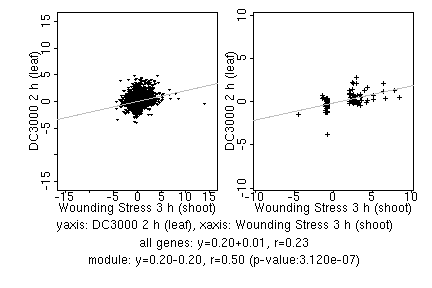
marker gene list
| Wounding Stress3 h (shoot) |
| DC3000 2 h (leaf) |
| probeID | AGICode | Annotation | gene expression |
|
|---|---|---|---|---|
| 1 | 261175_at | AT1G04800 | glycine-rich protein | -0.63 |
| 2 | 264436_at | AT1G10370 | ATGSTU17/ERD9/GST30/GST30B (EARLY-RESPONSIVE TO DEHYDRATION 9); glutathione transferase | 2.69 |
| 3 | 263210_at | AT1G10585 | transcription factor | 4.54 |
| 4 | 265188_at | AT1G23800 | ALDH2B7 (Aldehyde dehydrogenase 2B7); 3-chloroallyl aldehyde dehydrogenase/ aldehyde dehydrogenase (NAD) | 2.64 |
| 5 | 259789_at | AT1G29395 | COR414-TM1 (cold regulated 414 thylakoid membrane 1) | 2.78 |
| 6 | 261596_at | AT1G33080 | MATE efflux family protein | -0.79 |
| 7 | 264415_at | AT1G43160 | RAP2.6 (related to AP2 6); DNA binding / transcription factor | 8.62 |
| 8 | 245749_at | AT1G51090 | heavy-metal-associated domain-containing protein | 3.51 |
| 9 | 262127_at | AT1G52550 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G15780.1) | -0.58 |
| 10 | 259715_at | AT1G60990 | aminomethyltransferase | -0.70 |
| 11 | 255856_at | AT1G66940 | protein kinase-related | -0.92 |
| 12 | 256297_at | AT1G69500 | oxygen binding | -4.33 |
| 13 | 260450_at | AT1G72390 | similar to unnamed protein product [Vitis vinifera] (GB:CAO45587.1) | -1.26 |
| 14 | 262172_at | AT1G74970 | RPS9 (RIBOSOMAL PROTEIN S9); structural constituent of ribosome | -0.75 |
| 15 | 259878_at | AT1G76790 | O-methyltransferase family 2 protein | 2.92 |
| 16 | 261884_at | AT1G80910 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G16020.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G16020.2); similar to unnamed protein product [Vitis vinifera] (GB:CAO42386.1); contains domain UNCHARACTERIZED (PTHR13056) | -0.57 |
| 17 | 266743_at | AT2G02990 | RNS1 (RIBONUCLEASE 1); endoribonuclease | 7.92 |
| 18 | 264787_at | AT2G17840 | ERD7 (EARLY-RESPONSIVE TO DEHYDRATION 7) | 2.71 |
| 19 | 265433_at | AT2G20950 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G38550.1); contains InterPro domain Phospholipase-like, arabidopsis (InterPro:IPR007942) | -0.92 |
| 20 | 266246_at | AT2G27690 | CYP94C1 (cytochrome P450, family 94, subfamily C, polypeptide 1); oxygen binding | 2.54 |
| 21 | 267545_at | AT2G32690 | pseudogene, glycine-rich protein | -0.77 |
| 22 | 267411_at | AT2G34930 | disease resistance family protein | 3.17 |
| 23 | 263908_at | AT2G36480 | zinc finger (C2H2-type) family protein | -0.63 |
| 24 | 267147_at | AT2G38240 | oxidoreductase, 2OG-Fe(II) oxygenase family protein | 6.78 |
| 25 | 266415_at | AT2G38530 | LTP2 (LIPID TRANSFER PROTEIN 2); lipid binding | 3.56 |
| 26 | 263830_at | AT2G40260 | myb family transcription factor | -1.09 |
| 27 | 263497_at | AT2G42540 | COR15A (COLD-REGULATED 15A) | 5.26 |
| 28 | 260551_at | AT2G43510 | ATTI1 (ARABIDOPSIS THALIANA TRYPSIN INHIBITOR PROTEIN 1) | 3.04 |
| 29 | 263783_at | AT2G46400 | WRKY46 (WRKY DNA-binding protein 46); transcription factor | 2.15 |
| 30 | 245123_at | AT2G47450 | CAO (CHAOS); chromatin binding | -0.88 |
