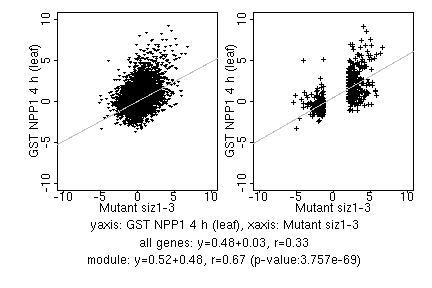
marker gene list
| Mutant siz1-3 |
| GST NPP1 4 h (leaf) |
| probeID | AGICode | Annotation | gene expression |
|
|---|---|---|---|---|
| 1 | 261027_at | AT1G01340 | ATCNGC10 (CYCLIC NUCLEOTIDE GATED CHANNEL 10); calmodulin binding / cyclic nucleotide binding / ion channel | 2.15 |
| 2 | 259428_at | AT1G01560 | ATMPK11 (Arabidopsis thaliana MAP kinase 11); MAP kinase/ kinase | 2.64 |
| 3 | 262118_at | AT1G02850 | glycosyl hydrolase family 1 protein | 2.14 |
| 4 | 265067_at | AT1G03850 | glutaredoxin family protein | 3.06 |
| 5 | 265066_at | AT1G03870 | FLA9 | -2.49 |
| 6 | 263656_at | AT1G04240 | SHY2 (SHORT HYPOCOTYL 2); transcription factor | -1.42 |
| 7 | 263177_at | AT1G05540 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G30160.2); contains InterPro domain Protein of unknown function DUF295 (InterPro:IPR005174) | -1.65 |
| 8 | 260957_at | AT1G06080 | ADS1 (DELTA 9 DESATURASE 1); oxidoreductase | -1.71 |
| 9 | 256066_at | AT1G06980 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G30230.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO21365.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN59819.1) | -3.41 |
| 10 | 260648_at | AT1G08050 | zinc finger (C3HC4-type RING finger) family protein | 2.81 |
| 11 | 261692_at | AT1G08450 | CRT3 (CALRETICULIN 3); calcium ion binding | 2.89 |
| 12 | 264648_at | AT1G09080 | BIP3; ATP binding | 4.75 |
| 13 | 264511_at | AT1G09350 | ATGOLS3 (ARABIDOPSIS THALIANA GALACTINOL SYNTHASE 3); transferase, transferring glycosyl groups / transferase, transferring hexosyl groups | -2.28 |
| 14 | 264434_at | AT1G10340 | ankyrin repeat family protein | 4.58 |
| 15 | 261825_at | AT1G11545 | xyloglucan:xyloglucosyl transferase, putative / xyloglucan endotransglycosylase, putative / endo-xyloglucan transferase, putative | -2.59 |
| 16 | 264346_at | AT1G12010 | 1-aminocyclopropane-1-carboxylate oxidase, putative / ACC oxidase, putative | -4.49 |
| 17 | 264342_at | AT1G12080 | contains domain PTHR22683 (PTHR22683) | -2.17 |
| 18 | 260996_at | AT1G12210 | RFL1 (RPS5-LIKE 1); ATP binding / protein binding | 2.91 |
| 19 | 259534_at | AT1G12290 | disease resistance protein (CC-NBS-LRR class), putative | 2.31 |
| 20 | 259410_at | AT1G13340 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G34220.2); similar to unknown [Carica papaya] (GB:ABS01355.1); contains InterPro domain Protein of unknown function DUF292, eukaryotic (InterPro:IPR005061) | 2.23 |
| 21 | 259385_at | AT1G13470 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G13520.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO42040.1); contains InterPro domain Protein of unknown function DUF1262 (InterPro:IPR010683) | 4.28 |
| 22 | 261480_at | AT1G14280 | PKS2 (PHYTOCHROME KINASE SUBSTRATE 2) | -1.37 |
| 23 | 260776_at | AT1G14580 | zinc finger (C2H2 type) family protein | -1.29 |
| 24 | 262830_at | AT1G14700 | ATPAP3/PAP3 (purple acid phosphatase 3); acid phosphatase/ protein serine/threonine phosphatase | -1.33 |
| 25 | 259502_at | AT1G15670 | kelch repeat-containing F-box family protein | 2.47 |
| 26 | 259489_at | AT1G15790 | similar to protein binding / transcription cofactor [Arabidopsis thaliana] (TAIR:AT1G15780.1); similar to Os08g0564800 [Oryza sativa (japonica cultivar-group)] (GB:NP_001062533.1); similar to hypothetical protein OsJ_027171 [Oryza sativa (japonica cultivar-group)] (GB:EAZ43688.1); similar to CTV.22 [Poncirus trifoliata] (GB:AAN62354.1) | 2.93 |
| 27 | 262731_at | AT1G16420 | ATMC8 (METACASPASE 8); caspase | 2.47 |
| 28 | 256114_at | AT1G16850 | unknown protein | -2.17 |
| 29 | 260734_at | AT1G17600 | disease resistance protein (TIR-NBS-LRR class), putative | 2.64 |
| 30 | 260735_at | AT1G17610 | disease resistance protein-related | 2.72 |
| 31 | 259398_at | AT1G17700 | prenylated rab acceptor (PRA1) family protein | -1.50 |
| 32 | 261661_at | AT1G18360 | hydrolase, alpha/beta fold family protein | -1.22 |
| 33 | 261717_at | AT1G18400 | BEE1 (BR ENHANCED EXPRESSION 1); transcription factor | -1.38 |
| 34 | 261431_at | AT1G18710 | AtMYB47 (myb domain protein 47); DNA binding / transcription factor | -3.23 |
| 35 | 256012_at | AT1G19250 | FMO1 (FLAVIN-DEPENDENT MONOOXYGENASE 1); monooxygenase | 3.99 |
| 36 | 261248_at | AT1G20030 | pathogenesis-related thaumatin family protein | -1.28 |
| 37 | 261226_at | AT1G20190 | ATEXPA11 (ARABIDOPSIS THALIANA EXPANSIN A11) | -2.63 |
| 38 | 255941_at | AT1G20350 | ATTIM17-1 (Arabidopsis thaliana translocase inner membrane subunit 17-1); P-P-bond-hydrolysis-driven protein transmembrane transporter | 3.14 |
| 39 | 259570_at | AT1G20440 | COR47 (cold regulated 47) | -1.90 |
| 40 | 259516_at | AT1G20450 | ERD10/LTI45 (EARLY RESPONSIVE TO DEHYDRATION 10) | -1.62 |
| 41 | 261449_at | AT1G21120 | O-methyltransferase, putative | 3.88 |
| 42 | 259559_at | AT1G21240 | WAK3 (WALL ASSOCIATED KINASE 3); kinase/ protein serine/threonine kinase | 5.28 |
| 43 | 259561_at | AT1G21250 | WAK1 (CELL WALL-ASSOCIATED KINASE); kinase | 3.06 |
| 44 | 255962_at | AT1G22330 | RNA binding | -1.22 |
| 45 | 255969_at | AT1G22330 | RNA binding | -1.80 |
| 46 | 264209_at | AT1G22740 | RAB7 (Ras-related protein 7); GTP binding | -1.27 |
| 47 | 264902_at | AT1G23060 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G70950.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO42267.1) | -1.70 |
| 48 | 264900_at | AT1G23080 | PIN7 (PIN-FORMED 7); auxin:hydrogen symporter/ transporter | -1.57 |
| 49 | 264866_at | AT1G24140 | matrixin family protein | 2.53 |
| 50 | 265028_at | AT1G24530 | transducin family protein / WD-40 repeat family protein | 2.25 |
| 51 | 245637_at | AT1G25230 | purple acid phosphatase family protein | -1.31 |
| 52 | 245816_at | AT1G26210 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G68870.1) | -1.23 |
| 53 | 261021_at | AT1G26380 | FAD-binding domain-containing protein | 4.21 |
| 54 | 261005_at | AT1G26420 | FAD-binding domain-containing protein | 5.10 |
| 55 | 263688_at | AT1G26920 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G69760.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO48418.1) | -1.44 |
| 56 | 264989_at | AT1G27200 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G40720.1); similar to unknown [Populus trichocarpa] (GB:ABK95005.1); contains InterPro domain Protein of unknown function DUF23 (InterPro:IPR008166) | -2.10 |
| 57 | 262748_at | AT1G28610 | GDSL-motif lipase, putative | -1.21 |
| 58 | 259789_at | AT1G29395 | COR414-TM1 (cold regulated 414 thylakoid membrane 1) | -1.41 |
| 59 | 259773_at | AT1G29500 | auxin-responsive protein, putative | -1.34 |
| 60 | 246633_at | AT1G29720 | protein kinase family protein | 2.38 |
| 61 | 265161_at | AT1G30900 | vacuolar sorting receptor, putative | 3.14 |
| 62 | 260706_at | AT1G32350 | AOX1D (ALTERNATIVE OXIDASE 1D); alternative oxidase | 2.40 |
| 63 | 261242_at | AT1G32960 | ATSBT3.3; subtilase | 5.70 |
| 64 | 261216_at | AT1G33030 | O-methyltransferase family 2 protein | 2.66 |
| 65 | 260119_at | AT1G33930 | avirulence-responsive family protein / avirulence induced gene (AIG1) family protein | -2.41 |
