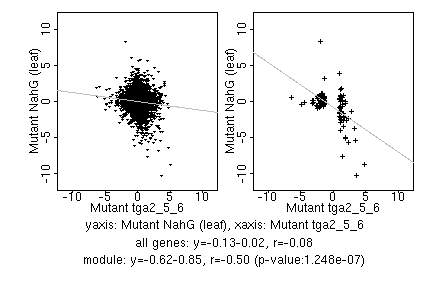
marker gene list
| Mutant tga2_5_6 |
| Mutant NahG(leaf) |
| probeID | AGICode | Annotation | gene expression |
|
|---|---|---|---|---|
| 1 | 264148_at | AT1G02220 | ANAC003 (Arabidopsis NAC domain containing protein 3); transcription factor | 1.06 |
| 2 | 262118_at | AT1G02850 | glycosyl hydrolase family 1 protein | -1.59 |
| 3 | 265067_at | AT1G03850 | glutaredoxin family protein | 1.39 |
| 4 | 263668_at | AT1G04350 | 2-oxoglutarate-dependent dioxygenase, putative | -1.63 |
| 5 | 264525_at | AT1G10060 | ATBCAT-1; branched-chain-amino-acid transaminase/ catalytic | -1.20 |
| 6 | 260475_at | AT1G11080 | SCPL31 (serine carboxypeptidase-like 31); serine carboxypeptidase | -2.08 |
| 7 | 262705_at | AT1G16260 | protein kinase family protein | 1.70 |
| 8 | 262518_at | AT1G17170 | ATGSTU24 (ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE (CLASS TAU) 24); glutathione transferase | -3.02 |
| 9 | 261221_at | AT1G19960 | similar to transmembrane receptor [Arabidopsis thaliana] (TAIR:AT2G32140.1) | 3.52 |
| 10 | 262796_at | AT1G20850 | XCP2 (XYLEM CYSTEINE PEPTIDASE 2); cysteine-type peptidase/ peptidase | 1.43 |
| 11 | 259561_at | AT1G21250 | WAK1 (CELL WALL-ASSOCIATED KINASE); kinase | 2.05 |
| 12 | 264872_at | AT1G24260 | SEP3 (SEPALLATA3); transcription factor | -1.88 |
| 13 | 260997_at | AT1G26610 | zinc finger (C2H2 type) family protein | 1.21 |
| 14 | 262743_at | AT1G29020 | calcium-binding EF hand family protein | 1.49 |
| 15 | 256497_at | AT1G31580 | ECS1 | 2.97 |
| 16 | 261339_at | AT1G35710 | leucine-rich repeat transmembrane protein kinase, putative | 1.65 |
| 17 | 260813_at | AT1G43700 | VIP1 (VIRE2-INTERACTING PROTEIN 1); transcription factor | -1.66 |
| 18 | 260867_at | AT1G43790 | similar to hydroxyproline-rich glycoprotein family protein [Arabidopsis thaliana] (TAIR:AT5G48920.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN71192.1) | 1.23 |
| 19 | 261684_at | AT1G47400 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G47395.1) | 1.45 |
| 20 | 261598_at | AT1G49750 | leucine-rich repeat family protein | 1.27 |
| 21 | 256321_at | AT1G55020 | LOX1 (Lipoxygenase 1); lipoxygenase | 1.18 |
| 22 | 260592_at | AT1G55850 | ATCSLE1 (Cellulose synthase-like E1); cellulose synthase/ transferase, transferring glycosyl groups | -1.80 |
| 23 | 255852_at | AT1G66970 | glycerophosphoryl diester phosphodiesterase family protein | 1.19 |
| 24 | 260189_at | AT1G67550 | URE (UREASE); urease | -3.10 |
| 25 | 260033_at | AT1G68760 | ATNUDT1 (Arabidopsis thaliana Nudix hydrolase homolog 1); dihydroneopterin triphosphate pyrophosphohydrolase/ hydrolase | -1.26 |
| 26 | 256299_at | AT1G69530 | ATEXPA1 (ARABIDOPSIS THALIANA EXPANSIN A1) | -1.22 |
| 27 | 260419_at | AT1G69730 | protein kinase family protein | 1.22 |
| 28 | 260179_at | AT1G70690 | kinase-related | 1.19 |
| 29 | 260211_at | AT1G74440 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G18720.1); similar to unknown [Populus trichocarpa] (GB:ABK94463.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN69808.1); contains InterPro domain Protein of unknown function DUF962 (InterPro:IPR009305) | 1.36 |
| 30 | 259705_at | AT1G77450 | ANAC032 (Arabidopsis NAC domain containing protein 32); transcription factor | -2.22 |
| 31 | 262935_at | AT1G79410 | ATOCT5 (ARABIDOPSIS THALIANA ORGANIC CATION/CARNITINE TRANSPORTER5); carbohydrate transmembrane transporter/ sugar:hydrogen ion symporter | -2.31 |
| 32 | 263402_at | AT2G04050 | MATE efflux family protein | 1.54 |
| 33 | 263046_at | AT2G05380 | GRP3S (GLYCINE-RICH PROTEIN 3 SHORT ISOFORM) | -6.23 |
| 34 | 263098_at | AT2G16005 | MD-2-related lipid recognition domain-containing protein / ML domain-containing protein | -2.92 |
