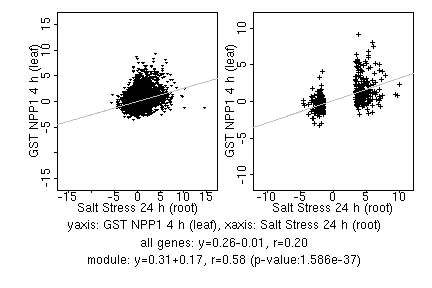
marker gene list
| Salt Stress 24 h (root) |
| GST NPP1 4 h (leaf) |
| probeID | AGICode | Annotation | gene expression |
|
|---|---|---|---|---|
| 1 | 261580_at | AT1G01110 | IQD18 (IQ-domain 18) | -1.79 |
| 2 | 259439_at | AT1G01480 | ACS2 (1-Amino-cyclopropane-1-carboxylate synthase 2) | 7.13 |
| 3 | 260910_at | AT1G02690 | importin alpha-2 subunit, putative | -1.42 |
| 4 | 264824_at | AT1G03420 | transposable element gene | -1.35 |
| 5 | 265085_at | AT1G03780 | targeting protein-related | -1.58 |
| 6 | 265083_at | AT1G03820 | similar to E6-4 [Gossypium hirsutum] (GB:AAY43793.1); similar to E6-3 [Gossypium hirsutum] (GB:AAY43792.1); similar to E6 (GB:AAB03081.1) | -1.90 |
| 7 | 264611_at | AT1G04680 | pectate lyase family protein | -1.33 |
| 8 | 265192_at | AT1G05060 | similar to hypothetical protein [Vitis vinifera] (GB:CAN75913.1) | 3.51 |
| 9 | 265216_at | AT1G05100 | MAPKKK18 (Mitogen-activated protein kinase kinase kinase 18); kinase | 5.37 |
| 10 | 264580_at | AT1G05340 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G32210.1) | 3.54 |
| 11 | 263231_at | AT1G05680 | UDP-glucoronosyl/UDP-glucosyl transferase family protein | 6.53 |
| 12 | 256062_at | AT1G07090 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G58500.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN60458.1); contains InterPro domain Protein of unknown function DUF640 (InterPro:IPR006936) | -1.87 |
| 13 | 261077_at | AT1G07430 | protein phosphatase 2C, putative / PP2C, putative | 7.04 |
| 14 | 261090_at | AT1G07560 | leucine-rich repeat protein kinase, putative | -1.37 |
| 15 | 261460_at | AT1G07880 | ATMPK13 (ARABIDOPSIS THALIANA MAP KINASE 13); MAP kinase/ kinase | -1.71 |
| 16 | 264647_at | AT1G09090 | ATRBOHB (RESPIRATORY BURST OXIDASE HOMOLOG B); FAD binding / calcium ion binding / iron ion binding / oxidoreductase | -1.62 |
| 17 | 264262_at | AT1G09200 | histone H3 | -1.66 |
| 18 | 264659_at | AT1G09930 | ATOPT2 (oligopeptide transporter 2); oligopeptide transporter | 7.35 |
| 19 | 264457_at | AT1G10400 | UDP-glycosyltransferase/ transferase, transferring glycosyl groups | -1.71 |
| 20 | 260475_at | AT1G11080 | SCPL31 (serine carboxypeptidase-like 31); serine carboxypeptidase | -2.66 |
| 21 | 255937_at | AT1G12610 | DDF1 (DWARF AND DELAYED FLOWERING 1); DNA binding / transcription factor | 6.10 |
| 22 | 259360_at | AT1G13310 | similar to unnamed protein product [Vitis vinifera] (GB:CAO42102.1); contains InterPro domain BRO1 (InterPro:IPR004328) | 6.49 |
| 23 | 262658_at | AT1G14220 | ribonuclease T2 family protein | -2.72 |
| 24 | 261474_at | AT1G14540 | anionic peroxidase, putative | 6.52 |
| 25 | 261763_at | AT1G15520 | ATPDR12/PDR12 (PLEIOTROPIC DRUG RESISTANCE 12); ATPase, coupled to transmembrane movement of substances | 4.27 |
| 26 | 257480_at | AT1G15640 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G15620.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN70531.1) | -1.33 |
| 27 | 261033_at | AT1G17380 | JAZ5/TIFY11A (JASMONATE-ZIM-DOMAIN PROTEIN 5) | 4.41 |
| 28 | 259464_at | AT1G18990 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G74830.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO62468.1); contains InterPro domain Protein of unknown function DUF593 (InterPro:IPR007656) | 4.14 |
| 29 | 256012_at | AT1G19250 | FMO1 (FLAVIN-DEPENDENT MONOOXYGENASE 1); monooxygenase | 4.67 |
| 30 | 261221_at | AT1G19960 | similar to transmembrane receptor [Arabidopsis thaliana] (TAIR:AT2G32140.1) | -1.87 |
| 31 | 261224_at | AT1G20160 | ATSBT5.2; subtilase | -2.00 |
| 32 | 261456_at | AT1G21050 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G76610.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN67637.1); contains InterPro domain Protein of unknown function DUF617, plant (InterPro:IPR006460) | -1.61 |
| 33 | 261454_at | AT1G21090 | hydroxyproline-rich glycoprotein family protein | -1.45 |
| 34 | 260902_at | AT1G21440 | mutase family protein | -2.63 |
| 35 | 263016_at | AT1G23410 | ubiquitin extension protein, putative / 40S ribosomal protein S27A (RPS27aA) | -1.45 |
| 36 | 265184_at | AT1G23710 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G70420.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO66069.1); contains InterPro domain Protein of unknown function DUF1645 (InterPro:IPR012442) | 4.67 |
| 37 | 265024_at | AT1G24600 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G67920.1) | 4.56 |
| 38 | 255733_at | AT1G25400 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G68440.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO42150.1) | 4.01 |
| 39 | 255727_at | AT1G25510 | aspartyl protease family protein | -2.75 |
| 40 | 261021_at | AT1G26380 | FAD-binding domain-containing protein | 3.94 |
| 41 | 261648_at | AT1G27730 | STZ (SALT TOLERANCE ZINC FINGER); nucleic acid binding / transcription factor/ zinc ion binding | 4.22 |
| 42 | 259579_at | AT1G28010 | PGP14 (P-GLYCOPROTEIN 14); ATPase, coupled to transmembrane movement of substances | -1.91 |
| 43 | 259599_at | AT1G28110 | SCPL45; serine carboxypeptidase | -1.57 |
| 44 | 261443_at | AT1G28480 | GRX480; thiol-disulfide exchange intermediate | 4.39 |
| 45 | 260028_at | AT1G29980 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G34510.1); similar to unknown [Populus trichocarpa] (GB:ABK95079.1); contains InterPro domain Protein of unknown function DUF642 (InterPro:IPR006946); contains InterPro domain Galactose-binding like (InterPro:IPR008979) | -1.49 |
| 46 | 256306_at | AT1G30370 | lipase class 3 family protein | 4.37 |
