marker gene list
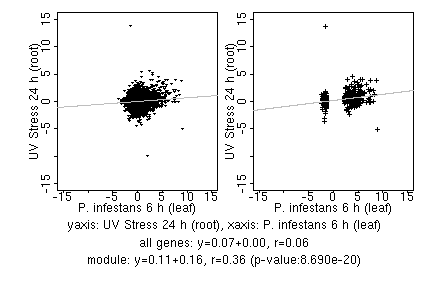
| P. infestans 6 h (leaf) |
| UV Stress 24 h (root) |
| probeID | AGICode | Annotation | gene expression |
|
|---|---|---|---|---|
| 1 | 261027_at | AT1G01340 | ATCNGC10 (CYCLIC NUCLEOTIDE GATED CHANNEL 10); calmodulin binding / cyclic nucleotide binding / ion channel | 2.76 |
| 2 | 259439_at | AT1G01480 | ACS2 (1-Amino-cyclopropane-1-carboxylate synthase 2) | 4.37 |
| 3 | 259445_at | AT1G02400 | ATGA2OX6/DTA1 (GIBBERELLIN 2-OXIDASE 6); gibberellin 2-beta-dioxygenase | 2.58 |
| 4 | 265067_at | AT1G03850 | glutaredoxin family protein | 3.05 |
| 5 | 265042_at | AT1G04040 | acid phosphatase class B family protein | -1.42 |
| 6 | 264580_at | AT1G05340 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G32210.1) | 3.82 |
| 7 | 263179_at | AT1G05710 | ethylene-responsive protein, putative | 2.49 |
| 8 | 260955_at | AT1G06000 | UDP-glucoronosyl/UDP-glucosyl transferase family protein | 2.96 |
| 9 | 260783_at | AT1G06160 | ethylene-responsive factor, putative | 3.39 |
| 10 | 261084_at | AT1G07440 | tropinone reductase, putative / tropine dehydrogenase, putative | -1.41 |
| 11 | 260626_at | AT1G08040 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G28310.2); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G28310.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO21860.1); contains InterPro domain Protein of unknown function DUF707 (InterPro:IPR007877) | -1.39 |
| 12 | 264645_at | AT1G08940 | phosphoglycerate/bisphosphoglycerate mutase family protein | 3.03 |
| 13 | 264648_at | AT1G09080 | BIP3; ATP binding | 4.75 |
| 14 | 264672_at | AT1G09750 | chloroplast nucleoid DNA-binding protein-related | -1.43 |
| 15 | 264663_at | AT1G09970 | leucine-rich repeat transmembrane protein kinase, putative | 2.82 |
| 16 | 264524_at | AT1G10070 | ATBCAT-2; branched-chain-amino-acid transaminase/ catalytic | 4.33 |
| 17 | 264467_at | AT1G10140 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G58420.1); contains InterPro domain Uncharacterised conserved protein UCP031279 (InterPro:IPR016972) | 2.63 |
| 18 | 264435_at | AT1G10360 | ATGSTU18 (GLUTATHIONE S-TRANSFERASE 29); glutathione transferase | -1.27 |
| 19 | 262762_at | AT1G10700 | ribose-phosphate pyrophosphokinase 3 / phosphoribosyl diphosphate synthetase 3 (PRS3) | 3.02 |
| 20 | 262815_at | AT1G11610 | CYP71A18 (cytochrome P450, family 71, subfamily A, polypeptide 18); oxygen binding | 5.75 |
| 21 | 261023_at | AT1G12200 | flavin-containing monooxygenase family protein / FMO family protein | 2.50 |
| 22 | 262826_at | AT1G13080 | CYP71B2 (CYTOCHROME P450 71B2); oxygen binding | -1.42 |
| 23 | 259358_at | AT1G13250 | GATL3 (Galacturonosyltransferase-like 3); polygalacturonate 4-alpha-galacturonosyltransferase/ transferase, transferring glycosyl groups / transferase, transferring hexosyl groups | -1.54 |
| 24 | 256096_at | AT1G13650 | similar to 18S pre-ribosomal assembly protein gar2-related [Arabidopsis thaliana] (TAIR:AT2G03810.4); similar to 18S pre-ribosomal assembly protein gar2-related [Arabidopsis thaliana] (TAIR:AT2G03810.2); similar to 18S pre-ribosomal assembly protein gar2-related [Arabidopsis thaliana] (TAIR:AT2G03810.3); similar to hypothetical protein Tc00.1047053503999.40 [Trypanosoma cruzi strain CL Brener] (GB:XP_813437.1) | -1.72 |
| 25 | 262612_at | AT1G14150 | oxygen evolving enhancer 3 (PsbQ) family protein | -1.29 |
| 26 | 261488_at | AT1G14345 | oxidoreductase | -1.61 |
| 27 | 261474_at | AT1G14540 | anionic peroxidase, putative | 5.92 |
| 28 | 262830_at | AT1G14700 | ATPAP3/PAP3 (purple acid phosphatase 3); acid phosphatase/ protein serine/threonine phosphatase | -1.39 |
| 29 | 262598_at | AT1G15260 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G16070.1) | -1.49 |
| 30 | 262577_at | AT1G15290 | binding | -1.38 |
| 31 | 261763_at | AT1G15520 | ATPDR12/PDR12 (PLEIOTROPIC DRUG RESISTANCE 12); ATPase, coupled to transmembrane movement of substances | 5.38 |
| 32 | 259502_at | AT1G15670 | kelch repeat-containing F-box family protein | 2.73 |
| 33 | 261836_at | AT1G16090 | WAKL7 (WALL ASSOCIATED KINASE-LIKE 7) | 3.54 |
| 34 | 262731_at | AT1G16420 | ATMC8 (METACASPASE 8); caspase | 4.09 |
| 35 | 262536_at | AT1G17100 | SOUL heme-binding family protein | -1.33 |
| 36 | 262518_at | AT1G17170 | ATGSTU24 (ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE (CLASS TAU) 24); glutathione transferase | 3.06 |
| 37 | 261033_at | AT1G17380 | JAZ5/TIFY11A (JASMONATE-ZIM-DOMAIN PROTEIN 5) | 2.70 |
| 38 | 259403_at | AT1G17745 | PGDH (3-PHOSPHOGLYCERATE DEHYDROGENASE); phosphoglycerate dehydrogenase | 4.00 |
| 39 | 255899_at | AT1G17970 | zinc finger (C3HC4-type RING finger) family protein | -1.38 |
| 40 | 256076_at | AT1G18060 | similar to unnamed protein product [Vitis vinifera] (GB:CAO68142.1) | -1.28 |
| 41 | 261663_at | AT1G18330 | EPR1 (EARLY-PHYTOCHROME-RESPONSIVE1); DNA binding / transcription factor | -1.35 |
| 42 | 261661_at | AT1G18360 | hydrolase, alpha/beta fold family protein | -1.35 |
| 43 | 261719_at | AT1G18380 | Identical to Uncharacterized protein At1g18380 precursor [Arabidopsis Thaliana] (GB:Q5BQ05;GB:Q5Q0G6;GB:Q8RX30;GB:Q9LPQ6); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G67025.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO68096.1) | 3.21 |
| 44 | 255753_at | AT1G18570 | MYB51 (MYB DOMAIN PROTEIN 51); DNA binding / transcription factor | 4.00 |
| 45 | 255773_at | AT1G18590 | sulfotransferase family protein | 2.64 |
| 46 | 259479_at | AT1G19020 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G48180.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO40966.1) | 3.44 |
| 47 | 256015_at | AT1G19150 | LHCA6 (Photosystem I light harvesting complex gene 6); chlorophyll binding | -1.35 |
| 48 | 256017_at | AT1G19180 | JAZ1/TIFY10A (JASMONATE-ZIM-DOMAIN PROTEIN 1); protein binding | 3.00 |
| 49 | 261247_at | AT1G20070 | unknown protein | -1.50 |
| 50 | 261449_at | AT1G21120 | O-methyltransferase, putative | 4.26 |
| 51 | 255923_at | AT1G22180 | SEC14 cytosolic factor family protein / phosphoglyceride transfer family protein | 3.03 |
| 52 | 261934_at | AT1G22400 | ATUGT85A1/UGT85A1 (UDP-GLUCOSYL TRANSFERASE 85A1); UDP-glycosyltransferase/ glucuronosyltransferase/ transferase, transferring glycosyl groups / transferase, transferring hexosyl groups | 2.59 |
| 53 | 264201_at | AT1G22630 | heat shock protein binding / unfolded protein binding | -1.52 |
| 54 | 264209_at | AT1G22740 | RAB7 (Ras-related protein 7); GTP binding | -1.25 |
| 55 | 264774_at | AT1G22890 | unknown protein | 2.62 |
| 56 | 264901_at | AT1G23090 | AST91 (SULFATE TRANSPORTER 91); sulfate transmembrane transporter | 2.46 |
| 57 | 264866_at | AT1G24140 | matrixin family protein | 3.35 |
| 58 | 245877_at | AT1G26220 | GCN5-related N-acetyltransferase (GNAT) family protein | -1.27 |
| 59 | 261021_at | AT1G26380 | FAD-binding domain-containing protein | 8.87 |
| 60 | 261020_at | AT1G26390 | FAD-binding domain-containing protein | 4.16 |
| 61 | 261005_at | AT1G26420 | FAD-binding domain-containing protein | 7.61 |
| 62 | 261016_at | AT1G26560 | glycosyl hydrolase family 1 protein | -1.27 |
| 63 | 259592_at | AT1G27950 | lipid transfer protein-related | -1.25 |
| 64 | 259599_at | AT1G28110 | SCPL45; serine carboxypeptidase | -2.16 |
| 65 | 245662_at | AT1G28190 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G12340.1); similar to unknown [Picea sitchensis] (GB:ABK21062.1) | 3.16 |
| 66 | 259787_at | AT1G29460 | auxin-responsive protein, putative | -1.96 |
| 67 | 259773_at | AT1G29500 | auxin-responsive protein, putative | -1.30 |
| 68 | 259783_at | AT1G29510 | SAUR68 (SMALL AUXIN UPREGULATED 68) | -1.90 |
| 69 | 259786_at | AT1G29660 | GDSL-motif lipase/hydrolase family protein | -1.47 |
| 70 | 259788_at | AT1G29670 | GDSL-motif lipase/hydrolase family protein | -1.33 |
| 71 | 246633_at | AT1G29720 | protein kinase family protein | -1.71 |
| 72 | 256159_at | AT1G30135 | JAZ8/TIFY5A (JASMONATE-ZIM-DOMAIN PROTEIN 8) | 5.03 |
| 73 | 245776_at | AT1G30260 | AGL79 (AGAMOUS-LIKE 79) | -1.36 |
