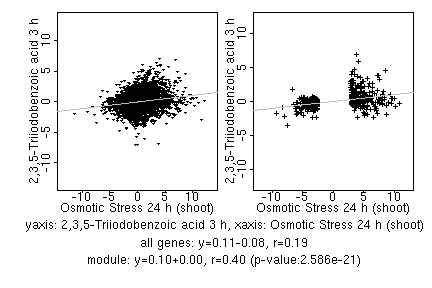
marker gene list
| Osmotic Stress 24 h (shoot) |
| 2,3,5-Triiodobenzoic acid3 h |
| probeID | AGICode | Annotation | gene expression |
|
|---|---|---|---|---|
| 1 | 259426_at | AT1G01470 | LEA14 (LATE EMBRYOGENESIS ABUNDANT 14) | 3.34 |
| 2 | 259417_at | AT1G02340 | HFR1 (LONG HYPOCOTYL IN FAR-RED); DNA binding / transcription factor | -2.86 |
| 3 | 260933_at | AT1G02470 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G02475.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO40177.1); contains InterPro domain Streptomyces cyclase/dehydrase (InterPro:IPR005031) | 3.37 |
| 4 | 262109_at | AT1G02730 | ATCSLD5 (CELLULOSE SYNTHASE-LIKE D5); 1,4-beta-D-xylan synthase/ cellulose synthase | -2.93 |
| 5 | 264822_at | AT1G03457 | RNA-binding protein, putative | -3.10 |
| 6 | 263677_at | AT1G04520 | 33 kDa secretory protein-related | -4.64 |
| 7 | 264611_at | AT1G04680 | pectate lyase family protein | -2.62 |
| 8 | 264580_at | AT1G05340 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G32210.1) | 7.86 |
| 9 | 263182_at | AT1G05575 | unknown protein | 3.14 |
| 10 | 263231_at | AT1G05680 | UDP-glucoronosyl/UDP-glucosyl transferase family protein | 3.95 |
| 11 | 260831_at | AT1G06830 | glutaredoxin family protein | -3.52 |
| 12 | 256061_at | AT1G07040 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G27030.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO41035.1) | 3.99 |
| 13 | 261080_at | AT1G07370 | PCNA1 (PROLIFERATING CELLULAR NUCLEAR ANTIGEN); DNA binding / DNA polymerase processivity factor | -3.26 |
| 14 | 261077_at | AT1G07430 | protein phosphatase 2C, putative / PP2C, putative | 7.71 |
| 15 | 261068_at | AT1G07450 | tropinone reductase, putative / tropine dehydrogenase, putative | -2.84 |
| 16 | 261420_at | AT1G07720 | beta-ketoacyl-CoA synthase family protein | 3.71 |
| 17 | 261785_at | AT1G08230 | amino acid transporter family protein | 3.53 |
| 18 | 264777_at | AT1G08630 | THA1 (THREONINE ALDOLASE 1); aldehyde-lyase/ threonine aldolase | 3.15 |
| 19 | 264266_at | AT1G09160 | protein phosphatase 2C-related / PP2C-related | -2.66 |
| 20 | 264514_at | AT1G09500 | cinnamyl-alcohol dehydrogenase family / CAD family | 4.98 |
| 21 | 264510_at | AT1G09530 | PAP3/PIF3/POC1 (PHYTOCHROME INTERACTING FACTOR 3); DNA binding / protein binding / transcription factor/ transcription regulator | 4.99 |
| 22 | 264672_at | AT1G09750 | chloroplast nucleoid DNA-binding protein-related | -4.27 |
| 23 | 264522_at | AT1G10050 | glycosyl hydrolase family 10 protein / carbohydrate-binding domain-containing protein | 3.14 |
| 24 | 264524_at | AT1G10070 | ATBCAT-2; branched-chain-amino-acid transaminase/ catalytic | 3.95 |
| 25 | 263236_at | AT1G10470 | ARR4 (RESPONSE REGULATOR 4); transcription regulator/ two-component response regulator | -3.22 |
| 26 | 261825_at | AT1G11545 | xyloglucan:xyloglucosyl transferase, putative / xyloglucan endotransglycosylase, putative / endo-xyloglucan transferase, putative | -3.32 |
| 27 | 259363_at | AT1G13270 | MAP1C (METHIONINE AMINOPEPTIDASE 1B); metalloexopeptidase | -2.63 |
| 28 | 256096_at | AT1G13650 | similar to 18S pre-ribosomal assembly protein gar2-related [Arabidopsis thaliana] (TAIR:AT2G03810.4); similar to 18S pre-ribosomal assembly protein gar2-related [Arabidopsis thaliana] (TAIR:AT2G03810.2); similar to 18S pre-ribosomal assembly protein gar2-related [Arabidopsis thaliana] (TAIR:AT2G03810.3); similar to hypothetical protein Tc00.1047053503999.40 [Trypanosoma cruzi strain CL Brener] (GB:XP_813437.1) | -3.33 |
| 29 | 262607_at | AT1G13990 | similar to unnamed protein product [Vitis vinifera] (GB:CAO68469.1) | 4.72 |
| 30 | 259502_at | AT1G15670 | kelch repeat-containing F-box family protein | 4.23 |
| 31 | 256114_at | AT1G16850 | unknown protein | 9.55 |
| 32 | 262482_at | AT1G17020 | SRG1 (SENESCENCE-RELATED GENE 1); oxidoreductase, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors | 9.87 |
| 33 | 259403_at | AT1G17745 | PGDH (3-PHOSPHOGLYCERATE DEHYDROGENASE); phosphoglycerate dehydrogenase | 4.04 |
| 34 | 255891_at | AT1G17870 | ATEGY3 | 4.79 |
| 35 | 256077_at | AT1G18090 | exonuclease, putative | -2.60 |
| 36 | 256125_at | AT1G18250 | ATLP-1 (Arabidopsis thaumatin-like protein 1) | -3.79 |
| 37 | 255774_at | AT1G18620 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G74160.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO49165.1) | -3.82 |
| 38 | 255779_at | AT1G18650 | glycosyl hydrolase family protein 17 | -2.51 |
| 39 | 261428_at | AT1G18870 | ICS2; isochorismate synthase | 3.17 |
| 40 | 259466_at | AT1G19050 | ARR7 (RESPONSE REGULATOR 7); transcription regulator/ two-component response regulator | -3.38 |
| 41 | 256015_at | AT1G19150 | LHCA6 (Photosystem I light harvesting complex gene 6); chlorophyll binding | -3.17 |
| 42 | 256017_at | AT1G19180 | JAZ1/TIFY10A (JASMONATE-ZIM-DOMAIN PROTEIN 1); protein binding | 5.17 |
| 43 | 255786_at | AT1G19670 | ATCLH1 (CORONATINE-INDUCED PROTEIN 1) | -2.42 |
| 44 | 261223_at | AT1G19950 | HVA22H (HVA22-LIKE PROTEIN H (ATHVA22H)) | -2.47 |
| 45 | 261230_at | AT1G20010 | TUB5 (tubulin beta-5 chain) | -2.90 |
| 46 | 259570_at | AT1G20440 | COR47 (cold regulated 47) | 4.63 |
| 47 | 259516_at | AT1G20450 | ERD10/LTI45 (EARLY RESPONSIVE TO DEHYDRATION 10) | 4.98 |
| 48 | 260877_at | AT1G21500 | similar to hypothetical protein OsI_030994 [Oryza sativa (indica cultivar-group)] (GB:EAZ09762.1); similar to Os09g0517000 [Oryza sativa (japonica cultivar-group)] (GB:NP_001063677.1); similar to unknown protein [Oryza sativa (japonica cultivar-group)] (GB:BAD33829.1) | -3.24 |
| 49 | 255962_at | AT1G22330 | RNA binding | -5.93 |
| 50 | 255969_at | AT1G22330 | RNA binding | -4.78 |
| 51 | 261942_at | AT1G22590 | AGL87; transcription factor | -2.73 |
| 52 | 264201_at | AT1G22630 | heat shock protein binding / unfolded protein binding | -2.70 |
| 53 | 264729_at | AT1G22990 | heavy-metal-associated domain-containing protein / copper chaperone (CCH)-related | 7.73 |
| 54 | 265175_at | AT1G23480 | ATCSLA03 (Cellulose synthase-like A3); transferase, transferring glycosyl groups | -3.55 |
| 55 | 263034_at | AT1G24020 | MLP423 (MLP-LIKE PROTEIN 423) | -2.53 |
| 56 | 255742_at | AT1G25560 | AP2 domain-containing transcription factor, putative | 3.62 |
| 57 | 261021_at | AT1G26380 | FAD-binding domain-containing protein | 7.55 |
| 58 | 261005_at | AT1G26420 | FAD-binding domain-containing protein | 5.88 |
| 59 | 259599_at | AT1G28110 | SCPL45; serine carboxypeptidase | -2.45 |
| 60 | 245688_at | AT1G28290 | AGP31 (ARABINOGALACTAN-PROTEIN 31); structural constituent of cell wall | -3.38 |
| 61 | 260898_at | AT1G29070 | ribosomal protein L34 family protein | -2.42 |
| 62 | 260884_at | AT1G29240 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G34170.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO48048.1); contains InterPro domain Protein of unknown function DUF688 (InterPro:IPR007789) | 3.80 |