| 47 | 264497_at | AT1G30840 | ATPUP4 (Arabidopsis thaliana purine permease 4); purine transmembrane transporter | -2.86 |
| 48 | 265101_at | AT1G30880 | unknown protein | -1.41 |
| 49 | 265160_at | AT1G31050 | transcription factor | -2.28 |
| 50 | 264415_at | AT1G43160 | RAP2.6 (related to AP2 6); DNA binding / transcription factor | 4.08 |
| 51 | 262434_at | AT1G47670 | amino acid transporter family protein | -1.79 |
| 52 | 262448_at | AT1G49450 | transducin family protein / WD-40 repeat family protein | 4.57 |
| 53 | 262396_at | AT1G49470 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G55230.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN63047.1); contains InterPro domain Protein of unknown function DUF716 (InterPro:IPR006904) | -3.13 |
| 54 | 261636_at | AT1G50110 | branched-chain amino acid aminotransferase 6 / branched-chain amino acid transaminase 6 (BCAT6) | -1.46 |
| 55 | 246375_at | AT1G51830 | ATP binding / kinase/ protein serine/threonine kinase | -2.05 |
| 56 | 246374_at | AT1G51840 | protein kinase-related | -1.74 |
| 57 | 265057_at | AT1G52140 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G16330.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN64915.1) | -1.52 |
| 58 | 262148_at | AT1G52560 | 26.5 kDa class I small heat shock protein-like (HSP26.5-P) | 4.93 |
| 59 | 260203_at | AT1G52890 | ANAC019 (Arabidopsis NAC domain containing protein 19); transcription factor | 5.48 |
| 60 | 264188_at | AT1G54690 | G-H2AX/GAMMA-H2AX/H2AXB/HTA3; DNA binding | -1.46 |
| 61 | 262085_at | AT1G56060 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G32190.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO68639.1); contains domain PD188784 (PD188784) | 5.11 |
| 62 | 256222_at | AT1G56210 | copper chaperone (CCH)-related | -1.88 |
| 63 | 245627_at | AT1G56600 | ATGOLS2 (ARABIDOPSIS THALIANA GALACTINOL SYNTHASE 2); transferase, transferring glycosyl groups / transferase, transferring hexosyl groups | 5.29 |
| 64 | 245674_at | AT1G56680 | glycoside hydrolase family 19 protein | -2.59 |
| 65 | 246405_at | AT1G57630 | disease resistance protein (TIR class), putative | 3.66 |
| 66 | 246406_at | AT1G57650 | disease resistance protein (NBS-LRR class), putative | 5.22 |
| 67 | 262072_at | AT1G59590 | ZCF37 | 3.63 |
| 68 | 264217_at | AT1G60190 | armadillo/beta-catenin repeat family protein / U-box domain-containing protein | 3.85 |
| 69 | 264941_at | AT1G60680 | AGD2 (ARF-GAP DOMAIN 2); aldo-keto reductase | -2.15 |
| 70 | 264760_at | AT1G61290 | SYP124 (syntaxin 124); SNAP receptor | 6.75 |
| 71 | 264758_at | AT1G61340 | F-box family protein | 4.19 |
| 72 | 264763_at | AT1G61450 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G61415.1); similar to Os05g0275600 [Oryza sativa (japonica cultivar-group)] (GB:NP_001055074.1) | -1.71 |
| 73 | 265063_at | AT1G61500 | S-locus protein kinase, putative | -2.08 |
| 74 | 265030_at | AT1G61610 | S-locus lectin protein kinase family protein | 4.85 |
| 75 | 264734_at | AT1G62280 | SLAH1 (SLAC1 HOMOLOGUE 1); transporter | -2.89 |
| 76 | 265117_at | AT1G62500 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | -2.35 |
| 77 | 262863_at | AT1G64910 | glycosyltransferase family protein | -1.79 |
| 78 | 264166_at | AT1G65370 | meprin and TRAF homology domain-containing protein / MATH domain-containing protein | -1.44 |
| 79 | 256526_at | AT1G66090 | disease resistance protein (TIR-NBS class), putative | 4.24 |
| 80 | 256528_at | AT1G66140 | ZFP4 (ZINC FINGER PROTEIN 4); nucleic acid binding / transcription factor/ zinc ion binding | -1.33 |
| 81 | 256525_at | AT1G66180 | aspartyl protease family protein | -1.34 |
| 82 | 245196_at | AT1G67750 | pectate lyase family protein | -1.99 |
| 83 | 260442_at | AT1G68220 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G13380.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN76984.1); contains InterPro domain Protein of unknown function DUF1218 (InterPro:IPR009606) | -1.39 |
| 84 | 260040_at | AT1G68765 | IDA (INFLORESCENCE DEFICIENT IN ABSCISSION) | 5.35 |
| 85 | 260181_at | AT1G70710 | AtGH9B1 (ARABIDOPSIS THALIANA GLYCOSYL HYDROLASE 9B1); hydrolase, hydrolyzing O-glycosyl compounds | -1.60 |
| 86 | 259947_at | AT1G71530 | protein kinase family protein | 5.13 |
| 87 | 259851_at | AT1G72250 | kinesin motor protein-related | -1.42 |
| 88 | 260451_at | AT1G72360 | ethylene-responsive element-binding protein, putative | -1.98 |
| 89 | 259912_at | AT1G72670 | IQD8 (IQ-domain 8); calmodulin binding | -4.37 |
| 90 | 245736_at | AT1G73330 | ATDR4 (Arabidopsis thaliana drought-repressed 4) | -2.06 |
| 91 | 260046_at | AT1G73805 | calmodulin binding | 4.08 |
| 92 | 260068_at | AT1G73805 | calmodulin binding | 3.69 |
| 93 | 260390_at | AT1G73940 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G49410.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G49410.2); similar to unnamed protein product [Vitis vinifera] (GB:CAO62865.1) | -1.38 |
| 94 | 262218_at | AT1G74770 | protein binding / zinc ion binding | -2.29 |
| 95 | 262213_at | AT1G74870 | protein binding / zinc ion binding | 9.60 |
| 96 | 262211_at | AT1G74930 | ORA47; DNA binding / transcription factor | 3.56 |
| 97 | 262978_at | AT1G75780 | TUB1 (tubulin beta-1 chain); structural molecule | -1.34 |
| 98 | 261777_at | AT1G76210 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G20520.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN69930.1); contains InterPro domain Protein of unknown function DUF241, plant (InterPro:IPR004320) | 5.45 |
