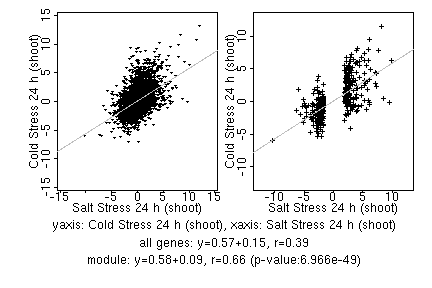
marker gene list
| Salt Stress 24 h (shoot) |
| Cold Stress 24 h (shoot) |
| probeID | AGICode | Annotation | gene expression |
|
|---|---|---|---|---|
| 1 | 261046_at | AT1G01390 | UDP-glucoronosyl/UDP-glucosyl transferase family protein | -2.19 |
| 2 | 259426_at | AT1G01470 | LEA14 (LATE EMBRYOGENESIS ABUNDANT 14) | 2.76 |
| 3 | 259417_at | AT1G02340 | HFR1 (LONG HYPOCOTYL IN FAR-RED); DNA binding / transcription factor | -2.38 |
| 4 | 260917_at | AT1G02700 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G02140.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN70483.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO40074.1) | 2.83 |
| 5 | 262118_at | AT1G02850 | glycosyl hydrolase family 1 protein | 3.37 |
| 6 | 265066_at | AT1G03870 | FLA9 | -1.84 |
| 7 | 264609_at | AT1G04530 | binding | -2.14 |
| 8 | 265216_at | AT1G05100 | MAPKKK18 (Mitogen-activated protein kinase kinase kinase 18); kinase | 4.66 |
| 9 | 264580_at | AT1G05340 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G32210.1) | 5.47 |
| 10 | 260831_at | AT1G06830 | glutaredoxin family protein | -2.52 |
| 11 | 256061_at | AT1G07040 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G27030.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO41035.1) | 2.17 |
| 12 | 261077_at | AT1G07430 | protein phosphatase 2C, putative / PP2C, putative | 5.88 |
| 13 | 261068_at | AT1G07450 | tropinone reductase, putative / tropine dehydrogenase, putative | -2.34 |
| 14 | 261065_at | AT1G07500 | unknown protein | 2.44 |
| 15 | 261420_at | AT1G07720 | beta-ketoacyl-CoA synthase family protein | 2.43 |
| 16 | 264783_at | AT1G08650 | PPCK1 (PHOSPHOENOLPYRUVATE CARBOXYLASE KINASE); kinase | 2.49 |
| 17 | 264264_at | AT1G09250 | transcription factor | -1.84 |
| 18 | 264510_at | AT1G09530 | PAP3/PIF3/POC1 (PHYTOCHROME INTERACTING FACTOR 3); DNA binding / protein binding / transcription factor/ transcription regulator | 3.09 |
| 19 | 264672_at | AT1G09750 | chloroplast nucleoid DNA-binding protein-related | -1.85 |
| 20 | 263236_at | AT1G10470 | ARR4 (RESPONSE REGULATOR 4); transcription regulator/ two-component response regulator | -2.50 |
| 21 | 262793_at | AT1G13110 | CYP71B7 (cytochrome P450, family 71, subfamily B, polypeptide 7); oxygen binding | -2.18 |
| 22 | 262607_at | AT1G13990 | similar to unnamed protein product [Vitis vinifera] (GB:CAO68469.1) | 2.57 |
| 23 | 261768_at | AT1G15550 | GA4 (GA REQUIRING 4); gibberellin 3-beta-dioxygenase | 3.07 |
| 24 | 256114_at | AT1G16850 | unknown protein | 8.30 |
| 25 | 262482_at | AT1G17020 | SRG1 (SENESCENCE-RELATED GENE 1); oxidoreductase, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors | 7.56 |
| 26 | 259403_at | AT1G17745 | PGDH (3-PHOSPHOGLYCERATE DEHYDROGENASE); phosphoglycerate dehydrogenase | 3.57 |
| 27 | 255891_at | AT1G17870 | ATEGY3 | 3.99 |
| 28 | 255774_at | AT1G18620 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G74160.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO49165.1) | -1.68 |
| 29 | 256017_at | AT1G19180 | JAZ1/TIFY10A (JASMONATE-ZIM-DOMAIN PROTEIN 1); protein binding | 3.07 |
| 30 | 260664_at | AT1G19510 | myb family transcription factor | -2.29 |
| 31 | 259570_at | AT1G20440 | COR47 (cold regulated 47) | 3.86 |
| 32 | 259516_at | AT1G20450 | ERD10/LTI45 (EARLY RESPONSIVE TO DEHYDRATION 10) | 4.69 |
| 33 | 259561_at | AT1G21250 | WAK1 (CELL WALL-ASSOCIATED KINASE); kinase | 2.19 |
| 34 | 260877_at | AT1G21500 | similar to hypothetical protein OsI_030994 [Oryza sativa (indica cultivar-group)] (GB:EAZ09762.1); similar to Os09g0517000 [Oryza sativa (japonica cultivar-group)] (GB:NP_001063677.1); similar to unknown protein [Oryza sativa (japonica cultivar-group)] (GB:BAD33829.1) | -1.84 |
| 35 | 255969_at | AT1G22330 | RNA binding | -1.79 |
| 36 | 264195_at | AT1G22690 | gibberellin-responsive protein, putative | -4.85 |
| 37 | 262986_at | AT1G23390 | kelch repeat-containing F-box family protein | -3.15 |
| 38 | 265175_at | AT1G23480 | ATCSLA03 (Cellulose synthase-like A3); transferase, transferring glycosyl groups | -2.09 |
| 39 | 265024_at | AT1G24600 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G67920.1) | 3.61 |
| 40 | 245816_at | AT1G26210 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G68870.1) | 2.40 |
| 41 | 259789_at | AT1G29395 | COR414-TM1 (cold regulated 414 thylakoid membrane 1) | 4.06 |
| 42 | 257506_at | AT1G29440 | auxin-responsive family protein | -2.06 |
| 43 | 259787_at | AT1G29460 | auxin-responsive protein, putative | -3.54 |
| 44 | 259783_at | AT1G29510 | SAUR68 (SMALL AUXIN UPREGULATED 68) | -2.67 |
| 45 | 257507_at | AT1G29600 | zinc finger (CCCH-type) family protein | -2.40 |
| 46 | 245769_at | AT1G30220 | ATINT2 (INOSITOL TRANSPORTER 2); carbohydrate transmembrane transporter/ sugar:hydrogen ion symporter | 2.49 |
| 47 | 261712_at | AT1G32780 | alcohol dehydrogenase, putative | -3.21 |
| 48 | 261285_at | AT1G35720 | ANNAT1 (ANNEXIN ARABIDOPSIS 1); calcium ion binding / calcium-dependent phospholipid binding | 2.53 |
| 49 | 265136_at | AT1G51270 | structural molecule | -3.38 |
| 50 | 259839_at | AT1G52190 | proton-dependent oligopeptide transport (POT) family protein | -1.89 |
| 51 | 262128_at | AT1G52690 | late embryogenesis abundant protein, putative / LEA protein, putative | 7.55 |