| 63 | 259789_at | AT1G29395 | COR414-TM1 (cold regulated 414 thylakoid membrane 1) | 5.13 |
| 64 | 257506_at | AT1G29440 | auxin-responsive family protein | -3.94 |
| 65 | 259784_at | AT1G29450 | auxin-responsive protein, putative | -7.79 |
| 66 | 259783_at | AT1G29510 | SAUR68 (SMALL AUXIN UPREGULATED 68) | -3.87 |
| 67 | 259786_at | AT1G29660 | GDSL-motif lipase/hydrolase family protein | -4.77 |
| 68 | 245769_at | AT1G30220 | ATINT2 (INOSITOL TRANSPORTER 2); carbohydrate transmembrane transporter/ sugar:hydrogen ion symporter | 3.72 |
| 69 | 261801_at | AT1G30520 | AAE14 (ACYL-ACTIVATING ENZYME 14); AMP binding | -3.73 |
| 70 | 246260_at | AT1G31820 | amino acid permease family protein | 3.30 |
| 71 | 261712_at | AT1G32780 | alcohol dehydrogenase, putative | -3.69 |
| 72 | 261981_at | AT1G33811 | GDSL-motif lipase/hydrolase family protein | -4.63 |
| 73 | 255980_at | AT1G33970 | avirulence-responsive protein, putative / avirulence induced gene protein, putative / AIG protein, putative | 3.22 |
| 74 | 262029_at | AT1G35680 | 50S ribosomal protein L21, chloroplast / CL21 (RPL21) | -2.70 |
| 75 | 260943_at | AT1G45145 | ATTRX5 (thioredoxin H-type 5); thiol-disulfide exchange intermediate | 3.94 |
| 76 | 259616_at | AT1G47960 | C/VIF1 (CELL WALL / VACUOLAR INHIBITOR OF FRUCTOSIDASE 1); pectinesterase inhibitor | 3.28 |
| 77 | 259618_at | AT1G48000 | MYB112 (myb domain protein 112); DNA binding / transcription factor | 4.66 |
| 78 | 262235_at | AT1G48350 | ribosomal protein L18 family protein | -2.46 |
| 79 | 260754_at | AT1G49000 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G18560.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN78728.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO68009.1) | 5.75 |
| 80 | 245743_at | AT1G51080 | similar to unnamed protein product [Vitis vinifera] (GB:CAO41687.1) | -2.48 |
| 81 | 245749_at | AT1G51090 | heavy-metal-associated domain-containing protein | 4.18 |
| 82 | 262151_at | AT1G52510 | hydrolase, alpha/beta fold family protein | -2.51 |
| 83 | 262128_at | AT1G52690 | late embryogenesis abundant protein, putative / LEA protein, putative | 8.61 |
| 84 | 260203_at | AT1G52890 | ANAC019 (Arabidopsis NAC domain containing protein 19); transcription factor | 5.18 |
| 85 | 260986_at | AT1G53580 | ETHE1/GLX2-3/GLY3 (GLYOXALASE 2-3); hydroxyacylglutathione hydrolase | 3.11 |
| 86 | 263157_at | AT1G54100 | ALDH7B4 (ALDEHYDE DEHYDROGENASE 7B4); 3-chloroallyl aldehyde dehydrogenase | 4.20 |
| 87 | 259664_at | AT1G55330 | AGP21 (ARABINOGALACTAN PROTEIN 21) | -2.57 |
| 88 | 265076_at | AT1G55490 | CPN60B (CHAPERONIN 60 BETA); ATP binding / protein binding / unfolded protein binding | -2.50 |
| 89 | 260603_at | AT1G55960 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G13062.2); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G13062.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO41766.1); contains InterPro domain Lipid-binding START (InterPro:IPR002913) | -3.01 |
| 90 | 262086_at | AT1G56050 | GTP-binding protein-related | -2.58 |
| 91 | 245627_at | AT1G56600 | ATGOLS2 (ARABIDOPSIS THALIANA GALACTINOL SYNTHASE 2); transferase, transferring glycosyl groups / transferase, transferring hexosyl groups | 5.41 |
| 92 | 246403_at | AT1G57590 | carboxylesterase | 6.20 |
| 93 | 264217_at | AT1G60190 | armadillo/beta-catenin repeat family protein / U-box domain-containing protein | 4.45 |
| 94 | 264920_at | AT1G60550 | naphthoate synthase, putative / dihydroxynaphthoic acid synthetase, putative / DHNA synthetase, putative | -2.64 |
| 95 | 264931_at | AT1G60590 | polygalacturonase, putative / pectinase, putative | -5.21 |
| 96 | 265117_at | AT1G62500 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | -2.72 |
| 97 | 265119_at | AT1G62570 | flavin-containing monooxygenase family protein / FMO family protein | 3.87 |
| 98 | 262644_at | AT1G62710 | BETA-VPE (vacuolar processing enzyme beta); cysteine-type endopeptidase | 3.33 |
| 99 | 262693_at | AT1G62780 | similar to hypothetical protein [Vitis vinifera] (GB:CAN83165.1) | -2.59 |
| 100 | 262667_at | AT1G62810 | copper amine oxidase, putative | 4.25 |
| 101 | 262347_at | AT1G64110 | AAA-type ATPase family protein | 6.82 |
| 102 | 259736_at | AT1G64390 | ATGH9C2 (ARABIDOPSIS THALIANA GLYCOSYL HYDROLASE 9C2); hydrolase, hydrolyzing O-glycosyl compounds | -3.05 |
| 103 | 261907_at | AT1G65060 | 4CL3 (4-coumarate:CoA ligase 3); 4-coumarate-CoA ligase | -3.29 |
| 104 | 263142_at | AT1G65230 | similar to hypothetical protein [Vitis vinifera] (GB:CAN76393.1) | -2.99 |
| 105 | 256527_at | AT1G66100 | thionin, putative | -5.67 |
| 106 | 259822_at | AT1G66230 | MYB20 (myb domain protein 20); DNA binding / transcription factor | -2.68 |
| 107 | 260140_at | AT1G66390 | PAP2 (PRODUCTION OF ANTHOCYANIN PIGMENT 2); DNA binding / transcription factor | 8.45 |
| 108 | 255856_at | AT1G66940 | protein kinase-related | -2.90 |
| 109 | 264968_at | AT1G67360 | rubber elongation factor (REF) family protein | 3.69 |
| 110 | 245196_at | AT1G67750 | pectate lyase family protein | -3.08 |
| 111 | 260005_at | AT1G67920 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G24600.1) | 6.08 |
| 112 | 260266_at | AT1G68520 | zinc finger (B-box type) family protein | -3.04 |
| 113 | 262229_at | AT1G68620 | hydrolase | 4.60 |
| 114 | 260041_at | AT1G68780 | leucine-rich repeat family protein | -2.62 |
| 115 | 260357_at | AT1G69260 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G13740.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO41856.1); contains InterPro domain Protein of unknown function DUF1675 (InterPro:IPR012463) | 5.48 |
| 116 | 256296_at | AT1G69480 | EXS family protein / ERD1/XPR1/SYG1 family protein | 3.32 |
| 117 | 256299_at | AT1G69530 | ATEXPA1 (ARABIDOPSIS THALIANA EXPANSIN A1) | -4.27 |
| 118 | 264340_at | AT1G70280 | NHL repeat-containing protein | -2.70 |
| 119 | 264338_at | AT1G70300 | KUP6 (K+ uptake permease 6); potassium ion transmembrane transporter | 4.35 |
| 120 | 262290_at | AT1G70985 | hydroxyproline-rich glycoprotein family protein | -6.42 |
| 121 | 260427_at | AT1G72430 | auxin-responsive protein-related | -3.39 |
| 122 | 259911_at | AT1G72680 | cinnamyl-alcohol dehydrogenase, putative | 3.13 |
| 123 | 262376_at | AT1G72970 | HTH (HOTHEAD); aldehyde-lyase | -3.36 |
| 124 | 262369_at | AT1G73010 | phosphoric monoester hydrolase | 5.24 |
| 125 | 260101_at | AT1G73260 | trypsin and protease inhibitor family protein / Kunitz family protein | 5.71 |
| 126 | 245731_at | AT1G73500 | ATMKK9 (Arabidopsis thaliana MAP kinase kinase 9); kinase | 3.84 |
| 127 | 260386_at | AT1G74010 | strictosidine synthase family protein | 4.51 |
| 128 | 260388_at | AT1G74070 | peptidyl-prolyl cis-trans isomerase cyclophilin-type family protein | -3.26 |
| 129 | 260221_at | AT1G74670 | gibberellin-responsive protein, putative | -5.96 |
| 130 | 259925_at | AT1G75040 | PR5 (PATHOGENESIS-RELATED GENE 5) | 5.01 |