| 99 | 259979_at | AT1G76600 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G21010.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN67638.1) | 5.31 |
| 100 | 259680_at | AT1G77690 | amino acid permease, putative | -1.38 |
| 101 | 260805_at | AT1G78320 | ATGSTU23 (Arabidopsis thaliana Glutathione S-transferase (class tau) 23); glutathione transferase | -2.23 |
| 102 | 262060_at | AT1G80170 | polygalacturonase, putative / pectinase, putative | -1.47 |
| 103 | 261893_at | AT1G80690 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G25170.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO45044.1); contains InterPro domain Protein of unknown function DUF862, eukaryotic (InterPro:IPR008580) | -1.51 |
| 104 | 261892_at | AT1G80840 | WRKY40 (WRKY DNA-binding protein 40); transcription factor | 3.65 |
| 105 | 265864_at | AT2G01750 | ATMAP70-3 (microtubule-associated proteins 70-3); microtubule binding | -1.44 |
| 106 | 263596_at | AT2G01900 | endonuclease/exonuclease/phosphatase family protein | -1.64 |
| 107 | 265250_at | AT2G01950 | BRL2 (BRI1-LIKE 2); ATP binding / protein serine/threonine kinase | -1.86 |
| 108 | 264042_at | AT2G03760 | ST (steroid sulfotransferase); sulfotransferase | 5.53 |
| 109 | 263402_at | AT2G04050 | MATE efflux family protein | 7.44 |
| 110 | 263401_at | AT2G04070 | transporter | 5.70 |
| 111 | 266613_at | AT2G14900 | gibberellin-regulated family protein | -2.19 |
| 112 | 265482_at | AT2G15780 | glycine-rich protein | 5.72 |
| 113 | 264790_at | AT2G17820 | ATHK1 (HISTIDINE KINASE 1) | -1.37 |
| 114 | 257456_at | AT2G18120 | SRS4 (SHI-RELATED SEQUENCE 4) | -2.11 |
| 115 | 263062_at | AT2G18180 | SEC14 cytosolic factor, putative / phosphoglyceride transfer protein, putative | 4.79 |
| 116 | 263061_at | AT2G18190 | AAA-type ATPase family protein | 5.30 |
| 117 | 265327_at | AT2G18210 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G36500.1); similar to hypothetical protein [Thellungiella halophila] (GB:ABB45855.1) | 4.23 |
| 118 | 266687_at | AT2G19670 | protein arginine N-methyltransferase, putative | -1.62 |
| 119 | 265588_at | AT2G19970 | pathogenesis-related protein, putative | -4.33 |
| 120 | 265586_at | AT2G19990 | PR-1-LIKE (PATHOGENESIS-RELATED PROTEIN-1-LIKE) | -4.17 |
| 121 | 265589_at | AT2G20170 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G23390.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO22094.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN72570.1); contains InterPro domain Protein of unknown function DUF239, plant (InterPro:IPR004314) | 4.90 |
| 122 | 263369_at | AT2G20480 | similar to unknown [Populus trichocarpa] (GB:ABK93010.1); similar to hypothetical protein OsI_030526 [Oryza sativa (indica cultivar-group)] (GB:EAZ09294.1); similar to Os09g0446000 [Oryza sativa (japonica cultivar-group)] (GB:NP_001063306.1) | -1.92 |
| 123 | 265428_at | AT2G20720 | pentatricopeptide (PPR) repeat-containing protein | 4.71 |
| 124 | 265422_at | AT2G20800 | NDB4 (NAD(P)H DEHYDROGENASE B4); NADH dehydrogenase | 7.36 |
| 125 | 264025_at | AT2G21050 | amino acid permease, putative | -2.24 |
| 126 | 263515_at | AT2G21640 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G05570.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G39235.1); similar to hypothetical protein OsI_027197 [Oryza sativa (indica cultivar-group)] (GB:EAZ05965.1) | 4.53 |
| 127 | 263431_at | AT2G22170 | lipid-associated family protein | -1.41 |
| 128 | 266799_at | AT2G22860 | ATPSK2 (PHYTOSULFOKINE 2 PRECURSOR); growth factor | 3.80 |
| 129 | 245082_at | AT2G23270 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G37290.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN62855.1) | 5.53 |
| 130 | 263539_at | AT2G24850 | TAT3 (TYROSINE AMINOTRANSFERASE 3); transaminase | 5.80 |
| 131 | 266841_at | AT2G26150 | ATHSFA2 (Arabidopsis thaliana heat shock transcription factor A2); DNA binding / transcription factor | 4.68 |
| 132 | 245033_at | AT2G26380 | disease resistance protein-related / LRR protein-related | 4.70 |
| 133 | 245052_at | AT2G26440 | pectinesterase family protein | -1.82 |
| 134 | 245041_at | AT2G26530 | AR781 | 4.13 |
| 135 | 245038_at | AT2G26560 | PLP2 (PHOSPHOLIPASE A 2A); nutrient reservoir | 4.45 |
| 136 | 267614_at | AT2G26710 | BAS1/CYP734A1 (PHYB ACTIVATION TAGGED SUPPRESSOR 1); oxygen binding / steroid hydroxylase | -1.55 |
| 137 | 264078_at | AT2G28470 | BGAL8 (BETA-GALACTOSIDASE 8); beta-galactosidase | -1.37 |
| 138 | 266222_at | AT2G28780 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G09450.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO21693.1) | -2.21 |
| 139 | 266223_at | AT2G28790 | osmotin-like protein, putative | -1.54 |
| 140 | 266274_at | AT2G29380 | protein phosphatase 2C, putative / PP2C, putative | 5.00 |
| 141 | 266267_at | AT2G29460 | ATGSTU4 (GLUTATHIONE S-TRANSFERASE 22); glutathione transferase | 4.24 |
| 142 | 266644_at | AT2G29660 | zinc finger (C2H2 type) family protein | -2.32 |
| 143 | 267246_at | AT2G30250 | WRKY25 (WRKY DNA-binding protein 25); transcription factor | 3.99 |
| 144 | 267573_at | AT2G30670 | tropinone reductase, putative / tropine dehydrogenase, putative | -2.37 |
| 145 | 267567_at | AT2G30770 | CYP71A13 (CYTOCHROME P450, FAMILY 71, SUBFAMILY A, POLYPEPTIDE 13); indoleacetaldoxime dehydratase/ oxygen binding | 4.01 |
| 146 | 264117_at | AT2G31210 | basic helix-loop-helix (bHLH) family protein | 5.19 |
| 147 | 265668_at | AT2G32020 | GCN5-related N-acetyltransferase (GNAT) family protein | 4.64 |
| 148 | 265723_at | AT2G32140 | transmembrane receptor | 3.60 |
| 149 | 267451_at | AT2G33710 | AP2 domain-containing transcription factor family protein | 3.71 |
