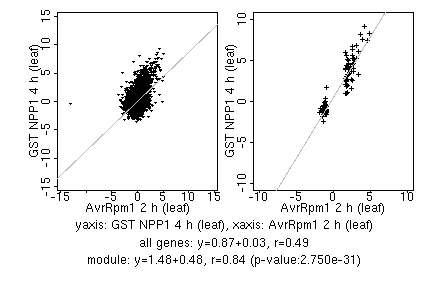
marker gene list
| AvrRpm1 2 h (leaf) |
| GST NPP1 4 h (leaf) |
| probeID | AGICode | Annotation | gene expression |
|
|---|---|---|---|---|
| 1 | 264574_at | AT1G05300 | ZIP5 (ZINC TRANSPORTER 5 PRECURSOR); cation transmembrane transporter | 2.54 |
| 2 | 260815_at | AT1G06950 | ATTIC110/TIC110 (TRANSLOCON AT THE INNER ENVELOPE MEMBRANE OF CHLOROPLASTS 110) | -1.02 |
| 3 | 256044_at | AT1G07160 | protein phosphatase 2C, putative / PP2C, putative | 1.98 |
| 4 | 263209_at | AT1G10522 | similar to unnamed protein product [Vitis vinifera] (GB:CAO40945.1) | -1.32 |
| 5 | 261198_at | AT1G12940 | ATNRT2.5 (NITRATE TRANSPORTER2.5); nitrate transmembrane transporter | 2.03 |
| 6 | 261474_at | AT1G14540 | anionic peroxidase, putative | 2.92 |
| 7 | 261836_at | AT1G16090 | WAKL7 (WALL ASSOCIATED KINASE-LIKE 7) | 2.22 |
| 8 | 260655_at | AT1G19320 | pathogenesis-related thaumatin family protein | 2.01 |
| 9 | 261449_at | AT1G21120 | O-methyltransferase, putative | 1.95 |
| 10 | 261937_at | AT1G22570 | proton-dependent oligopeptide transport (POT) family protein | -1.20 |
| 11 | 261021_at | AT1G26380 | FAD-binding domain-containing protein | 4.37 |
| 12 | 261005_at | AT1G26420 | FAD-binding domain-containing protein | 4.02 |
| 13 | 263228_at | AT1G30700 | FAD-binding domain-containing protein | 5.04 |
| 14 | 261191_at | AT1G32900 | starch synthase, putative | -1.20 |
| 15 | 261240_at | AT1G32940 | ATSBT3.5; subtilase | 1.85 |
| 16 | 246368_at | AT1G51890 | leucine-rich repeat protein kinase, putative | 3.02 |
| 17 | 265008_at | AT1G61560 | MLO6 (MILDEW RESISTANCE LOCUS O 6); calmodulin binding | 1.76 |
| 18 | 260005_at | AT1G67920 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G24600.1) | 2.23 |
| 19 | 262232_at | AT1G68600 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G25480.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G17470.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO42118.1); contains InterPro domain Protein of unknown function UPF0005 (InterPro:IPR006214) | -0.77 |
| 20 | 260362_at | AT1G70530 | protein kinase family protein | 1.94 |
| 21 | 256341_at | AT1G72040 | deoxynucleoside kinase family | -0.94 |
| 22 | 245777_at | AT1G73540 | ATNUDT21 (Arabidopsis thaliana Nudix hydrolase homolog 21); hydrolase | 1.98 |
| 23 | 262133_at | AT1G78000 | SULTR1;2 (SULFATE TRANSPORTER 1;2); sulfate transmembrane transporter | 1.79 |
| 24 | 260804_at | AT1G78410 | VQ motif-containing protein | 2.22 |
| 25 | 261899_at | AT1G80820 | CCR2 (CINNAMOYL COA REDUCTASE) | 2.18 |
| 26 | 265895_at | AT2G15000 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G34265.2); similar to unnamed protein product [Vitis vinifera] (GB:CAO70855.1) | -0.99 |
| 27 | 265475_at | AT2G15620 | NIR1 (NITRITE REDUCTASE); ferredoxin-nitrate reductase | -1.30 |