| 131 | 261782_at | AT1G76110 | high mobility group (HMG1/2) family protein / ARID/BRIGHT DNA-binding domain-containing protein | -3.06 |
| 132 | 261772_at | AT1G76240 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G17080.1); similar to Os06g0725500 [Oryza sativa (japonica cultivar-group)] (GB:NP_001058623.1); similar to hypothetical protein OsI_023643 [Oryza sativa (indica cultivar-group)] (GB:EAZ02411.1); contains InterPro domain Protein of unknown function DUF241, plant (InterPro:IPR004320) | -3.38 |
| 133 | 264958_at | AT1G76960 | unknown protein | 4.97 |
| 134 | 264954_at | AT1G77060 | mutase family protein | -2.39 |
| 135 | 264953_at | AT1G77120 | ADH1 (ALCOHOL DEHYDROGENASE 1) | 5.78 |
| 136 | 259705_at | AT1G77450 | ANAC032 (Arabidopsis NAC domain containing protein 32); transcription factor | 4.94 |
| 137 | 260804_at | AT1G78410 | VQ motif-containing protein | 7.76 |
| 138 | 262061_at | AT1G80110 | ATPP2-B11; carbohydrate binding | 6.11 |
| 139 | 262047_at | AT1G80160 | lactoylglutathione lyase family protein / glyoxalase I family protein | 6.43 |
| 140 | 260302_at | AT1G80310 | sulfate transmembrane transporter | 3.09 |
| 141 | 257474_at | AT1G80850 | methyladenine glycosylase family protein | -2.71 |
| 142 | 266329_at | AT2G01590 | similar to unnamed protein product [Vitis vinifera] (GB:CAO42320.1) | -2.54 |
| 143 | 266181_at | AT2G02390 | ATGSTZ1 (GLUTATHIONE S-TRANSFERASE 18); glutathione transferase | 3.76 |
| 144 | 266743_at | AT2G02990 | RNS1 (RIBONUCLEASE 1); endoribonuclease | 3.33 |
| 145 | 263674_at | AT2G04790 | similar to unnamed protein product [Vitis vinifera] (GB:CAO23994.1) | -6.08 |
| 146 | 263046_at | AT2G05380 | GRP3S (GLYCINE-RICH PROTEIN 3 SHORT ISOFORM) | 3.79 |
| 147 | 265377_at | AT2G05790 | glycosyl hydrolase family 17 protein | -3.20 |
| 148 | 266215_at | AT2G06850 | EXGT-A1 (ENDOXYLOGLUCAN TRANSFERASE); hydrolase, acting on glycosyl bonds | -2.96 |
| 149 | 266384_at | AT2G14660 | similar to hypothetical protein [Vitis vinifera] (GB:CAN83372.1); contains InterPro domain Protein of unknown function DUF55 (InterPro:IPR002740) | -2.42 |
| 150 | 265501_at | AT2G15490 | UGT73B4; UDP-glycosyltransferase/ transferase, transferring glycosyl groups | 4.20 |
| 151 | 265480_at | AT2G15970 | COR413-PM1 (cold regulated 413 plasma membrane 1) | 3.19 |
| 152 | 263607_at | AT2G16270 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G16630.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN75064.1) | -3.52 |
| 153 | 263612_at | AT2G16440 | DNA replication licensing factor, putative | -2.41 |
| 154 | 263073_at | AT2G17500 | auxin efflux carrier family protein | 3.45 |
| 155 | 264787_at | AT2G17840 | ERD7 (EARLY-RESPONSIVE TO DEHYDRATION 7) | 3.23 |
| 156 | 265817_at | AT2G18050 | HIS1-3 (HISTONE H1-3); DNA binding | 5.17 |
| 157 | 265342_at | AT2G18300 | basic helix-loop-helix (bHLH) family protein | -8.20 |
| 158 | 266687_at | AT2G19670 | protein arginine N-methyltransferase, putative | -2.68 |
| 159 | 266690_at | AT2G19900 | ATNADP-ME1 (NADP-MALIC ENZYME 1); malate dehydrogenase (oxaloacetate-decarboxylating) (NADP+)/ malic enzyme/ oxidoreductase, acting on NADH or NADPH, NAD or NADP as acceptor | 5.84 |
| 160 | 263374_at | AT2G20560 | DNAJ heat shock family protein | 4.08 |
| 161 | 263544_at | AT2G21590 | APL4 (large subunit of AGP 4); glucose-1-phosphate adenylyltransferase | 5.39 |
| 162 | 263517_at | AT2G21620 | RD2 (RESPONSIVE TO DESSICATION 2) | 3.47 |
| 163 | 263882_at | AT2G21790 | R1/RNR1 (RIBONUCLEOTIDE REDUCTASE 1); ribonucleoside-diphosphate reductase | -3.47 |
| 164 | 263881_at | AT2G21820 | similar to hypothetical protein MtrDRAFT_AC155884g16v2 [Medicago truncatula] (GB:ABN08202.1) | 7.31 |
| 165 | 267265_at | AT2G22980 | SCPL13; serine carboxypeptidase | -3.50 |
| 166 | 264377_at | AT2G25060 | plastocyanin-like domain-containing protein | -3.11 |
| 167 | 265913_at | AT2G25625 | similar to unnamed protein product [Vitis vinifera] (GB:CAO44157.1) | 4.77 |
| 168 | 245038_at | AT2G26560 | PLP2 (PHOSPHOLIPASE A 2A); nutrient reservoir | 5.05 |
| 169 | 267614_at | AT2G26710 | BAS1/CYP734A1 (PHYB ACTIVATION TAGGED SUPPRESSOR 1); oxygen binding / steroid hydroxylase | -4.13 |
| 170 | 265665_at | AT2G27420 | cysteine proteinase, putative | -5.00 |
| 171 | 264069_at | AT2G28000 | CPN60A (chloroplast / 60 kDa chaperonin alpha subunit); ATP binding / protein binding / unfolded protein binding | -2.60 |
| 172 | 265276_at | AT2G28400 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G60680.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO21845.1); contains InterPro domain Protein of unknown function DUF584 (InterPro:IPR007608) | 6.62 |
| 173 | 266229_at | AT2G28840 | ankyrin repeat family protein | 4.35 |
| 174 | 266790_at | AT2G28950 | ATEXPA6 (ARABIDOPSIS THALIANA EXPANSIN A6) | -3.91 |
| 175 | 266292_at | AT2G29350 | SAG13 (Senescence-associated gene 13); oxidoreductase | 6.42 |
| 176 | 266267_at | AT2G29460 | ATGSTU4 (GLUTATHIONE S-TRANSFERASE 22); glutathione transferase | 6.63 |
| 177 | 266270_at | AT2G29470 | ATGSTU3 (GLUTATHIONE S-TRANSFERASE 21); glutathione transferase | 5.69 |
| 178 | 266294_at | AT2G29500 | 17.6 kDa class I small heat shock protein (HSP17.6B-CI) | 3.65 |
| 179 | 267300_at | AT2G30140 | UDP-glucoronosyl/UDP-glucosyl transferase family protein | 3.31 |
| 180 | 267496_at | AT2G30550 | lipase class 3 family protein | 3.52 |
| 181 | 263475_at | AT2G31945 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G05575.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO22015.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN61524.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN71356.1) | 3.06 |
| 182 | 265724_at | AT2G32100 | ATOFP16/OFP16 (Arabidopsis thaliana ovate family protein 16) | -4.06 |
| 183 | 267116_at | AT2G32560 | F-box family protein | -2.88 |
| 184 | 267595_at | AT2G32990 | ATGH9B8 (ARABIDOPSIS THALIANA GLYCOSYL HYDROLASE 9B8); hydrolase, hydrolyzing O-glycosyl compounds | -2.75 |
| 185 | 255789_at | AT2G33370 | 60S ribosomal protein L23 (RPL23B) | -3.04 |
| 186 | 255795_at | AT2G33380 | RD20 (RESPONSIVE TO DESSICATION 20); calcium ion binding | 3.26 |
| 187 | 255787_at | AT2G33590 | cinnamoyl-CoA reductase family | 3.40 |
| 188 | 267459_at | AT2G33850 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G28400.1); similar to unknown [Brassica napus] (GB:AAC06020.1) | -3.52 |
| 189 | 266995_at | AT2G34500 | CYP710A1 (cytochrome P450, family 710, subfamily A, polypeptide 1); C-22 sterol desaturase/ oxygen binding | 3.43 |
| 190 | 266956_at | AT2G34510 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G29980.1); similar to unknown [Populus trichocarpa] (GB:ABK95079.1); contains InterPro domain Protein of unknown function DUF642 (InterPro:IPR006946); contains InterPro domain Galactose-binding like (InterPro:IPR008979) | -2.60 |
| 191 | 266901_at | AT2G34600 | JAZ7/TIFY5B (JASMONATE-ZIM-DOMAIN PROTEIN 7) | 5.72 |