| 66 | 260116_at | AT1G33960 | AIG1 (AVRRPT2-INDUCED GENE 1); GTP binding | 4.09 |
| 67 | 261161_at | AT1G34420 | leucine-rich repeat family protein / protein kinase family protein | 2.40 |
| 68 | 259550_at | AT1G35230 | AGP5 (ARABINOGALACTAN-PROTEIN 5) | 4.87 |
| 69 | 262010_at | AT1G35612 | transposable element gene | -1.24 |
| 70 | 261339_at | AT1G35710 | leucine-rich repeat transmembrane protein kinase, putative | 3.12 |
| 71 | 261292_at | AT1G36940 | unknown protein | -1.48 |
| 72 | 259507_at | AT1G43910 | AAA-type ATPase family protein | 2.85 |
| 73 | 245244_at | AT1G44350 | ILL6 (IAA-leucine resistant (ILR)-like gene 6); metallopeptidase | -2.18 |
| 74 | 262244_at | AT1G48260 | CIPK17 (SNF1-RELATED PROTEIN KINASE 3.21); kinase | -1.47 |
| 75 | 260754_at | AT1G49000 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G18560.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN78728.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO68009.1) | 3.54 |
| 76 | 260771_at | AT1G49160 | WNK7 (Arabidopsis WNK kinase 7); kinase | -1.55 |
| 77 | 245749_at | AT1G51090 | heavy-metal-associated domain-containing protein | -2.98 |
| 78 | 265136_at | AT1G51270 | structural molecule | 2.25 |
| 79 | 256169_at | AT1G51800 | leucine-rich repeat protein kinase, putative | 3.80 |
| 80 | 246366_at | AT1G51850 | leucine-rich repeat protein kinase, putative | 3.79 |
| 81 | 246373_at | AT1G51860 | leucine-rich repeat protein kinase, putative | 6.47 |
| 82 | 246368_at | AT1G51890 | leucine-rich repeat protein kinase, putative | 4.03 |
| 83 | 246370_at | AT1G51920 | unknown protein | 5.92 |
| 84 | 265053_at | AT1G52000 | jacalin lectin family protein | -1.46 |
| 85 | 259839_at | AT1G52190 | proton-dependent oligopeptide transport (POT) family protein | -1.57 |
| 86 | 259640_at | AT1G52400 | BGL1 (BETA-GLUCOSIDASE HOMOLOG 1); hydrolase, hydrolyzing O-glycosyl compounds | -1.32 |
| 87 | 259609_at | AT1G52410 | TSA1 (TSK-ASSOCIATING PROTEIN 1); calcium ion binding / protein binding | -2.00 |
| 88 | 260146_at | AT1G52770 | phototropic-responsive NPH3 family protein | 2.46 |
| 89 | 260152_at | AT1G52830 | IAA6 (indoleacetic acid-induced protein 6); transcription factor | -1.99 |
| 90 | 261315_at | AT1G53170 | ATERF-8/ATERF8 (ETHYLENE RESPONSE ELEMENT BINDING FACTOR 4); DNA binding / transcription factor/ transcription repressor | -1.28 |
| 91 | 263198_at | AT1G53990 | GLIP3 (GDSL-motif lipase 3); carboxylesterase/ lipase | 3.43 |
| 92 | 263174_at | AT1G54040 | ESP (EPITHIOSPECIFIER PROTEIN) | -2.05 |
| 93 | 264238_at | AT1G54740 | similar to structural constituent of ribosome [Arabidopsis thaliana] (TAIR:AT1G22110.1); similar to unknown [Populus trichocarpa x Populus deltoides] (GB:ABK96485.1) | -1.22 |
| 94 | 262085_at | AT1G56060 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G32190.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO68639.1); contains domain PD188784 (PD188784) | 3.11 |
| 95 | 245627_at | AT1G56600 | ATGOLS2 (ARABIDOPSIS THALIANA GALACTINOL SYNTHASE 2); transferase, transferring glycosyl groups / transferase, transferring hexosyl groups | -1.63 |
| 96 | 245657_at | AT1G56720 | protein kinase family protein | -1.26 |
| 97 | 246405_at | AT1G57630 | disease resistance protein (TIR class), putative | 4.36 |
| 98 | 246406_at | AT1G57650 | disease resistance protein (NBS-LRR class), putative | 5.28 |
| 99 | 245840_at | AT1G58420 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G10140.1); contains InterPro domain Uncharacterised conserved protein UCP031279 (InterPro:IPR016972) | 3.05 |
| 100 | 264289_at | AT1G61890 | MATE efflux family protein | -1.65 |
| 101 | 264746_at | AT1G62300 | WRKY6 (WRKY DNA-binding protein 6); transcription factor | 2.22 |
| 102 | 260637_at | AT1G62380 | ACO2 (ACC OXIDASE 2) | -1.25 |
| 103 | 265116_at | AT1G62480 | vacuolar calcium-binding protein-related | -1.22 |
| 104 | 265111_at | AT1G62510 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | -2.03 |
| 105 | 265118_at | AT1G62660 | beta-fructosidase (BFRUCT3) / beta-fructofuranosidase / invertase, vacuolar | -1.58 |
| 106 | 262643_at | AT1G62770 | invertase/pectin methylesterase inhibitor family protein | -1.41 |
| 107 | 259793_at | AT1G64380 | AP2 domain-containing transcription factor, putative | -1.83 |
| 108 | 261975_at | AT1G64640 | plastocyanin-like domain-containing protein | -2.09 |
| 109 | 264157_at | AT1G65310 | ATXTH17 (XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 17); hydrolase, acting on glycosyl bonds | -5.20 |
| 110 | 264635_at | AT1G65500 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G65486.1) | 2.74 |
| 111 | 264680_at | AT1G65510 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G65490.1) | 2.79 |
| 112 | 262930_at | AT1G65690 | harpin-induced protein-related / HIN1-related / harpin-responsive protein-related | 2.85 |
| 113 | 260141_at | AT1G66350 | RGL1 (RGA-LIKE 1); transcription factor | -1.74 |
| 114 | 256366_at | AT1G66880 | serine/threonine protein kinase family protein | 3.13 |
| 115 | 255912_at | AT1G66960 | lupeol synthase, putative / 2,3-oxidosqualene-triterpenoid cyclase, putative | 5.25 |
| 116 | 255879_at | AT1G67000 | kinase | 3.12 |
| 117 | 245193_at | AT1G67810 | Fe-S metabolism associated domain-containing protein | 2.42 |
| 118 | 260015_at | AT1G67980 | CCoAMT (caffeoyl-CoA 3-O-methyltransferase); caffeoyl-CoA O-methyltransferase | 3.63 |
| 119 | 259371_at | AT1G69080 | universal stress protein (USP) family protein | -1.21 |
| 120 | 259373_at | AT1G69160 | unknown protein | -1.72 |
| 121 | 260396_at | AT1G69720 | HO3 (HEME OXYGENASE 3); heme oxygenase (decyclizing) | 2.31 |
| 122 | 260419_at | AT1G69730 | protein kinase family protein | 2.55 |
| 123 | 264696_at | AT1G70230 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G01430.1); similar to Os09g0375300 [Oryza sativa (japonica cultivar-group)] (GB:NP_001063036.1); similar to Os06g0235200 [Oryza sativa (japonica cultivar-group)] (GB:NP_001057240.1); similar to leaf senescence protein-like [Oryza sativa (japonica cultivar-group)] (GB:BAD26032.1); contains InterPro domain Protein of unknown function DUF231, plant (InterPro:IPR004253) | -1.56 |
| 124 | 264314_at | AT1G70420 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G23710.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO66069.1); contains InterPro domain Protein of unknown function DUF1645 (InterPro:IPR012442) | -1.27 |
| 125 | 259751_at | AT1G71030 | ATMYBL2 (Arabidopsis myb-like 2); DNA binding / transcription factor | -1.32 |
| 126 | 259893_at | AT1G71390 | disease resistance family protein / LRR family protein | 3.47 |
| 127 | 256337_at | AT1G72060 | serine-type endopeptidase inhibitor | 3.11 |
| 128 | 259846_at | AT1G72140 | proton-dependent oligopeptide transport (POT) family protein | -2.01 |
| 129 | 259850_at | AT1G72240 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G22470.1) | 4.27 |
| 130 | 259852_at | AT1G72280 | AERO1 (ARABIDOPSIS ENDOPLASMIC RETICULUM OXIDOREDUCTINS 1); FAD binding / electron carrier/ oxidoreductase, acting on sulfur group of donors, disulfide as acceptor / protein binding | 2.28 |