| 28 | 264616_at | AT2G17740 | DC1 domain-containing protein | 3.50 |
| 29 | 267436_at | AT2G19190 | FRK1 (FLG22-INDUCED RECEPTOR-LIKE KINASE 1); kinase | 3.33 |
| 30 | 265596_at | AT2G20020 | Identical to CRS2-associated factor 1, chloroplast precursor [Arabidopsis Thaliana] (GB:Q9SL79;GB:Q94AQ2); similar to ATCAF2/CAF2 (ARABIDOPSIS THALIANA HOMOLOG OF MAIZE CAF2) [Arabidopsis thaliana] (TAIR:AT1G23400.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO65472.1); contains InterPro domain CRS1/YhbY (InterPro:IPR001890) | -1.04 |
| 31 | 265306_at | AT2G20320 | DENN (AEX-3) domain-containing protein | 1.79 |
| 32 | 263411_at | AT2G28710 | zinc finger (C2H2 type) family protein | 2.68 |
| 33 | 266270_at | AT2G29470 | ATGSTU3 (GLUTATHIONE S-TRANSFERASE 21); glutathione transferase | 2.43 |
| 34 | 265725_at | AT2G32030 | GCN5-related N-acetyltransferase (GNAT) family protein | 2.53 |
| 35 | 263948_at | AT2G35980 | YLS9 (YELLOW-LEAF-SPECIFIC GENE 9) | 3.31 |
| 36 | 263904_at | AT2G36380 | ATPDR6/PDR6 (PLEIOTROPIC DRUG RESISTANCE 6); ATPase, coupled to transmembrane movement of substances | -0.98 |
| 37 | 265208_at | AT2G36690 | oxidoreductase, 2OG-Fe(II) oxygenase family protein | 2.01 |
| 38 | 267174_at | AT2G37600 | 60S ribosomal protein L36 (RPL36A) | -1.00 |
| 39 | 266975_at | AT2G39380 | ATEXO70H2 (EXOCYST SUBUNIT EXO70 FAMILY PROTEIN H2); protein binding | 2.81 |
| 40 | 267562_at | AT2G39670 | radical SAM domain-containing protein | -1.36 |
| 41 | 267361_at | AT2G39920 | acid phosphatase class B family protein | -0.90 |
| 42 | 267384_at | AT2G44370 | DC1 domain-containing protein | 3.78 |
| 43 | 245148_at | AT2G45220 | pectinesterase family protein | 4.73 |
| 44 | 263783_at | AT2G46400 | WRKY46 (WRKY DNA-binding protein 46); transcription factor | 1.85 |
| 45 | 258483_at | AT3G02570 | MEE31 (maternal effect embryo arrest 31); mannose-6-phosphate isomerase | -0.91 |
| 46 | 259172_at | AT3G03630 | CS26 (cysteine synthase 26); cysteine synthase | -1.34 |
| 47 | 258534_at | AT3G06730 | thioredoxin family protein | -1.07 |
| 48 | 258696_at | AT3G09650 | HCF152 (HIGH CHLOROPHYLL FLUORESCENCE 152) | -0.85 |
| 49 | 257690_at | AT3G12830 | auxin-responsive family protein | 1.75 |
| 50 | 257856_at | AT3G12930 | similar to unknown [Populus trichocarpa] (GB:ABK94112.1); contains InterPro domain Iojap-related protein (InterPro:IPR004394) | -1.16 |
| 51 | 258252_at | AT3G15720 | glycoside hydrolase family 28 protein / polygalacturonase (pectinase) family protein | -1.26 |
| 52 | 258161_at | AT3G17930 | similar to unknown [Populus trichocarpa] (GB:ABK96142.1) | -0.82 |
| 53 | 257756_at | AT3G18680 | aspartate/glutamate/uridylate kinase family protein | -0.96 |
| 54 | 258025_at | AT3G19480 | D-3-phosphoglycerate dehydrogenase, putative / 3-PGDH, putative | -0.84 |