| 192 | 266899_at | AT2G34620 | mitochondrial transcription termination factor-related / mTERF-related | -3.10 |
| 193 | 263948_at | AT2G35980 | YLS9 (YELLOW-LEAF-SPECIFIC GENE 9) | 5.48 |
| 194 | 263963_at | AT2G36080 | DNA-binding protein, putative | 4.37 |
| 195 | 263931_at | AT2G36220 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G52710.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO15901.1) | 6.52 |
| 196 | 267181_at | AT2G37760 | aldo/keto reductase family protein | 3.45 |
| 197 | 267168_at | AT2G37770 | aldo/keto reductase family protein | 6.02 |
| 198 | 266098_at | AT2G37870 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | 6.48 |
| 199 | 267092_at | AT2G38120 | AUX1 (AUXIN RESISTANT 1); amino acid transmembrane transporter/ transporter | -3.15 |
| 200 | 267088_at | AT2G38140 | PSRP4 (PLASTID-SPECIFIC RIBOSOMAL PROTEIN 4); structural constituent of ribosome | -2.66 |
| 201 | 267035_at | AT2G38400 | AGT3 (ALANINE:GLYOXYLATE AMINOTRANSFERASE 3); alanine-glyoxylate transaminase | 4.62 |
| 202 | 267028_at | AT2G38470 | WRKY33 (WRKY DNA-binding protein 33); transcription factor | 3.49 |
| 203 | 266415_at | AT2G38530 | LTP2 (LIPID TRANSFER PROTEIN 2); lipid binding | 5.77 |
| 204 | 263264_at | AT2G38810 | HTA8; DNA binding | -5.88 |
| 205 | 266170_at | AT2G39050 | hydroxyproline-rich glycoprotein family protein | 3.97 |
| 206 | 266993_at | AT2G39210 | nodulin family protein | 3.19 |
| 207 | 255826_at | AT2G40490 | HEME2; uroporphyrinogen decarboxylase | -2.77 |
| 208 | 255822_at | AT2G40610 | ATEXPA8 (ARABIDOPSIS THALIANA EXPANSIN A8) | -9.12 |
| 209 | 267080_at | AT2G41190 | amino acid transporter family protein | 7.18 |
| 210 | 266368_at | AT2G41380 | embryo-abundant protein-related | 4.31 |
| 211 | 267635_at | AT2G42220 | rhodanese-like domain-containing protein | -3.08 |
| 212 | 263497_at | AT2G42540 | COR15A (COLD-REGULATED 15A) | 5.30 |
| 213 | 263492_at | AT2G42560 | late embryogenesis abundant domain-containing protein / LEA domain-containing protein | 10.29 |
| 214 | 266395_at | AT2G43100 | aconitase C-terminal domain-containing protein | -2.43 |
| 215 | 260551_at | AT2G43510 | ATTI1 (ARABIDOPSIS THALIANA TRYPSIN INHIBITOR PROTEIN 1) | 5.04 |
| 216 | 260568_at | AT2G43570 | chitinase, putative | 7.92 |
| 217 | 260561_at | AT2G43580 | chitinase, putative | 5.90 |
| 218 | 260560_at | AT2G43590 | chitinase, putative | 8.23 |
| 219 | 260556_at | AT2G43620 | chitinase, putative | 5.08 |
| 220 | 260567_at | AT2G43820 | GT/UGT74F2 (UDP-GLUCOSYLTRANSFERASE 74F2); UDP-glucosyltransferase/ UDP-glycosyltransferase/ transferase, transferring glycosyl groups / transferase, transferring hexosyl groups | 3.26 |
| 221 | 245136_at | AT2G45210 | auxin-responsive protein-related | 7.97 |
| 222 | 251395_at | AT2G45470 | FLA8 (Arabinogalactan protein 8) | -4.85 |
| 223 | 267559_at | AT2G45570 | CYP76C2 (cytochrome P450, family 76, subfamily C, polypeptide 2); oxygen binding | 8.20 |
| 224 | 266555_at | AT2G46270 | GBF3 (G-BOX BINDING FACTOR 3); transcription factor | 3.66 |
| 225 | 266327_at | AT2G46680 | ATHB-7 (ARABIDOPSIS THALIANA HOMEOBOX 7); transcription factor | 5.24 |
| 226 | 266462_at | AT2G47770 | benzodiazepine receptor-related | 7.68 |
| 227 | 258957_at | AT3G01420 | ALPHA-DOX1 (ALPHA-DIOXYGENASE 1) | 3.99 |
| 228 | 259193_at | AT3G01480 | CYP38 (CYCLOPHILIN 38); peptidyl-prolyl cis-trans isomerase | -3.15 |
| 229 | 259001_at | AT3G01960 | unknown protein | -2.64 |
| 230 | 258975_at | AT3G01970 | WRKY45 (WRKY DNA-binding protein 45); transcription factor | 4.82 |
| 231 | 258497_at | AT3G02380 | COL2 (CONSTANS-LIKE 2); transcription factor/ zinc ion binding | -2.62 |
| 232 | 258498_at | AT3G02480 | ABA-responsive protein-related | 11.00 |
| 233 | 258480_at | AT3G02640 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G16250.1); similar to unknown [Populus trichocarpa] (GB:ABK93711.1) | -2.83 |
| 234 | 259058_at | AT3G03470 | CYP89A9 (cytochrome P450, family 87, subfamily A, polypeptide 9); oxygen binding | 5.33 |
| 235 | 259172_at | AT3G03630 | CS26 (cysteine synthase 26); cysteine synthase | -2.40 |
| 236 | 259331_at | AT3G03840 | auxin-responsive protein, putative | -7.40 |
| 237 | 259340_at | AT3G03870 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G18130.2) | 3.46 |
| 238 | 258809_at | AT3G04070 | ANAC047 (Arabidopsis NAC domain containing protein 47); transcription factor | 5.33 |
| 239 | 258791_at | AT3G04720 | PR4 (PATHOGENESIS-RELATED 4) | 3.63 |
| 240 | 258895_at | AT3G05600 | epoxide hydrolase, putative | -3.27 |
| 241 | 258887_at | AT3G05630 | PLDP2 (PHOSPHOLIPASE D ZETA 2); phospholipase D | 5.59 |
| 242 | 258897_at | AT3G05730 | Encodes a defensin-like (DEFL) family protein. | -2.78 |
| 243 | 258880_at | AT3G06420 | ATG8H (AUTOPHAGY 8H); microtubule binding | 3.05 |
| 244 | 258535_at | AT3G06750 | hydroxyproline-rich glycoprotein family protein | -2.72 |
| 245 | 258983_at | AT3G08860 | alanine--glyoxylate aminotransferase, putative / beta-alanine-pyruvate aminotransferase, putative / AGT, putative | 4.39 |
| 246 | 258982_at | AT3G08870 | lectin protein kinase, putative | 3.30 |
| 247 | 258989_at | AT3G08920 | rhodanese-like domain-containing protein | -2.68 |
| 248 | 258708_at | AT3G09580 | amine oxidase family protein | -2.58 |
| 249 | 258695_at | AT3G09640 | APX2 (ASCORBATE PEROXIDASE 2); L-ascorbate peroxidase | 7.43 |
| 250 | 258939_at | AT3G10020 | similar to Os12g0147200 [Oryza sativa (japonica cultivar-group)] (GB:NP_001066153.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO15981.1); similar to Os11g0149200 [Oryza sativa (japonica cultivar-group)] (GB:NP_001065754.1) | 3.42 |
| 251 | 258884_at | AT3G10050 | OMR1 (L-O-METHYLTHREONINE RESISTANT 1); L-threonine ammonia-lyase | -2.55 |
| 252 | 258932_at | AT3G10150 | ATPAP16/PAP16 (purple acid phosphatase 16); acid phosphatase/ protein serine/threonine phosphatase | -2.79 |
| 253 | 259150_at | AT3G10320 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G41640.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO23616.1); contains InterPro domain Protein of unknown function DUF563 (InterPro:IPR007657) | 7.09 |
| 254 | 258920_at | AT3G10520 | AHB2 (NON-SYMBIOTIC HAEMOGLOBIN 2) | -2.82 |
| 255 | 259231_at | AT3G11410 | AHG3/ATPP2CA (ARABIDOPSIS THALIANA PROTEIN PHOSPHATASE 2CA); protein binding / protein serine/threonine phosphatase | 3.10 |
| 256 | 259286_at | AT3G11480 | BSMT1; S-adenosylmethionine-dependent methyltransferase | 8.22 |
| 257 | 256275_at | AT3G12110 | ACT11 (ACTIN-11); structural constituent of cytoskeleton | -2.74 |
| 258 | 256262_at | AT3G12150 | similar to hypothetical protein [Vitis vinifera] (GB:CAN81264.1); contains domain PTHR13617 (PTHR13617); contains domain SSF53474 (SSF53474); contains domain G3DSA:3.40.50.1820 (G3DSA:3.40.50.1820); contains domain PTHR13617:SF1 (PTHR13617:SF1) | -3.10 |
| 259 | 256245_at | AT3G12580 | HSP70 (heat shock protein 70); ATP binding | 4.56 |