| 35 | 265588_at | AT2G19970 | pathogenesis-related protein, putative | -1.86 |
| 36 | 264016_at | AT2G21220 | auxin-responsive protein, putative | -1.66 |
| 37 | 265611_at | AT2G25510 | unknown protein | 2.55 |
| 38 | 266296_at | AT2G29420 | ATGSTU7 (GLUTATHIONE S-TRANSFERASE 25); glutathione transferase | -2.49 |
| 39 | 266294_at | AT2G29500 | 17.6 kDa class I small heat shock protein (HSP17.6B-CI) | -1.58 |
| 40 | 263478_at | AT2G31880 | leucine-rich repeat transmembrane protein kinase, putative | 1.16 |
| 41 | 265698_at | AT2G32160 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G32170.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO64660.1); contains InterPro domain N2227-like (InterPro:IPR012901) | 1.75 |
| 42 | 267461_at | AT2G33830 | dormancy/auxin associated family protein | 1.68 |
| 43 | 263947_at | AT2G35820 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G35810.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO69007.1); contains domain G3DSA:2.60.120.480 (G3DSA:2.60.120.480) | -1.27 |
| 44 | 257375_at | AT2G38640 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G41590.1); similar to unknown [Populus trichocarpa] (GB:ABK94888.1); contains InterPro domain Protein of unknown function DUF567 (InterPro:IPR007612) | -1.21 |
| 45 | 257382_at | AT2G40750 | WRKY54 (WRKY DNA-binding protein 54); transcription factor | 5.02 |
| 46 | 260557_at | AT2G43610 | glycoside hydrolase family 19 protein | 1.17 |
| 47 | 260556_at | AT2G43620 | chitinase, putative | 1.38 |
| 48 | 258746_at | AT3G05950 | germin-like protein, putative | -2.33 |
| 49 | 258646_at | AT3G08040 | FRD3 (FERRIC REDUCTASE DEFECTIVE 3); antiporter | -1.76 |
| 50 | 259040_at | AT3G09270 | ATGSTU8 (Arabidopsis thaliana Glutathione S-transferase (class tau) 8); glutathione transferase | -1.75 |
| 51 | 258923_at | AT3G10450 | SCPL7; serine carboxypeptidase | -1.23 |
| 52 | 256269_at | AT3G12250 | TGA6 (TGA1a-related gene 6); DNA binding / transcription factor | -1.92 |
| 53 | 258114_at | AT3G14660 | CYP72A13 (cytochrome P450, family 72, subfamily A, polypeptide 13); oxygen binding | -1.85 |
| 54 | 257216_at | AT3G14990 | 4-methyl-5(b-hydroxyethyl)-thiazole monophosphate biosynthesis protein, putative | -2.94 |
| 55 | 258033_at | AT3G21250 | ATMRP6 (Arabidopsis thaliana multidrug resistance-associated protein 6) | -1.94 |
| 56 | 258322_at | AT3G22740 | HMT3 (Homocysteine S-methyltransferase 3); homocysteine S-methyltransferase | 2.08 |
| 57 | 257823_at | AT3G25190 | nodulin, putative | -4.32 |
| 58 | 257642_at | AT3G25710 | basic helix-loop-helix (bHLH) family protein | -1.20 |
| 59 | 252612_at | AT3G45160 | contains domain PROKAR_LIPOPROTEIN (PS51257) | -1.59 |
| 60 | 252170_at | AT3G50480 | HR4 (HOMOLOG OF RPW8 4) | 1.45 |
| 61 | 252126_at | AT3G50950 | disease resistance protein (CC-NBS-LRR class), putative | 1.06 |
| 62 | 252102_at | AT3G50970 | LTI30 (LOW TEMPERATURE-INDUCED 30) | -1.22 |
| 63 | 251704_at | AT3G56360 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G05250.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO41488.1) | 1.58 |
| 64 | 251705_at | AT3G56400 | WRKY70 (WRKY DNA-binding protein 70); transcription factor | 2.41 |
| 65 | 251673_at | AT3G57240 | BG3 (BETA-1,3-GLUCANASE 3); hydrolase, hydrolyzing O-glycosyl compounds | 2.20 |
| 66 | 255702_at | AT4G00230 | XSP1 (XYLEM SERINE PEPTIDASE 1); subtilase | 1.07 |
| 67 | 255666_at | AT4G00390 | transcription regulator | -1.72 |
| 68 | 254828_at | AT4G12550 | AIR1 (Auxin-Induced in Root cultures 1); lipid binding | 1.11 |
| 69 | 254759_at | AT4G13180 | short-chain dehydrogenase/reductase (SDR) family protein | -2.23 |
| 70 | 245265_at | AT4G14400 | ACD6 (ACCELERATED CELL DEATH 6); protein binding | 3.33 |
| 71 | 245615_at | AT4G14470 | transposable element gene | 1.49 |