| 131 | 245734_at | AT1G73480 | hydrolase, alpha/beta fold family protein | -1.70 |
| 132 | 260046_at | AT1G73805 | calmodulin binding | 3.12 |
| 133 | 260068_at | AT1G73805 | calmodulin binding | 2.81 |
| 134 | 260070_at | AT1G73830 | BEE3 (BR ENHANCED EXPRESSION 3); DNA binding / transcription factor | -1.91 |
| 135 | 260239_at | AT1G74360 | leucine-rich repeat transmembrane protein kinase, putative | 2.84 |
| 136 | 260225_at | AT1G74590 | ATGSTU10 (Arabidopsis thaliana Glutathione S-transferase (class tau) 10); glutathione transferase | 2.48 |
| 137 | 262177_at | AT1G74710 | ICS1 (ISOCHORISMATE SYNTHASEI); isochorismate synthase | 3.25 |
| 138 | 262211_at | AT1G74930 | ORA47; DNA binding / transcription factor | -4.24 |
| 139 | 259925_at | AT1G75040 | PR5 (PATHOGENESIS-RELATED GENE 5) | 4.43 |
| 140 | 262947_at | AT1G75750 | GASA1 (GAST1 PROTEIN HOMOLOG 1) | -1.25 |
| 141 | 262978_at | AT1G75780 | TUB1 (tubulin beta-1 chain); structural molecule | -1.42 |
| 142 | 262698_at | AT1G75960 | AMP-binding protein, putative | -2.54 |
| 143 | 264958_at | AT1G76960 | unknown protein | 3.06 |
| 144 | 262164_at | AT1G78070 | WD-40 repeat family protein | -2.08 |
| 145 | 260806_at | AT1G78260 | RNA recognition motif (RRM)-containing protein | -1.36 |
| 146 | 263133_at | AT1G78450 | SOUL heme-binding family protein | -1.69 |
| 147 | 264100_at | AT1G78970 | LUP1 (LUPEOL SYNTHASE 1); lupeol synthase | -1.43 |
| 148 | 262891_at | AT1G79460 | GA2 (GA REQUIRING 2); ent-kaurene synthase | -1.43 |
| 149 | 261394_at | AT1G79680 | wall-associated kinase, putative | 2.41 |
| 150 | 263401_at | AT2G04070 | transporter | 2.44 |
| 151 | 263854_at | AT2G04430 | ATNUDT5 (Arabidopsis thaliana Nudix hydrolase homolog 5); hydrolase | 2.37 |
| 152 | 263852_at | AT2G04450 | ATNUDT6 (Arabidopsis thaliana Nudix hydrolase homolog 6); ADP-ribose diphosphatase/ NAD binding / hydrolase | 3.52 |
| 153 | 266215_at | AT2G06850 | EXGT-A1 (ENDOXYLOGLUCAN TRANSFERASE); hydrolase, acting on glycosyl bonds | -1.66 |
| 154 | 265658_at | AT2G13810 | ALD1 (AGD2-LIKE DEFENSE RESPONSE PROTEIN1); transaminase | 3.82 |
| 155 | 265837_at | AT2G14560 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G33840.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO48825.1); contains InterPro domain Protein of unknown function DUF567 (InterPro:IPR007612) | 2.31 |
| 156 | 266385_at | AT2G14610 | PR1 (PATHOGENESIS-RELATED GENE 1) | 4.57 |
| 157 | 265499_at | AT2G15480 | UGT73B5 (UDP-GLUCOSYL TRANSFERASE 73B5); UDP-glycosyltransferase/ transferase, transferring glycosyl groups | 2.48 |
| 158 | 265480_at | AT2G15970 | COR413-PM1 (cold regulated 413 plasma membrane 1) | -1.37 |
| 159 | 263574_at | AT2G16990 | tetracycline transporter | -1.75 |
| 160 | 263584_at | AT2G17040 | ANAC036 (Arabidopsis NAC domain containing protein 36); transcription factor | 2.31 |
| 161 | 264788_at | AT2G17880 | DNAJ heat shock protein, putative | -2.17 |
| 162 | 265327_at | AT2G18210 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G36500.1); similar to hypothetical protein [Thellungiella halophila] (GB:ABB45855.1) | 2.71 |
| 163 | 266070_at | AT2G18660 | EXLB3 (EXPANSIN-LIKE B3 PRECURSOR) | 5.88 |
| 164 | 266017_at | AT2G18690 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G18680.1); similar to hypothetical protein OsI_029427 [Oryza sativa (indica cultivar-group)] (GB:EAZ08195.1) | 3.37 |
| 165 | 266946_at | AT2G18890 | protein kinase family protein | -1.29 |
| 166 | 267436_at | AT2G19190 | FRK1 (FLG22-INDUCED RECEPTOR-LIKE KINASE 1); kinase | 4.42 |
| 167 | 266693_at | AT2G19800 | MIOX2 (MYO-INOSITOL OXYGENASE 2) | -2.86 |
| 168 | 265597_at | AT2G20142 | transmembrane receptor | 3.27 |
| 169 | 265387_at | AT2G20670 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G32480.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO69754.1); contains InterPro domain Protein of unknown function DUF506, plant (InterPro:IPR006502) | -1.49 |
| 170 | 263765_at | AT2G21540 | SEC14 cytosolic factor, putative / phosphoglyceride transfer protein, putative | -1.27 |
| 171 | 263876_at | AT2G21880 | AtRABG2/AtRab7A (Arabidopsis Rab GTPase homolog G2); GTP binding | -1.56 |
| 172 | 263433_at | AT2G22240 | inositol-3-phosphate synthase isozyme 2 / myo-inositol-1-phosphate synthase 2 / MI-1-P synthase 2 / IPS 2 | -1.26 |
| 173 | 245082_at | AT2G23270 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G37290.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN62855.1) | 2.46 |
| 174 | 267288_at | AT2G23680 | stress-responsive protein, putative | 2.71 |
| 175 | 265993_at | AT2G24160 | pseudogene, leucine rich repeat protein family, contains leucine rich-repeat domains Pfam:PF00560, INTERPRO:IPR001611; contains some similarity to Cf-4 (Lycopersicon hirsutum) gi|2808683|emb|CAA05268; blastp match of 37% identity and 8.4e-98 P-value to GP|2808683|emb|CAA05268.1||AJ002235 Cf-4 {Lycopersicon hirsutum} | 2.89 |
| 176 | 263318_at | AT2G24762 | ATGDU4 (ARABIDOPSIS THALIANA GLUTAMINE DUMPER 4) | -1.51 |
| 177 | 265659_at | AT2G25440 | leucine-rich repeat family protein | 3.18 |
| 178 | 266841_at | AT2G26150 | ATHSFA2 (Arabidopsis thaliana heat shock transcription factor A2); DNA binding / transcription factor | 3.28 |
| 179 | 245035_at | AT2G26400 | ARD/ATARD3 (ACIREDUCTONE DIOXYGENASE); acireductone dioxygenase [iron(II)-requiring]/ heteroglycan binding / metal ion binding | 3.71 |
| 180 | 245052_at | AT2G26440 | pectinesterase family protein | 2.31 |
| 181 | 267612_at | AT2G26690 | nitrate transporter (NTP2) | -1.36 |
| 182 | 266247_at | AT2G27660 | DC1 domain-containing protein | 2.22 |
| 183 | 266780_at | AT2G29110 | ATGLR2.8 (Arabidopsis thaliana glutamate receptor 2.8) | 3.55 |
| 184 | 266782_at | AT2G29120 | ATGLR2.7 (Arabidopsis thaliana glutamate receptor 2.7) | 2.90 |
| 185 | 266267_at | AT2G29460 | ATGSTU4 (GLUTATHIONE S-TRANSFERASE 22); glutathione transferase | 3.62 |
| 186 | 267300_at | AT2G30140 | UDP-glucoronosyl/UDP-glucosyl transferase family protein | 3.08 |
| 187 | 267565_at | AT2G30750 | CYP71A12 (CYTOCHROME P450, FAMILY 71, SUBFAMILY A, POLYPEPTIDE 12); oxygen binding | 2.79 |
| 188 | 265723_at | AT2G32140 | transmembrane receptor | 2.99 |
| 189 | 267546_at | AT2G32680 | disease resistance family protein | 4.59 |
| 190 | 266996_at | AT2G34490 | CYP710A2 (cytochrome P450, family 710, subfamily A, polypeptide 2); C-22 sterol desaturase/ oxygen binding | -1.21 |
| 191 | 266901_at | AT2G34600 | JAZ7/TIFY5B (JASMONATE-ZIM-DOMAIN PROTEIN 7) | -3.48 |
| 192 | 263948_at | AT2G35980 | YLS9 (YELLOW-LEAF-SPECIFIC GENE 9) | 5.30 |
| 193 | 267158_at | AT2G37640 | ATEXPA3 (ARABIDOPSIS THALIANA EXPANSIN A3) | -1.71 |
| 194 | 266167_at | AT2G38860 | YLS5 (yellow-leaf-specific gene 5) | 2.26 |
| 195 | 266172_at | AT2G39010 | PIP2;6/PIP2E (plasma membrane intrinsic protein 2;6); water channel | -1.76 |
| 196 | 266993_at | AT2G39210 | nodulin family protein | 2.79 |
| 197 | 245088_at | AT2G39850 | subtilase | -1.51 |
| 198 | 265877_at | AT2G42380 | bZIP transcription factor family protein | -2.70 |
| 199 | 263495_at | AT2G42530 | COR15B | -2.03 |
| 200 | 263497_at | AT2G42540 | COR15A (COLD-REGULATED 15A) | -3.31 |
