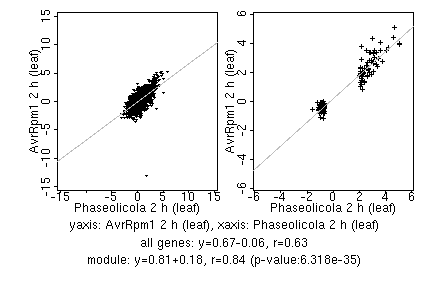
marker gene list
| Phaseolicola 2 h (leaf) |
| AvrRpm1 2 h (leaf) |
| probeID | AGICode | Annotation | gene expression |
|
|---|---|---|---|---|
| 1 | 261055_at | AT1G01300 | aspartyl protease family protein | -0.76 |
| 2 | 259443_at | AT1G02360 | chitinase, putative | 2.20 |
| 3 | 256044_at | AT1G07160 | protein phosphatase 2C, putative / PP2C, putative | 2.83 |
| 4 | 264348_at | AT1G12110 | NRT1.1 (NITRATE TRANSPORTER 1.1); transporter | -1.01 |
| 5 | 261474_at | AT1G14540 | anionic peroxidase, putative | 3.52 |
| 6 | 262598_at | AT1G15260 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G16070.1) | -1.13 |
| 7 | 261836_at | AT1G16090 | WAKL7 (WALL ASSOCIATED KINASE-LIKE 7) | 2.69 |
| 8 | 255899_at | AT1G17970 | zinc finger (C3HC4-type RING finger) family protein | -0.59 |
| 9 | 261937_at | AT1G22570 | proton-dependent oligopeptide transport (POT) family protein | -0.69 |
| 10 | 261021_at | AT1G26380 | FAD-binding domain-containing protein | 4.64 |
| 11 | 261005_at | AT1G26420 | FAD-binding domain-containing protein | 3.69 |
| 12 | 259775_at | AT1G29530 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G34310.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G34310.2); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G34310.3) | -1.00 |
| 13 | 263228_at | AT1G30700 | FAD-binding domain-containing protein | 4.76 |
| 14 | 256466_at | AT1G32580 | plastid developmental protein DAG, putative | -0.59 |
| 15 | 261594_at | AT1G33240 | AT-GTL1 (Arabidopsis thaliana GT2-like 1); transcription factor | -0.91 |
| 16 | 256447_at | AT1G33440 | proton-dependent oligopeptide transport (POT) family protein | -0.62 |
| 17 | 246368_at | AT1G51890 | leucine-rich repeat protein kinase, putative | 3.25 |
| 18 | 246370_at | AT1G51920 | unknown protein | 2.97 |
| 19 | 260645_at | AT1G53300 | TTL1 (TETRATRICOPETIDE-REPEAT THIOREDOXIN-LIKE 1); binding | -0.64 |
| 20 | 264757_at | AT1G61360 | S-locus lectin protein kinase family protein | 2.34 |
| 21 | 265008_at | AT1G61560 | MLO6 (MILDEW RESISTANCE LOCUS O 6); calmodulin binding | 2.67 |
| 22 | 264424_at | AT1G61740 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G11540.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO38814.1); contains InterPro domain Protein of unknown function DUF81; (InterPro:IPR002781) | -0.78 |
| 23 | 264746_at | AT1G62300 | WRKY6 (WRKY DNA-binding protein 6); transcription factor | 2.72 |
| 24 | 262871_at | AT1G65010 | hydrogen ion transmembrane transporter/ protein binding | -0.89 |
| 25 | 262930_at | AT1G65690 | harpin-induced protein-related / HIN1-related / harpin-responsive protein-related | 3.38 |
| 26 | 260005_at | AT1G67920 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G24600.1) | 2.73 |
| 27 | 260347_at | AT1G69420 | zinc finger (DHHC type) family protein | -0.82 |
| 28 | 264700_at | AT1G70100 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G24160.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO40971.1) | -0.77 |
| 29 | 264315_at | AT1G70370 | BURP domain-containing protein / polygalacturonase, putative | -0.98 |