| 72 | 245543_at | AT4G15260 | UDP-glucoronosyl/UDP-glucosyl transferase family protein | -1.29 |
| 73 | 254549_at | AT4G19880 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G45020.1); similar to Intracellular chloride channel [Medicago truncatula] (GB:ABC75353.2); contains InterPro domain Thioredoxin-like fold (InterPro:IPR012336); contains InterPro domain Glutathione S-transferase, C-terminal-like (InterPro:IPR010987); contains InterPro domain Glutathione S-transferase, C-terminal (InterPro:IPR004046); contains InterPro domain Glutathione S-transferase, predicted (InterPro:IPR016639) | -1.43 |
| 74 | 254255_at | AT4G23220 | protein kinase family protein | 1.32 |
| 75 | 253997_at | AT4G26090 | RPS2 (RESISTANT TO P. SYRINGAE 2) | 1.88 |
| 76 | 253936_at | AT4G26880 | stigma-specific Stig1 family protein | 1.26 |
| 77 | 253344_at | AT4G33550 | lipid binding | -1.90 |
| 78 | 253191_at | AT4G35350 | XCP1 (XYLEM CYSTEINE PEPTIDASE 1); cysteine-type peptidase | 1.70 |
| 79 | 251109_at | AT5G01600 | ATFER1 (FERRETIN 1); ferric iron binding | -1.19 |
| 80 | 250942_at | AT5G03350 | legume lectin family protein | 3.64 |
| 81 | 250764_at | AT5G05960 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | -1.59 |
| 82 | 250655_at | AT5G06960 | OBF5 (OCS-ELEMENT BINDING FACTOR 5); DNA binding / transcription factor | -4.76 |
| 83 | 250499_at | AT5G09730 | BXL3 (BETA-XYLOSIDASE 3); hydrolase, hydrolyzing O-glycosyl compounds | -1.50 |
| 84 | 250302_at | AT5G11920 | ATCWINV6 (6-&1-FRUCTAN EXOHYDROLASE); hydrolase, hydrolyzing O-glycosyl compounds / inulinase/ levanase | 1.79 |
| 85 | 246463_at | AT5G16970 | AT-AER (ALKENAL REDUCTASE); 2-alkenal reductase | -1.27 |
| 86 | 246464_at | AT5G16980 | NADP-dependent oxidoreductase, putative | -1.88 |
| 87 | 245947_at | AT5G19530 | ACL5 (ACAULIS 5) | -1.21 |
| 88 | 249754_at | AT5G24530 | oxidoreductase, 2OG-Fe(II) oxygenase family protein | 2.02 |
| 89 | 249485_at | AT5G39020 | protein kinase family protein | 1.20 |
| 90 | 249295_at | AT5G41280 | Identical to Cysteine-rich repeat secretory protein 57 precursor (CRRSP57) [Arabidopsis Thaliana] (GB:Q9FHD5;GB:Q5BPI3); similar to receptor-like protein kinase-related [Arabidopsis thaliana] (TAIR:AT5G41290.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO15206.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN62393.1); contains InterPro domain Protein of unknown function DUF26 (InterPro:IPR002902) | 1.96 |
| 91 | 249070_at | AT5G44030 | CESA4 (CELLULOSE SYNTHASE 4); transferase, transferring glycosyl groups | 1.18 |
| 92 | 254521_at | AT5G44820 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G19970.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO46707.1); contains domain PTHR10483:SF6 (PTHR10483:SF6); contains domain PTHR10483 (PTHR10483) | 1.18 |
| 93 | 248932_at | AT5G46050 | ATPTR3/PTR3 (PEPTIDE TRANSPORTER PROTEIN 3); transporter | 1.68 |
| 94 | 248782_at | AT5G47340 | palmitoyl protein thioesterase family protein | -2.17 |
| 95 | 248605_at | AT5G49410 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G73940.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO48310.1) | 1.06 |
| 96 | 248092_at | AT5G55170 | SUM3 (SMALL UBIQUITIN-LIKE MODIFIER 3) | 1.10 |
| 97 | 248062_at | AT5G55450 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | 1.57 |
| 98 | 247541_at | AT5G61660 | glycine-rich protein | -1.44 |
| 99 | 247488_at | AT5G61820 | similar to MtN19-like protein [Pisum sativum] (GB:AAU14999.2); contains InterPro domain Stress up-regulated Nod 19 (InterPro:IPR011692) | -1.24 |
| 100 | 247351_at | AT5G63790 | ANAC102 (Arabidopsis NAC domain containing protein 102); transcription factor | -1.41 |
| 101 | 247024_at | AT5G66985 | unknown protein | -1.95 |