| 201 | 263981_at | AT2G42870 | HLH1/PAR1 (PHY RAPIDLY REGULATED 1); transcription regulator | -1.38 |
| 202 | 265248_at | AT2G43010 | PIF4 (PHYTOCHROME INTERACTING FACTOR 4); DNA binding / transcription factor | -1.50 |
| 203 | 260568_at | AT2G43570 | chitinase, putative | 5.05 |
| 204 | 260556_at | AT2G43620 | chitinase, putative | 3.19 |
| 205 | 267345_at | AT2G44240 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G44220.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO67164.1); contains InterPro domain Protein of unknown function DUF239, plant (InterPro:IPR004314) | 3.52 |
| 206 | 267385_at | AT2G44380 | DC1 domain-containing protein | 4.73 |
| 207 | 263783_at | AT2G46400 | WRKY46 (WRKY DNA-binding protein 46); transcription factor | 2.75 |
| 208 | 266320_at | AT2G46640 | unknown protein | -1.95 |
| 209 | 266761_at | AT2G47130 | short-chain dehydrogenase/reductase (SDR) family protein | 2.96 |
| 210 | 266503_at | AT2G47780 | rubber elongation factor (REF) protein-related | -1.45 |
| 211 | 266464_at | AT2G47800 | ATMRP4 (Arabidopsis thaliana multidrug resistance-associated protein 4) | 2.52 |
| 212 | 266460_at | AT2G47930 | AGP26/ATAGP26 (ARABINOGALACTAN PROTEINS 26) | -1.72 |
| 213 | 259320_at | AT3G01080 | WRKY58 (WRKY DNA-binding protein 58); transcription factor | 4.01 |
| 214 | 258947_at | AT3G01830 | calmodulin-related protein, putative | 2.76 |
| 215 | 259095_at | AT3G05020 | ACP1 (ACYL CARRIER PROTEIN 1) | -1.27 |
| 216 | 259106_at | AT3G05490 | RALFL22 (RALF-LIKE 22) | -1.26 |
| 217 | 258742_at | AT3G05800 | transcription factor | -1.93 |
| 218 | 258751_at | AT3G05890 | RCI2B (RARE-COLD-INDUCIBLE 2B) | -2.28 |
| 219 | 258736_at | AT3G05900 | neurofilament protein-related | -2.40 |
| 220 | 258528_at | AT3G06770 | glycoside hydrolase family 28 protein / polygalacturonase (pectinase) family protein | -1.30 |
| 221 | 259015_at | AT3G07350 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G25240.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO38687.1); contains InterPro domain Protein of unknown function DUF506, plant (InterPro:IPR006502) | -1.40 |
| 222 | 259213_at | AT3G09010 | protein kinase family protein | 2.20 |
| 223 | 259033_at | AT3G09410 | pectinacetylesterase family protein | 2.63 |
| 224 | 256252_at | AT3G11340 | UDP-glucoronosyl/UDP-glucosyl transferase family protein | 3.00 |
| 225 | 256245_at | AT3G12580 | HSP70 (heat shock protein 70); ATP binding | 2.19 |
| 226 | 257185_at | AT3G13100 | ATMRP7 (Arabidopsis thaliana multidrug resistance-associated protein 7) | 4.78 |
| 227 | 256647_at | AT3G13610 | oxidoreductase, 2OG-Fe(II) oxygenase family protein | 4.27 |
| 228 | 256787_at | AT3G13790 | ATBFRUCT1/ATCWINV1 (ARABIDOPSIS THALIANA CELL WALL INVERTASE 1); beta-fructofuranosidase/ hydrolase, hydrolyzing O-glycosyl compounds | 2.78 |
| 229 | 258203_at | AT3G13950 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G13266.1); similar to Ankyrin [Medicago truncatula] (GB:ABN08906.1) | 4.00 |
| 230 | 258196_at | AT3G13980 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G54200.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN69469.1) | -1.44 |
| 231 | 257280_at | AT3G14440 | NCED3 (NINE-CIS-EPOXYCAROTENOID DIOXYGENASE3) | -1.98 |
| 232 | 264223_s_at | AT3G16030 | CES101 (CALLUS EXPRESSION OF RBCS 101); carbohydrate binding / kinase | 2.35 |
| 233 | 258054_at | AT3G16240 | DELTA-TIP (delta tonoplast integral protein); water channel | -1.23 |
| 234 | 258432_at | AT3G16570 | RALFL23 (RALF-LIKE 23) | -1.30 |
| 235 | 258421_at | AT3G16690 | nodulin MtN3 family protein | -1.71 |
| 236 | 258434_at | AT3G16770 | ATEBP/ERF72/RAP2.3 (RELATED TO AP2 3); DNA binding / protein binding / transcription activator/ transcription factor | -1.21 |
| 237 | 257651_at | AT3G16850 | glycoside hydrolase family 28 protein / polygalacturonase (pectinase) family protein | -1.30 |
| 238 | 257061_at | AT3G18250 | contains domain PROKAR_LIPOPROTEIN (PS51257) | 3.90 |
| 239 | 257671_at | AT3G20450 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G11905.1); similar to unknown [Populus trichocarpa] (GB:ABK95443.1); similar to Os05g0272900 [Oryza sativa (japonica cultivar-group)] (GB:NP_001055067.1); contains InterPro domain B-cell receptor-associated 31-like; (InterPro:IPR008417) | -1.32 |
| 240 | 256969_at | AT3G21080 | ABC transporter-related | 4.04 |
| 241 | 258181_at | AT3G21670 | nitrate transporter (NTP3) | -1.80 |
| 242 | 256933_at | AT3G22600 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | 3.12 |
| 243 | 258300_at | AT3G23340 | CKL10 (Casein Kinase I-like 10); casein kinase I/ kinase | -1.40 |
| 244 | 257591_at | AT3G24900 | disease resistance family protein / LRR family protein | 2.62 |
| 245 | 257100_at | AT3G25010 | disease resistance family protein | 3.81 |
| 246 | 256756_at | AT3G25610 | haloacid dehalogenase-like hydrolase family protein | 2.17 |
| 247 | 257623_at | AT3G26210 | CYP71B23 (cytochrome P450, family 71, subfamily B, polypeptide 23); oxygen binding | 3.42 |
| 248 | 257624_at | AT3G26220 | CYP71B3 (cytochrome P450, family 71, subfamily B, polypeptide 3); oxygen binding | 2.69 |
| 249 | 256874_at | AT3G26320 | CYP71B36 (cytochrome P450, family 71, subfamily B, polypeptide 36); oxygen binding | 2.41 |
| 250 | 256883_at | AT3G26440 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G13000.2); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G13000.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO42517.1); contains InterPro domain Protein of unknown function DUF707 (InterPro:IPR007877) | 2.25 |
| 251 | 256877_at | AT3G26470 | similar to ADR1-L1 (ADR1-LIKE 1), ATP binding / protein binding [Arabidopsis thaliana] (TAIR:AT4G33300.2); similar to ADR1-L1 (ADR1-LIKE 1), ATP binding / protein binding [Arabidopsis thaliana] (TAIR:AT4G33300.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO61278.1); contains InterPro domain Disease resistance, plant (InterPro:IPR014011) | 2.98 |
| 252 | 256872_at | AT3G26490 | phototropic-responsive NPH3 family protein | -1.32 |
| 253 | 258277_at | AT3G26830 | PAD3 (PHYTOALEXIN DEFICIENT 3); oxygen binding | 3.41 |
| 254 | 258239_at | AT3G27690 | LHCB2:4 (Photosystem II light harvesting complex gene 2.3); chlorophyll binding | -2.43 |
| 255 | 257271_at | AT3G28007 | nodulin MtN3 family protein | -1.96 |
| 256 | 256576_at | AT3G28210 | PMZ; zinc ion binding | 2.75 |
| 257 | 256577_at | AT3G28220 | meprin and TRAF homology domain-containing protein / MATH domain-containing protein | -3.19 |
| 258 | 256602_at | AT3G28310 | similar to AT14A [Arabidopsis thaliana] (TAIR:AT3G28290.1); similar to AT14A [Arabidopsis thaliana] (TAIR:AT3G28300.1); contains InterPro domain Protein of unknown function DUF677 (InterPro:IPR007749) | -2.90 |
| 259 | 256593_at | AT3G28510 | AAA-type ATPase family protein | 5.49 |
| 260 | 256596_at | AT3G28540 | AAA-type ATPase family protein | 3.25 |
| 261 | 256989_at | AT3G28580 | AAA-type ATPase family protein | 4.19 |
| 262 | 257137_at | AT3G28860 | ATMDR11/ATPGP19/MDR1/MDR11/PGP19 (P-GLYCOPROTEIN 19); ATPase, coupled to transmembrane movement of substances / auxin efflux transmembrane transporter | -1.49 |