| 30 | 260048_at | AT1G73750 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G15060.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO42199.1); contains InterPro domain Uncharacterised conserved protein UCP031088, alpha/beta hydrolase, At1g15070 (InterPro:IPR016969) | -0.71 |
| 31 | 260390_at | AT1G73940 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G49410.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G49410.2); similar to unnamed protein product [Vitis vinifera] (GB:CAO62865.1) | -0.71 |
| 32 | 262170_at | AT1G74940 | senescence-associated protein-related | -0.94 |
| 33 | 261772_at | AT1G76240 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G17080.1); similar to Os06g0725500 [Oryza sativa (japonica cultivar-group)] (GB:NP_001058623.1); similar to hypothetical protein OsI_023643 [Oryza sativa (indica cultivar-group)] (GB:EAZ02411.1); contains InterPro domain Protein of unknown function DUF241, plant (InterPro:IPR004320) | -0.96 |
| 34 | 264960_at | AT1G76930 | ATEXT4 (EXTENSIN 4) | 3.17 |
| 35 | 260804_at | AT1G78410 | VQ motif-containing protein | 2.47 |
| 36 | 261899_at | AT1G80820 | CCR2 (CINNAMOYL COA REDUCTASE) | 2.79 |
| 37 | 266331_at | AT2G01570 | RGA1 (REPRESSOR OF GA1-3 1); transcription factor | -0.75 |
| 38 | 264616_at | AT2G17740 | DC1 domain-containing protein | 3.23 |
| 39 | 267436_at | AT2G19190 | FRK1 (FLG22-INDUCED RECEPTOR-LIKE KINASE 1); kinase | 3.39 |
| 40 | 267136_at | AT2G23460 | XLG1 (EXTRA-LARGE G-PROTEIN 1); signal transducer | -0.64 |
| 41 | 266279_at | AT2G29290 | tropinone reductase, putative / tropine dehydrogenase, putative | -1.02 |
| 42 | 266270_at | AT2G29470 | ATGSTU3 (GLUTATHIONE S-TRANSFERASE 21); glutathione transferase | 2.62 |
| 43 | 266337_at | AT2G32390 | ATGLR3.5 (GLUTAMATE RECEPTOR 6) | -0.61 |
| 44 | 267116_at | AT2G32560 | F-box family protein | -0.58 |
| 45 | 263948_at | AT2G35980 | YLS9 (YELLOW-LEAF-SPECIFIC GENE 9) | 2.80 |
| 46 | 265208_at | AT2G36690 | oxidoreductase, 2OG-Fe(II) oxygenase family protein | 2.15 |
| 47 | 266010_at | AT2G37430 | zinc finger (C2H2 type) family protein (ZAT11) | 3.17 |
| 48 | 266975_at | AT2G39380 | ATEXO70H2 (EXOCYST SUBUNIT EXO70 FAMILY PROTEIN H2); protein binding | 3.01 |
| 49 | 263378_at | AT2G40180 | ATHPP2C5; protein serine/threonine phosphatase | 4.29 |
| 50 | 267384_at | AT2G44370 | DC1 domain-containing protein | 2.21 |
| 51 | 266131_at | AT2G45160 | scarecrow transcription factor family protein | -0.77 |
| 52 | 266896_at | AT2G45810 | DEAD/DEAH box helicase, putative | -0.63 |
| 53 | 266486_at | AT2G47950 | unknown protein | 3.14 |
| 54 | 265771_at | AT2G48030 | endonuclease/exonuclease/phosphatase family protein | -0.77 |
| 55 | 258975_at | AT3G01970 | WRKY45 (WRKY DNA-binding protein 45); transcription factor | 3.43 |
| 56 | 258606_at | AT3G02840 | immediate-early fungal elicitor family protein | 2.08 |
| 57 | 259033_at | AT3G09410 | pectinacetylesterase family protein | 2.39 |
| 58 | 258368_at | AT3G14240 | subtilase family protein | -1.05 |
| 59 | 258123_at | AT3G18040 | MPK9 (MAP KINASE 9); MAP kinase | -0.74 |
| 60 | 257964_at | AT3G19850 | phototropic-responsive NPH3 family protein | -0.84 |
| 61 | 256968_at | AT3G21070 | NADK1 (NAD kinase 1); NAD+ kinase/ NADH kinase/ calmodulin binding | 2.27 |