| 52 | 263157_at | AT1G54100 | ALDH7B4 (ALDEHYDE DEHYDROGENASE 7B4); 3-chloroallyl aldehyde dehydrogenase | 3.52 |
| 53 | 263158_at | AT1G54160 | CCAAT-binding transcription factor (CBF-B/NF-YA) family protein | 2.65 |
| 54 | 264562_at | AT1G55760 | BTB/POZ domain-containing protein | 2.19 |
| 55 | 260603_at | AT1G55960 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G13062.2); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G13062.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO41766.1); contains InterPro domain Lipid-binding START (InterPro:IPR002913) | -2.62 |
| 56 | 245627_at | AT1G56600 | ATGOLS2 (ARABIDOPSIS THALIANA GALACTINOL SYNTHASE 2); transferase, transferring glycosyl groups / transferase, transferring hexosyl groups | 4.27 |
| 57 | 245628_at | AT1G56650 | PAP1 (PRODUCTION OF ANTHOCYANIN PIGMENT 1); DNA binding / transcription factor | 3.18 |
| 58 | 246403_at | AT1G57590 | carboxylesterase | 5.04 |
| 59 | 256021_at | AT1G58270 | ZW9 | 2.47 |
| 60 | 264217_at | AT1G60190 | armadillo/beta-catenin repeat family protein / U-box domain-containing protein | 3.57 |
| 61 | 264741_at | AT1G62290 | aspartyl protease family protein | 2.15 |
| 62 | 265119_at | AT1G62570 | flavin-containing monooxygenase family protein / FMO family protein | 2.85 |
| 63 | 262644_at | AT1G62710 | BETA-VPE (vacuolar processing enzyme beta); cysteine-type endopeptidase | 2.15 |
| 64 | 262347_at | AT1G64110 | AAA-type ATPase family protein | 4.11 |
| 65 | 261957_at | AT1G64660 | ATMGL; catalytic/ methionine gamma-lyase | 3.14 |
| 66 | 261914_at | AT1G65870 | disease resistance-responsive family protein | -1.75 |
| 67 | 256527_at | AT1G66100 | thionin, putative | -5.08 |
| 68 | 256528_at | AT1G66140 | ZFP4 (ZINC FINGER PROTEIN 4); nucleic acid binding / transcription factor/ zinc ion binding | -1.50 |
| 69 | 259822_at | AT1G66230 | MYB20 (myb domain protein 20); DNA binding / transcription factor | -2.46 |
| 70 | 255856_at | AT1G66940 | protein kinase-related | -1.53 |
| 71 | 264968_at | AT1G67360 | rubber elongation factor (REF) family protein | 2.92 |
| 72 | 260012_at | AT1G67865 | unknown protein | -1.58 |
| 73 | 260005_at | AT1G67920 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G24600.1) | 3.43 |
| 74 | 260357_at | AT1G69260 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G13740.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO41856.1); contains InterPro domain Protein of unknown function DUF1675 (InterPro:IPR012463) | 4.05 |
| 75 | 256299_at | AT1G69530 | ATEXPA1 (ARABIDOPSIS THALIANA EXPANSIN A1) | -3.74 |
| 76 | 260410_at | AT1G69870 | proton-dependent oligopeptide transport (POT) family protein | 2.13 |
| 77 | 264339_at | AT1G70290 | ATTPS8 (Arabidopsis thaliana trehalose phosphatase/synthase 8); transferase, transferring glycosyl groups | -1.64 |
| 78 | 260181_at | AT1G70710 | AtGH9B1 (ARABIDOPSIS THALIANA GLYCOSYL HYDROLASE 9B1); hydrolase, hydrolyzing O-glycosyl compounds | -1.52 |
| 79 | 262309_at | AT1G70820 | phosphoglucomutase, putative / glucose phosphomutase, putative | -2.30 |
| 80 | 262290_at | AT1G70985 | hydroxyproline-rich glycoprotein family protein | -2.13 |
| 81 | 262369_at | AT1G73010 | phosphoric monoester hydrolase | 2.57 |
| 82 | 262357_at | AT1G73040 | jacalin lectin family protein | 3.08 |
| 83 | 260248_at | AT1G74310 | ATHSP101 (HEAT SHOCK PROTEIN 101); ATP binding / ATPase | 2.41 |
| 84 | 260221_at | AT1G74670 | gibberellin-responsive protein, putative | -4.92 |
| 85 | 256503_at | AT1G75250 | myb family transcription factor | -6.14 |
| 86 | 261754_at | AT1G76130 | AMY2/ATAMY2 (ALPHA-AMYLASE-LIKE 2); alpha-amylase | 2.12 |
| 87 | 261772_at | AT1G76240 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G17080.1); similar to Os06g0725500 [Oryza sativa (japonica cultivar-group)] (GB:NP_001058623.1); similar to hypothetical protein OsI_023643 [Oryza sativa (indica cultivar-group)] (GB:EAZ02411.1); contains InterPro domain Protein of unknown function DUF241, plant (InterPro:IPR004320) | -3.98 |
| 88 | 264958_at | AT1G76960 | unknown protein | 3.01 |
| 89 | 264953_at | AT1G77120 | ADH1 (ALCOHOL DEHYDROGENASE 1) | 4.09 |
| 90 | 259705_at | AT1G77450 | ANAC032 (Arabidopsis NAC domain containing protein 32); transcription factor | 2.62 |
| 91 | 260774_at | AT1G78290 | serine/threonine protein kinase, putative | -1.59 |
| 92 | 263127_at | AT1G78610 | mechanosensitive ion channel domain-containing protein / MS ion channel domain-containing protein | 2.47 |
| 93 | 262050_at | AT1G80130 | binding | 3.13 |
| 94 | 260297_at | AT1G80280 | hydrolase, alpha/beta fold family protein | -1.57 |
| 95 | 260287_at | AT1G80440 | kelch repeat-containing F-box family protein | -3.07 |
| 96 | 266181_at | AT2G02390 | ATGSTZ1 (GLUTATHIONE S-TRANSFERASE 18); glutathione transferase | 2.52 |
| 97 | 264040_at | AT2G03730 | ACR5 (ACT Domain Repeat 5) | -1.61 |
| 98 | 265478_at | AT2G15890 | MEE14 (maternal effect embryo arrest 14) | -2.47 |
| 99 | 265480_at | AT2G15970 | COR413-PM1 (cold regulated 413 plasma membrane 1) | 2.75 |
| 100 | 263558_at | AT2G16380 | SEC14 cytosolic factor family protein / phosphoglyceride transfer family protein | -1.59 |
| 101 | 264787_at | AT2G17840 | ERD7 (EARLY-RESPONSIVE TO DEHYDRATION 7) | 3.34 |
| 102 | 265342_at | AT2G18300 | basic helix-loop-helix (bHLH) family protein | -4.73 |
