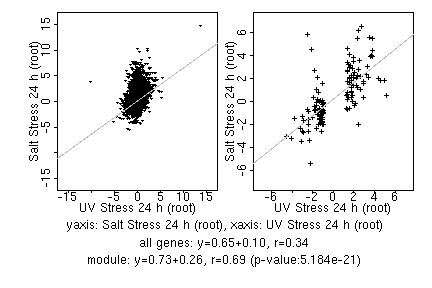
marker gene list
| UV Stress 24 h (root) |
| Salt Stress 24 h (root) |
| probeID | AGICode | Annotation | gene expression |
|
|---|---|---|---|---|
| 1 | 264580_at | AT1G05340 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G32210.1) | 2.66 |
| 2 | 261385_at | AT1G05450 | protease inhibitor/seed storage/lipid transfer protein (LTP)-related | 1.63 |
| 3 | 261249_at | AT1G05880 | zinc ion binding | 1.83 |
| 4 | 260783_at | AT1G06160 | ethylene-responsive factor, putative | 1.83 |
| 5 | 260623_at | AT1G08090 | ATNRT2:1 (Arabidopsis thaliana high affinity nitrate transporter 2.1); nitrate transmembrane transporter | -2.83 |
| 6 | 261474_at | AT1G14540 | anionic peroxidase, putative | 2.84 |
| 7 | 257480_at | AT1G15640 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G15620.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN70531.1) | -1.29 |
| 8 | 255889_at | AT1G17840 | ABCG11/COF1/DSO/WBC11 (DESPERADO); ATPase, coupled to transmembrane movement of substances | -1.27 |
| 9 | 255753_at | AT1G18570 | MYB51 (MYB DOMAIN PROTEIN 51); DNA binding / transcription factor | 1.47 |
| 10 | 261221_at | AT1G19960 | similar to transmembrane receptor [Arabidopsis thaliana] (TAIR:AT2G32140.1) | -1.01 |
| 11 | 261459_at | AT1G21100 | O-methyltransferase, putative | 3.48 |
| 12 | 261449_at | AT1G21120 | O-methyltransferase, putative | 1.96 |
| 13 | 264774_at | AT1G22890 | unknown protein | 2.30 |
| 14 | 264861_at | AT1G24320 | alpha-glucosidase, putative | -1.61 |
| 15 | 265024_at | AT1G24600 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G67920.1) | 1.46 |
| 16 | 245875_at | AT1G26240 | proline-rich extensin-like family protein | 1.93 |
| 17 | 261021_at | AT1G26380 | FAD-binding domain-containing protein | 3.74 |
| 18 | 261020_at | AT1G26390 | FAD-binding domain-containing protein | 4.44 |
| 19 | 261006_at | AT1G26410 | FAD-binding domain-containing protein | 3.38 |
| 20 | 261005_at | AT1G26420 | FAD-binding domain-containing protein | 2.05 |
| 21 | 262743_at | AT1G29020 | calcium-binding EF hand family protein | -1.35 |
| 22 | 259788_at | AT1G29670 | GDSL-motif lipase/hydrolase family protein | -2.27 |
| 23 | 245774_at | AT1G30210 | TCP24; transcription factor | -1.13 |
| 24 | 263228_at | AT1G30700 | FAD-binding domain-containing protein | 1.49 |
| 25 | 256447_at | AT1G33440 | proton-dependent oligopeptide transport (POT) family protein | -0.92 |
| 26 | 256423_at | AT1G33540 | SCPL18 (serine carboxypeptidase-like 18); serine carboxypeptidase | -1.44 |
| 27 | 245767_at | AT1G33610 | leucine-rich repeat family protein | -0.88 |
| 28 | 256542_at | AT1G42550 | PMI1 (PLASTID MOVEMENT IMPAIRED1) | 1.49 |
| 29 | 259454_at | AT1G44050 | DC1 domain-containing protein | -1.97 |
| 30 | 261606_at | AT1G49570 | peroxidase, putative | 2.99 |
| 31 | 259813_at | AT1G49860 | ATGSTF14 (Arabidopsis thaliana Glutathione S-transferase (class phi) 14); glutathione transferase | -2.92 |
| 32 | 261690_at | AT1G50090 | aminotransferase class IV family protein | -2.43 |
| 33 | 256405_at | AT1G50700 | CPK33 (calcium-dependent protein kinase 33); calmodulin-dependent protein kinase/ kinase | -0.89 |
| 34 | 265049_at | AT1G52060 | similar to jacalin lectin family protein [Arabidopsis thaliana] (TAIR:AT1G52070.1); similar to jasmonate inducible protein [Brassica napus] (GB:CAA72271.1); contains InterPro domain Mannose-binding lectin (InterPro:IPR001229) | 1.63 |
| 35 | 256222_at | AT1G56210 | copper chaperone (CCH)-related | -0.96 |
| 36 | 246393_at | AT1G58150 | unknown protein | -0.91 |
| 37 | 264278_at | AT1G60130 | jacalin lectin family protein | -1.69 |