| 260 | 256237_at | AT3G12610 | DRT100 (DNA-DAMAGE REPAIR/TOLERATION 100); protein binding | -2.96 |
| 261 | 257853_at | AT3G12960 | similar to seed maturation protein [Glycine tomentella] (GB:ABB72392.1) | 6.75 |
| 262 | 256983_at | AT3G13470 | chaperonin, putative | -2.46 |
| 263 | 256772_at | AT3G13750 | BGAL1 (BETA GALACTOSIDASE 1); beta-galactosidase | -3.86 |
| 264 | 258368_at | AT3G14240 | subtilase family protein | -2.68 |
| 265 | 258362_at | AT3G14280 | similar to hypothetical protein [Vitis vinifera] (GB:CAN62135.1) | 7.26 |
| 266 | 258091_at | AT3G14560 | unknown protein | 4.21 |
| 267 | 258063_at | AT3G14620 | CYP72A8 (cytochrome P450, family 72, subfamily A, polypeptide 8); oxygen binding | 3.05 |
| 268 | 258386_at | AT3G15520 | peptidyl-prolyl cis-trans isomerase TLP38, chloroplast / thylakoid lumen PPIase of 38 kDa / cyclophilin / rotamase | -2.63 |
| 269 | 258222_at | AT3G15680 | zinc finger (Ran-binding) family protein | -2.64 |
| 270 | 258250_at | AT3G15850 | FAD5 (FATTY ACID DESATURASE 5); oxidoreductase | -3.57 |
| 271 | 258338_at | AT3G16150 | L-asparaginase, putative / L-asparagine amidohydrolase, putative | -3.37 |
| 272 | 258054_at | AT3G16240 | DELTA-TIP (delta tonoplast integral protein); water channel | -2.79 |
| 273 | 258158_at | AT3G17790 | ATACP5 (acid phosphatase 5); acid phosphatase/ protein serine/threonine phosphatase | 5.05 |
| 274 | 258162_at | AT3G17810 | dihydroorotate dehydrogenase family protein / dihydroorotate oxidase family protein | 3.86 |
| 275 | 258156_at | AT3G18050 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G28100.1); similar to unknown [Populus trichocarpa] (GB:ABK95654.1) | -2.47 |
| 276 | 257122_at | AT3G20250 | APUM5 (ARABIDOPSIS PUMILIO 5); RNA binding | 3.33 |
| 277 | 256796_at | AT3G22210 | unknown protein | -4.75 |
| 278 | 256933_at | AT3G22600 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | 5.28 |
| 279 | 258321_at | AT3G22840 | ELIP1 (EARLY LIGHT-INDUCABLE PROTEIN); chlorophyll binding | 4.89 |
| 280 | 256914_at | AT3G23880 | F-box family protein | -4.68 |
| 281 | 256864_at | AT3G23890 | TOPII (TOPOISOMERASE II); ATP binding / DNA binding / DNA topoisomerase (ATP-hydrolyzing) | -5.17 |
| 282 | 256862_at | AT3G23940 | dehydratase family | -2.48 |
| 283 | 258133_at | AT3G24500 | ATMBF1C/MBF1C (MULTIPROTEIN BRIDGING FACTOR 1C); DNA binding / transcription coactivator/ transcription factor | 3.30 |
| 284 | 257595_at | AT3G24750 | unknown protein | 7.18 |
| 285 | 258277_at | AT3G26830 | PAD3 (PHYTOALEXIN DEFICIENT 3); oxygen binding | 3.69 |
| 286 | 257809_at | AT3G27060 | TSO2 (TSO MEANING 'UGLY' IN CHINESE); ribonucleoside-diphosphate reductase | -2.52 |
| 287 | 257714_at | AT3G27360 | histone H3 | -2.44 |
| 288 | 257297_at | AT3G28040 | leucine-rich repeat transmembrane protein kinase, putative | -3.01 |
| 289 | 257137_at | AT3G28860 | ATMDR11/ATPGP19/MDR1/MDR11/PGP19 (P-GLYCOPROTEIN 19); ATPase, coupled to transmembrane movement of substances / auxin efflux transmembrane transporter | -2.52 |
| 290 | 257774_at | AT3G29250 | oxidoreductase | 6.74 |
| 291 | 246335_at | AT3G44880 | ACD1 (ACCELERATED CELL DEATH 1) | 3.49 |
| 292 | 246339_at | AT3G44890 | RPL9 (ribosomal protein L9); structural constituent of ribosome | -2.54 |
| 293 | 252529_at | AT3G46490 | oxidoreductase, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors | -2.92 |
| 294 | 252442_at | AT3G46940 | deoxyuridine 5'-triphosphate nucleotidohydrolase family | -3.80 |
| 295 | 252400_at | AT3G48020 | similar to F-box family protein-related [Arabidopsis thaliana] (TAIR:AT5G62860.1); similar to unknown [Populus trichocarpa] (GB:ABK95323.1) | 4.13 |
| 296 | 252358_at | AT3G48425 | endonuclease/exonuclease/phosphatase family protein | -3.34 |
| 297 | 252317_at | AT3G48720 | transferase family protein | -2.96 |
| 298 | 252280_at | AT3G49260 | IQD21 (IQ-DOMAIN 21, IQ-domain 21); calmodulin binding | -2.67 |
| 299 | 252199_at | AT3G50270 | transferase family protein | -4.10 |
| 300 | 252133_at | AT3G50900 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G66490.1); similar to hypothetical protein [Thellungiella halophila] (GB:ABB45855.1) | 3.11 |
| 301 | 252102_at | AT3G50970 | LTI30 (LOW TEMPERATURE-INDUCED 30) | 8.44 |
| 302 | 252137_at | AT3G50980 | XERO1 (DEHYDRIN XERO 1) | 5.30 |
| 303 | 252117_at | AT3G51430 | YLS2 (yellow-leaf-specific gene 2); strictosidine synthase | 3.33 |
| 304 | 252068_at | AT3G51440 | strictosidine synthase family protein | 4.29 |
| 305 | 252076_at | AT3G51660 | macrophage migration inhibitory factor family protein / MIF family protein | 3.23 |
| 306 | 246302_at | AT3G51860 | CAX3 (cation exchanger 3); cation:cation antiporter | 3.99 |
| 307 | 256678_at | AT3G52380 | CP33 (PIGMENT DEFECTIVE 322); RNA binding | -2.67 |
| 308 | 252001_at | AT3G52750 | FTSZ2-2 (FtsZ2-2); structural molecule | -2.76 |
| 309 | 251982_at | AT3G53190 | pectate lyase family protein | -3.44 |
| 310 | 251920_at | AT3G53900 | uracil phosphoribosyltransferase, putative / UMP pyrophosphorylase, putative / UPRTase, putative | -2.72 |
| 311 | 251928_at | AT3G53980 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | 5.24 |
| 312 | 251895_at | AT3G54420 | ATEP3 (Arabidopsis thaliana chitinase class IV); chitinase | 3.21 |
| 313 | 251859_at | AT3G54680 | proteophosphoglycan-related | 3.16 |
| 314 | 251784_at | AT3G55330 | PPL1 (PSBP-LIKE PROTEIN 1); calcium ion binding | -2.48 |
| 315 | 251759_at | AT3G55630 | ATDFD (A. THALIANA DHFS-FPGS HOMOLOG D); tetrahydrofolylpolyglutamate synthase | -3.59 |
| 316 | 251714_at | AT3G56370 | leucine-rich repeat transmembrane protein kinase, putative | -2.94 |
| 317 | 246293_at | AT3G56710 | SIB1 (SIGMA FACTOR BINDING PROTEIN 1); binding | 3.41 |
| 318 | 251668_at | AT3G57010 | strictosidine synthase family protein | 8.82 |
| 319 | 251642_at | AT3G57520 | ATSIP2 (ARABIDOPSIS THALIANA SEED IMBIBITION 2); hydrolase, hydrolyzing O-glycosyl compounds | 3.76 |
| 320 | 251644_at | AT3G57540 | remorin family protein | 3.75 |
| 321 | 251479_at | AT3G59700 | ATHLECRK (ARABIDOPSIS THALIANA LECTIN-RECEPTOR KINASE); kinase | 4.07 |
| 322 | 251355_at | AT3G61100 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G61090.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO40196.1); contains InterPro domain Protein of unknown function DUF537 (InterPro:IPR007491) | -2.67 |
| 323 | 251329_at | AT3G61450 | SYP73 (SYNTAXIN OF PLANTS 73) | 4.31 |
| 324 | 251272_at | AT3G61890 | ATHB-12 (ARABIDOPSIS THALIANA HOMEOBOX PROTEIN 12); transcription factor | 4.01 |
| 325 | 251293_at | AT3G61930 | unknown protein | 4.66 |
| 326 | 251305_at | AT3G62030 | ROC4 (ROTAMASE CYP 4); peptidyl-prolyl cis-trans isomerase | -2.46 |
| 327 | 251219_at | AT3G62390 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G12060.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO40116.1); contains InterPro domain Protein of unknown function DUF231, plant (InterPro:IPR004253) | -2.80 |