| 74 | 245775_at | AT1G30270 | CIPK23 (CBL-INTERACTING PROTEIN KINASE 23); kinase | 2.72 |
| 75 | 261801_at | AT1G30520 | AAE14 (ACYL-ACTIVATING ENZYME 14); AMP binding | -1.79 |
| 76 | 263221_at | AT1G30620 | HSR8/MUR4/UXE1 (MURUS 4); UDP-arabinose 4-epimerase/ catalytic | 3.21 |
| 77 | 263228_at | AT1G30700 | FAD-binding domain-containing protein | 6.59 |
| 78 | 246313_at | AT1G31920 | pentatricopeptide (PPR) repeat-containing protein | -1.70 |
| 79 | 260706_at | AT1G32350 | AOX1D (ALTERNATIVE OXIDASE 1D); alternative oxidase | 5.64 |
| 80 | 260696_at | AT1G32520 | similar to unnamed protein product [Vitis vinifera] (GB:CAO63428.1) | -1.23 |
| 81 | 261242_at | AT1G32960 | ATSBT3.3; subtilase | 4.39 |
| 82 | 261216_at | AT1G33030 | O-methyltransferase family 2 protein | 3.92 |
| 83 | 245755_at | AT1G35210 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G22470.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN82663.1) | 2.81 |
| 84 | 259550_at | AT1G35230 | AGP5 (ARABINOGALACTAN-PROTEIN 5) | 2.55 |
| 85 | 256499_at | AT1G36640 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G36622.1) | 3.20 |
| 86 | 261363_at | AT1G41830 | SKS6 (SKU5 Similar 6); pectinesterase | -1.21 |
| 87 | 260943_at | AT1G45145 | ATTRX5 (thioredoxin H-type 5); thiol-disulfide exchange intermediate | 2.58 |
| 88 | 261638_at | AT1G49975 | similar to hypothetical protein [Vitis vinifera] (GB:CAN66219.1) | -1.33 |
| 89 | 245743_at | AT1G51080 | similar to unnamed protein product [Vitis vinifera] (GB:CAO41687.1) | -1.46 |
| 90 | 260490_at | AT1G51500 | CER5 (ECERIFERUM 5); ATPase, coupled to transmembrane movement of substances | -1.21 |
| 91 | 246368_at | AT1G51890 | leucine-rich repeat protein kinase, putative | 4.17 |
| 92 | 246370_at | AT1G51920 | unknown protein | 4.60 |
| 93 | 246371_at | AT1G51940 | protein kinase family protein / peptidoglycan-binding LysM domain-containing protein | -1.31 |
| 94 | 263198_at | AT1G53990 | GLIP3 (GDSL-motif lipase 3); carboxylesterase/ lipase | 4.45 |
| 95 | 264191_at | AT1G54730 | sugar transporter, putative | -1.28 |
| 96 | 264240_at | AT1G54820 | protein kinase family protein | -1.49 |
| 97 | 259658_at | AT1G55370 | carbohydrate binding / catalytic | -1.33 |
| 98 | 262085_at | AT1G56060 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G32190.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO68639.1); contains domain PD188784 (PD188784) | 3.27 |
| 99 | 245657_at | AT1G56720 | protein kinase family protein | -1.42 |
| 100 | 246405_at | AT1G57630 | disease resistance protein (TIR class), putative | 7.55 |
| 101 | 262903_at | AT1G59950 | aldo/keto reductase, putative | 4.25 |
| 102 | 264963_at | AT1G60600 | ABC4 (ABERRANT CHLOROPLAST DEVELOPMENT 4); 1,4-dihydroxy-2-naphthoate octaprenyltransferase/ prenyltransferase | -1.30 |
| 103 | 264929_at | AT1G60730 | aldo/keto reductase family protein | 3.87 |
| 104 | 264400_at | AT1G61800 | GPT2 (glucose-6-phosphate/phosphate translocator 2); antiporter/ glucose-6-phosphate transmembrane transporter | 6.28 |
| 105 | 264433_at | AT1G61810 | BGLU45; hydrolase, hydrolyzing O-glycosyl compounds | 5.57 |
| 106 | 264746_at | AT1G62300 | WRKY6 (WRKY DNA-binding protein 6); transcription factor | 3.37 |
| 107 | 260243_at | AT1G63720 | similar to hydroxyproline-rich glycoprotein family protein [Arabidopsis thaliana] (TAIR:AT5G52430.1); similar to unknown [Thellungiella halophila] (GB:ABJ98061.1) | 2.64 |
| 108 | 259793_at | AT1G64380 | AP2 domain-containing transcription factor, putative | -2.08 |
| 109 | 261973_at | AT1G64610 | WD-40 repeat family protein | 2.74 |
| 110 | 262870_at | AT1G64710 | alcohol dehydrogenase, putative | 2.98 |
| 111 | 263142_at | AT1G65230 | similar to hypothetical protein [Vitis vinifera] (GB:CAN76393.1) | -1.26 |
| 112 | 264161_at | AT1G65420 | Identical to Ycf20-like protein [Arabidopsis Thaliana] (GB:O80813); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G56830.2); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G56830.3); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G56830.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO44991.1); contains InterPro domain Protein of unknown function DUF565 (InterPro:IPR007572) | -1.44 |
| 113 | 262930_at | AT1G65690 | harpin-induced protein-related / HIN1-related / harpin-responsive protein-related | 3.78 |
| 114 | 261921_at | AT1G65900 | similar to unnamed protein product [Vitis vinifera] (GB:CAO22185.1); contains domain Helical backbone metal receptor (SSF53807) | -1.43 |
| 115 | 245193_at | AT1G67810 | Fe-S metabolism associated domain-containing protein | 5.63 |
| 116 | 245215_at | AT1G67830 | ATFXG1 (ALPHA-FUCOSIDASE 1); alpha-L-fucosidase/ carboxylesterase | -1.61 |
| 117 | 260015_at | AT1G67980 | CCoAMT (caffeoyl-CoA 3-O-methyltransferase); caffeoyl-CoA O-methyltransferase | 4.18 |
| 118 | 260431_at | AT1G68190 | zinc finger (B-box type) family protein | -1.40 |
| 119 | 259858_at | AT1G68400 | leucine-rich repeat transmembrane protein kinase, putative | -1.26 |
| 120 | 260260_at | AT1G68540 | oxidoreductase family protein | -1.67 |
| 121 | 262286_at | AT1G68585 | metal ion binding | -1.34 |
| 122 | 262229_at | AT1G68620 | hydrolase | 4.48 |
| 123 | 257516_at | AT1G69040 | ACR4 (ACT REPEAT 4); amino acid binding | -1.23 |
| 124 | 260347_at | AT1G69420 | zinc finger (DHHC type) family protein | -1.45 |
| 125 | 256304_at | AT1G69523 | UbiE/COQ5 methyltransferase family protein | -1.25 |
| 126 | 260371_at | AT1G69690 | TCP family transcription factor, putative | -1.39 |
| 127 | 260406_at | AT1G69920 | ATGSTU12 (Arabidopsis thaliana Glutathione S-transferase (class tau) 12); glutathione transferase | 5.12 |
| 128 | 260405_at | AT1G69930 | ATGSTU11 (Arabidopsis thaliana Glutathione S-transferase (class tau) 11); glutathione transferase | 4.12 |
| 129 | 262263_at | AT1G70940 | PIN3 (PIN-FORMED 3); auxin:hydrogen symporter/ transporter | -1.30 |
| 130 | 256336_at | AT1G72030 | GCN5-related N-acetyltransferase (GNAT) family protein | -1.45 |
| 131 | 260399_at | AT1G72520 | lipoxygenase, putative | 3.54 |
| 132 | 262381_at | AT1G72900 | disease resistance protein (TIR-NBS class), putative | 2.80 |
| 133 | 262370_at | AT1G73090 | similar to unnamed protein product [Vitis vinifera] (GB:CAO23197.1) | -1.29 |
| 134 | 260394_at | AT1G74080 | MYB122 (myb domain protein 122); DNA binding / transcription factor | 5.78 |
| 135 | 260239_at | AT1G74360 | leucine-rich repeat transmembrane protein kinase, putative | 5.11 |
| 136 | 260229_at | AT1G74370 | zinc finger (C3HC4-type RING finger) family protein | -1.25 |
| 137 | 260225_at | AT1G74590 | ATGSTU10 (Arabidopsis thaliana Glutathione S-transferase (class tau) 10); glutathione transferase | 5.61 |
| 138 | 260221_at | AT1G74670 | gibberellin-responsive protein, putative | -1.46 |
| 139 | 256456_at | AT1G75180 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G19400.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G19400.2); similar to unnamed protein product [Vitis vinifera] (GB:CAO47631.1); contains domain PTHR10996:SF1 (PTHR10996:SF1); contains domain PTHR10996 (PTHR10996) | -1.47 |
| 140 | 262679_at | AT1G75830 | LCR67/PDF1.1 (Low-molecular-weight cysteine-rich 67) | 9.05 |
| 141 | 262698_at | AT1G75960 | AMP-binding protein, putative | -1.30 |
| 142 | 261782_at | AT1G76110 | high mobility group (HMG1/2) family protein / ARID/BRIGHT DNA-binding domain-containing protein | -1.83 |
| 143 | 261772_at | AT1G76240 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G17080.1); similar to Os06g0725500 [Oryza sativa (japonica cultivar-group)] (GB:NP_001058623.1); similar to hypothetical protein OsI_023643 [Oryza sativa (indica cultivar-group)] (GB:EAZ02411.1); contains InterPro domain Protein of unknown function DUF241, plant (InterPro:IPR004320) | -1.35 |
| 144 | 259879_at | AT1G76650 | CML38; calcium ion binding | 3.17 |
| 145 | 264960_at | AT1G76930 | ATEXT4 (EXTENSIN 4) | 5.41 |
| 146 | 264959_at | AT1G77090 | thylakoid lumenal 29.8 kDa protein | -1.31 |