| 150 | 267006_at | AT2G34190 | xanthine/uracil permease family protein | -1.50 |
| 151 | 263948_at | AT2G35980 | YLS9 (YELLOW-LEAF-SPECIFIC GENE 9) | 5.42 |
| 152 | 263916_at | AT2G36440 | unknown protein | 7.08 |
| 153 | 266007_at | AT2G37380 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G39370.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO41570.1) | -2.14 |
| 154 | 266010_at | AT2G37430 | zinc finger (C2H2 type) family protein (ZAT11) | 5.41 |
| 155 | 265964_at | AT2G37510 | RNA-binding protein, putative | -1.36 |
| 156 | 266098_at | AT2G37870 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | 5.74 |
| 157 | 267147_at | AT2G38240 | oxidoreductase, 2OG-Fe(II) oxygenase family protein | 4.41 |
| 158 | 267140_at | AT2G38250 | DNA-binding protein-related | 4.77 |
| 159 | 267028_at | AT2G38470 | WRKY33 (WRKY DNA-binding protein 33); transcription factor | 3.90 |
| 160 | 266403_at | AT2G38600 | acid phosphatase class B family protein | -1.49 |
| 161 | 245063_at | AT2G39795 | mitochondrial glycoprotein family protein / MAM33 family protein | -1.40 |
| 162 | 263378_at | AT2G40180 | ATHPP2C5; protein serine/threonine phosphatase | 4.33 |
| 163 | 263836_at | AT2G40330 | Bet v I allergen family protein | -1.97 |
| 164 | 267101_at | AT2G41480 | peroxidase | -2.57 |
| 165 | 260522_x_at | AT2G41730 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G24640.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO14635.1) | 7.12 |
| 166 | 266395_at | AT2G43100 | aconitase C-terminal domain-containing protein | -2.73 |
| 167 | 257373_at | AT2G43140 | DNA binding / transcription factor | -1.98 |
| 168 | 260539_at | AT2G43480 | peroxidase, putative | -1.47 |
| 169 | 266821_at | AT2G44840 | ATERF13/EREBP (ETHYLENE-RESPONSIVE ELEMENT BINDING FACTOR 13); DNA binding / transcription factor | 6.41 |
| 170 | 267502_at | AT2G45550 | CYP76C4 (cytochrome P450, family 76, subfamily C, polypeptide 4); oxygen binding | -2.03 |
| 171 | 267559_at | AT2G45570 | CYP76C2 (cytochrome P450, family 76, subfamily C, polypeptide 2); oxygen binding | 6.38 |
| 172 | 260581_at | AT2G47190 | MYB2 (myb domain protein 2); DNA binding / transcription factor | 4.11 |
| 173 | 266462_at | AT2G47770 | benzodiazepine receptor-related | 4.75 |
| 174 | 266507_at | AT2G47860 | phototropic-responsive NPH3 family protein | -1.58 |
| 175 | 259264_at | AT3G01260 | aldose 1-epimerase family protein | -2.29 |
| 176 | 259000_at | AT3G01860 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G27210.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO14583.1) | -1.62 |
| 177 | 257536_at | AT3G02800 | phosphoprotein phosphatase | 4.10 |
| 178 | 258606_at | AT3G02840 | immediate-early fungal elicitor family protein | 3.91 |
| 179 | 258851_at | AT3G03190 | ATGSTF11 (GLUTATHIONE S-TRANSFERASE F11); glutathione transferase | -3.21 |
| 180 | 258815_at | AT3G04000 | short-chain dehydrogenase/reductase (SDR) family protein | 4.31 |
| 181 | 258840_at | AT3G04620 | nucleic acid binding | 3.76 |
| 182 | 258516_at | AT3G06490 | MYB108 (MYB DOMAIN PROTEIN 108); DNA binding / transcription factor | 3.91 |
| 183 | 258752_at | AT3G09520 | ATEXO70H4 (exocyst subunit EXO70 family protein H4); protein binding | 4.09 |
| 184 | 258695_at | AT3G09640 | APX2 (ASCORBATE PEROXIDASE 2); L-ascorbate peroxidase | 3.76 |
| 185 | 258920_at | AT3G10520 | AHB2 (NON-SYMBIOTIC HAEMOGLOBIN 2) | -1.45 |
| 186 | 258764_at | AT3G10720 | pectinesterase, putative | -1.41 |
| 187 | 257135_at | AT3G12900 | oxidoreductase, 2OG-Fe(II) oxygenase family protein | -3.54 |
| 188 | 256784_at | AT3G13674 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G55205.1) | -1.46 |
| 189 | 256774_at | AT3G13760 | DC1 domain-containing protein | -1.77 |
| 190 | 256779_at | AT3G13784 | ATCWINV5 (ARABIDOPSIS THALIANA CELL WALL INVERTASE 5); hydrolase, hydrolyzing O-glycosyl compounds | 4.36 |
| 191 | 258364_at | AT3G14225 | GLIP4; carboxylesterase | 4.00 |
| 192 | 258369_at | AT3G14310 | ATPME3 (Arabidopsis thaliana pectin methylesterase 3) | -2.00 |
| 193 | 257080_at | AT3G15240 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G53900.2); similar to unnamed protein product [Vitis vinifera] (GB:CAO39314.1); contains domain gb def: At3g15240 (PTHR13902:SF3); contains domain SERINE/THREONINE-PROTEIN KINASE WNK (WITH NO LYSINE)-RELATED (PTHR13902) | -1.50 |
| 194 | 257054_at | AT3G15353 | MT3 (METALLOTHIONEIN 3) | -1.86 |
| 195 | 258395_at | AT3G15500 | ATNAC3 (ARABIDOPSIS NAC DOMAIN CONTAINING PROTEIN 55); transcription factor | 4.72 |
| 196 | 258222_at | AT3G15680 | zinc finger (Ran-binding) family protein | -1.51 |
| 197 | 258413_at | AT3G17300 | similar to unnamed protein product [Vitis vinifera] (GB:CAO70895.1); contains InterPro domain Complex 1 LYR protein (InterPro:IPR008011) | -1.34 |
| 198 | 258158_at | AT3G17790 | ATACP5 (acid phosphatase 5); acid phosphatase/ protein serine/threonine phosphatase | 3.59 |
| 199 | 257720_at | AT3G18450 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G18460.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO68031.1); contains InterPro domain Protein of unknown function Cys-rich (InterPro:IPR006461) | -1.35 |
| 200 | 257035_at | AT3G19270 | CYP707A4 (cytochrome P450, family 707, subfamily A, polypeptide 4); oxygen binding | 4.40 |
| 201 | 257020_at | AT3G19590 | WD-40 repeat family protein / mitotic checkpoint protein, putative | -1.43 |
| 202 | 257115_at | AT3G20150 | kinesin motor family protein | -1.92 |