| 328 | 251221_at | AT3G62550 | universal stress protein (USP) family protein | 3.09 |
| 329 | 251174_at | AT3G63200 | PLA IIIB/PLP9 (Patatin-like protein 9); nutrient reservoir | -5.10 |
| 330 | 255662_at | AT4G00400 | GPAT8 (GLYCEROL-3-PHOSPHATE ACYLTRANSFERASE 8); acyltransferase/ glycerol-3-phosphate O-acyltransferase | -2.77 |
| 331 | 255692_at | AT4G00400 | GPAT8 (GLYCEROL-3-PHOSPHATE ACYLTRANSFERASE 8); acyltransferase/ glycerol-3-phosphate O-acyltransferase | -2.79 |
| 332 | 255675_at | AT4G00480 | ATMYC1 (Arabidopsis thaliana myc-related transcription factor 1); DNA binding / transcription factor | -2.50 |
| 333 | 255645_at | AT4G00880 | auxin-responsive family protein | -2.80 |
| 334 | 255604_at | AT4G01080 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G01430.1); similar to unknown protein Cr17 [Brassica napus] (GB:AAX51387.1); contains InterPro domain Protein of unknown function DUF231, plant (InterPro:IPR004253) | -4.04 |
| 335 | 255621_at | AT4G01390 | meprin and TRAF homology domain-containing protein / MATH domain-containing protein | -4.66 |
| 336 | 255590_at | AT4G01610 | cathepsin B-like cysteine protease, putative | 3.75 |
| 337 | 255543_at | AT4G01870 | tolB protein-related | 4.17 |
| 338 | 255513_at | AT4G02060 | PRL (PROLIFERA); ATP binding / DNA binding / DNA-dependent ATPase | -3.12 |
| 339 | 255521_at | AT4G02280 | SUS3; UDP-glycosyltransferase/ sucrose synthase/ transferase, transferring glycosyl groups | 6.31 |
| 340 | 255527_at | AT4G02360 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G02813.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO40037.1); contains InterPro domain Protein of unknown function DUF538 (InterPro:IPR007493) | 7.43 |
| 341 | 255479_at | AT4G02380 | SAG21 (SENESCENCE-ASSOCIATED GENE 21) | 7.54 |
| 342 | 255502_at | AT4G02410 | lectin protein kinase family protein | 3.56 |
| 343 | 255460_at | AT4G02800 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G30050.1); similar to Os04g0228100 [Oryza sativa (japonica cultivar-group)] (GB:NP_001052288.1); similar to H0209A05.2 [Oryza sativa (indica cultivar-group)] (GB:CAH66085.1) | -4.69 |
| 344 | 255433_at | AT4G03210 | XTH9 (XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 9); hydrolase, acting on glycosyl bonds | -3.55 |
| 345 | 255340_at | AT4G04490 | protein kinase family protein | 5.48 |
| 346 | 255298_at | AT4G04840 | methionine sulfoxide reductase domain-containing protein / SeIR domain-containing protein | -5.32 |
| 347 | 254917_at | AT4G11350 | transferase, transferring glycosyl groups | 4.65 |
| 348 | 254815_at | AT4G12420 | SKU5 (skewed 5); copper ion binding | -2.81 |
| 349 | 254831_at | AT4G12600 | ribosomal protein L7Ae/L30e/S12e/Gadd45 family protein | -2.42 |
| 350 | 254783_at | AT4G12830 | hydrolase, alpha/beta fold family protein | -4.38 |
| 351 | 254789_at | AT4G12880 | plastocyanin-like domain-containing protein | -2.69 |
| 352 | 254746_at | AT4G12980 | auxin-responsive protein, putative | -3.52 |
| 353 | 245347_at | AT4G14890 | ferredoxin family protein | -2.40 |
| 354 | 245352_at | AT4G15490 | UGT84A3; UDP-glycosyltransferase/ sinapate 1-glucosyltransferase/ transferase, transferring glycosyl groups | 3.04 |
| 355 | 245343_at | AT4G15830 | binding | -4.98 |
| 356 | 245393_at | AT4G16260 | glycosyl hydrolase family 17 protein | 6.88 |
| 357 | 245318_at | AT4G16980 | arabinogalactan-protein family | -3.53 |
| 358 | 245463_at | AT4G17030 | ATEXLB1 (ARABIDOPSIS THALIANA EXPANSIN-LIKE B1) | 3.67 |
| 359 | 245247_at | AT4G17230 | SCL13 (SCARECROW-LIKE 13); transcription factor | 3.23 |
| 360 | 245383_at | AT4G17810 | nucleic acid binding / transcription factor/ zinc ion binding | -3.77 |
| 361 | 254667_at | AT4G18280 | glycine-rich cell wall protein-related | 3.92 |
| 362 | 254669_at | AT4G18370 | DEG5/DEGP5/HHOA (DEGP PROTEASE 5); serine-type peptidase/ trypsin | -2.79 |
| 363 | 254609_at | AT4G18970 | GDSL-motif lipase/hydrolase family protein | -4.91 |
| 364 | 254580_at | AT4G19390 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G13720.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO63752.1); contains InterPro domain Uncharacterised conserved protein UCP022348 (InterPro:IPR016804); contains InterPro domain Protein of unknown function UPF0114 (InterPro:IPR005134) | 4.73 |
| 365 | 254466_at | AT4G20430 | subtilase family protein | -2.40 |
| 366 | 254393_at | AT4G21580 | oxidoreductase, zinc-binding dehydrogenase family protein | 3.50 |
| 367 | 254346_at | AT4G21980 | APG8A (autophagy 8A) | 3.21 |
| 368 | 254328_at | AT4G22570 | APT3 (ADENINE PHOSPHORIBOSYL TRANSFERASE 3); adenine phosphoribosyltransferase | -2.45 |
| 369 | 254276_at | AT4G22820 | zinc finger (AN1-like) family protein | 3.67 |
| 370 | 254241_at | AT4G23190 | CRK11 (CYSTEINE-RICH RLK11); kinase | 3.20 |
| 371 | 254239_at | AT4G23400 | PIP1;5/PIP1D (plasma membrane intrinsic protein 1;5); water channel | -2.48 |
| 372 | 254210_at | AT4G23450 | zinc finger (C3HC4-type RING finger) family protein | 3.27 |
| 373 | 254263_at | AT4G23493 | unknown protein | 3.12 |
| 374 | 254232_at | AT4G23600 | CORI3 (CORONATINE INDUCED 1, JASMONIC ACID RESPONSIVE 2); transaminase | 5.84 |
| 375 | 254233_at | AT4G23800 | high mobility group (HMG1/2) family protein | -3.68 |
| 376 | 254221_at | AT4G23820 | glycoside hydrolase family 28 protein / polygalacturonase (pectinase) family protein | -4.22 |
| 377 | 254178_at | AT4G23880 | unknown protein | 3.19 |
| 378 | 254157_at | AT4G24220 | VEP1 (VEIN PATTERNING 1); binding / catalytic | 3.20 |
| 379 | 254119_at | AT4G24780 | pectate lyase family protein | -3.90 |
| 380 | 254102_at | AT4G25050 | ACP4 (ACYL CARRIER PROTEIN 4) | -2.77 |
| 381 | 254105_at | AT4G25080 | CHLM (MAGNESIUM-PROTOPORPHYRIN IX METHYLTRANSFERASE); magnesium protoporphyrin IX methyltransferase | -3.17 |
| 382 | 253971_at | AT4G26530 | fructose-bisphosphate aldolase, putative | -5.13 |
| 383 | 253872_at | AT4G27410 | RD26 (RESPONSIVE TO DESSICATION 26); transcription factor | 7.29 |
| 384 | 253884_at | AT4G27670 | HSP21 (HEAT SHOCK PROTEIN 21) | 8.64 |
| 385 | 253829_at | AT4G28040 | nodulin MtN21 family protein | 5.18 |
| 386 | 253736_at | AT4G28780 | GDSL-motif lipase/hydrolase family protein | -4.07 |
| 387 | 253619_at | AT4G30460 | glycine-rich protein | 4.40 |
| 388 | 253579_at | AT4G30610 | BRS1 (BRI1 SUPPRESSOR 1) | -4.05 |
| 389 | 253480_at | AT4G31840 | plastocyanin-like domain-containing protein | -2.60 |
| 390 | 253453_at | AT4G31860 | protein phosphatase 2C, putative / PP2C, putative | 3.05 |
| 391 | 253440_at | AT4G32570 | TIFY8 | -2.95 |
| 392 | 253401_at | AT4G32870 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G25770.2); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G25770.1); similar to unknown [Populus trichocarpa x Populus deltoides] (GB:ABK96434.1); contains domain SSF55961 (SSF55961) | 5.21 |
| 393 | 253373_at | AT4G33150 | LKR (SACCHAROPINE DEHYDROGENASE) | 7.04 |