| 55 | 257964_at | AT3G19850 | phototropic-responsive NPH3 family protein | -0.82 |
| 56 | 257171_at | AT3G23760 | similar to transferase, transferring glycosyl groups [Arabidopsis thaliana] (TAIR:AT4G14100.1); similar to unknown [Populus trichocarpa] (GB:ABK94230.1) | -1.40 |
| 57 | 256862_at | AT3G23940 | dehydratase family | -1.03 |
| 58 | 257903_at | AT3G28460 | similar to hypothetical protein [Vitis vinifera] (GB:CAN74970.1); contains InterPro domain Protein of unknown function methylase putative (InterPro:IPR016065) | -1.17 |
| 59 | 252739_at | AT3G43250 | cell cycle control protein-related | 2.02 |
| 60 | 252612_at | AT3G45160 | contains domain PROKAR_LIPOPROTEIN (PS51257) | -0.83 |
| 61 | 252433_at | AT3G47560 | esterase/lipase/thioesterase family protein | -1.17 |
| 62 | 252362_at | AT3G48500 | PDE312/PTAC10 (PIGMENT DEFECTIVE 312); RNA binding / unfolded protein binding | -0.79 |
| 63 | 252346_at | AT3G48650 | pseudogene, At14a-related protein, similar to At14a (GI:11994571 and GI:11994573) (Arabidopsis thaliana) | 2.84 |
| 64 | 252245_at | AT3G49710 | pentatricopeptide (PPR) repeat-containing protein | -0.84 |
| 65 | 256678_at | AT3G52380 | CP33 (PIGMENT DEFECTIVE 322); RNA binding | -1.04 |
| 66 | 251958_at | AT3G53560 | chloroplast lumen common family protein | -0.94 |
| 67 | 251935_at | AT3G54090 | pfkB-type carbohydrate kinase family protein | -1.02 |
| 68 | 251832_at | AT3G55150 | ATEXO70H1 (exocyst subunit EXO70 family protein H1); protein binding | 3.33 |
| 69 | 251248_at | AT3G62150 | PGP21 (P-GLYCOPROTEIN 21); ATPase, coupled to transmembrane movement of substances | 1.77 |
| 70 | 255595_at | AT4G01700 | chitinase, putative | 2.33 |
| 71 | 255465_at | AT4G02990 | mitochondrial transcription termination factor family protein / mTERF family protein | -0.87 |
| 72 | 254869_at | AT4G11890 | protein kinase family protein | 2.06 |
| 73 | 254819_at | AT4G12500 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | 2.79 |
| 74 | 254755_at | AT4G13220 | similar to hypothetical protein [Vitis vinifera] (GB:CAN75300.1) | -0.78 |
| 75 | 245494_at | AT4G16390 | chloroplastic RNA-binding protein P67, putative | -0.95 |
| 76 | 245354_at | AT4G17600 | LIL3:1; transcription factor | -0.82 |
| 77 | 254645_at | AT4G18520 | binding | -0.83 |
| 78 | 254502_at | AT4G20130 | PTAC14 (PLASTID TRANSCRIPTIONALLY ACTIVE14) | -0.98 |
| 79 | 254372_at | AT4G21620 | glycine-rich protein | -0.82 |
| 80 | 254271_at | AT4G23150 | protein kinase family protein | 1.86 |
| 81 | 253956_at | AT4G26700 | ATFIM1 (Arabidopsis thaliana fimbrin 1); actin binding | -0.82 |
| 82 | 253607_at | AT4G30330 | small nuclear ribonucleoprotein E, putative / snRNP-E, putative / Sm protein E, putative | -1.29 |
| 83 | 253638_at | AT4G30470 | cinnamoyl-CoA reductase-related | -0.89 |
| 84 | 253286_at | AT4G34260 | catalytic | -0.81 |