| 203 | 257950_at | AT3G21780 | UGT71B6 (UDP-GLUCOSYL TRANSFERASE 71B6); UDP-glycosyltransferase/ abscisic acid glucosyltransferase/ transferase, transferring glycosyl groups | 4.98 |
| 204 | 258451_at | AT3G22360 | AOX1B (alternative oxidase 1B); alternative oxidase | 5.47 |
| 205 | 258452_at | AT3G22370 | AOX1A (alternative oxidase 1A); alternative oxidase | 3.64 |
| 206 | 258325_at | AT3G22830 | AT-HSFA6B (Arabidopsis thaliana heat shock transcription factor A6B); DNA binding / transcription factor | 8.07 |
| 207 | 256833_at | AT3G22910 | calcium-transporting ATPase, plasma membrane-type, putative / Ca(2+)-ATPase, putative (ACA13) | 4.95 |
| 208 | 257927_at | AT3G23240 | ATERF1/ERF1 (ETHYLENE RESPONSE FACTOR 1); DNA binding / transcription activator/ transcription factor | 4.21 |
| 209 | 257919_at | AT3G23250 | AtMYB15/AtY19/MYB15 (myb domain protein 15); DNA binding / transcription factor | 4.32 |
| 210 | 257163_at | AT3G24310 | MYB305 (myb domain protein 305); DNA binding / transcription factor | 4.17 |
| 211 | 257823_at | AT3G25190 | nodulin, putative | -3.02 |
| 212 | 257840_at | AT3G25250 | AGC2-1 (OXIDATIVE SIGNAL-INDUCIBLE1); kinase | 4.59 |
| 213 | 256762_at | AT3G25655 | unknown protein | 3.68 |
| 214 | 257613_at | AT3G26610 | polygalacturonase, putative / pectinase, putative | -2.08 |
| 215 | 258277_at | AT3G26830 | PAD3 (PHYTOALEXIN DEFICIENT 3); oxygen binding | 3.80 |
| 216 | 257810_at | AT3G27070 | TOM20-1 (TRANSLOCASE OUTER MEMBRANE 20-1) | -1.69 |
| 217 | 257735_at | AT3G27400 | pectate lyase family protein | -1.80 |
| 218 | 258028_at | AT3G27473 | DC1 domain-containing protein | -1.64 |
| 219 | 256847_at | AT3G27950 | early nodule-specific protein, putative | -2.44 |
| 220 | 256989_at | AT3G28580 | AAA-type ATPase family protein | 3.71 |
| 221 | 256563_at | AT3G29780 | RALFL27 (RALF-LIKE 27) | -1.82 |
| 222 | 257307_at | AT3G30210 | MYB121 (myb domain protein 121); DNA binding / transcription factor | 5.47 |
| 223 | 252679_at | AT3G44260 | CCR4-NOT transcription complex protein, putative | 4.03 |
| 224 | 252612_at | AT3G45160 | contains domain PROKAR_LIPOPROTEIN (PS51257) | -2.36 |
| 225 | 252557_at | AT3G45960 | ATEXLA3 (ARABIDOPSIS THALIANA EXPANSIN-LIKE A3) | 4.01 |
| 226 | 252383_at | AT3G47780 | ATATH6 (ABC2 homolog 6); ATPase, coupled to transmembrane movement of substances | 3.83 |
| 227 | 252368_at | AT3G48520 | CYP94B3 (cytochrome P450, family 94, subfamily B, polypeptide 3); oxygen binding | 4.16 |
| 228 | 252296_at | AT3G48970 | copper-binding family protein | -1.48 |
| 229 | 252265_at | AT3G49620 | DIN11 (DARK INDUCIBLE 11); oxidoreductase | 7.16 |
| 230 | 252224_at | AT3G49860 | ATARLA1B (ADP-ribosylation factor-like A1B); GTP binding | -1.35 |
| 231 | 252193_at | AT3G50060 | MYB77; DNA binding / transcription factor | 4.02 |
| 232 | 252195_at | AT3G50190 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G50140.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G50130.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO71911.1); contains InterPro domain Protein of unknown function DUF247, plant (InterPro:IPR004158) | -3.09 |
| 233 | 252196_at | AT3G50200 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G50170.2); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G50170.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN59797.1); contains InterPro domain Protein of unknown function DUF247, plant (InterPro:IPR004158) | -2.05 |
| 234 | 252205_at | AT3G50350 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G33985.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN61779.1); contains InterPro domain Protein of unknown function DUF1685 (InterPro:IPR012881) | -3.13 |
| 235 | 252131_at | AT3G50930 | AAA-type ATPase family protein | 6.07 |
| 236 | 252148_at | AT3G51280 | male sterility MS5, putative | -1.92 |
| 237 | 252149_at | AT3G51290 | proline-rich family protein | -1.66 |
| 238 | 252077_at | AT3G51720 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G38370.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO48842.1); contains InterPro domain Protein of unknown function DUF827, plant (InterPro:IPR008545) | -1.85 |
| 239 | 252073_at | AT3G51750 | unknown protein | 4.05 |
| 240 | 246310_at | AT3G51895 | SULTR3;1 (SULFATE TRANSPORTER 1); sulfate transmembrane transporter | 4.04 |
| 241 | 251982_at | AT3G53190 | pectate lyase family protein | -2.28 |
| 242 | 251950_at | AT3G53600 | zinc finger (C2H2 type) family protein | 10.20 |
| 243 | 251886_at | AT3G54260 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G42570.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO70127.1); contains InterPro domain Protein of unknown function DUF231, plant (InterPro:IPR004253) | -2.85 |
| 244 | 251899_at | AT3G54400 | aspartyl protease family protein | -1.51 |
| 245 | 251871_at | AT3G54520 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G54530.1) | 9.81 |
| 246 | 251846_at | AT3G54560 | HTA11; DNA binding | -2.08 |
| 247 | 251824_at | AT3G55090 | ATPase, coupled to transmembrane movement of substances | 3.97 |
| 248 | 251827_at | AT3G55120 | A11/CFI/TT5 (TRANSPARENT TESTA 5); chalcone isomerase | -1.41 |
| 249 | 251759_at | AT3G55630 | ATDFD (A. THALIANA DHFS-FPGS HOMOLOG D); tetrahydrofolylpolyglutamate synthase | 5.38 |
| 250 | 246293_at | AT3G56710 | SIB1 (SIGMA FACTOR BINDING PROTEIN 1); binding | 4.58 |
| 251 | 251545_at | AT3G58810 | MTPA2; efflux transmembrane transporter/ zinc ion transmembrane transporter | -2.23 |
| 252 | 251524_at | AT3G58990 | aconitase C-terminal domain-containing protein | -3.56 |
| 253 | 251517_at | AT3G59370 | contains domain PTHR22683 (PTHR22683) | -2.19 |