| 394 | 253344_at | AT4G33550 | lipid binding | 5.02 |
| 395 | 253293_at | AT4G33905 | peroxisomal membrane protein 22 kDa, putative | 3.96 |
| 396 | 253270_at | AT4G34160 | CYCD3/CYCD3;1/D3 (CYCLIN D3;1); cyclin-dependent protein kinase regulator/ protein binding | -2.94 |
| 397 | 253277_at | AT4G34230 | CAD5 (CINNAMYL ALCOHOL DEHYDROGENASE 5); cinnamyl-alcohol dehydrogenase | 3.72 |
| 398 | 253203_at | AT4G34710 | ADC2 (ARGININE DECARBOXYLASE 2) | 3.49 |
| 399 | 253224_at | AT4G34860 | beta-fructofuranosidase, putative / invertase, putative / saccharase, putative / beta-fructosidase, putative | 3.54 |
| 400 | 253161_at | AT4G35770 | SEN1 (DARK INDUCIBLE 1) | 3.32 |
| 401 | 253120_at | AT4G35790 | ATPLDDELTA (Arabidopsis thaliana phospholipase D delta); phospholipase D | 3.40 |
| 402 | 246231_at | AT4G37080 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G42690.2); similar to hypothetical protein [Populus tremula] (GB:CAM84232.1); contains InterPro domain Protein of unknown function DUF547 (InterPro:IPR006869) | -2.40 |
| 403 | 246272_at | AT4G37150 | esterase, putative | 3.39 |
| 404 | 253050_at | AT4G37450 | AGP18 (Arabinogalactan protein 18) | -2.70 |
| 405 | 252976_s_at | AT4G38550 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G20950.1); contains InterPro domain Phospholipase-like, arabidopsis (InterPro:IPR007942) | 3.27 |
| 406 | 252971_at | AT4G38770 | PRP4 (PROLINE-RICH PROTEIN 4) | -2.71 |
| 407 | 252970_at | AT4G38850 | SAUR_AC1 (SMALL AUXIN UP RNA 1 FROM ARABIDOPSIS THALIANA ECOTYPE COLUMBIA) | -3.21 |
| 408 | 252916_at | AT4G38950 | kinesin motor family protein | -3.04 |
| 409 | 252888_at | AT4G39210 | APL3 (large subunit of AGP 3) | 3.05 |
| 410 | 252876_at | AT4G39970 | haloacid dehalogenase-like hydrolase family protein | -2.49 |
| 411 | 251142_at | AT5G01015 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G65295.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN82885.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO45347.1) | -4.20 |
| 412 | 251137_at | AT5G01300 | phosphatidylethanolamine-binding family protein | 6.61 |
| 413 | 251083_at | AT5G01590 | similar to unnamed protein product [Vitis vinifera] (GB:CAO63840.1) | -2.48 |
| 414 | 251036_at | AT5G02160 | unknown protein | -3.48 |
| 415 | 250968_at | AT5G02890 | transferase family protein | -5.43 |
| 416 | 250955_at | AT5G03190 | CPuORF47 (Conserved peptide upstream open reading frame 47) | 4.28 |
| 417 | 250624_at | AT5G07330 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G63060.1); similar to unknown [Xerophyta humilis] (GB:AAT45004.1) | 6.87 |
| 418 | 250598_at | AT5G07690 | MYB29 (myb domain protein 29); DNA binding / transcription factor | -3.85 |
| 419 | 250496_at | AT5G09650 | ATPPA6 (ARABIDOPSIS THALIANA PYROPHOSPHORYLASE 6); inorganic diphosphatase/ pyrophosphatase | -2.43 |
| 420 | 250503_at | AT5G09820 | plastid-lipid associated protein PAP / fibrillin family protein | -2.47 |
| 421 | 250506_at | AT5G09930 | ATGCN2 (Arabidopsis thaliana general control non-repressible 2) | 3.84 |
| 422 | 250478_at | AT5G10250 | phototropic-responsive protein, putative | -5.94 |
| 423 | 250434_at | AT5G10390 | histone H3 | -3.06 |
| 424 | 250433_at | AT5G10400 | histone H3 | -2.97 |
| 425 | 245904_at | AT5G11110 | ATSPS2F/SPS1 (SUCROSE PHOSPHATE SYNTHASE 1); sucrose-phosphate synthase | 4.05 |
| 426 | 250413_at | AT5G11160 | APT5 (ADENINE PHOSPHORIBOSYLTRANSFERASE 5); adenine phosphoribosyltransferase | -2.41 |
| 427 | 250327_at | AT5G12050 | similar to unnamed protein product [Vitis vinifera] (GB:CAO45643.1) | -4.04 |
| 428 | 245981_at | AT5G13100 | oxidoreductase | -2.42 |
| 429 | 245980_at | AT5G13140 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G26960.1); similar to unknown [Euphorbia pulcherrima] (GB:ABO43960.1); contains InterPro domain Pollen Ole e 1 allergen and extensin (InterPro:IPR006041) | -3.32 |
| 430 | 245982_at | AT5G13170 | nodulin MtN3 family protein | 3.92 |
| 431 | 250279_at | AT5G13200 | GRAM domain-containing protein / ABA-responsive protein-related | 4.60 |
| 432 | 250228_at | AT5G13840 | WD-40 repeat family protein | -2.63 |
| 433 | 250212_at | AT5G13960 | SUVH4 (SU(VAR)3-9 HOMOLOG 4) | -2.71 |
| 434 | 250199_at | AT5G14180 | MPL1 (MYZUS PERSICAE-INDUCED LIPASE 1); catalytic | 4.91 |
| 435 | 246540_at | AT5G15600 | SP1L4 (SPIRAL1-LIKE4) | -2.65 |
| 436 | 246487_at | AT5G16030 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G02500.1); similar to hypothetical protein [Cleome spinosa] (GB:ABD96956.1) | -2.91 |
| 437 | 250012_x_at | AT5G18060 | auxin-responsive protein, putative | -3.04 |
| 438 | 250028_at | AT5G18130 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G03870.2); similar to unknown [Medicago truncatula] (GB:ABK28852.1) | 4.57 |
| 439 | 249920_at | AT5G19260 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G06020.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN75990.1) | -2.88 |
| 440 | 249917_at | AT5G22460 | esterase/lipase/thioesterase family protein | 6.11 |
| 441 | 249900_at | AT5G22640 | EMB1211 (EMBRYO DEFECTIVE 1211) | -2.54 |
| 442 | 249770_at | AT5G24110 | WRKY30 (WRKY DNA-binding protein 30); transcription factor | 7.45 |
| 443 | 249732_at | AT5G24420 | glucosamine/galactosamine-6-phosphate isomerase-related | -3.97 |
| 444 | 246919_at | AT5G25460 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G11420.1); similar to unknown [Ricinus communis] (GB:CAB02653.1); contains InterPro domain Protein of unknown function DUF642 (InterPro:IPR006946) | -3.30 |
| 445 | 246855_at | AT5G26280 | meprin and TRAF homology domain-containing protein / MATH domain-containing protein | -4.03 |
| 446 | 246831_at | AT5G26340 | MSS1 (SUGAR TRANSPORT PROTEIN 13); carbohydrate transmembrane transporter/ hexose:hydrogen symporter/ high-affinity hydrogen:glucose symporter/ sugar:hydrogen ion symporter | 3.89 |
| 447 | 246791_at | AT5G27280 | zinc finger (DNL type) family protein | 3.40 |
| 448 | 246792_at | AT5G27290 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G54680.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G54680.2); similar to unnamed protein product [Vitis vinifera] (GB:CAO68604.1) | -2.90 |
| 449 | 246777_at | AT5G27420 | zinc finger (C3HC4-type RING finger) family protein | 3.49 |
| 450 | 246779_at | AT5G27520 | mitochondrial substrate carrier family protein | 3.39 |
| 451 | 249645_at | AT5G36910 | THI2.2 (THIONIN 2.2); toxin receptor binding | -4.65 |
| 452 | 249575_at | AT5G37670 | 15.7 kDa class I-related small heat shock protein-like (HSP15.7-CI) | 4.56 |
| 453 | 249510_at | AT5G38510 | rhomboid family protein | -2.57 |
| 454 | 249494_at | AT5G39050 | transferase family protein | 3.25 |
| 455 | 249472_at | AT5G39210 | CRR7 (CHLORORESPIRATORY REDUCTION 7) | -3.40 |
| 456 | 249454_at | AT5G39520 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G39530.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO15021.1) | 8.15 |