| 263 | 256743_at | AT3G29370 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G39240.1) | -1.72 |
| 264 | 256598_at | AT3G30180 | BR6OX2/CYP85A2 (BRASSINOSTEROID-6-OXIDASE 2); monooxygenase/ oxygen binding | -1.33 |
| 265 | 252692_at | AT3G43960 | cysteine proteinase, putative | -1.52 |
| 266 | 252681_at | AT3G44350 | ANAC061 (Arabidopsis NAC domain containing protein 61); transcription factor | 3.30 |
| 267 | 252607_at | AT3G44990 | XTR8 (xyloglucan:xyloglucosyl transferase 8); hydrolase, acting on glycosyl bonds | -2.58 |
| 268 | 252549_at | AT3G45860 | receptor-like protein kinase, putative | 4.31 |
| 269 | 252563_at | AT3G45970 | ATEXLA1 (ARABIDOPSIS THALIANA EXPANSIN-LIKE A1) | -2.49 |
| 270 | 252533_at | AT3G46110 | signal transducer | 2.17 |
| 271 | 252520_at | AT3G46370 | leucine-rich repeat protein kinase, putative | -4.82 |
| 272 | 252529_at | AT3G46490 | oxidoreductase, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors | -1.77 |
| 273 | 252415_at | AT3G47340 | ASN1 (DARK INDUCIBLE 6) | -2.06 |
| 274 | 252417_at | AT3G47480 | calcium-binding EF hand family protein | 3.99 |
| 275 | 252421_at | AT3G47540 | chitinase, putative | 2.92 |
| 276 | 252373_at | AT3G48090 | EDS1 (ENHANCED DISEASE SUSCEPTIBILITY 1); signal transducer/ triacylglycerol lipase | 2.47 |
| 277 | 252345_at | AT3G48640 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G66670.2); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G66670.1) | 3.33 |
| 278 | 252346_at | AT3G48650 | pseudogene, At14a-related protein, similar to At14a (GI:11994571 and GI:11994573) (Arabidopsis thaliana) | 2.91 |
| 279 | 252296_at | AT3G48970 | copper-binding family protein | -1.75 |
| 280 | 252243_at | AT3G50120 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G50150.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G50130.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G50170.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO71911.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN59797.1); contains InterPro domain Protein of unknown function DUF247, plant (InterPro:IPR004158) | -1.57 |
| 281 | 252170_at | AT3G50480 | HR4 (HOMOLOG OF RPW8 4) | 2.48 |
| 282 | 252184_at | AT3G50660 | DWF4 (DWARF 4) | -1.27 |
| 283 | 252136_at | AT3G50770 | calmodulin-related protein, putative | 2.59 |
| 284 | 252129_at | AT3G50890 | ATHB28 (ARABIDOPSIS THALIANA HOMEOBOX PROTEIN 28); DNA binding / transcription factor | -1.42 |
| 285 | 252131_at | AT3G50930 | AAA-type ATPase family protein | 3.33 |
| 286 | 252102_at | AT3G50970 | LTI30 (LOW TEMPERATURE-INDUCED 30) | -1.38 |
| 287 | 252143_at | AT3G51150 | kinesin motor family protein | -1.57 |
| 288 | 252098_at | AT3G51330 | aspartyl protease family protein | 2.81 |
| 289 | 246302_at | AT3G51860 | CAX3 (cation exchanger 3); cation:cation antiporter | 2.70 |
| 290 | 252081_at | AT3G51910 | AT-HSFA7A (Arabidopsis thaliana heat shock transcription factor A7A); DNA binding / transcription factor | 2.13 |
| 291 | 252060_at | AT3G52430 | PAD4 (PHYTOALEXIN DEFICIENT 4); triacylglycerol lipase | 3.91 |
| 292 | 251996_at | AT3G52840 | BGAL2 (beta-galactosidase 2); beta-galactosidase | -1.26 |
| 293 | 251977_at | AT3G53250 | auxin-responsive family protein | -3.82 |
| 294 | 251884_at | AT3G54150 | embryo-abundant protein-related | 3.73 |
| 295 | 251895_at | AT3G54420 | ATEP3 (Arabidopsis thaliana chitinase class IV); chitinase | 2.14 |
| 296 | 251768_at | AT3G55940 | phosphoinositide-specific phospholipase C, putative | -1.36 |
| 297 | 251769_at | AT3G55950 | protein kinase family protein | 2.45 |
| 298 | 251673_at | AT3G57240 | BG3 (BETA-1,3-GLUCANASE 3); hydrolase, hydrolyzing O-glycosyl compounds | 3.87 |
| 299 | 251625_at | AT3G57260 | BGL2 (PATHOGENESIS-RELATED PROTEIN 2); glucan 1,3-beta-glucosidase/ hydrolase, hydrolyzing O-glycosyl compounds | 4.30 |
| 300 | 251633_at | AT3G57460 | catalytic/ metal ion binding / metalloendopeptidase/ zinc ion binding | 2.62 |
| 301 | 251647_at | AT3G57770 | protein kinase, putative | -1.46 |
| 302 | 251612_at | AT3G57950 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G42180.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN83225.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO23380.1) | 3.02 |
| 303 | 251575_at | AT3G58120 | bZIP transcription factor family protein | -1.83 |
| 304 | 251479_at | AT3G59700 | ATHLECRK (ARABIDOPSIS THALIANA LECTIN-RECEPTOR KINASE); kinase | 2.15 |
| 305 | 251402_at | AT3G60290 | oxidoreductase, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors | -2.32 |
| 306 | 251400_at | AT3G60420 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G60450.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO70569.1); contains InterPro domain Phosphoglycerate mutase (InterPro:IPR013078); contains InterPro domain PRIB5 (InterPro:IPR012398) | 2.95 |
| 307 | 251419_at | AT3G60470 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G44930.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN69719.1); contains InterPro domain Protein of unknown function DUF247, plant (InterPro:IPR004158) | 4.15 |
| 308 | 251306_at | AT3G61260 | DNA-binding family protein / remorin family protein | -1.37 |
| 309 | 251272_at | AT3G61890 | ATHB-12 (ARABIDOPSIS THALIANA HOMEOBOX PROTEIN 12); transcription factor | -1.34 |
| 310 | 255630_at | AT4G00700 | C2 domain-containing protein | 4.94 |
| 311 | 255599_at | AT4G01010 | ATCNGC13 (cyclic nucleotide gated channel 13); calmodulin binding / cyclic nucleotide binding / ion channel | 2.30 |
| 312 | 255568_at | AT4G01250 | WRKY22 (WRKY DNA-binding protein 22); transcription factor | -1.55 |
| 313 | 255617_at | AT4G01330 | protein kinase family protein | -1.27 |
| 314 | 255595_at | AT4G01700 | chitinase, putative | 2.36 |
| 315 | 255506_at | AT4G02130 | GATL6/LGT10; polygalacturonate 4-alpha-galacturonosyltransferase/ transferase, transferring glycosyl groups / transferase, transferring hexosyl groups | -1.32 |
| 316 | 255524_at | AT4G02330 | ATPMEPCRB; pectinesterase | 3.11 |
| 317 | 255430_at | AT4G03320 | TIC20-IV (TRANSLOCON AT THE INNER ENVELOPE MEMBRANE OF CHLOROPLASTS 20-IV); P-P-bond-hydrolysis-driven protein transmembrane transporter | 2.31 |
| 318 | 255406_at | AT4G03450 | ankyrin repeat family protein | 2.90 |
| 319 | 255381_at | AT4G03510 | RMA1 (Ring finger protein with Membrane Anchor 1); protein binding / ubiquitin-protein ligase/ zinc ion binding | -1.37 |
| 320 | 255319_at | AT4G04220 | disease resistance family protein | 2.21 |
| 321 | 255341_at | AT4G04500 | protein kinase family protein | 4.90 |
| 322 | 255342_at | AT4G04510 | protein kinase family protein | 3.29 |
| 323 | 255285_at | AT4G04630 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G21970.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO17943.1); contains InterPro domain Protein of unknown function DUF584 (InterPro:IPR007608) | -1.58 |
| 324 | 255028_at | AT4G09890 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G47480.1); similar to unknown [Populus trichocarpa] (GB:ABK92947.1) | -1.21 |
| 325 | 254975_at | AT4G10500 | oxidoreductase, 2OG-Fe(II) oxygenase family protein | 5.95 |