| 85 | 252908_at | AT4G39670 | glycolipid binding / glycolipid transporter | 2.79 |
| 86 | 250910_at | AT5G03720 | AT-HSFA3 (Arabidopsis thaliana heat shock transcription factor A3); DNA binding / transcription factor | -1.11 |
| 87 | 250529_at | AT5G08610 | DEAD box RNA helicase (RH26) | -0.82 |
| 88 | 245209_at | AT5G12340 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G28190.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO16848.1) | -0.80 |
| 89 | 246492_at | AT5G16140 | peptidyl-tRNA hydrolase family protein | -1.15 |
| 90 | 250018_at | AT5G18150 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G14602.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO23480.1); contains domain PTHR12133 (PTHR12133) | 2.25 |
| 91 | 249867_at | AT5G23020 | IMS2/MAM-L/MAM3 (METHYLTHIOALKYMALATE SYNTHASE-LIKE); 2-isopropylmalate synthase/ methylthioalkylmalate synthase | -1.55 |
| 92 | 249826_at | AT5G23310 | FSD3 (FE SUPEROXIDE DISMUTASE 3); iron superoxide dismutase | -1.68 |
| 93 | 249763_at | AT5G24010 | protein kinase family protein | -1.26 |
| 94 | 246821_at | AT5G26920 | calmodulin binding | 2.87 |
| 95 | 246777_at | AT5G27420 | zinc finger (C3HC4-type RING finger) family protein | 1.80 |
| 96 | 249727_at | AT5G35490 | unknown protein | -0.96 |
| 97 | 249354_at | AT5G40480 | EMB3012 (EMBRYO DEFECTIVE 3012) | -1.26 |
| 98 | 249197_at | AT5G42380 | CML37/CML39; calcium ion binding | 2.72 |
| 99 | 249188_at | AT5G42830 | transferase family protein | 2.27 |
| 100 | 249051_at | AT5G44390 | FAD-binding domain-containing protein | 1.92 |
| 101 | 248709_at | AT5G48470 | similar to unnamed protein product [Vitis vinifera] (GB:CAO17042.1) | -1.15 |
| 102 | 248448_at | AT5G51190 | AP2 domain-containing transcription factor, putative | 2.07 |
| 103 | 248327_at | AT5G52750 | heavy-metal-associated domain-containing protein | 1.78 |
| 104 | 248322_at | AT5G52760 | heavy-metal-associated domain-containing protein | 2.59 |
| 105 | 247949_at | AT5G57220 | CYP81F2 (cytochrome P450, family 81, subfamily F, polypeptide 2); oxygen binding | 2.67 |
| 106 | 247814_at | AT5G58310 | hydrolase, alpha/beta fold family protein | -1.30 |
| 107 | 247655_at | AT5G59820 | RHL41 (RESPONSIVE TO HIGH LIGHT 41); nucleic acid binding / transcription factor/ zinc ion binding | 1.89 |
| 108 | 247543_at | AT5G61600 | ethylene-responsive element-binding family protein | 1.86 |
| 109 | 247240_at | AT5G64660 | U-box domain-containing protein | 1.78 |
| 110 | 247214_at | AT5G64850 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G09960.1); similar to 80C09_15 [Brassica rapa subsp. pekinensis] (GB:AAZ41826.1) | -1.09 |
| 111 | 247215_at | AT5G64905 | PROPEP3 (Elicitor peptide 3 precursor) | 2.71 |
| 112 | 246993_at | AT5G67450 | AZF1 (ARABIDOPSIS ZINC-FINGER PROTEIN 1); nucleic acid binding / transcription factor/ zinc ion binding | 3.49 |