| 103 | 266072_at | AT2G18700 | ATTPS11 (Arabidopsis thaliana trehalose phosphatase/synthase 11); transferase, transferring glycosyl groups | -2.57 |
| 104 | 263374_at | AT2G20560 | DNAJ heat shock family protein | 2.73 |
| 105 | 265387_at | AT2G20670 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G32480.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO69754.1); contains InterPro domain Protein of unknown function DUF506, plant (InterPro:IPR006502) | -2.00 |
| 106 | 263517_at | AT2G21620 | RD2 (RESPONSIVE TO DESSICATION 2) | 2.48 |
| 107 | 263549_at | AT2G21650 | MEE3 (maternal effect embryo arrest 3); DNA binding / transcription factor | -4.55 |
| 108 | 263881_at | AT2G21820 | similar to hypothetical protein MtrDRAFT_AC155884g16v2 [Medicago truncatula] (GB:ABN08202.1) | 5.46 |
| 109 | 264005_at | AT2G22470 | AGP2 (ARABINOGALACTAN-PROTEIN 2) | 3.61 |
| 110 | 267265_at | AT2G22980 | SCPL13; serine carboxypeptidase | -2.32 |
| 111 | 267261_at | AT2G23120 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G23110.1) | 2.41 |
| 112 | 265913_at | AT2G25625 | similar to unnamed protein product [Vitis vinifera] (GB:CAO44157.1) | 2.60 |
| 113 | 266656_at | AT2G25900 | ATCTH (Arabidopsis thaliana Cys3His zinc finger protein); transcription factor | -1.74 |
| 114 | 265665_at | AT2G27420 | cysteine proteinase, putative | -1.89 |
| 115 | 266165_at | AT2G28190 | CSD2 (COPPER/ZINC SUPEROXIDE DISMUTASE 2); copper, zinc superoxide dismutase | -1.49 |
| 116 | 266229_at | AT2G28840 | ankyrin repeat family protein | 2.98 |
| 117 | 266225_at | AT2G28900 | OEP16 (OUTER ENVELOPE PROTEIN 16); P-P-bond-hydrolysis-driven protein transmembrane transporter | 2.40 |
| 118 | 266790_at | AT2G28950 | ATEXPA6 (ARABIDOPSIS THALIANA EXPANSIN A6) | -1.52 |
| 119 | 266279_at | AT2G29290 | tropinone reductase, putative / tropine dehydrogenase, putative | -2.10 |
| 120 | 266277_at | AT2G29310 | tropinone reductase, putative / tropine dehydrogenase, putative | -1.56 |
| 121 | 266805_at | AT2G30010 | similar to PMR5 (POWDERY MILDEW RESISTANT 5) [Arabidopsis thaliana] (TAIR:AT5G58600.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO21434.1); contains InterPro domain Protein of unknown function DUF231, plant (InterPro:IPR004253) | -3.32 |
| 122 | 267523_at | AT2G30600 | BTB/POZ domain-containing protein | -3.29 |
| 123 | 267524_at | AT2G30600 | BTB/POZ domain-containing protein | -1.74 |
| 124 | 265724_at | AT2G32100 | ATOFP16/OFP16 (Arabidopsis thaliana ovate family protein 16) | -2.73 |
| 125 | 255795_at | AT2G33380 | RD20 (RESPONSIVE TO DESSICATION 20); calcium ion binding | 2.98 |
| 126 | 255787_at | AT2G33590 | cinnamoyl-CoA reductase family | 2.54 |
| 127 | 266899_at | AT2G34620 | mitochondrial transcription termination factor-related / mTERF-related | -1.72 |
| 128 | 266544_at | AT2G35300 | late embryogenesis abundant group 1 domain-containing protein / LEA group 1 domain-containing protein | 6.38 |
| 129 | 263951_at | AT2G35960 | NHL12 (NDR1/HIN1-like 12) | -1.75 |
| 130 | 263931_at | AT2G36220 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G52710.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO15901.1) | 4.29 |
| 131 | 267181_at | AT2G37760 | aldo/keto reductase family protein | 3.05 |
| 132 | 267168_at | AT2G37770 | aldo/keto reductase family protein | 5.01 |
| 133 | 266098_at | AT2G37870 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | 6.14 |
| 134 | 267034_at | AT2G38310 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G05440.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO48777.1); contains InterPro domain Bet v I allergen; (InterPro:IPR000916); contains InterPro domain Streptomyces cyclase/dehydrase (InterPro:IPR005031) | -2.58 |
| 135 | 263264_at | AT2G38810 | HTA8; DNA binding | -1.73 |
| 136 | 266170_at | AT2G39050 | hydroxyproline-rich glycoprotein family protein | 2.94 |
| 137 | 267357_at | AT2G40000 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G55840.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO41329.1); contains InterPro domain Hs1pro-1, C-terminal (InterPro:IPR009743); contains InterPro domain Hs1pro-1, N-terminal (InterPro:IPR009869) | -1.53 |
| 138 | 255826_at | AT2G40490 | HEME2; uroporphyrinogen decarboxylase | -1.75 |
| 139 | 255822_at | AT2G40610 | ATEXPA8 (ARABIDOPSIS THALIANA EXPANSIN A8) | -10.27 |
| 140 | 267080_at | AT2G41190 | amino acid transporter family protein | 5.82 |
| 141 | 267635_at | AT2G42220 | rhodanese-like domain-containing protein | -1.80 |
| 142 | 263497_at | AT2G42540 | COR15A (COLD-REGULATED 15A) | 4.89 |
| 143 | 263492_at | AT2G42560 | late embryogenesis abundant domain-containing protein / LEA domain-containing protein | 6.04 |
| 144 | 263986_at | AT2G42790 | CSY3 (CITRATE SYNTHASE 3); citrate (SI)-synthase | 2.96 |
| 145 | 265265_at | AT2G42900 | similar to unnamed protein product [Vitis vinifera] (GB:CAO70018.1); contains InterPro domain Plant Basic Secretory Protein (InterPro:IPR007541) | -2.26 |
| 146 | 260546_at | AT2G43520 | ATTI2 (ARABIDOPSIS THALIANA TRYPSIN INHIBITOR PROTEIN 2); trypsin inhibitor | -2.60 |
| 147 | 266106_at | AT2G45170 | AtATG8e (AUTOPHAGY 8E); microtubule binding | -2.14 |
| 148 | 245138_at | AT2G45190 | AFO (ABNORMAL FLORAL ORGANS); transcription factor | -1.56 |