| 326 | 254938_at | AT4G10770 | ATOPT7 (oligopeptide transporter 7); oligopeptide transporter | -1.35 |
| 327 | 254954_at | AT4G10910 | unknown protein | -1.88 |
| 328 | 254897_at | AT4G11470 | protein kinase family protein | 3.48 |
| 329 | 254869_at | AT4G11890 | protein kinase family protein | 3.69 |
| 330 | 254785_at | AT4G12730 | FLA2 | -1.40 |
| 331 | 254746_at | AT4G12980 | auxin-responsive protein, putative | -1.74 |
| 332 | 254716_at | AT4G13560 | UNE15 (unfertilized embryo sac 15) | -1.26 |
| 333 | 254685_at | AT4G13790 | auxin-responsive protein, putative | -2.15 |
| 334 | 245325_at | AT4G14130 | XTR7 (XYLOGLUCAN ENDOTRANSGLYCOSYLASE 7); hydrolase, acting on glycosyl bonds | -2.17 |
| 335 | 245329_at | AT4G14365 | zinc finger (C3HC4-type RING finger) family protein / ankyrin repeat family protein | 2.80 |
| 336 | 245610_at | AT4G14380 | similar to hypothetical protein [Vitis vinifera] (GB:CAN63630.1) | -1.83 |
| 337 | 245566_at | AT4G14610 | pseudogene, disease resistance protein (CC-NBS-LRR class), putative, domain signature CC-NBS-LRR exists, suggestive of a disease resistance protein.; blastp match of 45% identity and 2.2e-162 P-value to GP|24461866|gb|AAN62353.1|AF506028_20|AF506028 NBS-LRR type disease resistance protein {Poncirus trifoliata} | 2.41 |
| 338 | 245336_at | AT4G16515 | unknown protein | -1.69 |
| 339 | 245449_at | AT4G16870 | transposable element gene | -1.48 |
| 340 | 245346_at | AT4G17090 | CT-BMY (BETA-AMYLASE 3, BETA-AMYLASE 8); beta-amylase | -1.27 |
| 341 | 245427_at | AT4G17550 | transporter-related | -1.86 |
| 342 | 254660_at | AT4G18250 | receptor serine/threonine kinase, putative | 4.68 |
| 343 | 254667_at | AT4G18280 | glycine-rich cell wall protein-related | -1.30 |
| 344 | 254609_at | AT4G18970 | GDSL-motif lipase/hydrolase family protein | -1.28 |
| 345 | 254563_at | AT4G19120 | ERD3 (EARLY-RESPONSIVE TO DEHYDRATION 3) | -1.41 |
| 346 | 254564_at | AT4G19170 | NCED4 (NINE-CIS-EPOXYCAROTENOID DIOXYGENASE 4) | -1.30 |
| 347 | 254571_at | AT4G19370 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G31720.1); similar to unknown [Picea sitchensis] (GB:ABK21519.1); similar to Os02g0703300 [Oryza sativa (japonica cultivar-group)] (GB:NP_001047854.1); contains InterPro domain Protein of unknown function DUF1218 (InterPro:IPR009606) | 2.52 |
| 348 | 254524_at | AT4G20000 | VQ motif-containing protein | 6.82 |
| 349 | 254453_at | AT4G21120 | AAT1 (CATIONIC AMINO ACID TRANSPORTER 1); cationic amino acid transmembrane transporter | 2.99 |
| 350 | 254413_at | AT4G21440 | ATM4/ATMYB102 (ARABIDOPSIS MYB-LIKE 102); DNA binding / transcription factor | -1.33 |
| 351 | 254387_at | AT4G21850 | methionine sulfoxide reductase domain-containing protein / SeIR domain-containing protein | 2.50 |
| 352 | 254318_at | AT4G22530 | embryo-abundant protein-related | 2.21 |
| 353 | 254271_at | AT4G23150 | protein kinase family protein | 5.13 |
| 354 | 254243_at | AT4G23210 | CRK13/HIG1; kinase | 2.59 |
| 355 | 254255_at | AT4G23220 | protein kinase family protein | 3.19 |
| 356 | 254252_at | AT4G23310 | receptor-like protein kinase, putative | 5.41 |
| 357 | 254253_at | AT4G23320 | protein kinase family protein | 3.25 |
| 358 | 254240_at | AT4G23496 | SP1L5 (SPIRAL1-LIKE5) | -1.31 |
| 359 | 254232_at | AT4G23600 | CORI3 (CORONATINE INDUCED 1, JASMONIC ACID RESPONSIVE 2); transaminase | -1.80 |
| 360 | 254229_at | AT4G23610 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G54200.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO39657.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN73280.1) | 2.93 |
| 361 | 254093_at | AT4G25110 | ATMC2 (METACASPASE 2); caspase | 3.58 |
| 362 | 254110_at | AT4G25260 | invertase/pectin methylesterase inhibitor family protein | -1.66 |
| 363 | 254075_at | AT4G25470 | CBF2 (FREEZING TOLERANCE QTL 4); DNA binding / transcription activator/ transcription factor | -3.96 |
| 364 | 254066_at | AT4G25480 | DREB1A (DEHYDRATION RESPONSE ELEMENT B1A); DNA binding / transcription activator/ transcription factor | -2.71 |
| 365 | 254074_at | AT4G25490 | CBF1 (C-REPEAT/DRE BINDING FACTOR 1); DNA binding / transcription activator/ transcription factor | -3.38 |
| 366 | 254024_at | AT4G25780 | pathogenesis-related protein, putative | -3.93 |
| 367 | 253832_at | AT4G27654 | unknown protein | -4.18 |
| 368 | 253776_at | AT4G28390 | AAC3 (ADP/ATP CARRIER 3); ATP:ADP antiporter/ binding | 2.35 |
| 369 | 253737_at | AT4G28703 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G04300.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO44490.1); similar to Os04g0509400 [Oryza sativa (japonica cultivar-group)] (GB:NP_001053279.1); similar to OSJNBb0065L13.6 [Oryza sativa (japonica cultivar-group)] (GB:CAE03363.1); contains InterPro domain RmlC-like jelly roll fold (InterPro:IPR014710); contains InterPro domain Protein of unknown function DUF861, cupin-3 (InterPro:IPR008579); contains InterPro domain Cupin, RmlC-type (InterPro:IPR011051) | -1.26 |
| 370 | 253679_at | AT4G29610 | cytidine deaminase, putative / cytidine aminohydrolase, putative | -1.85 |
| 371 | 253697_at | AT4G29700 | type I phosphodiesterase/nucleotide pyrophosphatase family protein | -1.85 |
| 372 | 253660_at | AT4G30140 | GDSL-motif lipase/hydrolase family protein | -1.75 |
| 373 | 253666_at | AT4G30270 | MERI5B (MERISTEM-5); hydrolase, acting on glycosyl bonds / xyloglucan:xyloglucosyl transferase | -1.80 |
| 374 | 253406_at | AT4G32890 | zinc finger (GATA type) family protein | -1.96 |
| 375 | 253414_at | AT4G33050 | EDA39 (embryo sac development arrest 39); calmodulin binding | 2.19 |
| 376 | 253259_at | AT4G34410 | AP2 domain-containing transcription factor, putative | -4.98 |
| 377 | 253207_at | AT4G34770 | auxin-responsive family protein | -1.36 |
| 378 | 253172_at | AT4G35060 | heavy-metal-associated domain-containing protein / copper chaperone (CCH)-related | -2.08 |
| 379 | 253181_at | AT4G35180 | LHT7 (LYS/HIS TRANSPORTER 7); amino acid transmembrane transporter | 4.35 |
| 380 | 253165_at | AT4G35320 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G17300.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN78386.1) | -1.53 |
| 381 | 253161_at | AT4G35770 | SEN1 (DARK INDUCIBLE 1) | -1.25 |
| 382 | 253046_at | AT4G37370 | CYP81D8 (cytochrome P450, family 81, subfamily D, polypeptide 8); oxygen binding | 2.90 |
| 383 | 252983_at | AT4G37980 | ELI3-1 (ELICITOR-ACTIVATED GENE 3); binding / catalytic/ oxidoreductase/ zinc ion binding | -1.55 |
| 384 | 252997_at | AT4G38400 | ATEXLA2 (ARABIDOPSIS THALIANA EXPANSIN-LIKE A2) | -1.57 |
| 385 | 252977_at | AT4G38560 | similar to pEARLI4 [Arabidopsis thaliana] (TAIR:AT2G20960.1); contains InterPro domain Phospholipase-like, arabidopsis (InterPro:IPR007942) | 3.87 |
| 386 | 252950_at | AT4G38690 | 1-phosphatidylinositol phosphodiesterase-related | -1.77 |
| 387 | 252921_at | AT4G39030 | EDS5 (ENHANCED DISEASE SUSCEPTIBILITY 5); antiporter/ transporter | 2.81 |
| 388 | 252908_at | AT4G39670 | glycolipid binding / glycolipid transporter | 3.97 |
| 389 | 252863_at | AT4G39800 | MI-1-P SYNTHASE (Myo-inositol-1-phosphate synthase); inositol-3-phosphate synthase | -1.55 |