| 38 | 264433_at | AT1G61810 | BGLU45; hydrolase, hydrolyzing O-glycosyl compounds | 1.43 |
| 39 | 261949_at | AT1G64670 | BDG1 (BODYGUARD1); hydrolase | -1.69 |
| 40 | 264157_at | AT1G65310 | ATXTH17 (XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 17); hydrolase, acting on glycosyl bonds | 1.44 |
| 41 | 245193_at | AT1G67810 | Fe-S metabolism associated domain-containing protein | 1.41 |
| 42 | 260015_at | AT1G67980 | CCoAMT (caffeoyl-CoA 3-O-methyltransferase); caffeoyl-CoA O-methyltransferase | 1.53 |
| 43 | 260267_at | AT1G68530 | CUT1 (CUTICULAR 1); acyltransferase/ catalytic | -1.49 |
| 44 | 262229_at | AT1G68620 | hydrolase | 2.38 |
| 45 | 260030_at | AT1G68880 | ATBZIP (ARABIDOPSIS THALIANA BASIC LEUCINE-ZIPPER 8); DNA binding / transcription factor | -1.17 |
| 46 | 256299_at | AT1G69530 | ATEXPA1 (ARABIDOPSIS THALIANA EXPANSIN A1) | -1.51 |
| 47 | 262303_at | AT1G70920 | homeobox-leucine zipper protein, putative / HD-ZIP transcription factor, putative | 1.62 |
| 48 | 260394_at | AT1G74080 | MYB122 (myb domain protein 122); DNA binding / transcription factor | 1.89 |
| 49 | 260239_at | AT1G74360 | leucine-rich repeat transmembrane protein kinase, putative | 1.45 |
| 50 | 264953_at | AT1G77120 | ADH1 (ALCOHOL DEHYDROGENASE 1) | -1.68 |
| 51 | 266233_at | AT2G02340 | ATPP2-B8 (Phloem protein 2-B8); carbohydrate binding | -0.99 |
| 52 | 265329_at | AT2G18450 | SDH1-2 (Succinate dehydrogenase 1-2) | -1.14 |
| 53 | 266017_at | AT2G18690 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G18680.1); similar to hypothetical protein OsI_029427 [Oryza sativa (indica cultivar-group)] (GB:EAZ08195.1) | 1.62 |
| 54 | 267265_at | AT2G22980 | SCPL13; serine carboxypeptidase | 1.88 |
| 55 | 245082_at | AT2G23270 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G37290.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN62855.1) | 3.57 |
| 56 | 263800_at | AT2G24600 | ankyrin repeat family protein | -1.46 |
| 57 | 263539_at | AT2G24850 | TAT3 (TYROSINE AMINOTRANSFERASE 3); transaminase | -2.41 |
| 58 | 265611_at | AT2G25510 | unknown protein | -1.31 |
| 59 | 245035_at | AT2G26400 | ARD/ATARD3 (ACIREDUCTONE DIOXYGENASE); acireductone dioxygenase [iron(II)-requiring]/ heteroglycan binding / metal ion binding | -4.48 |
| 60 | 245038_at | AT2G26560 | PLP2 (PHOSPHOLIPASE A 2A); nutrient reservoir | 3.86 |
| 61 | 267573_at | AT2G30670 | tropinone reductase, putative / tropine dehydrogenase, putative | -3.08 |
| 62 | 267565_at | AT2G30750 | CYP71A12 (CYTOCHROME P450, FAMILY 71, SUBFAMILY A, POLYPEPTIDE 12); oxygen binding | 3.03 |
| 63 | 267567_at | AT2G30770 | CYP71A13 (CYTOCHROME P450, FAMILY 71, SUBFAMILY A, POLYPEPTIDE 13); indoleacetaldoxime dehydratase/ oxygen binding | 3.83 |
| 64 | 267457_at | AT2G33790 | pollen Ole e 1 allergen and extensin family protein | 2.07 |
| 65 | 266415_at | AT2G38530 | LTP2 (LIPID TRANSFER PROTEIN 2); lipid binding | -1.11 |
| 66 | 266168_at | AT2G38870 | protease inhibitor, putative | 2.68 |
| 67 | 266191_at | AT2G39040 | peroxidase, putative | 2.25 |
| 68 | 267337_at | AT2G39980 | transferase family protein | 2.47 |
| 69 | 263979_at | AT2G42840 | PDF1 (PROTODERMAL FACTOR 1) | -1.09 |
| 70 | 263970_at | AT2G42850 | CYP718 (cytochrome P450, family 718); oxygen binding | 2.49 |
| 71 | 265260_at | AT2G43000 | ANAC042 (Arabidopsis NAC domain containing protein 42); transcription factor | 2.02 |
| 72 | 257373_at | AT2G43140 | DNA binding / transcription factor | -1.19 |
| 73 | 260551_at | AT2G43510 | ATTI1 (ARABIDOPSIS THALIANA TRYPSIN INHIBITOR PROTEIN 1) | 1.71 |
| 74 | 260556_at | AT2G43620 | chitinase, putative | -1.74 |
