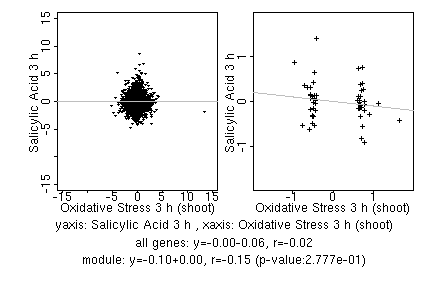
marker gene list
| Oxidative Stress 3 h (shoot) |
| Salicylic Acid 3 h |
| probeID | AGICode | Annotation | gene expression |
|
|---|---|---|---|---|
| 1 | 264505_at | AT1G09380 | integral membrane family protein / nodulin MtN21-related | -0.70 |
| 2 | 260685_at | AT1G17650 | phosphogluconate dehydrogenase (decarboxylating) | 0.69 |
| 3 | 265175_at | AT1G23480 | ATCSLA03 (Cellulose synthase-like A3); transferase, transferring glycosyl groups | -0.44 |
| 4 | 263024_at | AT1G23980 | zinc finger (C3HC4-type RING finger) family protein | -0.73 |
| 5 | 259606_at | AT1G27920 | microtubule associated protein (MAP65/ASE1) family protein | -0.49 |
| 6 | 259789_at | AT1G29395 | COR414-TM1 (cold regulated 414 thylakoid membrane 1) | 0.70 |
| 7 | 245790_at | AT1G32200 | ATS1 (ACYLTRANSFERASE 1) | 0.70 |
| 8 | 259646_at | AT1G55350 | DEK1 (DEFECTIVE KERNEL 1); calcium-dependent cysteine-type endopeptidase/ cysteine-type endopeptidase | 0.80 |
| 9 | 262909_at | AT1G59830 | PP2A-1 (protein phosphatase 2A-2); protein serine/threonine phosphatase | 0.66 |
| 10 | 261953_at | AT1G64440 | RHD1 (ROOT HAIR DEFECTIVE 1); UDP-glucose 4-epimerase | 0.70 |
| 11 | 260442_at | AT1G68220 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G13380.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN76984.1); contains InterPro domain Protein of unknown function DUF1218 (InterPro:IPR009606) | -0.49 |
| 12 | 264314_at | AT1G70420 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G23710.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO66069.1); contains InterPro domain Protein of unknown function DUF1645 (InterPro:IPR012442) | 0.76 |
| 13 | 263129_at | AT1G78620 | integral membrane family protein | 0.66 |
| 14 | 265875_at | AT2G01690 | binding | -0.49 |
| 15 | 265778_at | AT2G07360 | SH3 domain-containing protein | 0.63 |
| 16 | 265400_at | AT2G10940 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | 0.68 |
| 17 | 265392_at | AT2G20860 | LIP1 (LIPOIC ACID SYNTHASE 1); lipoic acid synthase | 0.70 |
| 18 | 265442_at | AT2G20940 | similar to unknown [Populus trichocarpa] (GB:ABK95988.1); contains InterPro domain Protein of unknown function DUF1279 (InterPro:IPR009688) | -0.55 |
| 19 | 264018_at | AT2G21170 | TIM (TRIOSEPHOSPHATE ISOMERASE); triose-phosphate isomerase | 0.79 |
| 20 | 265913_at | AT2G25625 | similar to unnamed protein product [Vitis vinifera] (GB:CAO44157.1) | -0.40 |
| 21 | 267247_at | AT2G30170 | catalytic | 0.77 |
| 22 | 267519_at | AT2G30470 | HSI2 (HIGH-LEVEL EXPRESSION OF SUGAR-INDUCIBLE GENE 2); transcription factor/ transcription repressor | 0.66 |
| 23 | 263957_at | AT2G35880 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G32330.3); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G32330.2); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G32330.1); similar to Os06g0606800 [Oryza sativa (japonica cultivar-group)] (GB:NP_001058026.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO44238.1); similar to Os11g0571900 [Oryza sativa (japonica cultivar-group)] (GB:NP_001068122.1); contains InterPro domain Targeting for Xklp2 (InterPro:IPR009675) | 0.81 |