| 147 | 262162_at | AT1G78020 | senescence-associated protein-related | -1.24 |
| 148 | 260081_at | AT1G78170 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G22250.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO61724.1) | -1.24 |
| 149 | 260804_at | AT1G78410 | VQ motif-containing protein | 3.79 |
| 150 | 264096_at | AT1G78995 | similar to unnamed protein product [Vitis vinifera] (GB:CAO41778.1) | -1.77 |
| 151 | 261394_at | AT1G79680 | wall-associated kinase, putative | 3.02 |
| 152 | 259860_at | AT1G80640 | protein kinase family protein | -1.22 |
| 153 | 261899_at | AT1G80820 | CCR2 (CINNAMOYL COA REDUCTASE) | 6.17 |
| 154 | 261892_at | AT1G80840 | WRKY40 (WRKY DNA-binding protein 40); transcription factor | 3.49 |
| 155 | 266329_at | AT2G01590 | similar to unnamed protein product [Vitis vinifera] (GB:CAO42320.1) | -1.31 |
| 156 | 265869_at | AT2G01760 | ARR14 (ARABIDOPSIS RESPONSE REGULATOR 14); transcription factor/ two-component response regulator | -1.72 |
| 157 | 263597_at | AT2G01870 | similar to unnamed protein product [Vitis vinifera] (GB:CAO21902.1) | -1.22 |
| 158 | 265705_at | AT2G03410 | Mo25 family protein | 4.50 |
| 159 | 265704_at | AT2G03420 | similar to expressed protein [Oryza sativa (japonica cultivar-group)] (GB:ABF94594.1); similar to hypothetical protein OsI_010237 [Oryza sativa (indica cultivar-group)] (GB:EAY89004.1) | -1.25 |
| 160 | 264037_at | AT2G03750 | sulfotransferase family protein | -1.26 |
| 161 | 263410_at | AT2G04039 | similar to unnamed protein product [Vitis vinifera] (GB:CAO41070.1) | -1.29 |
| 162 | 263403_at | AT2G04040 | ATDTX1; antiporter/ multidrug efflux pump/ multidrug transporter/ transporter | 5.71 |
| 163 | 263322_at | AT2G04270 | glycoside hydrolase starch-binding domain-containing protein | -1.43 |
| 164 | 263807_at | AT2G04400 | indole-3-glycerol phosphate synthase (IGPS) | 3.36 |
| 165 | 265377_at | AT2G05790 | glycosyl hydrolase family 17 protein | -1.36 |
| 166 | 265892_at | AT2G15020 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G64190.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN67723.1) | -1.63 |
| 167 | 263565_at | AT2G15390 | FUT4 (fucosyltransferase 4); fucosyltransferase/ transferase, transferring glycosyl groups | 3.58 |
| 168 | 265499_at | AT2G15480 | UGT73B5 (UDP-GLUCOSYL TRANSFERASE 73B5); UDP-glycosyltransferase/ transferase, transferring glycosyl groups | 5.25 |
| 169 | 265501_at | AT2G15490 | UGT73B4; UDP-glycosyltransferase/ transferase, transferring glycosyl groups | 5.68 |
| 170 | 265479_at | AT2G15760 | calmodulin-binding protein | 2.48 |
| 171 | 266536_at | AT2G16900 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G35110.2); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G35110.3); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G35110.4); similar to hypothetical protein [Vitis vinifera] (GB:CAN73143.1); contains InterPro domain Phospholipase-like, arabidopsis (InterPro:IPR007942) | 2.63 |
| 172 | 264616_at | AT2G17740 | DC1 domain-containing protein | 2.84 |
| 173 | 266017_at | AT2G18690 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G18680.1); similar to hypothetical protein OsI_029427 [Oryza sativa (indica cultivar-group)] (GB:EAZ08195.1) | 3.72 |
| 174 | 266946_at | AT2G18890 | protein kinase family protein | -1.36 |
| 175 | 267436_at | AT2G19190 | FRK1 (FLG22-INDUCED RECEPTOR-LIKE KINASE 1); kinase | 3.55 |
| 176 | 265939_at | AT2G19650 | DC1 domain-containing protein | -1.26 |
| 177 | 264014_at | AT2G21210 | auxin-responsive protein, putative | -1.62 |
| 178 | 263549_at | AT2G21650 | MEE3 (maternal effect embryo arrest 3); DNA binding / transcription factor | -1.39 |
| 179 | 266831_at | AT2G22830 | SQE2 (SQUALENE EPOXIDASE 2); oxidoreductase | -1.38 |
| 180 | 245076_at | AT2G23170 | GH3.3; indole-3-acetic acid amido synthetase | 3.03 |
| 181 | 245082_at | AT2G23270 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G37290.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN62855.1) | 4.48 |
| 182 | 267288_at | AT2G23680 | stress-responsive protein, putative | 2.72 |
| 183 | 266000_at | AT2G24180 | CYP71B6 (CYTOCHROME P450 71B6); oxygen binding | 3.13 |
| 184 | 263793_at | AT2G24630 | ATCSLC08 (Cellulose synthase-like C8); transferase, transferring glycosyl groups | -1.21 |
| 185 | 266662_at | AT2G25830 | YebC-related | -1.24 |
| 186 | 266648_at | AT2G25840 | OVA4 (OVULE ABORTION 4); ATP binding / aminoacyl-tRNA ligase | -1.43 |
| 187 | 266656_at | AT2G25900 | ATCTH (Arabidopsis thaliana Cys3His zinc finger protein); transcription factor | -1.24 |
| 188 | 257365_x_at | AT2G26020 | PDF1.2b (plant defensin 1.2b) | 3.86 |
| 189 | 267376_at | AT2G26330 | ER (ERECTA) | -1.64 |
| 190 | 245038_at | AT2G26560 | PLP2 (PHOSPHOLIPASE A 2A); nutrient reservoir | 3.11 |
| 191 | 267606_at | AT2G26640 | beta-ketoacyl-CoA synthase, putative | -1.21 |
| 192 | 267610_at | AT2G26650 | AKT1 (ARABIDOPSIS K TRANSPORTER 1); cyclic nucleotide binding / inward rectifier potassium channel | 4.45 |
| 193 | 266303_at | AT2G27060 | ATP binding / protein serine/threonine kinase | -1.29 |
| 194 | 266254_at | AT2G27810 | xanthine/uracil permease family protein | -1.37 |
| 195 | 266157_at | AT2G28100 | ATFUC1 (ALPHA-L-FUCOSIDASE 1); alpha-L-fucosidase | -1.54 |
| 196 | 266790_at | AT2G28950 | ATEXPA6 (ARABIDOPSIS THALIANA EXPANSIN A6) | -1.69 |
| 197 | 266279_at | AT2G29290 | tropinone reductase, putative / tropine dehydrogenase, putative | -1.24 |
| 198 | 266291_at | AT2G29320 | tropinone reductase, putative / tropine dehydrogenase, putative | -1.24 |
| 199 | 266292_at | AT2G29350 | SAG13 (Senescence-associated gene 13); oxidoreductase | 5.24 |
| 200 | 266273_at | AT2G29410 | MTPB1; efflux transmembrane transporter/ zinc ion transmembrane transporter | 3.79 |
| 201 | 266267_at | AT2G29460 | ATGSTU4 (GLUTATHIONE S-TRANSFERASE 22); glutathione transferase | 5.77 |
| 202 | 266270_at | AT2G29470 | ATGSTU3 (GLUTATHIONE S-TRANSFERASE 21); glutathione transferase | 3.72 |
| 203 | 266670_at | AT2G29740 | UDP-glucoronosyl/UDP-glucosyl transferase family protein | 3.48 |
| 204 | 267300_at | AT2G30140 | UDP-glucoronosyl/UDP-glucosyl transferase family protein | 2.55 |
| 205 | 267471_at | AT2G30390 | ferrochelatase II | -1.36 |
| 206 | 267565_at | AT2G30750 | CYP71A12 (CYTOCHROME P450, FAMILY 71, SUBFAMILY A, POLYPEPTIDE 12); oxygen binding | 6.83 |
| 207 | 267567_at | AT2G30770 | CYP71A13 (CYTOCHROME P450, FAMILY 71, SUBFAMILY A, POLYPEPTIDE 13); indoleacetaldoxime dehydratase/ oxygen binding | 7.68 |
| 208 | 265723_at | AT2G32140 | transmembrane receptor | 2.56 |
| 209 | 265679_at | AT2G32240 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G05320.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G05320.3); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G05320.2); similar to unnamed protein product [Vitis vinifera] (GB:CAO63724.1); contains domain PD023692 (PD023692); contains domain PTHR23160 (PTHR23160) | 3.28 |
| 210 | 267116_at | AT2G32560 | F-box family protein | -1.24 |
| 211 | 267549_at | AT2G32640 | similar to unnamed protein product [Vitis vinifera] (GB:CAO24043.1); contains domain G3DSA:3.50.50.60 (G3DSA:3.50.50.60); contains domain SSF51905 (SSF51905) | -1.32 |
| 212 | 255817_at | AT2G33330 | 33 kDa secretory protein-related | -1.43 |
| 213 | 255842_at | AT2G33530 | SCPL46 (serine carboxypeptidase-like 46); serine carboxypeptidase | -1.31 |
| 214 | 267451_at | AT2G33710 | AP2 domain-containing transcription factor family protein | 2.73 |
| 215 | 266995_at | AT2G34500 | CYP710A1 (cytochrome P450, family 710, subfamily A, polypeptide 1); C-22 sterol desaturase/ oxygen binding | 5.21 |
| 216 | 266956_at | AT2G34510 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G29980.1); similar to unknown [Populus trichocarpa] (GB:ABK95079.1); contains InterPro domain Protein of unknown function DUF642 (InterPro:IPR006946); contains InterPro domain Galactose-binding like (InterPro:IPR008979) | -1.57 |
| 217 | 266899_at | AT2G34620 | mitochondrial transcription termination factor-related / mTERF-related | -1.47 |