| 62 | 257540_at | AT3G21520 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G21550.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO71058.1); contains InterPro domain Protein of unknown function DUF679 (InterPro:IPR007770) | 2.46 |
| 63 | 258277_at | AT3G26830 | PAD3 (PHYTOALEXIN DEFICIENT 3); oxygen binding | 2.27 |
| 64 | 258244_at | AT3G27770 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G62960.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO14740.1) | -0.59 |
| 65 | 252737_at | AT3G43220 | phosphoinositide phosphatase family protein | -0.74 |
| 66 | 252443_at | AT3G46960 | ATP-dependent helicase | -0.71 |
| 67 | 252346_at | AT3G48650 | pseudogene, At14a-related protein, similar to At14a (GI:11994571 and GI:11994573) (Arabidopsis thaliana) | 2.20 |
| 68 | 252255_at | AT3G49220 | pectinesterase family protein | -0.68 |
| 69 | 252272_at | AT3G49670 | BAM2 (big apical meristem 2); ATP binding / protein serine/threonine kinase | -1.02 |
| 70 | 252098_at | AT3G51330 | aspartyl protease family protein | 2.13 |
| 71 | 251950_at | AT3G53600 | zinc finger (C2H2 type) family protein | 2.99 |
| 72 | 251832_at | AT3G55150 | ATEXO70H1 (exocyst subunit EXO70 family protein H1); protein binding | 2.92 |
| 73 | 251790_at | AT3G55470 | C2 domain-containing protein | 2.30 |
| 74 | 251479_at | AT3G59700 | ATHLECRK (ARABIDOPSIS THALIANA LECTIN-RECEPTOR KINASE); kinase | 2.19 |
| 75 | 255595_at | AT4G01700 | chitinase, putative | 2.13 |
| 76 | 245592_at | AT4G14540 | CCAAT-box binding transcription factor subunit B (NF-YB) (HAP3 ) (AHAP3) family | -0.83 |
| 77 | 245460_at | AT4G16990 | disease resistance protein (TIR-NBS class), putative | -0.97 |
| 78 | 254693_at | AT4G17880 | basic helix-loop-helix (bHLH) family protein | -0.97 |
| 79 | 254658_at | AT4G18230 | similar to unnamed protein product [Vitis vinifera] (GB:CAO66912.1); contains InterPro domain Oligosaccharide biosynthesis protein Alg14 like (InterPro:IPR013969) | -0.62 |
| 80 | 254660_at | AT4G18250 | receptor serine/threonine kinase, putative | 2.14 |
| 81 | 254645_at | AT4G18520 | binding | -0.59 |
| 82 | 254649_at | AT4G18570 | proline-rich family protein | -0.78 |
| 83 | 254564_at | AT4G19170 | NCED4 (NINE-CIS-EPOXYCAROTENOID DIOXYGENASE 4) | -0.71 |
| 84 | 254575_at | AT4G19460 | glycosyl transferase family 1 protein | 2.25 |
| 85 | 254543_at | AT4G19810 | glycosyl hydrolase family 18 protein | 2.19 |
| 86 | 253855_at | AT4G28050 | TET7 (TETRASPANIN7) | -0.69 |
| 87 | 253215_at | AT4G34950 | nodulin family protein | -0.74 |
| 88 | 252908_at | AT4G39670 | glycolipid binding / glycolipid transporter | 2.87 |
| 89 | 251097_at | AT5G01560 | lectin protein kinase, putative | 2.27 |
| 90 | 250983_at | AT5G02780 | In2-1 protein, putative | 5.14 |
| 91 | 250918_at | AT5G03610 | GDSL-motif lipase/hydrolase family protein | 2.26 |
| 92 | 250649_at | AT5G06690 | (THIOREDOXIN-LIKE 5); thiol-disulfide exchange intermediate | -1.00 |
| 93 | 250503_at | AT5G09820 | plastid-lipid associated protein PAP / fibrillin family protein | -0.93 |
| 94 | 245906_at | AT5G11070 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G35090.1) | -0.75 |
| 95 | 250368_at | AT5G11280 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G80200.1); similar to unknown [Populus trichocarpa] (GB:ABK96125.1) | -0.61 |