| 149 | 267559_at | AT2G45570 | CYP76C2 (cytochrome P450, family 76, subfamily C, polypeptide 2); oxygen binding | 5.93 |
| 150 | 266555_at | AT2G46270 | GBF3 (G-BOX BINDING FACTOR 3); transcription factor | 2.37 |
| 151 | 266327_at | AT2G46680 | ATHB-7 (ARABIDOPSIS THALIANA HOMEOBOX 7); transcription factor | 3.91 |
| 152 | 266462_at | AT2G47770 | benzodiazepine receptor-related | 6.53 |
| 153 | 266503_at | AT2G47780 | rubber elongation factor (REF) protein-related | 2.64 |
| 154 | 259104_at | AT3G02170 | LNG2 (LONGIFOLIA2) | -1.70 |
| 155 | 258498_at | AT3G02480 | ABA-responsive protein-related | 10.13 |
| 156 | 258480_at | AT3G02640 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G16250.1); similar to unknown [Populus trichocarpa] (GB:ABK93711.1) | -1.66 |
| 157 | 259058_at | AT3G03470 | CYP89A9 (cytochrome P450, family 87, subfamily A, polypeptide 9); oxygen binding | 3.66 |
| 158 | 259172_at | AT3G03630 | CS26 (cysteine synthase 26); cysteine synthase | -1.76 |
| 159 | 258815_at | AT3G04000 | short-chain dehydrogenase/reductase (SDR) family protein | 2.53 |
| 160 | 258901_at | AT3G05640 | protein phosphatase 2C, putative / PP2C, putative | 3.04 |
| 161 | 258897_at | AT3G05730 | Encodes a defensin-like (DEFL) family protein. | -2.12 |
| 162 | 258751_at | AT3G05890 | RCI2B (RARE-COLD-INDUCIBLE 2B) | 2.34 |
| 163 | 258552_at | AT3G07010 | pectate lyase family protein | -1.53 |
| 164 | 258983_at | AT3G08860 | alanine--glyoxylate aminotransferase, putative / beta-alanine-pyruvate aminotransferase, putative / AGT, putative | 4.29 |
| 165 | 258695_at | AT3G09640 | APX2 (ASCORBATE PEROXIDASE 2); L-ascorbate peroxidase | 7.32 |
| 166 | 258932_at | AT3G10150 | ATPAP16/PAP16 (purple acid phosphatase 16); acid phosphatase/ protein serine/threonine phosphatase | -2.33 |
| 167 | 258966_at | AT3G10690 | DNA gyrase subunit A family protein | -1.53 |
| 168 | 259231_at | AT3G11410 | AHG3/ATPP2CA (ARABIDOPSIS THALIANA PROTEIN PHOSPHATASE 2CA); protein binding / protein serine/threonine phosphatase | 2.15 |
| 169 | 259286_at | AT3G11480 | BSMT1; S-adenosylmethionine-dependent methyltransferase | 3.58 |
| 170 | 256262_at | AT3G12150 | similar to hypothetical protein [Vitis vinifera] (GB:CAN81264.1); contains domain PTHR13617 (PTHR13617); contains domain SSF53474 (SSF53474); contains domain G3DSA:3.40.50.1820 (G3DSA:3.40.50.1820); contains domain PTHR13617:SF1 (PTHR13617:SF1) | -2.63 |
| 171 | 256245_at | AT3G12580 | HSP70 (heat shock protein 70); ATP binding | 3.59 |
| 172 | 256237_at | AT3G12610 | DRT100 (DNA-DAMAGE REPAIR/TOLERATION 100); protein binding | -1.51 |
| 173 | 256772_at | AT3G13750 | BGAL1 (BETA GALACTOSIDASE 1); beta-galactosidase | -1.91 |
| 174 | 258362_at | AT3G14280 | similar to hypothetical protein [Vitis vinifera] (GB:CAN62135.1) | 5.80 |
| 175 | 257280_at | AT3G14440 | NCED3 (NINE-CIS-EPOXYCAROTENOID DIOXYGENASE3) | 3.40 |
| 176 | 258091_at | AT3G14560 | unknown protein | 2.53 |
| 177 | 258063_at | AT3G14620 | CYP72A8 (cytochrome P450, family 72, subfamily A, polypeptide 8); oxygen binding | 2.30 |
| 178 | 256599_at | AT3G14760 | similar to unnamed protein product [Vitis vinifera] (GB:CAO39487.1) | -1.81 |
| 179 | 257056_at | AT3G15350 | glycosyltransferase family 14 protein / core-2/I-branching enzyme family protein | 2.59 |
| 180 | 258402_at | AT3G15450 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G27450.1); similar to unknown [Populus trichocarpa] (GB:ABK93866.1); contains domain PTHR11772 (PTHR11772); contains domain G3DSA:3.60.20.10 (G3DSA:3.60.20.10); contains domain SSF56235 (SSF56235) | -2.59 |
| 181 | 258270_at | AT3G15650 | phospholipase/carboxylesterase family protein | 4.93 |
| 182 | 258250_at | AT3G15850 | FAD5 (FATTY ACID DESATURASE 5); oxidoreductase | -1.61 |
| 183 | 258339_at | AT3G16120 | dynein light chain, putative | -1.77 |
| 184 | 258158_at | AT3G17790 | ATACP5 (acid phosphatase 5); acid phosphatase/ protein serine/threonine phosphatase | 4.11 |
| 185 | 258162_at | AT3G17810 | dihydroorotate dehydrogenase family protein / dihydroorotate oxidase family protein | 2.61 |
| 186 | 257066_at | AT3G18280 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | 2.45 |
| 187 | 256655_at | AT3G18890 | binding / catalytic/ coenzyme binding | -1.84 |
| 188 | 258025_at | AT3G19480 | D-3-phosphoglycerate dehydrogenase, putative / 3-PGDH, putative | -1.63 |
| 189 | 257115_at | AT3G20150 | kinesin motor family protein | -1.67 |
| 190 | 257264_at | AT3G22060 | receptor protein kinase-related | -2.11 |
| 191 | 256765_at | AT3G22200 | POP2 (POLLEN-PISTIL INCOMPATIBILITY 2); 4-aminobutyrate transaminase | 2.29 |
| 192 | 256926_at | AT3G22540 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G14819.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO71190.1); contains InterPro domain Protein of unknown function DUF1677, plant (InterPro:IPR012876) | -1.61 |
| 193 | 258321_at | AT3G22840 | ELIP1 (EARLY LIGHT-INDUCABLE PROTEIN); chlorophyll binding | 3.51 |
| 194 | 256914_at | AT3G23880 | F-box family protein | -2.45 |
| 195 | 257623_at | AT3G26210 | CYP71B23 (cytochrome P450, family 71, subfamily B, polypeptide 23); oxygen binding | 2.20 |
| 196 | 256874_at | AT3G26320 | CYP71B36 (cytochrome P450, family 71, subfamily B, polypeptide 36); oxygen binding | -1.67 |
| 197 | 256980_at | AT3G26932 | DRB3 (DSRNA-BINDING PROTEIN 3); double-stranded RNA binding | -1.50 |