| 218 | 266620_at | AT2G35450 | hydrolase | -1.32 |
| 219 | 266608_at | AT2G35500 | shikimate kinase-related | -1.29 |
| 220 | 265796_at | AT2G35730 | heavy-metal-associated domain-containing protein | 4.77 |
| 221 | 265823_at | AT2G35760 | integral membrane family protein | -1.48 |
| 222 | 263935_at | AT2G35930 | U-box domain-containing protein | 5.39 |
| 223 | 263951_at | AT2G35960 | NHL12 (NDR1/HIN1-like 12) | -1.25 |
| 224 | 263948_at | AT2G35980 | YLS9 (YELLOW-LEAF-SPECIFIC GENE 9) | 8.22 |
| 225 | 263287_at | AT2G36145 | similar to unnamed protein product [Vitis vinifera] (GB:CAO15876.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN74693.1) | -1.61 |
| 226 | 263915_at | AT2G36430 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G22540.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO15956.1); contains InterPro domain Protein of unknown function DUF247, plant (InterPro:IPR004158) | -1.39 |
| 227 | 267092_at | AT2G38120 | AUX1 (AUXIN RESISTANT 1); amino acid transmembrane transporter/ transporter | -1.56 |
| 228 | 266167_at | AT2G38860 | YLS5 (yellow-leaf-specific gene 5) | 4.45 |
| 229 | 266168_at | AT2G38870 | protease inhibitor, putative | 3.37 |
| 230 | 266142_at | AT2G39030 | GCN5-related N-acetyltransferase (GNAT) family protein | 4.28 |
| 231 | 267356_at | AT2G39930 | ATISA1/ISA1 (ISOAMYLASE 1); alpha-amylase/ isoamylase | -1.22 |
| 232 | 255822_at | AT2G40610 | ATEXPA8 (ARABIDOPSIS THALIANA EXPANSIN A8) | -1.56 |
| 233 | 267078_at | AT2G40960 | nucleic acid binding | -1.31 |
| 234 | 266364_at | AT2G41230 | unknown protein | 4.32 |
| 235 | 266368_at | AT2G41380 | embryo-abundant protein-related | 5.75 |
| 236 | 266371_at | AT2G41410 | calmodulin, putative | 2.58 |
| 237 | 267537_at | AT2G41880 | GK-1 (GUANYLATE KINASE 1); guanylate kinase | 2.92 |
| 238 | 267586_at | AT2G41950 | similar to unnamed protein product [Vitis vinifera] (GB:CAO23423.1); contains domain S-adenosyl-L-methionine-dependent methyltransferases (SSF53335) | -1.22 |
| 239 | 265262_at | AT2G42980 | aspartyl protease family protein | 2.51 |
| 240 | 265260_at | AT2G43000 | ANAC042 (Arabidopsis NAC domain containing protein 42); transcription factor | 4.38 |
| 241 | 260568_at | AT2G43570 | chitinase, putative | 6.32 |
| 242 | 260560_at | AT2G43590 | chitinase, putative | 4.11 |
| 243 | 260556_at | AT2G43620 | chitinase, putative | 2.93 |
| 244 | 266873_at | AT2G44740 | CYCP4;1 (cyclin p4;1); cyclin-dependent protein kinase | -1.43 |
| 245 | 245148_at | AT2G45220 | pectinesterase family protein | 7.49 |
| 246 | 245130_at | AT2G45340 | leucine-rich repeat transmembrane protein kinase, putative | -1.56 |
| 247 | 267561_at | AT2G45590 | protein kinase family protein | -1.38 |
| 248 | 266581_at | AT2G46140 | late embryogenesis abundant protein, putative / LEA protein, putative | 2.49 |
| 249 | 266326_at | AT2G46650 | B5 #1 (cytochrome b5 family protein #1); heme binding / transition metal ion binding | 2.95 |
| 250 | 266324_at | AT2G46710 | rac GTPase activating protein, putative | -1.31 |
| 251 | 263319_at | AT2G47160 | BOR1 (REQUIRES HIGH BORON 1); anion exchanger | -1.52 |
| 252 | 266460_at | AT2G47930 | AGP26/ATAGP26 (ARABINOGALACTAN PROTEINS 26) | -1.70 |
| 253 | 258956_at | AT3G01440 | oxygen evolving enhancer 3 (PsbQ) family protein | -1.34 |
| 254 | 259185_at | AT3G01550 | triose phosphate/phosphate translocator, putative | -1.66 |
| 255 | 258975_at | AT3G01970 | WRKY45 (WRKY DNA-binding protein 45); transcription factor | 5.36 |
| 256 | 259132_at | AT3G02250 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G15740.1); similar to hypothetical protein [Cleome spinosa] (GB:ABD96934.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO49221.1); similar to hypothetical protein [Cleome spinosa] (GB:ABD96844.1); contains InterPro domain Protein of unknown function DUF246, plant (InterPro:IPR004348) | -1.24 |
| 257 | 257536_at | AT3G02800 | phosphoprotein phosphatase | 3.77 |
| 258 | 258606_at | AT3G02840 | immediate-early fungal elicitor family protein | 4.93 |
| 259 | 258613_at | AT3G02870 | VTC4; 3'(2'),5'-bisphosphate nucleotidase/ L-galactose-1-phosphate phosphatase/ inositol or phosphatidylinositol phosphatase | -1.30 |
| 260 | 258791_at | AT3G04720 | PR4 (PATHOGENESIS-RELATED 4) | 3.12 |
| 261 | 258804_at | AT3G04760 | pentatricopeptide (PPR) repeat-containing protein | -1.31 |
| 262 | 258534_at | AT3G06730 | thioredoxin family protein | -1.23 |
| 263 | 258528_at | AT3G06770 | glycoside hydrolase family 28 protein / polygalacturonase (pectinase) family protein | -1.61 |
| 264 | 258554_at | AT3G06980 | DEAD/DEAH box helicase, putative | -1.27 |
| 265 | 259010_at | AT3G07340 | basic helix-loop-helix (bHLH) family protein | -1.76 |
| 266 | 258692_at | AT3G08640 | alphavirus core protein family | 3.06 |
| 267 | 259213_at | AT3G09010 | protein kinase family protein | 2.67 |
| 268 | 259207_at | AT3G09050 | similar to unknown [Populus trichocarpa x Populus deltoides] (GB:ABK96465.1) | -1.25 |
| 269 | 259033_at | AT3G09410 | pectinacetylesterase family protein | 4.64 |
| 270 | 258927_at | AT3G10160 | ATDFC (A. THALIANA DHFS-FPGS HOMOLOG C); dihydrofolate synthase | -1.45 |
| 271 | 259150_at | AT3G10320 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G41640.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO23616.1); contains InterPro domain Protein of unknown function DUF563 (InterPro:IPR007657) | 4.93 |
| 272 | 256441_at | AT3G10940 | protein phosphatase-related | -1.69 |
| 273 | 256252_at | AT3G11340 | UDP-glucoronosyl/UDP-glucosyl transferase family protein | 5.57 |
| 274 | 256245_at | AT3G12580 | HSP70 (heat shock protein 70); ATP binding | 2.75 |
| 275 | 256237_at | AT3G12610 | DRT100 (DNA-DAMAGE REPAIR/TOLERATION 100); protein binding | -1.49 |
| 276 | 257706_at | AT3G12685 | similar to catalytic [Arabidopsis thaliana] (TAIR:AT1G24350.1); similar to unknown [Populus trichocarpa] (GB:ABK94924.1); contains InterPro domain Acid phosphatase/vanadium-dependent haloperoxidase related (InterPro:IPR003832) | -1.23 |
| 277 | 257846_at | AT3G12910 | transcription factor | 4.76 |
| 278 | 257856_at | AT3G12930 | similar to unknown [Populus trichocarpa] (GB:ABK94112.1); contains InterPro domain Iojap-related protein (InterPro:IPR004394) | -1.30 |
| 279 | 257180_at | AT3G13180 | NOL1/NOP2/sun family protein / antitermination NusB domain-containing protein | -1.78 |
| 280 | 256787_at | AT3G13790 | ATBFRUCT1/ATCWINV1 (ARABIDOPSIS THALIANA CELL WALL INVERTASE 1); beta-fructofuranosidase/ hydrolase, hydrolyzing O-glycosyl compounds | 3.21 |
| 281 | 258203_at | AT3G13950 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G13266.1); similar to Ankyrin [Medicago truncatula] (GB:ABN08906.1) | 3.69 |
| 282 | 257216_at | AT3G14990 | 4-methyl-5(b-hydroxyethyl)-thiazole monophosphate biosynthesis protein, putative | 2.90 |
| 283 | 257058_at | AT3G15352 | ATCOX17 (Arabidopsis thaliana cytochrome c oxidase 17) | 2.78 |
| 284 | 258395_at | AT3G15500 | ATNAC3 (ARABIDOPSIS NAC DOMAIN CONTAINING PROTEIN 55); transcription factor | 3.71 |
| 285 | 257294_at | AT3G15570 | phototropic-responsive NPH3 family protein | -1.47 |
| 286 | 258250_at | AT3G15850 | FAD5 (FATTY ACID DESATURASE 5); oxidoreductase | -1.23 |
| 287 | 258055_at | AT3G16250 | ferredoxin-related | -1.55 |
| 288 | 257206_at | AT3G16530 | legume lectin family protein | 3.26 |
| 289 | 257891_at | AT3G17170 | RFC3 (REGULATOR OF FATTY-ACID COMPOSITION 3); structural constituent of ribosome | -1.24 |
| 290 | 258156_at | AT3G18050 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G28100.1); similar to unknown [Populus trichocarpa] (GB:ABK95654.1) | -1.82 |
| 291 | 257061_at | AT3G18250 | contains domain PROKAR_LIPOPROTEIN (PS51257) | 3.11 |
| 292 | 257038_at | AT3G19260 | LAG1 HOMOLOG 2 (LONGEVITY ASSURANCE GENE1 HOMOLOG 2) | 2.80 |
| 293 | 257132_at | AT3G20230 | 50S ribosomal protein L18 family | -1.27 |
| 294 | 257974_at | AT3G20820 | leucine-rich repeat family protein | -1.31 |
| 295 | 257978_at | AT3G20860 | ATNEK5; kinase | 3.55 |