| 457 | 249377_at | AT5G40690 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G41730.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO14635.1) | 3.73 |
| 458 | 249237_at | AT5G42050 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G27090.1); similar to unknown [Populus trichocarpa] (GB:ABK95892.1); contains InterPro domain Kelch related (InterPro:IPR013089); contains InterPro domain Development and cell death (InterPro:IPR013989) | 3.71 |
| 459 | 249197_at | AT5G42380 | CML37/CML39; calcium ion binding | 3.26 |
| 460 | 249191_at | AT5G42760 | similar to unknown [Populus trichocarpa] (GB:ABK95091.1); contains InterPro domain O-methyltransferase, N-terminal (InterPro:IPR003455); contains InterPro domain Conserved hypothetical protein CHP00027, methylltransferase (InterPro:IPR011610) | -2.63 |
| 461 | 249117_at | AT5G43840 | AT-HSFA6A (Arabidopsis thaliana heat shock transcription factor A6A); DNA binding / transcription factor | 9.67 |
| 462 | 249073_at | AT5G44020 | acid phosphatase class B family protein | -4.43 |
| 463 | 249060_at | AT5G44560 | VPS2.2 | -4.01 |
| 464 | 249008_at | AT5G44680 | methyladenine glycosylase family protein | -3.09 |
| 465 | 248969_at | AT5G45310 | similar to unnamed protein product [Vitis vinifera] (GB:CAO63757.1) | 4.54 |
| 466 | 248959_at | AT5G45630 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G18980.1); similar to unknown [Cucumis sativus] (GB:ABY56081.1); similar to unknown [Cucumis melo] (GB:ABQ53634.1); contains InterPro domain Protein of unknown function DUF584 (InterPro:IPR007608) | 5.20 |
| 467 | 248963_at | AT5G45700 | NLI interacting factor (NIF) family protein | -5.74 |
| 468 | 248867_at | AT5G46830 | basic helix-loop-helix (bHLH) family protein | -5.11 |
| 469 | 248784_at | AT5G47380 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G66600.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G66600.3); similar to hypothetical protein 24.t00022 [Brassica oleracea] (GB:ABD64944.1); contains InterPro domain Protein of unknown function DUF547 (InterPro:IPR006869) | -4.90 |
| 470 | 248807_at | AT5G47500 | pectinesterase family protein | -3.67 |
| 471 | 248696_at | AT5G48360 | formin homology 2 domain-containing protein / FH2 domain-containing protein | -2.50 |
| 472 | 248656_at | AT5G48460 | fimbrin-like protein, putative | -3.52 |
| 473 | 248619_at | AT5G49630 | AAP6 (AMINO ACID PERMEASE 6); amino acid transmembrane transporter | -2.81 |
| 474 | 248505_at | AT5G50360 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G63350.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO68256.1) | 9.63 |
| 475 | 248526_at | AT5G50740 | metal ion binding | -3.83 |
| 476 | 248419_at | AT5G51550 | phosphate-responsive 1 family protein | -2.49 |
| 477 | 248377_at | AT5G51720 | similar to Os07g0467200 [Oryza sativa (japonica cultivar-group)] (GB:NP_001059590.1); similar to hypothetical protein OsI_025030 [Oryza sativa (indica cultivar-group)] (GB:EAZ03798.1); contains domain PTHR13680 (PTHR13680); contains domain PTHR13680:SF1 (PTHR13680:SF1) | -3.97 |
| 478 | 248372_at | AT5G51850 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G25430.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO17701.1) | -5.29 |
| 479 | 248352_at | AT5G52300 | LTI65/RD29B (RESPONSIVE TO DESSICATION 29B) | 8.80 |
| 480 | 248337_at | AT5G52310 | COR78 (COLD REGULATED 78) | 5.81 |
| 481 | 248332_at | AT5G52640 | HSP81-1 (HEAT SHOCK PROTEIN 81-1); ATP binding / unfolded protein binding | 3.04 |
| 482 | 248246_at | AT5G53200 | TRY (TRIPTYCHON); DNA binding / transcription factor | -3.23 |
| 483 | 248236_at | AT5G53870 | plastocyanin-like domain-containing protein | 5.78 |
| 484 | 248193_at | AT5G54080 | HGO (HOMOGENTISATE 1,2-DIOXYGENASE); homogentisate 1,2-dioxygenase | 3.60 |
| 485 | 248085_at | AT5G55480 | glycerophosphoryl diester phosphodiesterase family protein | -2.73 |
| 486 | 248055_at | AT5G55830 | lectin protein kinase, putative | -4.05 |
| 487 | 247977_at | AT5G56850 | similar to unknown [Populus trichocarpa x Populus deltoides] (GB:ABK96358.1) | -2.86 |
| 488 | 247943_at | AT5G57170 | macrophage migration inhibitory factor family protein / MIF family protein | -3.41 |
| 489 | 247863_at | AT5G57900 | SKIP1 (SKP1 INTERACTING PARTNER 1) | 3.66 |
| 490 | 247816_at | AT5G58260 | Encodes subunit NDH-N of NAD(P)H:plastoquinone dehydrogenase complex (Ndh complex) present in the thylakoid membrane of chloroplasts. This subunit is thought to be required for Ndh complex assembly. | -2.73 |
| 491 | 247754_at | AT5G59080 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G02020.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO15731.1) | -2.82 |
| 492 | 247723_at | AT5G59220 | protein phosphatase 2C, putative / PP2C, putative | 6.04 |
| 493 | 247718_at | AT5G59310 | LTP4 (LIPID TRANSFER PROTEIN 4); lipid binding | 10.12 |
| 494 | 247717_at | AT5G59320 | LTP3 (LIPID TRANSFER PROTEIN 3); lipid binding | 9.14 |
| 495 | 247655_at | AT5G59820 | RHL41 (RESPONSIVE TO HIGH LIGHT 41); nucleic acid binding / transcription factor/ zinc ion binding | 3.28 |
| 496 | 247651_at | AT5G59870 | HTA6; DNA binding | -2.91 |
| 497 | 247563_at | AT5G61130 | glycosyl hydrolase family protein 17 | -2.74 |
| 498 | 247488_at | AT5G61820 | similar to MtN19-like protein [Pisum sativum] (GB:AAU14999.2); contains InterPro domain Stress up-regulated Nod 19 (InterPro:IPR011692) | 3.06 |
| 499 | 247511_at | AT5G62040 | brother of FT and TFL1 protein (BFT) | 3.18 |
| 500 | 247474_at | AT5G62280 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G45360.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO49726.1); contains InterPro domain Protein of unknown function DUF1442 (InterPro:IPR009902) | -4.51 |
| 501 | 247376_at | AT5G63310 | NDPK2 (NUCLEOSIDE DIPHOSPHATE KINASE 2); ATP binding / nucleoside diphosphate kinase | -2.49 |
| 502 | 247347_at | AT5G63780 | SHA1 (SHOOT APICAL MERISTEM ARREST 1); protein binding / zinc ion binding | -2.82 |
| 503 | 247195_at | AT5G65500 | protein kinase family protein | 3.58 |
| 504 | 247162_at | AT5G65730 | xyloglucan:xyloglucosyl transferase, putative / xyloglucan endotransglycosylase, putative / endo-xyloglucan transferase, putative | -5.88 |
| 505 | 247128_at | AT5G66110 | metal ion binding | 5.56 |
| 506 | 247095_at | AT5G66400 | RAB18 (RESPONSIVE TO ABA 18) | 6.38 |
| 507 | 247101_at | AT5G66530 | aldose 1-epimerase family protein | -2.41 |
| 508 | 247055_at | AT5G66740 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G75160.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN74294.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO22871.1); contains InterPro domain Protein of unknown function DUF620 (InterPro:IPR006873) | -6.44 |
| 509 | 247026_at | AT5G67080 | MAPKKK19 (Mitogen-activated protein kinase kinase kinase 19); kinase | 5.58 |
| 510 | 247028_at | AT5G67100 | ICU2 (INCURVATA2); DNA-directed DNA polymerase | -4.49 |