| 31 | 266483_at | AT2G47910 | CRR6 (CHLORORESPIRATORY REDUCTION 6) | -0.72 |
| 32 | 259173_at | AT3G03640 | GLUC (Beta-glucosidase homolog); hydrolase, hydrolyzing O-glycosyl compounds | 2.32 |
| 33 | 258805_at | AT3G04010 | glycosyl hydrolase family 17 protein | 2.62 |
| 34 | 258893_at | AT3G05660 | kinase/ protein binding | 2.16 |
| 35 | 258554_at | AT3G06980 | DEAD/DEAH box helicase, putative | -0.74 |
| 36 | 258708_at | AT3G09580 | amine oxidase family protein | -0.93 |
| 37 | 256281_at | AT3G12560 | TRFL9 (TRF-LIKE 9); DNA binding | -0.67 |
| 38 | 256245_at | AT3G12580 | HSP70 (heat shock protein 70); ATP binding | 2.58 |
| 39 | 257547_at | AT3G13000 | transcription factor | -0.60 |
| 40 | 257028_at | AT3G19230 | leucine-rich repeat family protein | -0.58 |
| 41 | 256833_at | AT3G22910 | calcium-transporting ATPase, plasma membrane-type, putative / Ca(2+)-ATPase, putative (ACA13) | 2.71 |
| 42 | 257171_at | AT3G23760 | similar to transferase, transferring glycosyl groups [Arabidopsis thaliana] (TAIR:AT4G14100.1); similar to unknown [Populus trichocarpa] (GB:ABK94230.1) | -0.69 |
| 43 | 257644_at | AT3G25780 | AOC3 (ALLENE OXIDE CYCLASE 3) | 2.63 |
| 44 | 256577_at | AT3G28220 | meprin and TRAF homology domain-containing protein / MATH domain-containing protein | 2.35 |
| 45 | 252417_at | AT3G47480 | calcium-binding EF hand family protein | 3.54 |
| 46 | 252215_at | AT3G50240 | KICP-02; microtubule motor | -0.64 |
| 47 | 252102_at | AT3G50970 | LTI30 (LOW TEMPERATURE-INDUCED 30) | 5.25 |
| 48 | 252114_at | AT3G51450 | strictosidine synthase family protein | 2.61 |
| 49 | 246308_at | AT3G51820 | ATG4/CHLG/G4 (CHLOROPHYLL SYNTHASE); chlorophyll synthetase | -0.57 |
| 50 | 251864_at | AT3G54920 | PMR6 (POWDERY MILDEW RESISTANT 6); lyase/ pectate lyase | -0.65 |
| 51 | 251770_at | AT3G55970 | oxidoreductase, 2OG-Fe(II) oxygenase family protein | 6.60 |
| 52 | 246294_at | AT3G56910 | PSRP5 (PLASTID-SPECIFIC 50S RIBOSOMAL PROTEIN 5) | -0.58 |
| 53 | 251575_at | AT3G58120 | bZIP transcription factor family protein | -0.65 |
| 54 | 251513_at | AT3G59220 | PRN (PIRIN); calmodulin binding | 4.10 |
| 55 | 251400_at | AT3G60420 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G60450.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO70569.1); contains InterPro domain Phosphoglycerate mutase (InterPro:IPR013078); contains InterPro domain PRIB5 (InterPro:IPR012398) | 4.46 |
| 56 | 251262_at | AT3G62080 | SNF7 family protein | -0.66 |
| 57 | 255513_at | AT4G02060 | PRL (PROLIFERA); ATP binding / DNA binding / DNA-dependent ATPase | -0.67 |
| 58 | 245329_at | AT4G14365 | zinc finger (C3HC4-type RING finger) family protein / ankyrin repeat family protein | 2.97 |
| 59 | 245584_at | AT4G14940 | ATAO1 (Arabidosis thaliana amine oxidase 1); copper ion binding | -0.74 |
| 60 | 245275_at | AT4G15210 | ATBETA-AMY (BETA-AMYLASE); beta-amylase | 3.74 |
| 61 | 245291_at | AT4G16155 | dihydrolipoamide dehydrogenase 2, plastidic / lipoamide dehydrogenase 2 (PTLPD2) | -0.64 |