| 96 | 250267_at | AT5G12930 | similar to unnamed protein product [Vitis vinifera] (GB:CAO21945.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN80883.1) | 3.70 |
| 97 | 245976_at | AT5G13080 | WRKY75 (WRKY DNA-BINDING PROTEIN 75); transcription factor | 4.10 |
| 98 | 246549_at | AT5G15050 | glycosyltransferase family 14 protein / core-2/I-branching enzyme family protein | -0.75 |
| 99 | 250018_at | AT5G18150 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G14602.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO23480.1); contains domain PTHR12133 (PTHR12133) | 2.59 |
| 100 | 250001_at | AT5G18680 | AtTLP11 (TUBBY LIKE PROTEIN 11); phosphoric diester hydrolase/ transcription factor | -0.60 |
| 101 | 249964_at | AT5G19010 | MPK16 (mitogen-activated protein kinase 16); MAP kinase | -0.79 |
| 102 | 249770_at | AT5G24110 | WRKY30 (WRKY DNA-binding protein 30); transcription factor | 5.13 |
| 103 | 249730_at | AT5G24430 | calcium-dependent protein kinase, putative / CDPK, putative | 2.13 |
| 104 | 249750_at | AT5G24570 | unknown protein | -0.83 |
| 105 | 246959_at | AT5G24690 | similar to RER1 (RETICULATA-RELATED 1) [Arabidopsis thaliana] (TAIR:AT5G22790.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO24736.1) | -0.80 |
| 106 | 246902_at | AT5G25640 | similar to rhomboid family protein [Arabidopsis thaliana] (TAIR:AT5G25752.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO44068.1); contains InterPro domain Peptidase S54, rhomboid; (InterPro:IPR002610) | -0.62 |
| 107 | 246821_at | AT5G26920 | calmodulin binding | 2.21 |
| 108 | 246777_at | AT5G27420 | zinc finger (C3HC4-type RING finger) family protein | 2.28 |
| 109 | 246051_at | AT5G28840 | GME (GDP-D-MANNOSE 3',5'-EPIMERASE); GDP-mannose 3,5-epimerase/ NAD binding / catalytic | -0.71 |
| 110 | 246622_at | AT5G36250 | protein phosphatase 2C, putative / PP2C, putative | -0.61 |
| 111 | 249493_at | AT5G39080 | transferase family protein | -1.52 |
| 112 | 249224_at | AT5G42130 | mitochondrial substrate carrier family protein | -0.62 |
| 113 | 249188_at | AT5G42830 | transferase family protein | 2.28 |
| 114 | 248703_at | AT5G48430 | aspartic-type endopeptidase/ pepsin A | 3.09 |
| 115 | 248557_at | AT5G49980 | AFB5 (AUXIN F-BOX PROTEIN 5); ubiquitin-protein ligase | -0.72 |
| 116 | 248502_at | AT5G50450 | zinc finger (MYND type) family protein | -0.71 |
| 117 | 248046_at | AT5G56040 | leucine-rich repeat protein kinase, putative | -0.67 |
| 118 | 247940_at | AT5G57190 | PSD2 (PHOSPHATIDYLSERINE DECARBOXYLASE 2); phosphatidylserine decarboxylase | 2.13 |
| 119 | 247949_at | AT5G57220 | CYP81F2 (cytochrome P450, family 81, subfamily F, polypeptide 2); oxygen binding | 2.65 |
| 120 | 247610_at | AT5G60630 | unknown protein | 2.47 |
| 121 | 247246_at | AT5G64620 | C/VIF2 (CELL WALL / VACUOLAR INHIBITOR OF FRUCTOSIDASE 2); pectinesterase inhibitor | -0.87 |
| 122 | 247215_at | AT5G64905 | PROPEP3 (Elicitor peptide 3 precursor) | 3.84 |
| 123 | 247145_at | AT5G65600 | legume lectin family protein / protein kinase family protein | 3.17 |
| 124 | 246993_at | AT5G67450 | AZF1 (ARABIDOPSIS ZINC-FINGER PROTEIN 1); nucleic acid binding / transcription factor/ zinc ion binding | 2.58 |