| 75 | 267502_at | AT2G45550 | CYP76C4 (cytochrome P450, family 76, subfamily C, polypeptide 4); oxygen binding | -2.34 |
| 76 | 258791_at | AT3G04720 | PR4 (PATHOGENESIS-RELATED 4) | 1.61 |
| 77 | 258675_at | AT3G08770 | LTP6 (Lipid transfer protein 6); lipid binding | -0.99 |
| 78 | 258765_at | AT3G10710 | pectinesterase family protein | 2.06 |
| 79 | 258764_at | AT3G10720 | pectinesterase, putative | -0.94 |
| 80 | 259260_at | AT3G11370 | DC1 domain-containing protein | -1.28 |
| 81 | 259261_at | AT3G11390 | DC1 domain-containing protein | -1.32 |
| 82 | 257135_at | AT3G12900 | oxidoreductase, 2OG-Fe(II) oxygenase family protein | -3.03 |
| 83 | 257184_at | AT3G13090 | ATMRP8 (Arabidopsis thaliana multidrug resistance-associated protein 8) | -1.50 |
| 84 | 258008_at | AT3G19430 | late embryogenesis abundant protein-related / LEA protein-related | 1.90 |
| 85 | 257142_at | AT3G20090 | CYP705A18 (cytochrome P450, family 705, subfamily A, polypeptide 18); oxygen binding | -1.01 |
| 86 | 256818_at | AT3G21420 | oxidoreductase, 2OG-Fe(II) oxygenase family protein | -0.93 |
| 87 | 257540_at | AT3G21520 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G21550.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO71058.1); contains InterPro domain Protein of unknown function DUF679 (InterPro:IPR007770) | 1.83 |
| 88 | 257924_at | AT3G23190 | lesion inducing protein-related | -0.96 |
| 89 | 258130_at | AT3G24510 | Encodes a defensin-like (DEFL) family protein. | 5.09 |
| 90 | 257625_at | AT3G26230 | CYP71B24 (cytochrome P450, family 71, subfamily B, polypeptide 24); oxygen binding | 2.40 |
| 91 | 258277_at | AT3G26830 | PAD3 (PHYTOALEXIN DEFICIENT 3); oxygen binding | 2.01 |
| 92 | 258028_at | AT3G27473 | DC1 domain-containing protein | -1.26 |
| 93 | 256847_at | AT3G27950 | early nodule-specific protein, putative | -1.51 |
| 94 | 256593_at | AT3G28510 | AAA-type ATPase family protein | -2.29 |
| 95 | 252234_at | AT3G49780 | ATPSK4 (PHYTOSULFOKINE 4 PRECURSOR); growth factor | 1.84 |
| 96 | 252238_at | AT3G49960 | peroxidase, putative | 1.60 |
| 97 | 252196_at | AT3G50200 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G50170.2); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G50170.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN59797.1); contains InterPro domain Protein of unknown function DUF247, plant (InterPro:IPR004158) | -1.32 |
| 98 | 251419_at | AT3G60470 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G44930.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN69719.1); contains InterPro domain Protein of unknown function DUF247, plant (InterPro:IPR004158) | -2.86 |
| 99 | 251195_at | AT3G62930 | glutaredoxin family protein | -0.88 |
| 100 | 255252_at | AT4G04990 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G61260.1); contains InterPro domain Protein of unknown function DUF761, plant (InterPro:IPR008480) | 2.13 |
| 101 | 254916_at | AT4G11340 | Toll-Interleukin-Resistance (TIR) domain-containing protein | 2.41 |
| 102 | 245576_at | AT4G14770 | tesmin/TSO1-like CXC domain-containing protein | -0.98 |
| 103 | 245393_at | AT4G16260 | glycosyl hydrolase family 17 protein | 1.48 |
| 104 | 254522_at | AT4G19980 | unknown protein | -1.31 |
| 105 | 254247_at | AT4G23260 | protein kinase | -1.43 |
| 106 | 254233_at | AT4G23800 | high mobility group (HMG1/2) family protein | -0.89 |
| 107 | 253827_at | AT4G28085 | unknown protein | 1.47 |
| 108 | 253819_at | AT4G28350 | lectin protein kinase family protein | 2.51 |
| 109 | 253796_at | AT4G28460 | unknown protein | 2.48 |
| 110 | 253763_at | AT4G28850 | xyloglucan:xyloglucosyl transferase, putative / xyloglucan endotransglycosylase, putative / endo-xyloglucan transferase, putative | 5.23 |