| 198 | 257714_at | AT3G27360 | histone H3 | -1.52 |
| 199 | 258239_at | AT3G27690 | LHCB2:4 (Photosystem II light harvesting complex gene 2.3); chlorophyll binding | -1.86 |
| 200 | 257271_at | AT3G28007 | nodulin MtN3 family protein | 4.04 |
| 201 | 256603_at | AT3G28270 | Identical to UPF0496 protein At3g28270 [Arabidopsis Thaliana] (GB:Q9LHD9;GB:Q8H7D0;GB:Q8H7E9;GB:Q944H3); similar to AT14A [Arabidopsis thaliana] (TAIR:AT3G28290.1); similar to AT14A [Arabidopsis thaliana] (TAIR:AT3G28300.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO22906.1); contains InterPro domain Protein of unknown function DUF677 (InterPro:IPR007749) | 2.15 |
| 202 | 255723_at | AT3G29575 | similar to TMAC2 (TWO OR MORE ABRES-CONTAINING GENE 2) [Arabidopsis thaliana] (TAIR:AT3G02140.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO49169.1); contains InterPro domain Protein of unknown function DUF1675 (InterPro:IPR012463) | 2.96 |
| 203 | 252515_at | AT3G46230 | ATHSP17.4 (Arabidopsis thaliana heat shock protein 17.4) | 6.40 |
| 204 | 252442_at | AT3G46940 | deoxyuridine 5'-triphosphate nucleotidohydrolase family | -1.99 |
| 205 | 252415_at | AT3G47340 | ASN1 (DARK INDUCIBLE 6) | -2.64 |
| 206 | 252400_at | AT3G48020 | similar to F-box family protein-related [Arabidopsis thaliana] (TAIR:AT5G62860.1); similar to unknown [Populus trichocarpa] (GB:ABK95323.1) | 2.35 |
| 207 | 252300_at | AT3G49160 | pyruvate kinase family protein | -4.47 |
| 208 | 252167_at | AT3G50560 | short-chain dehydrogenase/reductase (SDR) family protein | -1.76 |
| 209 | 252178_at | AT3G50750 | brassinosteroid signalling positive regulator-related | -1.57 |
| 210 | 252102_at | AT3G50970 | LTI30 (LOW TEMPERATURE-INDUCED 30) | 6.30 |
| 211 | 252011_at | AT3G52720 | carbonic anhydrase family protein | -2.22 |
| 212 | 251982_at | AT3G53190 | pectate lyase family protein | -1.72 |
| 213 | 251975_at | AT3G53230 | cell division cycle protein 48, putative / CDC48, putative | 2.79 |
| 214 | 251741_at | AT3G56040 | similar to hypothetical protein OsJ_018104 [Oryza sativa (japonica cultivar-group)] (GB:EAZ34621.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN60362.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO41406.1); contains domain Nucleotide-diphospho-sugar transferases (SSF53448); contains domain no description (G3DSA:3.90.550.10) | -1.51 |
| 215 | 251722_at | AT3G56200 | amino acid transporter family protein | 2.39 |
| 216 | 251668_at | AT3G57010 | strictosidine synthase family protein | 6.36 |
| 217 | 251665_at | AT3G57040 | ARR9 (RESPONSE REACTOR 4); transcription regulator | -2.02 |
| 218 | 251642_at | AT3G57520 | ATSIP2 (ARABIDOPSIS THALIANA SEED IMBIBITION 2); hydrolase, hydrolyzing O-glycosyl compounds | 2.27 |
| 219 | 251272_at | AT3G61890 | ATHB-12 (ARABIDOPSIS THALIANA HOMEOBOX PROTEIN 12); transcription factor | 2.45 |
| 220 | 255626_at | AT4G00780 | meprin and TRAF homology domain-containing protein / MATH domain-containing protein | -1.87 |
| 221 | 255621_at | AT4G01390 | meprin and TRAF homology domain-containing protein / MATH domain-containing protein | -5.36 |
| 222 | 255590_at | AT4G01610 | cathepsin B-like cysteine protease, putative | 2.27 |
| 223 | 255521_at | AT4G02280 | SUS3; UDP-glycosyltransferase/ sucrose synthase/ transferase, transferring glycosyl groups | 4.03 |
| 224 | 255527_at | AT4G02360 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G02813.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO40037.1); contains InterPro domain Protein of unknown function DUF538 (InterPro:IPR007493) | 5.42 |
| 225 | 255479_at | AT4G02380 | SAG21 (SENESCENCE-ASSOCIATED GENE 21) | 6.11 |
| 226 | 255460_at | AT4G02800 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G30050.1); similar to Os04g0228100 [Oryza sativa (japonica cultivar-group)] (GB:NP_001052288.1); similar to H0209A05.2 [Oryza sativa (indica cultivar-group)] (GB:CAH66085.1) | -1.63 |
| 227 | 255298_at | AT4G04840 | methionine sulfoxide reductase domain-containing protein / SeIR domain-containing protein | -3.38 |
| 228 | 255259_at | AT4G05020 | NDB2 (NAD(P)H DEHYDROGENASE B2); disulfide oxidoreductase | 2.46 |
| 229 | 255250_at | AT4G05100 | AtMYB74 (myb domain protein 74); DNA binding / transcription factor | 2.50 |
| 230 | 254746_at | AT4G12980 | auxin-responsive protein, putative | -2.53 |
| 231 | 245624_at | AT4G14090 | UDP-glucoronosyl/UDP-glucosyl transferase family protein | 2.67 |
| 232 | 245352_at | AT4G15490 | UGT84A3; UDP-glycosyltransferase/ sinapate 1-glucosyltransferase/ transferase, transferring glycosyl groups | 2.25 |
| 233 | 245448_at | AT4G16860 | RPP4 (RECOGNITION OF PERONOSPORA PARASITICA 4) | -1.80 |
| 234 | 245463_at | AT4G17030 | ATEXLB1 (ARABIDOPSIS THALIANA EXPANSIN-LIKE B1) | 2.79 |
| 235 | 245362_at | AT4G17460 | HAT1 (homeobox-leucine zipper protein 1); DNA binding / transcription factor | -2.40 |
| 236 | 245427_at | AT4G17550 | transporter-related | 2.38 |
| 237 | 254667_at | AT4G18280 | glycine-rich cell wall protein-related | 3.16 |
| 238 | 254564_at | AT4G19170 | NCED4 (NINE-CIS-EPOXYCAROTENOID DIOXYGENASE 4) | -1.59 |
| 239 | 254580_at | AT4G19390 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G13720.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO63752.1); contains InterPro domain Uncharacterised conserved protein UCP022348 (InterPro:IPR016804); contains InterPro domain Protein of unknown function UPF0114 (InterPro:IPR005134) | 3.17 |