| 24 | 266055_at | AT2G40650 | pre-mRNA splicing factor PRP38 family protein | 0.74 |
| 25 | 267625_at | AT2G42240 | nucleic acid binding | -0.54 |
| 26 | 258510_at | AT3G06600 | unknown protein | -0.42 |
| 27 | 259008_at | AT3G09390 | MT2A (METALLOTHIONEIN 2A) | -0.47 |
| 28 | 257707_at | AT3G12650 | similar to unnamed protein product [Vitis vinifera] (GB:CAO15069.1) | -0.49 |
| 29 | 257291_at | AT3G15590 | DNA-binding protein, putative | -0.53 |
| 30 | 252685_at | AT3G44410 | pseudogene, disease resistence protein, putative, similar to disease resistance protein RPP1-WsB (Arabidopsis thaliana) gi|3860165|gb|AAC72978 | -0.45 |
| 31 | 256677_at | AT3G52190 | PHF1 (PHOSPHATE TRANSPORTER TRAFFIC FACILITATOR1); nucleotide binding | -0.40 |
| 32 | 251935_at | AT3G54090 | pfkB-type carbohydrate kinase family protein | -0.47 |
| 33 | 251727_at | AT3G56290 | similar to hypothetical protein [Vitis vinifera] (GB:CAN75527.1) | -0.41 |
| 34 | 251573_at | AT3G58140 | phenylalanyl-tRNA synthetase class IIc family protein | 0.78 |
| 35 | 254433_at | AT4G20850 | TPP2 (TRIPEPTIDYL PEPTIDASE II); subtilase | 0.67 |
| 36 | 254085_at | AT4G24960 | ATHVA22D (Arabidopsis thaliana HVA22 homologue D) | -0.56 |
| 37 | 253406_at | AT4G32890 | zinc finger (GATA type) family protein | 0.74 |
| 38 | 253207_at | AT4G34770 | auxin-responsive family protein | -0.43 |
| 39 | 250723_at | AT5G06300 | carboxy-lyase | -0.46 |
| 40 | 250225_at | AT5G14105 | similar to hypothetical protein [Vitis vinifera] (GB:CAN83318.1) | -0.54 |
| 41 | 249969_at | AT5G19090 | heavy-metal-associated domain-containing protein | 0.72 |
| 42 | 245950_at | AT5G19570 | similar to hypothetical protein OsI_018061 [Oryza sativa (indica cultivar-group)] (GB:EAY96828.1); similar to Os05g0188300 [Oryza sativa (japonica cultivar-group)] (GB:NP_001054840.1) | -0.62 |
| 43 | 246924_at | AT5G25060 | RNA recognition motif (RRM)-containing protein | 0.65 |
| 44 | 249612_at | AT5G37290 | armadillo/beta-catenin repeat family protein | -0.45 |
| 45 | 249206_at | AT5G42630 | ATS/KAN4 (ABERRANT TESTA SHAPE); DNA binding / transcription factor | 1.14 |
| 46 | 248741_at | AT5G48170 | SLY2 (SLEEPY2) | -0.55 |
| 47 | 248059_at | AT5G55540 | TRN1 (TORNADO 1) | 0.72 |
| 48 | 248004_at | AT5G56230 | prenylated rab acceptor (PRA1) family protein | -0.94 |
| 49 | 247896_at | AT5G57950 | 26S proteasome regulatory subunit, putative | -0.47 |
| 50 | 247838_at | AT5G57990 | UBP23 (UBIQUITIN-SPECIFIC PROTEASE 23); ubiquitin-specific protease | 0.65 |
| 51 | 247445_at | AT5G62640 | ELF5 (EARLY FLOWERING 5) | -0.45 |
| 52 | 247319_at | AT5G64050 | ATERS/ERS/OVA3 (OVULE ABORTION 3); glutamate-tRNA ligase | 0.92 |
| 53 | 247140_at | AT5G66250 | kinectin-related | 0.72 |
| 54 | 244923_s_at | ATMG01020 | hypothetical protein | 1.67 |