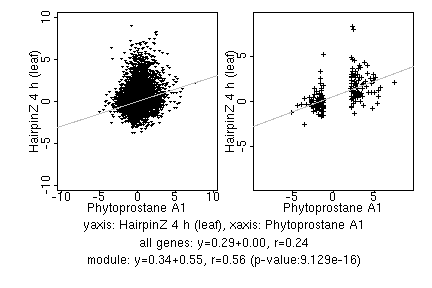
marker gene list
| Phytoprostane A1 4 h |
| HairpinZ 4 h (leaf) |
| probeID | AGICode | Annotation | gene expression |
|
|---|---|---|---|---|
| 1 | 261564_at | AT1G01720 | ATAF1 (Arabidopsis NAC domain containing protein 2); transcription factor | 2.67 |
| 2 | 261562_at | AT1G01750 | actin-depolymerizing factor, putative | -1.96 |
| 3 | 262105_at | AT1G02810 | pectinesterase family protein | -2.55 |
| 4 | 264611_at | AT1G04680 | pectate lyase family protein | -1.12 |
| 5 | 264580_at | AT1G05340 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G32210.1) | 2.33 |
| 6 | 263184_at | AT1G05560 | UGT1 (UDP-glucosyl transferase 75B1); UDP-glycosyltransferase/ transferase, transferring glycosyl groups | 3.50 |
| 7 | 263231_at | AT1G05680 | UDP-glucoronosyl/UDP-glucosyl transferase family protein | 4.75 |
| 8 | 263210_at | AT1G10585 | transcription factor | 3.71 |
| 9 | 259358_at | AT1G13250 | GATL3 (Galacturonosyltransferase-like 3); polygalacturonate 4-alpha-galacturonosyltransferase/ transferase, transferring glycosyl groups / transferase, transferring hexosyl groups | -1.33 |
| 10 | 262658_at | AT1G14220 | ribonuclease T2 family protein | -1.36 |
| 11 | 261491_at | AT1G14350 | FLP (FOUR LIPS); DNA binding / transcription factor | -1.39 |
| 12 | 261763_at | AT1G15520 | ATPDR12/PDR12 (PLEIOTROPIC DRUG RESISTANCE 12); ATPase, coupled to transmembrane movement of substances | 4.29 |
| 13 | 261838_at | AT1G16030 | HSP70B (heat shock protein 70B); ATP binding | 5.90 |
| 14 | 262518_at | AT1G17170 | ATGSTU24 (ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE (CLASS TAU) 24); glutathione transferase | 4.19 |
| 15 | 262517_at | AT1G17180 | ATGSTU25 (Arabidopsis thaliana Glutathione S-transferase (class tau) 25); glutathione transferase | 4.77 |
| 16 | 259479_at | AT1G19020 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G48180.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO40966.1) | 2.47 |
| 17 | 255962_at | AT1G22330 | RNA binding | -1.45 |
| 18 | 264775_at | AT1G22880 | ATCEL5/ATGH9B4 (ARABIDOPSIS THALIANA GLYCOSYL HYDROLASE 9B4); hydrolase, hydrolyzing O-glycosyl compounds | -1.85 |
| 19 | 264988_at | AT1G27140 | ATGSTU14 (GLUTATHIONE S-TRANSFERASE 13); glutathione transferase | -1.35 |
| 20 | 260883_at | AT1G29270 | similar to transcription regulator [Arabidopsis thaliana] (TAIR:AT2G40435.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN79056.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO48025.1) | -1.36 |
| 21 | 263228_at | AT1G30700 | FAD-binding domain-containing protein | 2.51 |
| 22 | 265102_at | AT1G30870 | cationic peroxidase, putative | -2.26 |
| 23 | 265161_at | AT1G30900 | vacuolar sorting receptor, putative | -1.09 |
| 24 | 261981_at | AT1G33811 | GDSL-motif lipase/hydrolase family protein | -1.23 |
| 25 | 261157_at | AT1G34510 | peroxidase, putative | -2.29 |
| 26 | 259454_at | AT1G44050 | DC1 domain-containing protein | -1.25 |
| 27 | 260758_at | AT1G48930 | ATGH9C1 (ARABIDOPSIS THALIANA GLYCOSYL HYDROLASE 9C1); hydrolase, hydrolyzing O-glycosyl compounds | -2.01 |
| 28 | 261691_at | AT1G50060 | pathogenesis-related protein, putative | -2.44 |
| 29 | 246375_at | AT1G51830 | ATP binding / kinase/ protein serine/threonine kinase | -1.76 |
| 30 | 262148_at | AT1G52560 | 26.5 kDa class I small heat shock protein-like (HSP26.5-P) | 7.64 |
| 31 | 262138_at | AT1G52660 | disease resistance protein, putative | -3.45 |
| 32 | 260643_at | AT1G53270 | ABC transporter family protein | 2.53 |
| 33 | 260978_at | AT1G53540 | 17.6 kDa class I small heat shock protein (HSP17.6C-CI) (AA 1-156) | 5.20 |