| 296 | 256967_at | AT3G21060 | transducin family protein / WD-40 repeat family protein | -1.31 |
| 297 | 257540_at | AT3G21520 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G21550.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO71058.1); contains InterPro domain Protein of unknown function DUF679 (InterPro:IPR007770) | 4.69 |
| 298 | 258181_at | AT3G21670 | nitrate transporter (NTP3) | -1.44 |
| 299 | 257950_at | AT3G21780 | UGT71B6 (UDP-GLUCOSYL TRANSFERASE 71B6); UDP-glycosyltransferase/ abscisic acid glucosyltransferase/ transferase, transferring glycosyl groups | 2.94 |
| 300 | 258456_at | AT3G22420 | WNK2 (WITH NO K 2); kinase | -1.26 |
| 301 | 256933_at | AT3G22600 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | 7.79 |
| 302 | 257765_at | AT3G23020 | pentatricopeptide (PPR) repeat-containing protein | -1.55 |
| 303 | 258100_at | AT3G23550 | MATE efflux family protein | 3.72 |
| 304 | 258108_at | AT3G23570 | dienelactone hydrolase family protein | 2.57 |
| 305 | 257204_at | AT3G23805 | RALFL24 (RALF-LIKE 24) | -1.54 |
| 306 | 257592_at | AT3G24982 | protein binding | 3.03 |
| 307 | 256756_at | AT3G25610 | haloacid dehalogenase-like hydrolase family protein | 4.50 |
| 308 | 257644_at | AT3G25780 | AOC3 (ALLENE OXIDE CYCLASE 3) | 2.99 |
| 309 | 257636_at | AT3G26200 | CYP71B22 (cytochrome P450, family 71, subfamily B, polypeptide 22); oxygen binding | 3.36 |
| 310 | 256883_at | AT3G26440 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G13000.2); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G13000.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO42517.1); contains InterPro domain Protein of unknown function DUF707 (InterPro:IPR007877) | 2.85 |
| 311 | 257831_at | AT3G26710 | similar to unnamed protein product [Vitis vinifera] (GB:CAO49107.1) | -1.23 |
| 312 | 258277_at | AT3G26830 | PAD3 (PHYTOALEXIN DEFICIENT 3); oxygen binding | 7.27 |
| 313 | 257297_at | AT3G28040 | leucine-rich repeat transmembrane protein kinase, putative | -1.71 |
| 314 | 257272_at | AT3G28130 | nodulin MtN21 family protein | -1.51 |
| 315 | 257903_at | AT3G28460 | similar to hypothetical protein [Vitis vinifera] (GB:CAN74970.1); contains InterPro domain Protein of unknown function methylase putative (InterPro:IPR016065) | -1.22 |
| 316 | 256989_at | AT3G28580 | AAA-type ATPase family protein | 6.99 |
| 317 | 256585_at | AT3G28760 | similar to unnamed protein product [Vitis vinifera] (GB:CAO14940.1); contains InterPro domain 3-dehydroquinate synthase, prokaryotic-type (InterPro:IPR002812) | -1.25 |
| 318 | 258002_at | AT3G28930 | AIG2 (AVRRPT2-INDUCED GENE 2) | 3.70 |
| 319 | 252739_at | AT3G43250 | cell cycle control protein-related | 4.60 |
| 320 | 252639_at | AT3G44550 | oxidoreductase, acting on the CH-CH group of donors | 4.07 |
| 321 | 252652_at | AT3G44720 | ADT4 (AROGENATE DEHYDRATASE 4); arogenate dehydratase/ prephenate dehydratase | 3.19 |
| 322 | 246332_at | AT3G44830 | lecithin:cholesterol acyltransferase family protein / LACT family protein | 2.59 |
| 323 | 252604_at | AT3G45060 | ATNRT2.6 (Arabidopsis thaliana high affinity nitrate transporter 2.6); nitrate transmembrane transporter | 3.86 |
| 324 | 252534_at | AT3G46130 | MYB111 (myb domain protein 111); DNA binding / transcription factor | -1.43 |
| 325 | 252487_at | AT3G46660 | UDP-glucoronosyl/UDP-glucosyl transferase family protein | 3.48 |
| 326 | 252353_at | AT3G48200 | similar to hypothetical protein OsI_020499 [Oryza sativa (indica cultivar-group)] (GB:EAY99266.1); similar to hypothetical protein OsJ_018975 [Oryza sativa (japonica cultivar-group)] (GB:EAZ35492.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO64300.1) | -1.32 |
| 327 | 252317_at | AT3G48720 | transferase family protein | -1.25 |
| 328 | 252334_at | AT3G48850 | mitochondrial phosphate transporter, putative | 5.52 |
| 329 | 252338_at | AT3G48890 | ATMP2 (MEMBRANE STEROID BINDING PROTEIN 2); heme binding / transition metal ion binding | 3.35 |
| 330 | 252265_at | AT3G49620 | DIN11 (DARK INDUCIBLE 11); oxidoreductase | 7.71 |
| 331 | 252272_at | AT3G49670 | BAM2 (big apical meristem 2); ATP binding / protein serine/threonine kinase | -1.44 |
| 332 | 252189_at | AT3G50070 | CYCD3;3 (CYCLIN D3;3); cyclin-dependent protein kinase | -1.96 |
| 333 | 252136_at | AT3G50770 | calmodulin-related protein, putative | 4.86 |
| 334 | 252133_at | AT3G50900 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G66490.1); similar to hypothetical protein [Thellungiella halophila] (GB:ABB45855.1) | 3.27 |
| 335 | 252131_at | AT3G50930 | AAA-type ATPase family protein | 3.88 |
| 336 | 252073_at | AT3G51750 | unknown protein | -1.46 |
| 337 | 256674_at | AT3G52360 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G35850.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO39911.1) | -1.23 |
| 338 | 252010_at | AT3G52740 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G44450.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO70571.1) | -1.55 |
| 339 | 251996_at | AT3G52840 | BGAL2 (beta-galactosidase 2); beta-galactosidase | -1.37 |
| 340 | 252023_at | AT3G52920 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G36410.1); similar to unknown [Populus trichocarpa] (GB:ABK95581.1); similar to unknown [Populus trichocarpa] (GB:ABK93998.1); contains InterPro domain Protein of unknown function DUF662 (InterPro:IPR007033) | -1.24 |
| 341 | 251971_at | AT3G53160 | UGT73C7 (UDP-GLUCOSYL TRANSFERASE 73C7); UDP-glycosyltransferase/ transferase, transferring glycosyl groups | 3.65 |
| 342 | 251884_at | AT3G54150 | embryo-abundant protein-related | 4.30 |
| 343 | 251895_at | AT3G54420 | ATEP3 (Arabidopsis thaliana chitinase class IV); chitinase | 4.94 |
| 344 | 251847_at | AT3G54640 | TSA1 (TRYPTOPHAN SYNTHASE ALPHA CHAIN); tryptophan synthase | 4.11 |
| 345 | 251853_at | AT3G54790 | armadillo/beta-catenin repeat family protein / U-box domain-containing protein | -1.36 |
| 346 | 251790_at | AT3G55470 | C2 domain-containing protein | 2.84 |
| 347 | 251759_at | AT3G55630 | ATDFD (A. THALIANA DHFS-FPGS HOMOLOG D); tetrahydrofolylpolyglutamate synthase | -1.28 |
| 348 | 251769_at | AT3G55950 | protein kinase family protein | 3.06 |
| 349 | 251713_at | AT3G56080 | dehydration-responsive protein-related | -1.62 |
| 350 | 251727_at | AT3G56290 | similar to hypothetical protein [Vitis vinifera] (GB:CAN75527.1) | -1.22 |
| 351 | 251665_at | AT3G57040 | ARR9 (RESPONSE REACTOR 4); transcription regulator | -1.29 |
| 352 | 251641_at | AT3G57470 | peptidase M16 family protein / insulinase family protein | -1.25 |
| 353 | 251586_at | AT3G58070 | GIS (GLABROUS INFLORESCENCE STEMS); nucleic acid binding / transcription factor/ zinc ion binding | -1.94 |
| 354 | 251575_at | AT3G58120 | bZIP transcription factor family protein | -1.83 |
| 355 | 251524_at | AT3G58990 | aconitase C-terminal domain-containing protein | -1.34 |
| 356 | 251500_at | AT3G59110 | protein kinase family protein | -1.31 |
| 357 | 251519_at | AT3G59400 | GUN4 (Genomes uncoupled 4) | -1.32 |
| 358 | 251479_at | AT3G59700 | ATHLECRK (ARABIDOPSIS THALIANA LECTIN-RECEPTOR KINASE); kinase | 2.50 |
| 359 | 251428_at | AT3G60140 | DIN2 (DARK INDUCIBLE 2); hydrolase, hydrolyzing O-glycosyl compounds | 2.72 |
| 360 | 251402_at | AT3G60290 | oxidoreductase, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors | -1.88 |
| 361 | 251422_at | AT3G60540 | sec61beta family protein | 3.00 |
| 362 | 251336_at | AT3G61190 | BAP1 (BON ASSOCIATION PROTEIN 1) | 3.08 |
| 363 | 251293_at | AT3G61930 | unknown protein | 3.80 |
| 364 | 251183_at | AT3G62630 | similar to calmodulin-binding protein [Arabidopsis thaliana] (TAIR:AT2G15760.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO40086.1); contains InterPro domain Protein of unknown function DUF1645 (InterPro:IPR012442) | -1.51 |
| 365 | 251174_at | AT3G63200 | PLA IIIB/PLP9 (Patatin-like protein 9); nutrient reservoir | -1.39 |
| 366 | 251176_at | AT3G63380 | calcium-transporting ATPase, plasma membrane-type, putative / Ca(2+)-ATPase, putative (ACA12) | 6.53 |