| 111 | 253660_at | AT4G30140 | GDSL-motif lipase/hydrolase family protein | -2.31 |
| 112 | 253502_at | AT4G31940 | CYP82C4 (cytochrome P450, family 82, subfamily C, polypeptide 4); oxygen binding | -2.13 |
| 113 | 253505_at | AT4G31970 | CYP82C2 (cytochrome P450, family 82, subfamily C, polypeptide 2); oxygen binding | 4.66 |
| 114 | 253044_at | AT4G37290 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G23270.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN62855.1) | 3.31 |
| 115 | 251039_at | AT5G02020 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G55646.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G59080.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO15731.1) | 1.49 |
| 116 | 250719_at | AT5G06250 | transcription factor | -0.96 |
| 117 | 250464_at | AT5G10040 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G65207.1); similar to 80A08_3 [Brassica rapa subsp. pekinensis] (GB:AAZ67588.1) | -3.96 |
| 118 | 250302_at | AT5G11920 | ATCWINV6 (6-&1-FRUCTAN EXOHYDROLASE); hydrolase, hydrolyzing O-glycosyl compounds / inulinase/ levanase | -1.32 |
| 119 | 250142_at | AT5G14650 | polygalacturonase, putative / pectinase, putative | -2.23 |
| 120 | 246459_at | AT5G16900 | leucine-rich repeat protein kinase, putative | -0.90 |
| 121 | 249775_at | AT5G24160 | squalene monooxygenase 1,2 / squalene epoxidase 1,2 (SQP1,2) | 2.10 |
| 122 | 246821_at | AT5G26920 | calmodulin binding | 1.66 |
| 123 | 246807_at | AT5G27100 | ATGLR2.1 (Arabidopsis thaliana glutamate receptor 2.1) | -0.96 |
| 124 | 249476_at | AT5G38910 | germin-like protein, putative | 1.42 |
| 125 | 249333_at | AT5G40990 | GLIP1 (GDSL LIPASE1); carboxylesterase | 3.88 |
| 126 | 249125_at | AT5G43450 | 2-oxoglutarate-dependent dioxygenase, putative | 2.52 |
| 127 | 248979_at | AT5G45080 | ATPP2-A6 (Phloem protein 2-A6); transmembrane receptor | -1.29 |
| 128 | 248942_at | AT5G45480 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G45470.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G45530.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G45540.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO39193.1); contains InterPro domain Protein of unknown function DUF594 (InterPro:IPR007658) | -0.89 |
| 129 | 248855_at | AT5G46590 | ANAC096 (Arabidopsis NAC domain containing protein 96); transcription factor | -1.86 |
| 130 | 248763_at | AT5G47550 | cysteine protease inhibitor, putative / cystatin, putative | -1.13 |
| 131 | 248727_at | AT5G47990 | CYP705A5 (cytochrome P450, family 705, subfamily A, polypeptide 5); oxygen binding | 1.41 |
| 132 | 248548_at | AT5G50300 | xanthine/uracil/vitamin C permease family protein | -1.12 |
| 133 | 248319_at | AT5G52710 | heavy-metal-associated domain-containing protein | -2.83 |
| 134 | 248322_at | AT5G52760 | heavy-metal-associated domain-containing protein | -2.04 |
| 135 | 248048_at | AT5G56080 | nicotianamine synthase, putative | -3.66 |
| 136 | 247949_at | AT5G57220 | CYP81F2 (cytochrome P450, family 81, subfamily F, polypeptide 2); oxygen binding | 3.03 |
| 137 | 247871_at | AT5G57530 | xyloglucan:xyloglucosyl transferase, putative / xyloglucan endotransglycosylase, putative / endo-xyloglucan transferase, putative | 2.69 |
| 138 | 247914_at | AT5G57540 | xyloglucan:xyloglucosyl transferase, putative / xyloglucan endotransglycosylase, putative / endo-xyloglucan transferase, putative | 1.92 |
| 139 | 247215_at | AT5G64905 | PROPEP3 (Elicitor peptide 3 precursor) | 1.80 |
| 140 | 247145_at | AT5G65600 | legume lectin family protein / protein kinase family protein | 3.86 |