| 34 | 263150_at | AT1G54050 | 17.4 kDa class III heat shock protein (HSP17.4-CIII) | 3.51 |
| 35 | 256352_at | AT1G54970 | ATPRP1 (PROLINE-RICH PROTEIN 1); structural constituent of cell wall | -2.53 |
| 36 | 260602_at | AT1G55920 | AtSerat2;1 (SERINE ACETYLTRANSFERASE 1) | 2.40 |
| 37 | 264929_at | AT1G60730 | aldo/keto reductase family protein | 2.71 |
| 38 | 264912_at | AT1G60750 | pseudogene, aldo/keto reductase family, contains Pfam profile PF00248: oxidoreductase, aldo/keto reductase family; blastp match of 71% identity and 8.2e-127 P-value to GP|2606077|gb|AAB84222.1||AF030301 auxin-induced protein {Helianthus annuus} | 5.55 |
| 39 | 261099_at | AT1G62980 | ATEXPA18 (ARABIDOPSIS THALIANA EXPANSIN A18) | -1.92 |
| 40 | 262325_at | AT1G64160 | disease resistance-responsive family protein / dirigent family protein | 3.02 |
| 41 | 259736_at | AT1G64390 | ATGH9C2 (ARABIDOPSIS THALIANA GLYCOSYL HYDROLASE 9C2); hydrolase, hydrolyzing O-glycosyl compounds | -1.31 |
| 42 | 262930_at | AT1G65690 | harpin-induced protein-related / HIN1-related / harpin-responsive protein-related | 2.37 |
| 43 | 245193_at | AT1G67810 | Fe-S metabolism associated domain-containing protein | 2.78 |
| 44 | 260405_at | AT1G69930 | ATGSTU11 (Arabidopsis thaliana Glutathione S-transferase (class tau) 11); glutathione transferase | 3.04 |
| 45 | 260248_at | AT1G74310 | ATHSP101 (HEAT SHOCK PROTEIN 101); ATP binding / ATPase | 5.01 |
| 46 | 259980_at | AT1G76520 | auxin efflux carrier family protein | 2.57 |
| 47 | 259979_at | AT1G76600 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G21010.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN67638.1) | 2.88 |
| 48 | 264481_at | AT1G77200 | AP2 domain-containing transcription factor TINY, putative | -2.74 |
| 49 | 259705_at | AT1G77450 | ANAC032 (Arabidopsis NAC domain containing protein 32); transcription factor | 2.78 |
| 50 | 260058_at | AT1G78100 | F-box family protein | -1.09 |
| 51 | 260803_at | AT1G78340 | ATGSTU22 (Arabidopsis thaliana Glutathione S-transferase (class tau) 22); glutathione transferase | 3.79 |
| 52 | 262045_at | AT1G80240 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G25460.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO44183.1); contains InterPro domain Protein of unknown function DUF642 (InterPro:IPR006946) | -5.04 |
| 53 | 263594_at | AT2G01880 | ATPAP7/PAP7 (purple acid phosphatase 7); acid phosphatase/ protein serine/threonine phosphatase | -1.71 |
| 54 | 263481_at | AT2G04025 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G13620.1) | -1.13 |
| 55 | 263809_at | AT2G04570 | GDSL-motif lipase/hydrolase family protein | -1.11 |
| 56 | 265499_at | AT2G15480 | UGT73B5 (UDP-GLUCOSYL TRANSFERASE 73B5); UDP-glycosyltransferase/ transferase, transferring glycosyl groups | 3.93 |
| 57 | 265501_at | AT2G15490 | UGT73B4; UDP-glycosyltransferase/ transferase, transferring glycosyl groups | 5.26 |
| 58 | 263073_at | AT2G17500 | auxin efflux carrier family protein | 3.46 |
| 59 | 265948_at | AT2G19590 | ACO1 (ACC OXIDASE 1); 1-aminocyclopropane-1-carboxylate oxidase | -1.11 |
| 60 | 265588_at | AT2G19970 | pathogenesis-related protein, putative | -2.20 |
| 61 | 263374_at | AT2G20560 | DNAJ heat shock family protein | 3.13 |
| 62 | 267287_at | AT2G23630 | SKS16 (SKU5 Similar 16); copper ion binding / pectinesterase | -1.97 |
| 63 | 266841_at | AT2G26150 | ATHSFA2 (Arabidopsis thaliana heat shock transcription factor A2); DNA binding / transcription factor | 3.72 |
| 64 | 266276_at | AT2G29330 | TRI (TROPINONE REDUCTASE); oxidoreductase | -1.58 |
| 65 | 266296_at | AT2G29420 | ATGSTU7 (GLUTATHIONE S-TRANSFERASE 25); glutathione transferase | 3.00 |