| 254 | 251456_at | AT3G60120 | glycosyl hydrolase family 1 protein | 4.59 |
| 255 | 251428_at | AT3G60140 | DIN2 (DARK INDUCIBLE 2); hydrolase, hydrolyzing O-glycosyl compounds | 3.60 |
| 256 | 251420_at | AT3G60490 | AP2 domain-containing transcription factor TINY, putative | -1.65 |
| 257 | 251282_at | AT3G61630 | CRF6 (CYTOKININ RESPONSE FACTOR 6); DNA binding / transcription factor | 3.66 |
| 258 | 251293_at | AT3G61930 | unknown protein | -1.93 |
| 259 | 251298_at | AT3G62040 | hydrolase | -2.11 |
| 260 | 251259_at | AT3G62260 | protein phosphatase 2C, putative / PP2C, putative | 3.96 |
| 261 | 251207_at | AT3G63050 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G48075.1) | 4.60 |
| 262 | 251166_at | AT3G63350 | AT-HSFA7B (Arabidopsis thaliana heat shock transcription factor A7B); DNA binding / transcription factor | 5.83 |
| 263 | 255645_at | AT4G00880 | auxin-responsive family protein | -1.50 |
| 264 | 255597_at | AT4G01730 | zinc finger (DHHC type) family protein | -1.74 |
| 265 | 255543_at | AT4G01870 | tolB protein-related | 3.95 |
| 266 | 255517_at | AT4G02290 | ATGH9B13 (ARABIDOPSIS THALIANA GLYCOSYL HYDROLASE 9B13); hydrolase, hydrolyzing O-glycosyl compounds | -2.80 |
| 267 | 255494_at | AT4G02710 | kinase interacting family protein | -1.37 |
| 268 | 255433_at | AT4G03210 | XTH9 (XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 9); hydrolase, acting on glycosyl bonds | -2.26 |
| 269 | 255340_at | AT4G04490 | protein kinase family protein | 7.57 |
| 270 | 255111_at | AT4G08780 | peroxidase, putative | -1.67 |
| 271 | 255807_at | AT4G10270 | wound-responsive family protein | -2.13 |
| 272 | 254905_at | AT4G11170 | disease resistance protein (TIR-NBS-LRR class), putative | 4.84 |
| 273 | 254926_at | AT4G11280 | ACS6 (1-AMINOCYCLOPROPANE-1-CARBOXYLIC ACID (ACC) SYNTHASE 6) | 4.78 |
| 274 | 254914_at | AT4G11290 | peroxidase, putative | -2.09 |
| 275 | 254846_at | AT4G11830 | PLDGAMMA2; phospholipase D | -1.47 |
| 276 | 254862_at | AT4G12030 | bile acid:sodium symporter family protein | -2.60 |
| 277 | 254805_at | AT4G12480 | pEARLI 1; lipid binding | 5.31 |
| 278 | 254823_at | AT4G12580 | unknown protein | 4.25 |
| 279 | 245325_at | AT4G14130 | XTR7 (XYLOGLUCAN ENDOTRANSGLYCOSYLASE 7); hydrolase, acting on glycosyl bonds | -2.15 |
| 280 | 245569_at | AT4G14660 | RNA polymerase Rpb7 N-terminal domain-containing protein | -1.60 |
| 281 | 245552_at | AT4G15360 | CYP705A3 (cytochrome P450, family 705, subfamily A, polypeptide 3); oxygen binding | -2.01 |
| 282 | 245343_at | AT4G15830 | binding | -1.85 |
| 283 | 245440_at | AT4G16680 | RNA helicase, putative | 6.70 |
| 284 | 245447_at | AT4G16820 | lipase class 3 family protein | 6.48 |
| 285 | 245399_at | AT4G17340 | DELTA-TIP2/TIP2;2 (tonoplast intrinsic protein 2;2); water channel | -2.58 |
| 286 | 245252_at | AT4G17500 | ATERF-1 (ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 1); DNA binding / transcription activator/ transcription factor | 3.62 |
| 287 | 245348_at | AT4G17770 | ATTPS5 (Arabidopsis thaliana trehalose phosphatase/synthase 5); transferase, transferring glycosyl groups / trehalose-phosphatase | -1.99 |
| 288 | 254705_at | AT4G17870 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G46790.1); similar to Streptomyces cyclase/dehydrase family protein [Brassica oleracea] (GB:ABD65175.1); contains InterPro domain Streptomyces cyclase/dehydrase (InterPro:IPR005031) | -1.71 |
| 289 | 254652_at | AT4G18170 | WRKY28 (WRKY DNA-binding protein 28); transcription factor | 3.49 |
| 290 | 254562_at | AT4G19230 | CYP707A1 (cytochrome P450, family 707, subfamily A, polypeptide 1); oxygen binding | 3.84 |
| 291 | 254571_at | AT4G19370 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G31720.1); similar to unknown [Picea sitchensis] (GB:ABK21519.1); similar to Os02g0703300 [Oryza sativa (japonica cultivar-group)] (GB:NP_001047854.1); contains InterPro domain Protein of unknown function DUF1218 (InterPro:IPR009606) | -1.43 |
| 292 | 254522_at | AT4G19980 | unknown protein | -2.02 |
| 293 | 254391_at | AT4G21590 | ENDO3 (ENDONUCLEASE 3); T/G mismatch-specific endonuclease/ endonuclease/ nucleic acid binding / single-stranded DNA specific endodeoxyribonuclease | -1.75 |
| 294 | 254394_at | AT4G21630 | subtilase family protein | 4.73 |
| 295 | 254377_at | AT4G21650 | subtilase family protein | 4.24 |
| 296 | 254301_at | AT4G22790 | MATE efflux family protein | -1.40 |
| 297 | 254241_at | AT4G23190 | CRK11 (CYSTEINE-RICH RLK11); kinase | 3.75 |
| 298 | 254233_at | AT4G23800 | high mobility group (HMG1/2) family protein | -2.04 |
| 299 | 254158_at | AT4G24380 | hydrolase, acting on ester bonds | 5.52 |
| 300 | 254119_at | AT4G24780 | pectate lyase family protein | -2.62 |
| 301 | 254059_at | AT4G25200 | ATHSP23.6-MITO (MITOCHONDRION-LOCALIZED SMALL HEAT SHOCK PROTEIN 23.6) | 3.62 |
| 302 | 254075_at | AT4G25470 | CBF2 (FREEZING TOLERANCE QTL 4); DNA binding / transcription activator/ transcription factor | 4.41 |
| 303 | 254042_at | AT4G25810 | XTR6 (XYLOGLUCAN ENDOTRANSGLYCOSYLASE 6); hydrolase, acting on glycosyl bonds | 4.27 |
| 304 | 253999_at | AT4G26200 | ACS7 (1-Amino-cyclopropane-1-carboxylate synthase 7); 1-aminocyclopropane-1-carboxylate synthase | 4.53 |
| 305 | 253815_at | AT4G28250 | ATEXPB3 (ARABIDOPSIS THALIANA EXPANSIN B3) | -2.60 |
| 306 | 253796_at | AT4G28460 | unknown protein | 5.50 |
| 307 | 253729_at | AT4G29360 | glycosyl hydrolase family 17 protein | -1.58 |
| 308 | 253643_at | AT4G29780 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G12010.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO43835.1); contains domain PTHR22930 (PTHR22930) | 4.33 |