| 367 | 255691_at | AT4G00370 | ANTR2 (anion transporter 2); organic anion transmembrane transporter | -1.51 |
| 368 | 255662_at | AT4G00400 | GPAT8 (GLYCEROL-3-PHOSPHATE ACYLTRANSFERASE 8); acyltransferase/ glycerol-3-phosphate O-acyltransferase | -1.45 |
| 369 | 255674_at | AT4G00430 | TMP-C (PLASMA MEMBRANE INTRINSIC PROTEIN 1;4); water channel | -1.36 |
| 370 | 255676_at | AT4G00490 | BAM2/BMY9 (BETA-AMYLASE 2); beta-amylase | -1.28 |
| 371 | 255660_at | AT4G00755 | F-box family protein | -1.28 |
| 372 | 255595_at | AT4G01700 | chitinase, putative | 2.78 |
| 373 | 255543_at | AT4G01870 | tolB protein-related | 2.60 |
| 374 | 255479_at | AT4G02380 | SAG21 (SENESCENCE-ASSOCIATED GENE 21) | 4.20 |
| 375 | 255488_at | AT4G02630 | protein kinase family protein | -1.69 |
| 376 | 255438_at | AT4G03070 | AOP1 (2-oxoglutarate?dependent dioxygenase 1.1); oxidoreductase, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors | -1.55 |
| 377 | 255406_at | AT4G03450 | ankyrin repeat family protein | 2.86 |
| 378 | 255340_at | AT4G04490 | protein kinase family protein | 4.85 |
| 379 | 255259_at | AT4G05020 | NDB2 (NAD(P)H DEHYDROGENASE B2); disulfide oxidoreductase | 2.83 |
| 380 | 255230_at | AT4G05390 | ATRFNR1 (ROOT FNR 1); oxidoreductase | 2.48 |
| 381 | 255110_at | AT4G08770 | peroxidase, putative | 7.48 |
| 382 | 255111_at | AT4G08780 | peroxidase, putative | 4.00 |
| 383 | 255088_at | AT4G09350 | DNAJ heat shock N-terminal domain-containing protein | -1.40 |
| 384 | 255025_at | AT4G09900 | hydrolase, alpha/beta fold family protein | -1.24 |
| 385 | 255016_at | AT4G10120 | ATSPS4F; sucrose-phosphate synthase/ transferase, transferring glycosyl groups | -1.37 |
| 386 | 254926_at | AT4G11280 | ACS6 (1-AMINOCYCLOPROPANE-1-CARBOXYLIC ACID (ACC) SYNTHASE 6) | 3.91 |
| 387 | 254922_at | AT4G11370 | RHA1A (RING-H2 finger A1A); protein binding / zinc ion binding | 4.34 |
| 388 | 254857_at | AT4G12120 | SEC1B; protein transporter | 2.58 |
| 389 | 254783_at | AT4G12830 | hydrolase, alpha/beta fold family protein | -1.96 |
| 390 | 254746_at | AT4G12980 | auxin-responsive protein, putative | -1.26 |
| 391 | 245592_at | AT4G14540 | CCAAT-box binding transcription factor subunit B (NF-YB) (HAP3 ) (AHAP3) family | -1.46 |
| 392 | 245368_at | AT4G15510 | photosystem II reaction center PsbP family protein | -1.50 |
| 393 | 245528_at | AT4G15530 | PPDK (PYRUVATE ORTHOPHOSPHATE DIKINASE); kinase/ pyruvate, phosphate dikinase | 2.67 |
| 394 | 245317_at | AT4G15610 | integral membrane family protein | 3.44 |
| 395 | 245393_at | AT4G16260 | glycosyl hydrolase family 17 protein | 6.14 |
| 396 | 245296_at | AT4G16370 | ATOPT3 (OLIGOPEPTIDE TRANSPORTER); oligopeptide transporter | -1.21 |
| 397 | 245442_at | AT4G16710 | glycosyltransferase family protein 28 | -1.45 |
| 398 | 245447_at | AT4G16820 | lipase class 3 family protein | 5.35 |
| 399 | 245252_at | AT4G17500 | ATERF-1 (ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 1); DNA binding / transcription activator/ transcription factor | 4.47 |
| 400 | 245383_at | AT4G17810 | nucleic acid binding / transcription factor/ zinc ion binding | -1.82 |
| 401 | 254693_at | AT4G17880 | basic helix-loop-helix (bHLH) family protein | -1.68 |
| 402 | 254660_at | AT4G18250 | receptor serine/threonine kinase, putative | 4.20 |
| 403 | 254673_at | AT4G18430 | AtRABA1e (Arabidopsis Rab GTPase homolog A1e); GTP binding | 3.38 |
| 404 | 254571_at | AT4G19370 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G31720.1); similar to unknown [Picea sitchensis] (GB:ABK21519.1); similar to Os02g0703300 [Oryza sativa (japonica cultivar-group)] (GB:NP_001047854.1); contains InterPro domain Protein of unknown function DUF1218 (InterPro:IPR009606) | 5.55 |
| 405 | 254432_at | AT4G20830 | FAD-binding domain-containing protein | 2.78 |
| 406 | 254408_at | AT4G21390 | B120; protein kinase/ sugar binding | 4.26 |
| 407 | 254396_at | AT4G21680 | proton-dependent oligopeptide transport (POT) family protein | 4.79 |
| 408 | 254387_at | AT4G21850 | methionine sulfoxide reductase domain-containing protein / SeIR domain-containing protein | 2.77 |
| 409 | 254318_at | AT4G22530 | embryo-abundant protein-related | 3.63 |
| 410 | 254328_at | AT4G22570 | APT3 (ADENINE PHOSPHORIBOSYL TRANSFERASE 3); adenine phosphoribosyltransferase | -2.08 |
| 411 | 254271_at | AT4G23150 | protein kinase family protein | 3.17 |
| 412 | 254241_at | AT4G23190 | CRK11 (CYSTEINE-RICH RLK11); kinase | 3.72 |
| 413 | 254250_at | AT4G23290 | protein kinase family protein | -1.60 |
| 414 | 254215_at | AT4G23700 | ATCHX17 (CATION/H+ EXCHANGER 17); monovalent cation:proton antiporter | 5.14 |
| 415 | 254181_at | AT4G23940 | FtsH protease, putative | -1.25 |
| 416 | 254200_at | AT4G24110 | similar to unnamed protein product [Vitis vinifera] (GB:CAO62924.1) | 4.74 |
| 417 | 254150_at | AT4G24350 | phosphorylase family protein | 2.60 |
| 418 | 254122_at | AT4G24510 | CER2 (ECERIFERUM 2); transferase | -1.39 |
| 419 | 254119_at | AT4G24780 | pectate lyase family protein | -1.66 |
| 420 | 254042_at | AT4G25810 | XTR6 (XYLOGLUCAN ENDOTRANSGLYCOSYLASE 6); hydrolase, acting on glycosyl bonds | 3.73 |
| 421 | 254041_at | AT4G25830 | integral membrane family protein | -1.21 |
| 422 | 254038_at | AT4G25910 | NFU3 (NFU domain protein 3) | -1.34 |
| 423 | 254011_at | AT4G26370 | antitermination NusB domain-containing protein | -1.28 |
| 424 | 253943_at | AT4G27030 | small conjugating protein ligase | -1.43 |
| 425 | 253891_at | AT4G27720 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G64650.2); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G64650.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G49310.1); similar to unknown [Populus trichocarpa] (GB:ABK93582.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO48607.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO62932.1); contains InterPro domain Protein of unknown function DUF791 (InterPro:IPR008509); contains InterPro domain MFS general substrate transporter (InterPro:IPR016196) | -1.46 |
| 426 | 253849_at | AT4G28080 | binding | -1.27 |
| 427 | 253827_at | AT4G28085 | unknown protein | 3.19 |
| 428 | 253796_at | AT4G28460 | unknown protein | 5.33 |
| 429 | 253768_at | AT4G28550 | RabGAP/TBC domain-containing protein | 4.74 |
| 430 | 253791_at | AT4G28640 | IAA11 (indoleacetic acid-induced protein 11); transcription factor | -1.26 |
| 431 | 253758_at | AT4G29060 | EMB2726 (EMBRYO DEFECTIVE 2726); translation elongation factor | -1.22 |
| 432 | 253657_at | AT4G30110 | HMA2 (Heavy metal ATPase 2); cadmium-transporting ATPase | -1.46 |
| 433 | 253628_at | AT4G30280 | ATXTH18/XTH18 (XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 18); hydrolase, acting on glycosyl bonds | 2.70 |
| 434 | 253589_at | AT4G30825 | pentatricopeptide (PPR) repeat-containing protein | -1.51 |
| 435 | 253503_at | AT4G31950 | CYP82C3 (cytochrome P450, family 82, subfamily C, polypeptide 3); oxygen binding | 7.37 |
| 436 | 253440_at | AT4G32570 | TIFY8 | -1.58 |
| 437 | 253411_at | AT4G32980 | ATH1 (ARABIDOPSIS THALIANA HOMEOBOX GENE 1); transcription factor | -1.38 |
| 438 | 253412_at | AT4G33000 | CBL10 (CALCINEURIN B-LIKE10); calcium ion binding | -1.36 |
| 439 | 253414_at | AT4G33050 | EDA39 (embryo sac development arrest 39); calmodulin binding | 3.10 |
| 440 | 253343_at | AT4G33540 | metallo-beta-lactamase family protein | 2.69 |
| 441 | 253274_at | AT4G34200 | EDA9 (embryo sac development arrest 9); NAD binding / amino acid binding / binding / catalytic/ cofactor binding / oxidoreductase, acting on the CH-OH group of donors, NAD or NADP as acceptor / phosphoglycerate dehydrogenase | 3.94 |
| 442 | 253278_at | AT4G34220 | leucine-rich repeat transmembrane protein kinase, putative | -1.49 |
| 443 | 253277_at | AT4G34230 | CAD5 (CINNAMYL ALCOHOL DEHYDROGENASE 5); cinnamyl-alcohol dehydrogenase | 2.62 |
| 444 | 253255_at | AT4G34760 | auxin-responsive family protein | -1.67 |
| 445 | 253173_at | AT4G35110 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G16900.1); contains InterPro domain Phospholipase-like, arabidopsis (InterPro:IPR007942) | 3.10 |