| 62 | 245422_at | AT4G17470 | palmitoyl protein thioesterase family protein | 3.11 |
| 63 | 245354_at | AT4G17600 | LIL3:1; transcription factor | -0.78 |
| 64 | 254243_at | AT4G23210 | CRK13/HIG1; kinase | 2.65 |
| 65 | 254232_at | AT4G23600 | CORI3 (CORONATINE INDUCED 1, JASMONIC ACID RESPONSIVE 2); transaminase | 3.76 |
| 66 | 254011_at | AT4G26370 | antitermination NusB domain-containing protein | -0.73 |
| 67 | 253788_at | AT4G28680 | tyrosine decarboxylase, putative | -0.69 |
| 68 | 253277_at | AT4G34230 | CAD5 (CINNAMYL ALCOHOL DEHYDROGENASE 5); cinnamyl-alcohol dehydrogenase | 2.72 |
| 69 | 253073_at | AT4G37410 | CYP81F4 (cytochrome P450, family 81, subfamily F, polypeptide 4); oxygen binding | 3.04 |
| 70 | 252989_at | AT4G38420 | SKS9 (SKU5 Similar 9); copper ion binding / oxidoreductase | -0.61 |
| 71 | 250662_at | AT5G07010 | sulfotransferase family protein | 6.61 |
| 72 | 250279_at | AT5G13200 | GRAM domain-containing protein / ABA-responsive protein-related | 2.15 |
| 73 | 250180_at | AT5G14450 | GDSL-motif lipase/hydrolase family protein | -0.58 |
| 74 | 250133_at | AT5G16400 | ATF2/TRXF2 (THIOREDOXIN F2); thiol-disulfide exchange intermediate | -0.60 |
| 75 | 250061_at | AT5G17710 | EMB1241 (EMBRYO DEFECTIVE 1241); adenyl-nucleotide exchange factor/ chaperone binding / protein binding / protein homodimerization | -0.58 |
| 76 | 246004_at | AT5G20630 | GLP3 (GERMIN-LIKE PROTEIN 3); manganese ion binding / metal ion binding / nutrient reservoir | -0.59 |
| 77 | 249836_at | AT5G23420 | HMGB6 (High mobility group B 6); transcription factor | -0.66 |
| 78 | 249656_at | AT5G37130 | binding | -0.62 |
| 79 | 249417_at | AT5G39670 | calcium-binding EF hand family protein | 2.94 |
| 80 | 248812_at | AT5G47330 | palmitoyl protein thioesterase family protein | 3.34 |
| 81 | 248686_at | AT5G48540 | 33 kDa secretory protein-related | 2.20 |
| 82 | 248597_at | AT5G49160 | MET1 (DECREASED METHYLATION 2DNA) | -0.77 |
| 83 | 248337_at | AT5G52310 | COR78 (COLD REGULATED 78) | 4.35 |
| 84 | 248332_at | AT5G52640 | HSP81-1 (HEAT SHOCK PROTEIN 81-1); ATP binding / unfolded protein binding | 2.24 |
| 85 | 248118_at | AT5G55050 | GDSL-motif lipase/hydrolase family protein | 3.18 |
| 86 | 247478_at | AT5G62360 | invertase/pectin methylesterase inhibitor family protein | 3.54 |
| 87 | 247386_at | AT5G63420 | EMB2746 (EMBRYO DEFECTIVE 2746); catalytic | -0.84 |
| 88 | 247340_at | AT5G63700 | zinc finger (C3HC4 type RING finger) family protein | -0.59 |
| 89 | 247109_at | AT5G65870 | ATPSK5 (PHYTOSULFOKINE 5 PRECURSOR); growth factor | 2.88 |
| 90 | 244973_at | ATCG00690 | Encodes photosystem II 5 kD protein subunit PSII-T. This is a plastid-encoded gene (PsbTc) which also has a nuclear-encoded paralog (PsbTn). | -0.78 |
| 91 | 244985_at | ATCG00810 | encodes a chloroplast ribosomal protein L22, a constituent of the large subunit of the ribosomal complex | -0.81 |
| 92 | 244938_at | ATCG01120 | encodes a chloroplast ribosomal protein S15, a constituent of the small subunit of the ribosomal complex | -1.32 |