| 309 | 253607_at | AT4G30330 | small nuclear ribonucleoprotein E, putative / snRNP-E, putative / Sm protein E, putative | -1.55 |
| 310 | 253598_at | AT4G30800 | 40S ribosomal protein S11 (RPS11B) | -1.43 |
| 311 | 253516_at | AT4G31360 | selenium binding | -1.51 |
| 312 | 253483_at | AT4G31910 | transferase family protein | -2.10 |
| 313 | 253424_at | AT4G32330 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G25480.1); similar to unknown [Populus trichocarpa] (GB:ABK95344.1); contains InterPro domain Targeting for Xklp2 (InterPro:IPR009675) | -1.46 |
| 314 | 253375_at | AT4G33280 | DNA binding / transcription factor | 3.90 |
| 315 | 253344_at | AT4G33550 | lipid binding | 3.63 |
| 316 | 253322_at | AT4G33980 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G42900.2); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G42900.3); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G42900.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN64989.1) | 3.78 |
| 317 | 253259_at | AT4G34410 | AP2 domain-containing transcription factor, putative | 5.38 |
| 318 | 253218_at | AT4G34980 | SLP2 (subtilisin-like serine protease 2); subtilase | -1.78 |
| 319 | 253225_at | AT4G35020 | ARAC3/ATROP6/RHO1PS/ROP6 (rho-related protein from plants 6); GTP binding / GTPase | -1.70 |
| 320 | 253046_at | AT4G37370 | CYP81D8 (cytochrome P450, family 81, subfamily D, polypeptide 8); oxygen binding | 6.53 |
| 321 | 253060_at | AT4G37710 | VQ motif-containing protein | 5.02 |
| 322 | 253012_at | AT4G37900 | glycine-rich protein | 4.58 |
| 323 | 251114_at | AT5G01380 | transcription factor | 4.73 |
| 324 | 251021_at | AT5G02140 | thaumatin-like protein, putative | -1.45 |
| 325 | 251014_at | AT5G02520 | unknown protein | -1.65 |
| 326 | 250798_at | AT5G05340 | peroxidase, putative | 3.54 |
| 327 | 250781_at | AT5G05410 | DREB2A (DRE-BINDING PROTEIN 2A); DNA binding / transcription activator/ transcription factor | 4.98 |
| 328 | 250661_at | AT5G07030 | pepsin A | -1.40 |
| 329 | 250622_at | AT5G07310 | AP2 domain-containing transcription factor, putative | 5.00 |
| 330 | 250471_at | AT5G10170 | inositol-3-phosphate synthase, putative / myo-inositol-1-phosphate synthase, putative / MI-1-P synthase, putative | -1.57 |
| 331 | 250434_at | AT5G10390 | histone H3 | -2.05 |
| 332 | 250277_at | AT5G12940 | leucine-rich repeat family protein | -1.41 |
| 333 | 245982_at | AT5G13170 | nodulin MtN3 family protein | 4.08 |
| 334 | 250286_at | AT5G13320 | PBS3 (AVRPPHB SUSCEPTIBLE 3) | 4.36 |
| 335 | 250228_at | AT5G13840 | WD-40 repeat family protein | -1.80 |
| 336 | 250182_at | AT5G14470 | GHMP kinase-related | 5.42 |
| 337 | 246584_at | AT5G14730 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G01513.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO64332.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN65788.1) | 3.87 |
| 338 | 246596_at | AT5G14740 | CA2 (BETA CARBONIC ANHYDRASE 2); carbonate dehydratase/ zinc ion binding | 6.12 |
| 339 | 246597_at | AT5G14760 | AO (L-ASPARTATE OXIDASE); L-aspartate oxidase | -1.71 |
| 340 | 246595_at | AT5G14780 | FDH (FORMATE DEHYDROGENASE); NAD binding / binding / catalytic/ cofactor binding / oxidoreductase, acting on the CH-OH group of donors, NAD or NADP as acceptor | 3.56 |
| 341 | 246562_at | AT5G15580 | LNG1 (LONGIFOLIA1) | -1.36 |
| 342 | 246518_at | AT5G15770 | ATGNA1 (ARABIDOPSIS THALIANA GLUCOSE-6-PHOSPHATE ACETYLTRANSFERASE 1); N-acetyltransferase/ glucosamine 6-phosphate N-acetyltransferase | -1.46 |
| 343 | 246522_at | AT5G15830 | ATBZIP3 (ARABIDOPSIS THALIANA BASIC LEUCINE-ZIPPER 3); DNA binding / transcription factor | -1.70 |
| 344 | 249971_at | AT5G19110 | extracellular dermal glycoprotein-related / EDGP-related | 5.58 |
| 345 | 245947_at | AT5G19530 | ACL5 (ACAULIS 5) | -2.83 |
| 346 | 246145_at | AT5G19880 | peroxidase, putative | 6.10 |
| 347 | 246099_at | AT5G20230 | ATBCB (ARABIDOPSIS BLUE-COPPER-BINDING PROTEIN); copper ion binding | 5.72 |
| 348 | 249860_at | AT5G22860 | serine carboxypeptidase S28 family protein | 3.54 |
| 349 | 249916_at | AT5G22880 | H2B/HTB2 (HISTONE H2B); DNA binding | -1.78 |
| 350 | 249770_at | AT5G24110 | WRKY30 (WRKY DNA-binding protein 30); transcription factor | 6.30 |
| 351 | 249743_at | AT5G24540 | glycosyl hydrolase family 1 protein | 3.48 |
| 352 | 246933_at | AT5G25160 | ZFP3 (ZINC FINGER PROTEIN 3); nucleic acid binding / transcription factor/ zinc ion binding | -1.42 |
| 353 | 246831_at | AT5G26340 | MSS1 (SUGAR TRANSPORT PROTEIN 13); carbohydrate transmembrane transporter/ hexose:hydrogen symporter/ high-affinity hydrogen:glucose symporter/ sugar:hydrogen ion symporter | 3.62 |
| 354 | 246821_at | AT5G26920 | calmodulin binding | 4.03 |
| 355 | 245955_at | AT5G28510 | glycosyl hydrolase family 1 protein | 6.14 |
| 356 | 245953_at | AT5G28520 | similar to jacalin lectin family protein [Arabidopsis thaliana] (TAIR:AT1G33790.1); similar to jasmonate inducible protein [Brassica napus] (GB:CAA72271.1); contains InterPro domain Mannose-binding lectin (InterPro:IPR001229) | 7.57 |
| 357 | 246108_at | AT5G28630 | glycine-rich protein | 4.07 |
| 358 | 249617_at | AT5G37450 | leucine-rich repeat transmembrane protein kinase, putative | -2.08 |
| 359 | 249535_at | AT5G38820 | amino acid transporter family protein | -1.48 |
| 360 | 249333_at | AT5G40990 | GLIP1 (GDSL LIPASE1); carboxylesterase | 3.92 |
| 361 | 249197_at | AT5G42380 | CML37/CML39; calcium ion binding | 6.50 |