| 390 | 252862_at | AT4G39830 | L-ascorbate oxidase, putative | 3.30 |
| 391 | 251084_at | AT5G01520 | zinc finger (C3HC4-type RING finger) family protein | -2.10 |
| 392 | 251072_at | AT5G01740 | similar to SAG20 (WOUND-INDUCED PROTEIN 12) [Arabidopsis thaliana] (TAIR:AT3G10985.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO48816.1); contains InterPro domain Wound-induced protein, Wun1 (InterPro:IPR009798) | -2.37 |
| 393 | 251026_at | AT5G02200 | FHL (FAR-RED-ELONGATED HYPOCOTYL1-LIKE); protein binding | -1.87 |
| 394 | 251028_at | AT5G02230 | haloacid dehalogenase-like hydrolase family protein | -1.70 |
| 395 | 250994_at | AT5G02490 | heat shock cognate 70 kDa protein 2 (HSC70-2) (HSP70-2) | 2.91 |
| 396 | 251017_at | AT5G02760 | protein phosphatase 2C family protein / PP2C family protein | -1.70 |
| 397 | 250983_at | AT5G02780 | In2-1 protein, putative | 4.65 |
| 398 | 250936_at | AT5G03120 | similar to hypothetical protein [Vitis vinifera] (GB:CAN68657.1) | -2.43 |
| 399 | 250980_at | AT5G03130 | unknown protein | -1.35 |
| 400 | 250937_at | AT5G03230 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G60680.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO21845.1); contains InterPro domain Protein of unknown function DUF584 (InterPro:IPR007608) | -1.48 |
| 401 | 250818_at | AT5G04930 | ALA1 (AMINOPHOSPHOLIPID ATPASE1); ATPase, coupled to transmembrane movement of ions, phosphorylative mechanism | 2.15 |
| 402 | 250775_at | AT5G05460 | hydrolase, acting on glycosyl bonds | 2.20 |
| 403 | 250665_at | AT5G06980 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G12320.1) | -1.37 |
| 404 | 250657_at | AT5G07000 | sulfotransferase family protein | -2.14 |
| 405 | 250575_at | AT5G08240 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G23160.1) | 2.42 |
| 406 | 245932_at | AT5G09290 | 3'(2'),5'-bisphosphate nucleotidase, putative / inositol polyphosphate 1-phosphatase, putative | 2.53 |
| 407 | 250476_at | AT5G10140 | FLC (FLOWERING LOCUS C); transcription factor | -2.35 |
| 408 | 250435_at | AT5G10380 | zinc finger (C3HC4-type RING finger) family protein | 2.61 |
| 409 | 250437_at | AT5G10430 | AGP4 (ARABINOGALACTAN-PROTEIN 4) | -1.51 |
| 410 | 250445_at | AT5G10760 | aspartyl protease family protein | 3.96 |
| 411 | 250302_at | AT5G11920 | ATCWINV6 (6-&1-FRUCTAN EXOHYDROLASE); hydrolase, hydrolyzing O-glycosyl compounds / inulinase/ levanase | 3.31 |
| 412 | 250327_at | AT5G12050 | similar to unnamed protein product [Vitis vinifera] (GB:CAO45643.1) | -1.52 |
| 413 | 245976_at | AT5G13080 | WRKY75 (WRKY DNA-BINDING PROTEIN 75); transcription factor | 3.42 |
| 414 | 250281_at | AT5G13240 | transcription regulator | -1.23 |
| 415 | 250286_at | AT5G13320 | PBS3 (AVRPPHB SUSCEPTIBLE 3) | 4.42 |
| 416 | 250155_at | AT5G15160 | bHLH family protein | -1.34 |
| 417 | 246487_at | AT5G16030 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G02500.1); similar to hypothetical protein [Cleome spinosa] (GB:ABD96956.1) | -1.27 |
| 418 | 246475_at | AT5G16720 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G70750.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO17158.1); contains InterPro domain Protein of unknown function DUF593 (InterPro:IPR007656) | -1.33 |
| 419 | 250062_at | AT5G17760 | AAA-type ATPase family protein | 2.88 |
| 420 | 250012_x_at | AT5G18060 | auxin-responsive protein, putative | -1.60 |
| 421 | 249983_at | AT5G18470 | curculin-like (mannose-binding) lectin family protein | 3.10 |
| 422 | 249940_at | AT5G22380 | ANAC090 (Arabidopsis NAC domain containing protein 90); transcription factor | 3.48 |
| 423 | 249917_at | AT5G22460 | esterase/lipase/thioesterase family protein | -2.63 |
| 424 | 249895_at | AT5G22500 | acyl CoA reductase, putative / male-sterility protein, putative | -1.32 |
| 425 | 249896_at | AT5G22530 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G22520.1) | 5.05 |
| 426 | 249889_at | AT5G22540 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G22550.2); similar to unnamed protein product [Vitis vinifera] (GB:CAO15956.1); contains InterPro domain Protein of unknown function DUF247, plant (InterPro:IPR004158) | 2.82 |
| 427 | 249890_at | AT5G22570 | WRKY38 (WRKY DNA-binding protein 38); transcription factor | 2.80 |
| 428 | 249894_at | AT5G22580 | Identical to Uncharacterized protein At5g22580 [Arabidopsis Thaliana] (GB:Q9FK81); similar to stable protein 1-related [Arabidopsis thaliana] (TAIR:AT3G17210.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN68720.1); contains InterPro domain Dimeric alpha-beta barrel (InterPro:IPR011008); contains InterPro domain Stress responsive alpha-beta barrel (InterPro:IPR013097) | -2.20 |
| 429 | 249797_at | AT5G23750 | remorin family protein | -1.43 |
| 430 | 249818_at | AT5G23860 | TUB8 (tubulin beta-8) | -1.23 |
| 431 | 249774_at | AT5G24150 | SQP1 (Squalene monooxygenase 1) | -1.64 |
| 432 | 249777_at | AT5G24210 | lipase class 3 family protein | 2.45 |
| 433 | 249741_at | AT5G24470 | APRR5 (PSEUDO-RESPONSE REGULATOR 5); transcription regulator | -1.22 |
| 434 | 249754_at | AT5G24530 | oxidoreductase, 2OG-Fe(II) oxygenase family protein | 3.66 |
| 435 | 249743_at | AT5G24540 | glycosyl hydrolase family 1 protein | 5.90 |
| 436 | 249755_at | AT5G24580 | copper-binding family protein | -1.90 |
| 437 | 246919_at | AT5G25460 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G11420.1); similar to unknown [Ricinus communis] (GB:CAB02653.1); contains InterPro domain Protein of unknown function DUF642 (InterPro:IPR006946) | -1.37 |
| 438 | 246821_at | AT5G26920 | calmodulin binding | 2.37 |
| 439 | 246783_at | AT5G27360 | SFP2; carbohydrate transmembrane transporter/ sugar:hydrogen ion symporter | -1.52 |
| 440 | 249677_at | AT5G35970 | DNA-binding protein, putative | -1.43 |
| 441 | 249645_at | AT5G36910 | THI2.2 (THIONIN 2.2); toxin receptor binding | -2.60 |
| 442 | 249581_at | AT5G37600 | ATGSR1 (Arabidopsis thaliana glutamine synthase clone R1); glutamate-ammonia ligase | 2.22 |
| 443 | 249481_at | AT5G38900 | DSBA oxidoreductase family protein | 3.65 |
| 444 | 249494_at | AT5G39050 | transferase family protein | 2.31 |
| 445 | 249417_at | AT5G39670 | calcium-binding EF hand family protein | 3.50 |
| 446 | 249383_at | AT5G39860 | PRE1 (PACLOBUTRAZOL RESISTANCE1); DNA binding / transcription factor | -1.91 |
| 447 | 249377_at | AT5G40690 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G41730.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO14635.1) | 2.29 |
| 448 | 249346_at | AT5G40780 | LHT1 (LYSINE HISTIDINE TRANSPORTER 1); amino acid transmembrane transporter | 2.44 |
| 449 | 249340_at | AT5G41140 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G63300.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO62788.1); contains domain PTHR23160 (PTHR23160) | -1.47 |
| 450 | 249197_at | AT5G42380 | CML37/CML39; calcium ion binding | 2.73 |
| 451 | 249208_at | AT5G42650 | AOS (ALLENE OXIDE SYNTHASE); hydro-lyase/ oxygen binding | -1.79 |
| 452 | 249188_at | AT5G42830 | transferase family protein | 3.43 |
| 453 | 249134_at | AT5G43150 | similar to unnamed protein product [Vitis vinifera] (GB:CAO61459.1) | -1.45 |