| 446 | 253181_at | AT4G35180 | LHT7 (LYS/HIS TRANSPORTER 7); amino acid transmembrane transporter | 4.26 |
| 447 | 253188_at | AT4G35300 | TMT2 (TONOPLAST MONOSACCHARIDE TRANSPORTER2); carbohydrate transmembrane transporter/ nucleoside transmembrane transporter/ sugar:hydrogen ion symporter | -1.33 |
| 448 | 253165_at | AT4G35320 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G17300.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN78386.1) | -1.48 |
| 449 | 253162_at | AT4G35630 | PSAT (phosphoserine aminotransferase); phosphoserine transaminase | 3.27 |
| 450 | 246275_at | AT4G36540 | BEE2 (BR ENHANCED EXPRESSION 2); DNA binding / transcription factor | -1.44 |
| 451 | 246231_at | AT4G37080 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G42690.2); similar to hypothetical protein [Populus tremula] (GB:CAM84232.1); contains InterPro domain Protein of unknown function DUF547 (InterPro:IPR006869) | -1.67 |
| 452 | 246200_at | AT4G37240 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G23690.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO45438.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN61825.1) | -1.54 |
| 453 | 253044_at | AT4G37290 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G23270.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN62855.1) | 5.08 |
| 454 | 253046_at | AT4G37370 | CYP81D8 (cytochrome P450, family 81, subfamily D, polypeptide 8); oxygen binding | 4.06 |
| 455 | 253040_at | AT4G37800 | xyloglucan:xyloglucosyl transferase, putative / xyloglucan endotransglycosylase, putative / endo-xyloglucan transferase, putative | -1.21 |
| 456 | 253028_at | AT4G38160 | PDE191 (PIGMENT DEFECTIVE 191) | -1.32 |
| 457 | 252993_at | AT4G38540 | monooxygenase, putative (MO2) | 3.21 |
| 458 | 252950_at | AT4G38690 | 1-phosphatidylinositol phosphodiesterase-related | -1.46 |
| 459 | 252972_at | AT4G38840 | auxin-responsive protein, putative | -1.21 |
| 460 | 252970_at | AT4G38850 | SAUR_AC1 (SMALL AUXIN UP RNA 1 FROM ARABIDOPSIS THALIANA ECOTYPE COLUMBIA) | -2.02 |
| 461 | 252921_at | AT4G39030 | EDS5 (ENHANCED DISEASE SUSCEPTIBILITY 5); antiporter/ transporter | 2.64 |
| 462 | 252934_at | AT4G39120 | inositol monophosphatase family protein | -1.29 |
| 463 | 252908_at | AT4G39670 | glycolipid binding / glycolipid transporter | 5.40 |
| 464 | 252870_at | AT4G39940 | AKN2 (APS-KINASE 2); ATP binding / kinase/ transferase, transferring phosphorus-containing groups | 2.51 |
| 465 | 252827_at | AT4G39950 | CYP79B2 (cytochrome P450, family 79, subfamily B, polypeptide 2); oxygen binding | 4.04 |
| 466 | 252831_at | AT4G39980 | DHS1 (3-DEOXY-D-ARABINO-HEPTULOSONATE 7-PHOSPHATE SYNTHASE 1); 3-deoxy-7-phosphoheptulonate synthase | 2.81 |
| 467 | 251142_at | AT5G01015 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G65295.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN82885.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO45347.1) | -1.53 |
| 468 | 251106_at | AT5G01500 | mitochondrial substrate carrier family protein | 2.63 |
| 469 | 251037_at | AT5G02100 | UNE18 (unfertilized embryo sac 18); oxysterol binding | 2.68 |
| 470 | 251008_at | AT5G02710 | similar to unnamed protein product [Vitis vinifera] (GB:CAO18074.1); contains InterPro domain Protein of unknown function UPF0153 (InterPro:IPR005358) | -1.49 |
| 471 | 250983_at | AT5G02780 | In2-1 protein, putative | 8.23 |
| 472 | 250975_at | AT5G03050 | similar to hypothetical protein [Vitis vinifera] (GB:CAN67127.1) | -1.25 |
| 473 | 250918_at | AT5G03610 | GDSL-motif lipase/hydrolase family protein | 2.71 |
| 474 | 250738_at | AT5G05730 | ASA1 (ANTHRANILATE SYNTHASE ALPHA SUBUNIT 1); anthranilate synthase | 2.99 |
| 475 | 250739_at | AT5G05740 | ATEGY2; metalloendopeptidase | -1.23 |
| 476 | 250690_at | AT5G06530 | ABC transporter family protein | -1.45 |
| 477 | 250598_at | AT5G07690 | MYB29 (myb domain protein 29); DNA binding / transcription factor | -1.48 |
| 478 | 250575_at | AT5G08240 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G23160.1) | 3.59 |
| 479 | 246036_at | AT5G08370 | ATAGAL2 (ARABIDOPSIS THALIANA ALPHA-GALACTOSIDASE 2); alpha-galactosidase | -1.51 |
| 480 | 250443_at | AT5G10520 | RBK1 (ROP BINDING PROTEIN KINASES 1); kinase | 5.53 |
| 481 | 246019_at | AT5G10690 | pentatricopeptide (PPR) repeat-containing protein / CBS domain-containing protein | -1.46 |
| 482 | 250366_at | AT5G11420 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G25460.1); similar to unknown [Ricinus communis] (GB:CAB02653.1); contains InterPro domain Protein of unknown function DUF642 (InterPro:IPR006946); contains InterPro domain Galactose-binding like (InterPro:IPR008979) | -1.41 |
| 483 | 250267_at | AT5G12930 | similar to unnamed protein product [Vitis vinifera] (GB:CAO21945.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN80883.1) | 7.51 |
| 484 | 245976_at | AT5G13080 | WRKY75 (WRKY DNA-BINDING PROTEIN 75); transcription factor | 8.56 |
| 485 | 245854_at | AT5G13490 | AAC2 (ADP/ATP CARRIER 2); ATP:ADP antiporter/ binding | 3.46 |
| 486 | 250255_at | AT5G13730 | SIG4 (SIGMA FACTOR 4); DNA binding / DNA-directed RNA polymerase/ transcription factor | -1.26 |
| 487 | 246483_at | AT5G16000 | NIK1 (NSP-INTERACTING KINASE 1); kinase | -1.58 |
| 488 | 246487_at | AT5G16030 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G02500.1); similar to hypothetical protein [Cleome spinosa] (GB:ABD96956.1) | -1.22 |
| 489 | 250094_at | AT5G17380 | pyruvate decarboxylase family protein | 3.50 |
| 490 | 250054_at | AT5G17860 | CAX7 (CALCIUM EXCHANGER 7); calcium:sodium antiporter/ cation:cation antiporter | 2.54 |
| 491 | 250014_at | AT5G17990 | TRP1 (TRYPTOPHAN BIOSYNTHESIS 1); anthranilate phosphoribosyltransferase | 3.08 |
| 492 | 249983_at | AT5G18470 | curculin-like (mannose-binding) lectin family protein | 4.18 |
| 493 | 246099_at | AT5G20230 | ATBCB (ARABIDOPSIS BLUE-COPPER-BINDING PROTEIN); copper ion binding | 3.29 |
| 494 | 245993_at | AT5G20700 | senescence-associated protein-related | -1.37 |
| 495 | 246159_at | AT5G20935 | similar to unnamed protein product [Vitis vinifera] (GB:CAO67410.1) | -1.33 |
| 496 | 246133_at | AT5G20960 | AAO1 (ALDEHYDE OXIDASE 1) | 3.27 |
| 497 | 249942_at | AT5G22300 | NIT4 (NITRILASE 4) | 3.45 |
| 498 | 249900_at | AT5G22640 | EMB1211 (EMBRYO DEFECTIVE 1211) | -1.32 |
| 499 | 249770_at | AT5G24110 | WRKY30 (WRKY DNA-binding protein 30); transcription factor | 4.42 |
| 500 | 249774_at | AT5G24150 | SQP1 (Squalene monooxygenase 1) | -1.64 |
| 501 | 246947_at | AT5G25120 | CYP71B11 (cytochrome P450, family 71, subfamily B, polypeptide 11); oxygen binding | -1.23 |
| 502 | 246931_at | AT5G25170 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G25190.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO44286.1); contains InterPro domain Protein of unknown function DUF862, eukaryotic (InterPro:IPR008580) | 2.50 |
| 503 | 246858_at | AT5G25930 | leucine-rich repeat family protein / protein kinase family protein | 3.42 |
| 504 | 246870_at | AT5G26030 | FC1 (FERROCHELATASE 1); ferrochelatase | 2.53 |
| 505 | 246777_at | AT5G27420 | zinc finger (C3HC4-type RING finger) family protein | 3.60 |
| 506 | 246779_at | AT5G27520 | mitochondrial substrate carrier family protein | 2.92 |
| 507 | 246744_at | AT5G27760 | hypoxia-responsive family protein | 3.48 |
| 508 | 249726_at | AT5G35480 | unknown protein | -1.36 |
| 509 | 249727_at | AT5G35490 | unknown protein | -1.82 |
| 510 | 249693_at | AT5G35750 | AHK2 (ARABIDOPSIS HISTIDINE KINASE 2) | -1.36 |
| 511 | 249694_at | AT5G35790 | G6PD1 (GLUCOSE-6-PHOSPHATE DEHYDROGENASE 1); glucose-6-phosphate dehydrogenase | -1.24 |
| 512 | 249638_at | AT5G36880 | acetyl-CoA synthetase, putative / acetate-CoA ligase, putative | 2.68 |
| 513 | 249527_at | AT5G38710 | proline oxidase, putative / osmotic stress-responsive proline dehydrogenase, putative | 3.05 |
| 514 | 249481_at | AT5G38900 | DSBA oxidoreductase family protein | 5.63 |