| 362 | 249212_at | AT5G42690 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G37080.2); similar to unnamed protein product [Vitis vinifera] (GB:CAO61290.1); contains InterPro domain Protein of unknown function DUF547 (InterPro:IPR006869) | -1.36 |
| 363 | 249188_at | AT5G42830 | transferase family protein | 3.48 |
| 364 | 249174_at | AT5G42900 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G33980.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN75496.1) | 7.18 |
| 365 | 249134_at | AT5G43150 | similar to unnamed protein product [Vitis vinifera] (GB:CAO61459.1) | -1.42 |
| 366 | 249128_at | AT5G43440 | 2-oxoglutarate-dependent dioxygenase, putative | 4.54 |
| 367 | 249125_at | AT5G43450 | 2-oxoglutarate-dependent dioxygenase, putative | 6.21 |
| 368 | 249003_at | AT5G44500 | small nuclear ribonucleoprotein associated protein B, putative / snRNP-B, putative / Sm protein B, putative | -1.65 |
| 369 | 249001_at | AT5G44990 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G19880.1); similar to Intracellular chloride channel [Medicago truncatula] (GB:ABC75353.2); contains InterPro domain Thioredoxin-like fold (InterPro:IPR012336); contains InterPro domain Glutathione S-transferase, C-terminal-like (InterPro:IPR010987); contains InterPro domain Glutathione S-transferase, predicted (InterPro:IPR016639) | 3.75 |
| 370 | 248990_at | AT5G45210 | disease resistance protein (TIR-NBS-LRR class), putative | -1.83 |
| 371 | 248963_at | AT5G45700 | NLI interacting factor (NIF) family protein | -2.97 |
| 372 | 248878_at | AT5G46160 | ribosomal protein L14 family protein / huellenlos paralog (HLP) | -1.38 |
| 373 | 248895_at | AT5G46330 | FLS2 (FLAGELLIN-SENSITIVE 2); ATP binding / kinase/ protein binding / protein serine/threonine kinase/ transmembrane receptor protein serine/threonine kinase | 4.91 |
| 374 | 248896_at | AT5G46350 | WRKY8 (WRKY DNA-binding protein 8); transcription factor | 4.45 |
| 375 | 248794_at | AT5G47220 | ATERF-2/ATERF2/ERF2 (ETHYLENE RESPONSE FACTOR 2); DNA binding / transcription activator/ transcription factor | 3.96 |
| 376 | 248793_at | AT5G47240 | ATNUDT8 (Arabidopsis thaliana Nudix hydrolase homolog 8); hydrolase | -2.55 |
| 377 | 248775_at | AT5G47850 | protein kinase, putative | 6.05 |
| 378 | 248625_at | AT5G48880 | KAT5/PKT1/PKT2 (PEROXISOMAL 3-KETO-ACYL-COA THIOLASE 1, PEROXISOMAL 3-KETO-ACYL-COA THIOLASE 2); acetyl-CoA C-acyltransferase | -1.80 |
| 379 | 248597_at | AT5G49160 | MET1 (DECREASED METHYLATION 2DNA) | -1.39 |
| 380 | 248585_at | AT5G49640 | unknown protein | -3.15 |
| 381 | 248548_at | AT5G50300 | xanthine/uracil/vitamin C permease family protein | -1.85 |
| 382 | 248511_at | AT5G50375 | CPI1 (CYCLOPROPYL ISOMERASE) | -1.64 |
| 383 | 248419_at | AT5G51550 | phosphate-responsive 1 family protein | -1.99 |
| 384 | 248420_at | AT5G51560 | leucine-rich repeat transmembrane protein kinase, putative | -1.43 |
| 385 | 248337_at | AT5G52310 | COR78 (COLD REGULATED 78) | 5.31 |
| 386 | 248333_at | AT5G52390 | photoassimilate-responsive protein, putative | 3.67 |
| 387 | 248358_at | AT5G52400 | CYP715A1 (cytochrome P450, family 715, subfamily A, polypeptide 1); oxygen binding | 5.12 |
| 388 | 248327_at | AT5G52750 | heavy-metal-associated domain-containing protein | 3.67 |
| 389 | 248322_at | AT5G52760 | heavy-metal-associated domain-containing protein | 4.52 |
| 390 | 247913_at | AT5G57510 | unknown protein | 5.17 |
| 391 | 247925_at | AT5G57560 | TCH4 (TOUCH 4); hydrolase, acting on glycosyl bonds / xyloglucan:xyloglucosyl transferase | 3.57 |
| 392 | 247848_at | AT5G58120 | disease resistance protein (TIR-NBS-LRR class), putative | 3.80 |
| 393 | 247789_at | AT5G58680 | armadillo/beta-catenin repeat family protein | 4.29 |
| 394 | 247723_at | AT5G59220 | protein phosphatase 2C, putative / PP2C, putative | 4.01 |
| 395 | 247718_at | AT5G59310 | LTP4 (LIPID TRANSFER PROTEIN 4); lipid binding | 4.14 |
| 396 | 247655_at | AT5G59820 | RHL41 (RESPONSIVE TO HIGH LIGHT 41); nucleic acid binding / transcription factor/ zinc ion binding | 4.71 |
| 397 | 247604_at | AT5G60950 | COBL5 (COBRA-LIKE PROTEIN 5 PRECURSOR) | -1.59 |
| 398 | 247435_at | AT5G62480 | ATGSTU9 (GLUTATHIONE S-TRANSFERASE TAU 9); glutathione transferase | 3.53 |
| 399 | 247347_at | AT5G63780 | SHA1 (SHOOT APICAL MERISTEM ARREST 1); protein binding / zinc ion binding | -1.64 |
| 400 | 247208_at | AT5G64870 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G25250.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G25260.1); similar to 80C09_16 [Brassica rapa subsp. pekinensis] (GB:AAZ41827.1); contains domain PTHR13806:SF3 (PTHR13806:SF3); contains domain PTHR13806 (PTHR13806) | 4.81 |
| 401 | 247145_at | AT5G65600 | legume lectin family protein / protein kinase family protein | 5.44 |
| 402 | 247161_at | AT5G65810 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G49720.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G49720.2); similar to hypothetical protein [Vitis vinifera] (GB:CAN74732.1); contains domain S-adenosyl-L-methionine-dependent methyltransferases (SSF53335) | -1.40 |
| 403 | 247116_at | AT5G65970 | MLO10 (MILDEW RESISTANCE LOCUS O 10); calmodulin binding | -2.27 |
| 404 | 247071_at | AT5G66640 | LIM domain-containing protein-related | 6.08 |
| 405 | 247056_at | AT5G66750 | CHR01/DDM1 (DECREASED DNA METHYLATION 1); helicase | -1.70 |
| 406 | 247024_at | AT5G66985 | unknown protein | -2.99 |
| 407 | 247037_at | AT5G67070 | RALFL34 (RALF-LIKE 34) | -1.43 |
| 408 | 247039_at | AT5G67270 | ATEB1C (MICROTUBULE END BINDING PROTEIN 1); microtubule binding | -1.88 |