| 454 | 249096_at | AT5G43910 | pfkB-type carbohydrate kinase family protein | 2.24 |
| 455 | 249071_at | AT5G44050 | MATE efflux family protein | -1.52 |
| 456 | 249037_at | AT5G44130 | FLA13 (FASCICLIN-LIKE ARABINOGALACTAN PROTEIN 13 PRECURSOR) | -1.25 |
| 457 | 249065_at | AT5G44260 | zinc finger (CCCH-type) family protein | -3.35 |
| 458 | 249004_at | AT5G44570 | unknown protein | 3.22 |
| 459 | 248976_at | AT5G44930 | ARAD2 (ARABINAN DEFICIENT 2); catalytic | -1.26 |
| 460 | 249001_at | AT5G44990 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G19880.1); similar to Intracellular chloride channel [Medicago truncatula] (GB:ABC75353.2); contains InterPro domain Thioredoxin-like fold (InterPro:IPR012336); contains InterPro domain Glutathione S-transferase, C-terminal-like (InterPro:IPR010987); contains InterPro domain Glutathione S-transferase, predicted (InterPro:IPR016639) | 4.93 |
| 461 | 248971_at | AT5G45000 | transmembrane receptor | 2.23 |
| 462 | 248969_at | AT5G45310 | similar to unnamed protein product [Vitis vinifera] (GB:CAO63757.1) | -1.42 |
| 463 | 248970_at | AT5G45380 | sodium:solute symporter family protein | 2.60 |
| 464 | 248910_at | AT5G45820 | CIPK20 (CBL-INTERACTING PROTEIN KINASE 20); kinase | -1.72 |
| 465 | 248932_at | AT5G46050 | ATPTR3/PTR3 (PEPTIDE TRANSPORTER PROTEIN 3); transporter | 3.90 |
| 466 | 248889_at | AT5G46230 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G09310.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO14438.1); contains InterPro domain Protein of unknown function DUF538 (InterPro:IPR007493) | 2.37 |
| 467 | 248846_at | AT5G46500 | similar to disease resistance protein (TIR-NBS-LRR class), putative [Arabidopsis thaliana] (TAIR:AT5G46260.1) | 2.61 |
| 468 | 248793_at | AT5G47240 | ATNUDT8 (Arabidopsis thaliana Nudix hydrolase homolog 8); hydrolase | -1.36 |
| 469 | 248763_at | AT5G47550 | cysteine protease inhibitor, putative / cystatin, putative | -1.48 |
| 470 | 248765_at | AT5G47650 | ATNUDT2 (Arabidopsis thaliana Nudix hydrolase homolog 2); ADP-ribose diphosphatase/ NAD binding / hydrolase | -1.31 |
| 471 | 248772_at | AT5G47800 | phototropic-responsive NPH3 family protein | -1.24 |
| 472 | 248775_at | AT5G47850 | protein kinase, putative | 4.77 |
| 473 | 248656_at | AT5G48460 | fimbrin-like protein, putative | -1.27 |
| 474 | 248681_at | AT5G48900 | pectate lyase family protein | -1.72 |
| 475 | 248623_at | AT5G49170 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G06840.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO44815.1) | -1.73 |
| 476 | 248622_at | AT5G49360 | BXL1 (BETA-XYLOSIDASE 1); hydrolase, hydrolyzing O-glycosyl compounds | -1.49 |
| 477 | 248551_at | AT5G50200 | WR3 (WOUND-RESPONSIVE 3); nitrate transmembrane transporter | 2.42 |
| 478 | 248467_at | AT5G50800 | nodulin MtN3 family protein | -1.57 |
| 479 | 248395_at | AT5G52120 | ATPP2-A14 (Phloem protein 2-A14); carbohydrate binding | -1.41 |
| 480 | 248337_at | AT5G52310 | COR78 (COLD REGULATED 78) | -2.77 |
| 481 | 248332_at | AT5G52640 | HSP81-1 (HEAT SHOCK PROTEIN 81-1); ATP binding / unfolded protein binding | 2.74 |
| 482 | 248321_at | AT5G52740 | heavy-metal-associated domain-containing protein | 3.22 |
| 483 | 248327_at | AT5G52750 | heavy-metal-associated domain-containing protein | 2.42 |
| 484 | 248322_at | AT5G52760 | heavy-metal-associated domain-containing protein | 3.85 |
| 485 | 248330_at | AT5G52810 | ornithine cyclodeaminase/mu-crystallin family protein | 2.54 |
| 486 | 248197_at | AT5G54190 | PORA (Protochlorophyllide reductase A); oxidoreductase/ protochlorophyllide reductase | -2.38 |
| 487 | 248169_at | AT5G54610 | ANK (ANKYRIN); protein binding | 3.11 |
| 488 | 248092_at | AT5G55170 | SUM3 (SMALL UBIQUITIN-LIKE MODIFIER 3) | 2.38 |
| 489 | 248062_at | AT5G55450 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | 2.57 |
| 490 | 248028_at | AT5G55620 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G09950.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO39940.1) | -1.41 |
| 491 | 248050_at | AT5G56100 | glycine-rich protein / oleosin | -1.28 |
| 492 | 247965_at | AT5G56540 | AGP14 (ARABINOGALACTAN PROTEIN 14) | -1.25 |
| 493 | 247949_at | AT5G57220 | CYP81F2 (cytochrome P450, family 81, subfamily F, polypeptide 2); oxygen binding | 2.86 |
| 494 | 247899_at | AT5G57345 | similar to unknown [Populus trichocarpa x Populus deltoides] (GB:ABK96592.1) | -1.36 |
| 495 | 247878_at | AT5G57760 | unknown protein | -1.55 |
| 496 | 247740_at | AT5G58940 | CRCK1 (CALMODULIN-BINDING RECEPTOR-LIKE CYTOPLASMIC KINASE 1); kinase | 2.30 |
| 497 | 247760_at | AT5G59130 | subtilase family protein | -1.35 |
| 498 | 247684_at | AT5G59670 | leucine-rich repeat protein kinase, putative | 3.44 |
| 499 | 247618_at | AT5G60280 | lectin protein kinase family protein | 3.41 |
| 500 | 247629_at | AT5G60410 | ATSIZ1/SIZ1; DNA binding / SUMO ligase | 2.31 |
| 501 | 247630_at | AT5G60410 | ATSIZ1/SIZ1; DNA binding / SUMO ligase | -1.22 |
| 502 | 247597_at | AT5G60860 | AtRABA1f (Arabidopsis Rab GTPase homolog A1f); GTP binding | -1.27 |
| 503 | 247600_at | AT5G60890 | ATMYB34/ATR1/MYB34 (ALTERED TRYPTOPHAN REGULATION, MYB DOMAIN PROTEIN 34); DNA binding / kinase/ transcription activator/ transcription factor | -1.55 |
| 504 | 247602_at | AT5G60900 | RLK1 (RECEPTOR-LIKE PROTEIN KINASE 1); carbohydrate binding / kinase | 2.85 |
| 505 | 247604_at | AT5G60950 | COBL5 (COBRA-LIKE PROTEIN 5 PRECURSOR) | 3.61 |
| 506 | 247554_at | AT5G61010 | ATEXO70E2 (EXOCYST SUBUNIT EXO70 FAMILY PROTEIN E2); protein binding | 2.22 |
| 507 | 247573_at | AT5G61160 | AACT1 (ANTHOCYANIN 5-AROMATIC ACYLTRANSFERASE 1); transferase | -3.95 |
| 508 | 247474_at | AT5G62280 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G45360.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO49726.1); contains InterPro domain Protein of unknown function DUF1442 (InterPro:IPR009902) | -2.92 |
| 509 | 247429_at | AT5G62620 | galactosyltransferase family protein | 2.27 |
| 510 | 247439_at | AT5G62670 | AHA11 (ARABIDOPSIS H(+)-ATPASE 11); ATPase | -1.96 |
| 511 | 247348_at | AT5G63810 | BGAL10 (beta-galactosidase 10); beta-galactosidase | -1.27 |
| 512 | 247314_at | AT5G64000 | SAL2; 3'(2'),5'-bisphosphate nucleotidase/ inositol or phosphatidylinositol phosphatase | 4.12 |
| 513 | 247327_at | AT5G64120 | peroxidase, putative | 2.67 |
| 514 | 247293_at | AT5G64510 | similar to unnamed protein product [Vitis vinifera] (GB:CAO49799.1) | 3.35 |
| 515 | 247215_at | AT5G64905 | PROPEP3 (Elicitor peptide 3 precursor) | 2.78 |
| 516 | 247126_at | AT5G66080 | protein phosphatase 2C family protein / PP2C family protein | -2.29 |
| 517 | 247071_at | AT5G66640 | LIM domain-containing protein-related | 2.92 |
| 518 | 247064_at | AT5G66890 | disease resistance protein (CC-NBS-LRR class), putative | 3.47 |
| 519 | 246988_at | AT5G67340 | armadillo/beta-catenin repeat family protein / U-box domain-containing protein | 2.38 |
| 520 | 245010_at | ATCG00420 | Encodes NADH dehydrogenase subunit J. Its transcription is increased upon sulfur depletion. | -1.75 |