| 515 | 249494_at | AT5G39050 | transferase family protein | 2.83 |
| 516 | 249472_at | AT5G39210 | CRR7 (CHLORORESPIRATORY REDUCTION 7) | -1.41 |
| 517 | 249459_at | AT5G39580 | peroxidase, putative | 3.51 |
| 518 | 249383_at | AT5G39860 | PRE1 (PACLOBUTRAZOL RESISTANCE1); DNA binding / transcription factor | -1.30 |
| 519 | 249372_at | AT5G40760 | G6PD6 (GLUCOSE-6-PHOSPHATE DEHYDROGENASE 6); glucose-6-phosphate dehydrogenase | 2.57 |
| 520 | 249333_at | AT5G40990 | GLIP1 (GDSL LIPASE1); carboxylesterase | 6.14 |
| 521 | 249288_at | AT5G41050 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G26960.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO49052.1) | -1.46 |
| 522 | 249230_at | AT5G42070 | similar to unnamed protein product [Vitis vinifera] (GB:CAO69519.1) | -1.22 |
| 523 | 249216_at | AT5G42240 | SCPL42 (serine carboxypeptidase-like 42); serine carboxypeptidase | -1.32 |
| 524 | 249242_at | AT5G42250 | alcohol dehydrogenase, putative | -1.22 |
| 525 | 249197_at | AT5G42380 | CML37/CML39; calcium ion binding | 3.89 |
| 526 | 249109_at | AT5G43700 | ATAUX2-11 (indoleacetic acid-induced protein 4); transcription factor | -1.38 |
| 527 | 249045_at | AT5G44380 | FAD-binding domain-containing protein | 4.27 |
| 528 | 249052_at | AT5G44420 | PDF1.2 (Low-molecular-weight cysteine-rich 77) | 4.62 |
| 529 | 249057_at | AT5G44480 | DUR (DEFECTIVE UGE IN ROOT); catalytic | 2.80 |
| 530 | 249059_at | AT5G44530 | subtilase family protein | -1.21 |
| 531 | 249011_at | AT5G44670 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G20170.1); similar to Os06g0328800 [Oryza sativa (japonica cultivar-group)] (GB:NP_001057533.1); similar to Os02g0712500 [Oryza sativa (japonica cultivar-group)] (GB:NP_001047907.1); similar to unknown protein [Oryza sativa (japonica cultivar-group)] (GB:BAD72474.1); contains InterPro domain Protein of unknown function DUF23 (InterPro:IPR008166) | -1.44 |
| 532 | 249008_at | AT5G44680 | methyladenine glycosylase family protein | -1.50 |
| 533 | 249001_at | AT5G44990 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G19880.1); similar to Intracellular chloride channel [Medicago truncatula] (GB:ABC75353.2); contains InterPro domain Thioredoxin-like fold (InterPro:IPR012336); contains InterPro domain Glutathione S-transferase, C-terminal-like (InterPro:IPR010987); contains InterPro domain Glutathione S-transferase, predicted (InterPro:IPR016639) | 5.44 |
| 534 | 248962_at | AT5G45680 | FK506-binding protein 1 (FKBP13) | -1.22 |
| 535 | 248920_at | AT5G45930 | CHLI2; magnesium chelatase | -1.23 |
| 536 | 248921_at | AT5G45950 | GDSL-motif lipase/hydrolase family protein | -1.45 |
| 537 | 248853_at | AT5G46570 | protein kinase family protein | -1.56 |
| 538 | 248794_at | AT5G47220 | ATERF-2/ATERF2/ERF2 (ETHYLENE RESPONSE FACTOR 2); DNA binding / transcription activator/ transcription factor | 3.13 |
| 539 | 248799_at | AT5G47230 | ERF5 (ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 5); DNA binding / transcription activator/ transcription factor | 3.10 |
| 540 | 248764_at | AT5G47640 | CCAAT-box binding transcription factor subunit B (NF-YB) (HAP3 ) (AHAP3) family (Hap3b) | -1.28 |
| 541 | 248656_at | AT5G48460 | fimbrin-like protein, putative | -1.22 |
| 542 | 248683_at | AT5G48490 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | -1.24 |
| 543 | 248646_at | AT5G49100 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G06868.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO44794.1) | -1.50 |
| 544 | 248564_at | AT5G49700 | DNA-binding protein-related | 5.18 |
| 545 | 248507_at | AT5G50420 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G53770.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G17270.1); similar to Os07g0572600 [Oryza sativa (japonica cultivar-group)] (GB:NP_001060060.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN80219.1); contains domain PTHR13398 (PTHR13398) | -1.25 |
| 546 | 248526_at | AT5G50740 | metal ion binding | -1.75 |
| 547 | 248419_at | AT5G51550 | phosphate-responsive 1 family protein | -1.50 |
| 548 | 248381_at | AT5G51830 | pfkB-type carbohydrate kinase family protein | 3.70 |
| 549 | 248358_at | AT5G52400 | CYP715A1 (cytochrome P450, family 715, subfamily A, polypeptide 1); oxygen binding | 2.83 |
| 550 | 248332_at | AT5G52640 | HSP81-1 (HEAT SHOCK PROTEIN 81-1); ATP binding / unfolded protein binding | 3.29 |
| 551 | 248287_at | AT5G52970 | thylakoid lumen 15.0 kDa protein | -1.37 |
| 552 | 248094_at | AT5G55220 | trigger factor type chaperone family protein | -1.33 |
| 553 | 248060_at | AT5G55560 | protein kinase family protein | 2.50 |
| 554 | 248028_at | AT5G55620 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G09950.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO39940.1) | -1.24 |
| 555 | 248075_at | AT5G55740 | CRR21 (CHLORORESPIRATORY REDUCTION 21) | -1.58 |
| 556 | 247964_at | AT5G56600 | PFN3/PRF3 (PROFILIN 3); actin binding | -1.29 |
| 557 | 247977_at | AT5G56850 | similar to unknown [Populus trichocarpa x Populus deltoides] (GB:ABK96358.1) | -1.61 |
| 558 | 247943_at | AT5G57170 | macrophage migration inhibitory factor family protein / MIF family protein | -1.25 |
| 559 | 247946_at | AT5G57180 | CIA2 (CHLOROPLAST IMPORT APPARATUS 2) | -1.30 |
| 560 | 247949_at | AT5G57220 | CYP81F2 (cytochrome P450, family 81, subfamily F, polypeptide 2); oxygen binding | 5.00 |
| 561 | 247913_at | AT5G57510 | unknown protein | 2.51 |
| 562 | 247880_at | AT5G57780 | transcription regulator | -1.24 |
| 563 | 247889_at | AT5G57930 | APO2 (ACCUMULATION OF PHOTOSYSTEM ONE 2) | -1.25 |
| 564 | 247816_at | AT5G58260 | Encodes subunit NDH-N of NAD(P)H:plastoquinone dehydrogenase complex (Ndh complex) present in the thylakoid membrane of chloroplasts. This subunit is thought to be required for Ndh complex assembly. | -1.47 |
| 565 | 247814_at | AT5G58310 | hydrolase, alpha/beta fold family protein | -2.11 |
| 566 | 247655_at | AT5G59820 | RHL41 (RESPONSIVE TO HIGH LIGHT 41); nucleic acid binding / transcription factor/ zinc ion binding | 3.03 |
| 567 | 247610_at | AT5G60630 | unknown protein | 2.88 |
| 568 | 247596_at | AT5G60840 | unknown protein | -1.28 |
| 569 | 247601_at | AT5G60850 | OBP4 (OBF BINDING PROTEIN 4); DNA binding / transcription factor | -1.29 |
| 570 | 247573_at | AT5G61160 | AACT1 (ANTHOCYANIN 5-AROMATIC ACYLTRANSFERASE 1); transferase | 5.08 |
| 571 | 247486_at | AT5G62140 | similar to unknown [Populus trichocarpa] (GB:ABK94834.1) | -1.32 |
| 572 | 247487_at | AT5G62150 | peptidoglycan-binding LysM domain-containing protein | 2.79 |
| 573 | 247435_at | AT5G62480 | ATGSTU9 (GLUTATHIONE S-TRANSFERASE TAU 9); glutathione transferase | 3.48 |
| 574 | 247439_at | AT5G62670 | AHA11 (ARABIDOPSIS H(+)-ATPASE 11); ATPase | -1.28 |
| 575 | 247347_at | AT5G63780 | SHA1 (SHOOT APICAL MERISTEM ARREST 1); protein binding / zinc ion binding | -1.31 |
| 576 | 247327_at | AT5G64120 | peroxidase, putative | 3.11 |
| 577 | 247216_at | AT5G64860 | DPE1 (DISPROPORTIONATING ENZYME); 4-alpha-glucanotransferase | -1.33 |
| 578 | 247205_at | AT5G64890 | PROPEP2 (Elicitor peptide 2 precursor) | 4.96 |
| 579 | 247215_at | AT5G64905 | PROPEP3 (Elicitor peptide 3 precursor) | 5.97 |
| 580 | 247189_at | AT5G65390 | AGP7 (Arabinogalactan protein 7) | -1.45 |
| 581 | 247145_at | AT5G65600 | legume lectin family protein / protein kinase family protein | 5.71 |
| 582 | 247074_at | AT5G66590 | allergen V5/Tpx-1-related family protein | -1.41 |
| 583 | 247026_at | AT5G67080 | MAPKKK19 (Mitogen-activated protein kinase kinase kinase 19); kinase | 4.96 |
| 584 | 246986_at | AT5G67280 | RLK (RECEPTOR-LIKE KINASE); ATP binding / kinase/ protein serine/threonine kinase | -1.25 |
| 585 | 246984_at | AT5G67310 | CYP81G1 (cytochrome P450, family 81, subfamily G, polypeptide 1); oxygen binding | 4.79 |
| 586 | 246998_at | AT5G67370 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G11840.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN75014.1); contains InterPro domain Protein of unknown function DUF1230 (InterPro:IPR009631) | -1.45 |