| 240 | 254544_at | AT4G19820 | glycosyl hydrolase family 18 protein | -2.39 |
| 241 | 254545_at | AT4G19830 | immunophilin / FKBP-type peptidyl-prolyl cis-trans isomerase family protein | -2.52 |
| 242 | 254276_at | AT4G22820 | zinc finger (AN1-like) family protein | 2.62 |
| 243 | 254247_at | AT4G23260 | protein kinase | -1.96 |
| 244 | 254250_at | AT4G23290 | protein kinase family protein | -2.42 |
| 245 | 254263_at | AT4G23493 | unknown protein | 2.31 |
| 246 | 254232_at | AT4G23600 | CORI3 (CORONATINE INDUCED 1, JASMONIC ACID RESPONSIVE 2); transaminase | 5.50 |
| 247 | 254119_at | AT4G24780 | pectate lyase family protein | -2.08 |
| 248 | 253994_at | AT4G26080 | ABI1 (ABA INSENSITIVE 1); calcium ion binding / protein serine/threonine phosphatase | 2.40 |
| 249 | 253971_at | AT4G26530 | fructose-bisphosphate aldolase, putative | -4.33 |
| 250 | 253973_at | AT4G26555 | immunophilin / FKBP-type peptidyl-prolyl cis-trans isomerase family protein | -1.70 |
| 251 | 253951_at | AT4G26860 | alanine racemase family protein | -1.52 |
| 252 | 253872_at | AT4G27410 | RD26 (RESPONSIVE TO DESSICATION 26); transcription factor | 5.53 |
| 253 | 253874_at | AT4G27450 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G15450.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO39242.1); contains domain G3DSA:3.60.20.10 (G3DSA:3.60.20.10); contains domain SSF56235 (SSF56235) | -2.82 |
| 254 | 253619_at | AT4G30460 | glycine-rich protein | 3.17 |
| 255 | 253638_at | AT4G30470 | cinnamoyl-CoA reductase-related | 2.31 |
| 256 | 253579_at | AT4G30610 | BRS1 (BRI1 SUPPRESSOR 1) | -1.86 |
| 257 | 253627_at | AT4G30650 | hydrophobic protein, putative / low temperature and salt responsive protein, putative | 2.42 |
| 258 | 253421_at | AT4G32340 | binding | -2.42 |
| 259 | 253440_at | AT4G32570 | TIFY8 | -2.12 |
| 260 | 253373_at | AT4G33150 | LKR (SACCHAROPINE DEHYDROGENASE) | 4.48 |
| 261 | 253343_at | AT4G33540 | metallo-beta-lactamase family protein | 2.41 |
| 262 | 253344_at | AT4G33550 | lipid binding | 4.13 |
| 263 | 253305_at | AT4G33666 | unknown protein | -2.53 |
| 264 | 253293_at | AT4G33905 | peroxisomal membrane protein 22 kDa, putative | 3.27 |
| 265 | 253278_at | AT4G34220 | leucine-rich repeat transmembrane protein kinase, putative | -1.78 |
| 266 | 253277_at | AT4G34230 | CAD5 (CINNAMYL ALCOHOL DEHYDROGENASE 5); cinnamyl-alcohol dehydrogenase | 2.78 |
| 267 | 253203_at | AT4G34710 | ADC2 (ARGININE DECARBOXYLASE 2) | 2.60 |
| 268 | 253161_at | AT4G35770 | SEN1 (DARK INDUCIBLE 1) | -3.22 |
| 269 | 246275_at | AT4G36540 | BEE2 (BR ENHANCED EXPRESSION 2); DNA binding / transcription factor | -2.60 |
| 270 | 246219_at | AT4G36760 | ATAPP1 (aminopeptidase P1) | 2.22 |
| 271 | 253061_at | AT4G37610 | BT5 (BTB and TAZ domain protein 5); protein binding / transcription regulator | -4.44 |
| 272 | 252989_at | AT4G38420 | SKS9 (SKU5 Similar 9); copper ion binding / oxidoreductase | -1.70 |
| 273 | 252970_at | AT4G38850 | SAUR_AC1 (SMALL AUXIN UP RNA 1 FROM ARABIDOPSIS THALIANA ECOTYPE COLUMBIA) | -3.60 |
| 274 | 252965_at | AT4G38860 | auxin-responsive protein, putative | -2.58 |
| 275 | 252888_at | AT4G39210 | APL3 (large subunit of AGP 3) | 3.55 |
| 276 | 252880_at | AT4G39730 | lipid-associated family protein | 2.36 |
| 277 | 252858_at | AT4G39770 | trehalose-6-phosphate phosphatase, putative | -2.03 |
| 278 | 252876_at | AT4G39970 | haloacid dehalogenase-like hydrolase family protein | -1.58 |
| 279 | 251142_at | AT5G01015 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G65295.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN82885.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO45347.1) | -1.95 |
| 280 | 251133_at | AT5G01240 | amino acid permease, putative | -1.49 |
| 281 | 251137_at | AT5G01300 | phosphatidylethanolamine-binding family protein | 3.95 |
| 282 | 251084_at | AT5G01520 | zinc finger (C3HC4-type RING finger) family protein | 3.06 |
| 283 | 251107_at | AT5G01610 | Identical to Uncharacterized protein At5g01610 [Arabidopsis Thaliana] (GB:Q9M015;GB:Q0WT00;GB:Q84VV4); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G08890.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G08890.2); similar to hypothetical protein [Vitis vinifera] (GB:CAN63135.1); contains InterPro domain Protein of unknown function DUF538 (InterPro:IPR007493) | -1.51 |
| 284 | 251104_at | AT5G01720 | F-box family protein (FBL3) | 2.54 |
| 285 | 251036_at | AT5G02160 | unknown protein | -2.40 |
| 286 | 251017_at | AT5G02760 | protein phosphatase 2C family protein / PP2C family protein | -2.92 |
| 287 | 250942_at | AT5G03350 | legume lectin family protein | -2.60 |
| 288 | 245711_at | AT5G04340 | C2H2 (ZINC FINGER OF ARABIDOPSIS THALIANA 6); nucleic acid binding / transcription factor/ zinc ion binding | 2.29 |
| 289 | 250891_at | AT5G04530 | beta-ketoacyl-CoA synthase family protein | 2.20 |
| 290 | 250753_at | AT5G05860 | UGT76C2 (UDP-glucosyl transferase 76C2); UDP-glycosyltransferase/ transferase, transferring glycosyl groups | -2.73 |
| 291 | 250624_at | AT5G07330 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G63060.1); similar to unknown [Xerophyta humilis] (GB:AAT45004.1) | 4.12 |
