEIN 12); transcription factor | -0.88 |
| 66 | 255457_at | AT4G02770 | PSAD-1 (photosystem I subunit D-1) | 1.96 |
| 67 | 255298_at | AT4G04840 | methionine sulfoxide reductase domain-containing protein / SeIR domain-containing protein | 2.47 |
| 68 | 255248_at | AT4G05180 | PSBQ/PSBQ-2/PSII-Q (PHOTOSYSTEM II SUBUNIT Q-2); calcium ion binding | 2.10 |
| 69 | 255046_at | AT4G09650 | ATP synthase delta chain, chloroplast, putative / H(+)-transporting two-sector ATPase, delta (OSCP) subunit, putative | 1.32 |
| 70 | 254970_at | AT4G10340 | LHCB5 (LIGHT HARVESTING COMPLEX OF PHOTOSYSTEM II 5); chlorophyll binding | 1.50 |
| 71 | 254790_at | AT4G12800 | PSAL (photosystem I subunit L) | 1.68 |
| 72 | 254754_at | AT4G13210 | pectate lyase family protein | -0.83 |
| 73 | 245501_at | AT4G15620 | integral membrane family protein | 1.61 |
| 74 | 254431_at | AT4G20840 | FAD-binding domain-containing protein | -0.72 |
| 75 | 254398_at | AT4G21280 | PSBQ/PSBQ-1/PSBQA; calcium ion binding | 1.49 |
| 76 | 245233_at | AT4G25580 | stress-responsive protein-related | -0.94 |
| 77 | 253738_at | AT4G28750 | PSAE-1 (PSA E1 KNOCKOUT); catalytic | 2.20 |
| 78 | 253522_at | AT4G31290 | ChaC-like family protein | -1.24 |
| 79 | 253421_at | AT4G32340 | binding | -1.26 |
| 80 | 253434_at | AT4G32500 | AKT5 (Arabidopsis K+ transporter 5); cyclic nucleotide binding / inward rectifier potassium channel | -0.73 |
| 81 | 253387_at | AT4G33010 | ATGLDP1 (ARABIDOPSIS THALIANA GLYCINE DECARBOXYLASE P-PROTEIN 1); glycine dehydrogenase (decarboxylating) | 1.39 |
| 82 | 253322_at | AT4G33980 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G42900.2); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G42900.3); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G42900.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN64989.1) | -1.22 |
| 83 | 253039_at | AT4G37760 | SQE3 (SQUALENE EPOXIDASE 3); oxidoreductase | 1.53 |
| 84 | 253009_at | AT4G37930 | SHM1 (SERINE HYDROXYMETHYLTRANSFERASE 1); glycine hydroxymethyltransferase/ poly(U) binding | 1.70 |
| 85 | 252906_at | AT4G39640 | GGT1 (GAMMA-GLUTAMYL TRANSPEPTIDASE 1); gamma-glutamyltransferase/ glutathione gamma-glutamylcysteinyltransferase | -0.79 |
| 86 | 252872_at | AT4G40010 | SNRK2-7/SNRK2.7/SRK2F (SNF1-RELATED PROTEIN KINASE 2.7); kinase | -0.92 |
| 87 | 251012_at | AT5G02580 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G55240.1); similar to hypothetical protein OsI_019565 [Oryza sativa (indica cultivar-group)] (GB:EAY98332.1); similar to Os05g0462000 [Oryza sativa (japonica cultivar-group)] (GB:NP_001055765.1); contains InterPro domain Conserved hypothetical protein CHP01589, plant (InterPro:IPR006476) | -1.17 |
| 88 | 250907_at | AT5G03670 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G36420.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO15955.1) | -1.01 |
| 89 | 250912_at | AT5G03740 | HD2C (HISTONE DEACETYLASE 2C); nucleic acid binding / zinc ion binding | -0.79 |
| 90 | 250714_at | AT5G06140 | phox (PX) domain-containing protein | -0.84 |
| 91 | 250665_at | AT5G06980 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G12320.1) | -1.13 |
| 92 | 250661_at | AT5G07030 | pepsin A | -0.78 |
| 93 | 250243_at | AT5G13630 | GUN5 (GENOMES UNCOUPLED 5) | 1.40 |
| 94 | 250214_at | AT5G13870 | EXGT-A4 (ENDOXYLOGLUCAN TRANSFERASE A4); hydrolase, acting on glycosyl bonds | -0.81 |
| 95 | 250053_at | AT5G17850 | cation exchanger, putative (CAX8) | -0.75 |
| 96 | 245970_at | AT5G20710 | BGAL7 (beta-galactosidase 7); beta-galactosidase | -0.85 |
| 97 | 249876_at | AT5G23060 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G59780.1); similar to extracellular Ca2+ sensing receptor [Glycine max] (GB:ABY57763.1); contains InterPro domain Rhodanese-like (InterPro:IPR001763) | 1.54 |
| 98 | 249645_at | AT5G36910 | THI2.2 (THIONIN 2.2); toxin receptor binding | 4.96 |
| 99 | 249606_at | AT5G37260 | CIR1/RVE2 (CIRCADIAN 1); DNA binding / transcription factor | -0.92 |
| 100 | 249415_at | AT5G39660 | CDF2 (CYCLING DOF FACTOR 2); DNA binding / protein binding / transcription factor | -0.74 |
| 101 | 249174_at | AT5G42900 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G33980.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN75496.1) | -1.41 |
| 102 | 248886_at | AT5G46110 | APE2 (ACCLIMATION OF PHOTOSYNTHESIS TO ENVIRONMENT); antiporter/ triose-phosphate transmembrane transporter | 1.32 |
| 103 | 248684_at | AT5G48485 | DIR1 (DEFECTIVE IN INDUCED RESISTANCE 1); lipid binding | 1.98 |
| 104 | 248683_at | AT5G48490 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | 3.10 |
| 105 | 248435_at | AT5G51210 | OLEO3 (OLEOSIN3) | -0.99 |
| 106 | 248193_at | AT5G54080 | HGO (HOMOGENTISATE 1,2-DIOXYGENASE); homogentisate 1,2-dioxygenase | -0.83 |
| 107 | 248151_at | AT5G54270 | LHCB3 (LIGHT-HARVESTING CHLOROPHYLL BINDING PROTEIN 3) | 2.41 |
| 108 | 248160_at | AT5G54470 | zinc finger (B-box type) family protein | -1.21 |
| 109 | 247320_at | AT5G64040 | PSAN (photosystem I reaction center subunit PSI-N); calmodulin binding | 1.43 |
| 110 | 247073_at | AT5G66570 | OE33/OEE1/OEE33/PSBO-1/PSBO1 (OXYGEN-EVOLVING ENHANCER 33); oxygen evolving/ poly(U) binding | 1.35 |