| 292 | 250502_at | AT5G09590 | mtHSC70-2 (HEAT SHOCK PROTEIN 70); ATP binding / unfolded protein binding | 2.37 |
| 293 | 245904_at | AT5G11110 | ATSPS2F/SPS1 (SUCROSE PHOSPHATE SYNTHASE 1); sucrose-phosphate synthase | 2.95 |
| 294 | 250327_at | AT5G12050 | similar to unnamed protein product [Vitis vinifera] (GB:CAO45643.1) | -2.96 |
| 295 | 245984_at | AT5G13090 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G24270.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO68319.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN73358.1) | -1.64 |
| 296 | 245982_at | AT5G13170 | nodulin MtN3 family protein | 3.48 |
| 297 | 250279_at | AT5G13200 | GRAM domain-containing protein / ABA-responsive protein-related | 3.25 |
| 298 | 250228_at | AT5G13840 | WD-40 repeat family protein | -1.54 |
| 299 | 246595_at | AT5G14780 | FDH (FORMATE DEHYDROGENASE); NAD binding / binding / catalytic/ cofactor binding / oxidoreductase, acting on the CH-OH group of donors, NAD or NADP as acceptor | 2.28 |
| 300 | 246522_at | AT5G15830 | ATBZIP3 (ARABIDOPSIS THALIANA BASIC LEUCINE-ZIPPER 3); DNA binding / transcription factor | -1.92 |
| 301 | 246487_at | AT5G16030 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G02500.1); similar to hypothetical protein [Cleome spinosa] (GB:ABD96956.1) | -1.91 |
| 302 | 250083_at | AT5G17220 | ATGSTF12 (GLUTATHIONE S-TRANSFERASE 26); glutathione transferase | 2.44 |
| 303 | 250062_at | AT5G17760 | AAA-type ATPase family protein | 2.52 |
| 304 | 250012_x_at | AT5G18060 | auxin-responsive protein, putative | -2.43 |
| 305 | 250028_at | AT5G18130 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G03870.2); similar to unknown [Medicago truncatula] (GB:ABK28852.1) | 2.34 |
| 306 | 250007_at | AT5G18670 | BMY3 (BETA-AMYLASE 9); beta-amylase | -1.65 |
| 307 | 249923_at | AT5G19120 | aspartic-type endopeptidase/ pepsin A | -2.92 |
| 308 | 246114_at | AT5G20250 | DIN10 (DARK INDUCIBLE 10); hydrolase, hydrolyzing O-glycosyl compounds | -4.61 |
| 309 | 245998_at | AT5G20830 | SUS1 (SUCROSE SYNTHASE 1); UDP-glycosyltransferase/ sucrose synthase | 2.94 |
| 310 | 249932_at | AT5G22390 | similar to unnamed protein product [Vitis vinifera] (GB:CAO48728.1) | -1.54 |
| 311 | 249917_at | AT5G22460 | esterase/lipase/thioesterase family protein | 4.92 |
| 312 | 249894_at | AT5G22580 | Identical to Uncharacterized protein At5g22580 [Arabidopsis Thaliana] (GB:Q9FK81); similar to stable protein 1-related [Arabidopsis thaliana] (TAIR:AT3G17210.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN68720.1); contains InterPro domain Dimeric alpha-beta barrel (InterPro:IPR011008); contains InterPro domain Stress responsive alpha-beta barrel (InterPro:IPR013097) | -1.50 |
| 313 | 249862_at | AT5G22920 | zinc finger (C3HC4-type RING finger) family protein | -3.78 |
| 314 | 249765_at | AT5G24030 | SLAH3 (SLAC1 HOMOLOGUE 3); transporter | 2.42 |
| 315 | 249732_at | AT5G24420 | glucosamine/galactosamine-6-phosphate isomerase-related | -3.20 |
| 316 | 246949_at | AT5G25140 | CYP71B13 (cytochrome P450, family 71, subfamily B, polypeptide 13); oxygen binding | -1.49 |
| 317 | 246908_at | AT5G25610 | RD22 (RESPONSIVE TO DESSICATION 22) | 2.16 |
| 318 | 246792_at | AT5G27290 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G54680.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G54680.2); similar to unnamed protein product [Vitis vinifera] (GB:CAO68604.1) | -1.56 |
| 319 | 246779_at | AT5G27520 | mitochondrial substrate carrier family protein | 2.34 |
| 320 | 245925_at | AT5G28770 | BZO2H3 (ARABIDOPSIS THALIANA BASIC LEUCINE ZIPPER 63); DNA binding / protein heterodimerization/ transcription factor | -2.72 |
| 321 | 249645_at | AT5G36910 | THI2.2 (THIONIN 2.2); toxin receptor binding | -4.71 |
| 322 | 249626_at | AT5G37540 | aspartyl protease family protein | 2.53 |
| 323 | 249383_at | AT5G39860 | PRE1 (PACLOBUTRAZOL RESISTANCE1); DNA binding / transcription factor | -3.45 |
| 324 | 249378_at | AT5G40450 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G28770.1); similar to hypothetical protein Kpol_1058p4 [Vanderwaltozyma polyspora DSM 70294] (GB:XP_001645325.1) | -2.10 |
| 325 | 249372_at | AT5G40760 | G6PD6 (GLUCOSE-6-PHOSPHATE DEHYDROGENASE 6); glucose-6-phosphate dehydrogenase | 2.25 |
| 326 | 249237_at | AT5G42050 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G27090.1); similar to unknown [Populus trichocarpa] (GB:ABK95892.1); contains InterPro domain Kelch related (InterPro:IPR013089); contains InterPro domain Development and cell death (InterPro:IPR013989) | 2.37 |
| 327 | 249215_at | AT5G42800 | DFR (DIHYDROFLAVONOL 4-REDUCTASE); dihydrokaempferol 4-reductase | 2.50 |
| 328 | 249126_at | AT5G43380 | TOPP6 (Type one serine/threonine protein phosphatase 6); protein serine/threonine phosphatase | 3.56 |
| 329 | 249117_at | AT5G43840 | AT-HSFA6A (Arabidopsis thaliana heat shock transcription factor A6A); DNA binding / transcription factor | 7.39 |
| 330 | 249073_at | AT5G44020 | acid phosphatase class B family protein | -1.78 |
| 331 | 249008_at | AT5G44680 | methyladenine glycosylase family protein | -1.87 |
| 332 | 248975_at | AT5G45040 | cytochrome c6 (ATC6) | -1.49 |
| 333 | 248964_at | AT5G45340 | CYP707A3 (cytochrome P450, family 707, subfamily A, polypeptide 3); oxygen binding | 2.12 |
| 334 | 248959_at | AT5G45630 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G18980.1); similar to unknown [Cucumis sativus] (GB:ABY56081.1); similar to unknown [Cucumis melo] (GB:ABQ53634.1); contains InterPro domain Protein of unknown function DUF584 (InterPro:IPR007608) | 3.25 |
| 335 | 248962_at | AT5G45680 | FK506-binding protein 1 (FKBP13) | -1.51 |
| 336 | 248910_at | AT5G45820 | CIPK20 (CBL-INTERACTING PROTEIN KINASE 20); kinase | -2.06 |
| 337 | 248807_at | AT5G47500 | pectinesterase family protein | -2.46 |
| 338 | 248763_at | AT5G47550 | cysteine protease inhibitor, putative / cystatin, putative | 2.15 |
| 339 | 248696_at | AT5G48360 | formin homology 2 domain-containing protein / FH2 domain-containing protein | -1.53 |
| 340 | 248684_at | AT5G48485 | DIR1 (DEFECTIVE IN INDUCED RESISTANCE 1); lipid binding | -1.64 |
| 341 | 248683_at | AT5G48490 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | -2.68 |
| 342 | 248657_at | AT5G48570 | peptidyl-prolyl cis-trans isomerase, putative / FK506-binding protein, putative | 2.36 |
| 343 | 248622_at | AT5G49360 | BXL1 (BETA-XYLOSIDASE 1); hydrolase, hydrolyzing O-glycosyl compounds | -2.42 |
| 344 | 248505_at | AT5G50360 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G63350.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO68256.1) | 6.08 |
| 345 | 248352_at | AT5G52300 | LTI65/RD29B (RESPONSIVE TO DESSICATION 29B) | 7.93 |
| 346 | 248337_at | AT5G52310 | COR78 (COLD REGULATED 78) | 5.52 |
| 347 | 248335_at | AT5G52450 | MATE efflux protein-related | 2.27 |
| 348 | 248332_at | AT5G52640 | HSP81-1 (HEAT SHOCK PROTEIN 81-1); ATP binding / unfolded protein binding | 2.24 |
| 349 | 248248_at | AT5G53120 | SPDS3 (SPERMIDINE SYNTHASE 3) | 2.88 |
| 350 | 248246_at | AT5G53200 | TRY (TRIPTYCHON); DNA binding / transcription factor | -1.96 |
| 351 | 248236_at | AT5G53870 | plastocyanin-like domain-containing protein | 4.65 |
| 352 | 248190_at | AT5G54130 | calcium-binding EF hand family protein | -1.94 |
| 353 | 248191_at | AT5G54130 | calcium-binding EF hand family protein | -2.01 |
| 354 | 248028_at | AT5G55620 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G09950.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO39940.1) | -2.05 |
| 355 | 247954_at | AT5G56870 | BGAL4 (beta-galactosidase 4); beta-galactosidase | -2.49 |
| 356 | 247957_at | AT5G57050 | ABI2 (ABA INSENSITIVE 2); protein serine/threonine phosphatase | 2.29 |
| 357 | 247903_at | AT5G57340 | similar to hypothetical protein MtrDRAFT_AC155282g59v2 [Medicago truncatula] (GB:ABN08100.1) | -1.58 |
| 358 | 247921_at | AT5G57660 | zinc finger (B-box type) family protein | -1.58 |
| 359 | 247880_at | AT5G57780 | transcription regulator | -3.20 |
| 360 | 247754_at | AT5G59080 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G02020.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO15731.1) | -3.33 |
| 361 | 247723_at | AT5G59220 | protein phosphatase 2C, putative / PP2C, putative | 4.27 |
| 362 | 247718_at | AT5G59310 | LTP4 (LIPID TRANSFER PROTEIN 4); lipid binding | 9.74 |
| 363 | 247717_at | AT5G59320 | LTP3 (LIPID TRANSFER PROTEIN 3); lipid binding | 8.71 |
| 364 | 247726_at | AT5G59430 | ATTRP1 (TELOMERE REPEAT BINDING PROTEIN 1); DNA binding | -1.50 |
| 365 | 247655_at | AT5G59820 | RHL41 (RESPONSIVE TO HIGH LIGHT 41); nucleic acid binding / transcription factor/ zinc ion binding | 2.79 |
| 366 | 247488_at | AT5G61820 | similar to MtN19-like protein [Pisum sativum] (GB:AAU14999.2); contains InterPro domain Stress up-regulated Nod 19 (InterPro:IPR011692) | 2.37 |
| 367 | 247474_at | AT5G62280 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G45360.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO49726.1); contains InterPro domain Protein of unknown function DUF1442 (InterPro:IPR009902) | -5.77 |
| 368 | 247447_at | AT5G62730 | proton-dependent oligopeptide transport (POT) family protein | -1.85 |
| 369 | 247377_at | AT5G63180 | pectate lyase family protein | -3.08 |
| 370 | 247347_at | AT5G63780 | SHA1 (SHOOT APICAL MERISTEM ARREST 1); protein binding / zinc ion binding | -2.02 |
| 371 | 247304_at | AT5G63850 | AAP4 (amino acid permease 4); amino acid transmembrane transporter | -1.54 |
| 372 | 247252_at | AT5G64770 | similar to 80C09_10 [Brassica rapa subsp. pekinensis] (GB:AAZ41821.1) | -1.67 |
| 373 | 247120_at | AT5G65990 | amino acid transporter family protein | 2.49 |
| 374 | 247095_at | AT5G66400 | RAB18 (RESPONSIVE TO ABA 18) | 5.59 |
| 375 | 247074_at | AT5G66590 | allergen V5/Tpx-1-related family protein | -2.29 |
| 376 | 247055_at | AT5G66740 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G75160.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN74294.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO22871.1); contains InterPro domain Protein of unknown function DUF620 (InterPro:IPR006873) | -2.39 |
| 377 | 247060_at | AT5G66760 | SDH1-1 (Succinate dehydrogenase 1-1) | 2.25 |
| 378 | 246987_at | AT5G67300 | ATMYB44/ATMYBR1/MYBR1 (MYB DOMAIN PROTEIN 44); DNA binding / transcription factor | 3.01 |
| 379 | 246997_at | AT5G67390 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G69360.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO71983.1) | -2.53 |
| 380 | 247013_at | AT5G67480 | BT4 (BTB AND TAZ DOMAIN PROTEIN 4); protein binding / transcription regulator | 3.49 |