| 66 | 266267_at | AT2G29460 | ATGSTU4 (GLUTATHIONE S-TRANSFERASE 22); glutathione transferase | 3.41 |
| 67 | 266269_at | AT2G29480 | ATGSTU2 (GLUTATHIONE S-TRANSFERASE 20); glutathione transferase | 2.82 |
| 68 | 266290_at | AT2G29490 | ATGSTU1 (GLUTATHIONE S-TRANSFERASE 19); glutathione transferase | 2.33 |
| 69 | 266294_at | AT2G29500 | 17.6 kDa class I small heat shock protein (HSP17.6B-CI) | 3.17 |
| 70 | 266669_at | AT2G29750 | UDP-glucoronosyl/UDP-glucosyl transferase family protein | -1.49 |
| 71 | 256712_at | AT2G34020 | calcium ion binding | -1.27 |
| 72 | 267319_at | AT2G34660 | ATMRP2 (MULTIDRUG RESISTANCE-ASSOCIATED PROTEIN 2); ATPase, coupled to transmembrane movement of substances | 2.33 |
| 73 | 265197_at | AT2G36750 | UGT72C1 (UDP-GLUCOSYL TRANSFERASE 72C1); UDP-glycosyltransferase/ transferase, transferring glycosyl groups | 2.61 |
| 74 | 266975_at | AT2G39380 | ATEXO70H2 (EXOCYST SUBUNIT EXO70 FAMILY PROTEIN H2); protein binding | -1.12 |
| 75 | 260540_at | AT2G43500 | RWP-RK domain-containing protein | 2.80 |
| 76 | 260558_at | AT2G43600 | glycoside hydrolase family 19 protein | -2.64 |
| 77 | 267385_at | AT2G44380 | DC1 domain-containing protein | -1.45 |
| 78 | 266590_at | AT2G46240 | BAG6 (ARABIDOPSIS THALIANA BCL-2-ASSOCIATED ATHANOGENE 6); calmodulin binding / protein binding | 3.66 |
| 79 | 266461_at | AT2G47730 | ATGSTF8 (GLUTATHIONE S-TRANSFERASE 8); glutathione transferase | 2.28 |
| 80 | 259264_at | AT3G01260 | aldose 1-epimerase family protein | -2.27 |
| 81 | 258912_at | AT3G06460 | GNS1/SUR4 membrane family protein | -1.12 |
| 82 | 259037_at | AT3G09350 | armadillo/beta-catenin repeat family protein | 3.02 |
| 83 | 258939_at | AT3G10020 | similar to Os12g0147200 [Oryza sativa (japonica cultivar-group)] (GB:NP_001066153.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO15981.1); similar to Os11g0149200 [Oryza sativa (japonica cultivar-group)] (GB:NP_001065754.1) | 3.05 |
| 84 | 256245_at | AT3G12580 | HSP70 (heat shock protein 70); ATP binding | 2.35 |
| 85 | 258143_at | AT3G18170 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G18180.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO68130.1); contains InterPro domain Protein of unknown function DUF563 (InterPro:IPR007657) | -1.76 |
| 86 | 258133_at | AT3G24500 | ATMBF1C/MBF1C (MULTIPROTEIN BRIDGING FACTOR 1C); DNA binding / transcription coactivator/ transcription factor | 2.93 |
| 87 | 257823_at | AT3G25190 | nodulin, putative | 2.46 |
| 88 | 258277_at | AT3G26830 | PAD3 (PHYTOALEXIN DEFICIENT 3); oxygen binding | 2.71 |
| 89 | 257153_at | AT3G27220 | kelch repeat-containing protein | -1.56 |
| 90 | 256576_at | AT3G28210 | PMZ; zinc ion binding | 4.25 |
| 91 | 256589_at | AT3G28740 | cytochrome P450 family protein | 5.67 |
| 92 | 256736_at | AT3G29410 | terpene synthase/cyclase family protein | -1.37 |
| 93 | 252671_at | AT3G44190 | pyridine nucleotide-disulphide oxidoreductase family protein | 2.33 |
| 94 | 252568_at | AT3G45410 | lectin protein kinase family protein | -1.44 |
| 95 | 252537_at | AT3G45710 | proton-dependent oligopeptide transport (POT) family protein | -3.42 |
| 96 | 252515_at | AT3G46230 | ATHSP17.4 (Arabidopsis thaliana heat shock protein 17.4) | 4.70 |
| 97 | 252490_at | AT3G46720 | UDP-glucoronosyl/UDP-glucosyl transferase family protein | -1.36 |
| 98 | 252238_at | AT3G49960 | peroxidase, putative | -2.34 |
| 99 | 252102_at | AT3G50970 | LTI30 (LOW TEMPERATURE-INDUCED 30) | 3.17 |
| 100 | 256673_at | AT3G52370 | FLA15 (FASCICLIN-LIKE ARABINOGALACTAN PROTEIN 15 PRECURSOR) | -1.29 |
| 101 | 251824_at | AT3G55090 | ATPase, coupled to transmembrane movement of substances | 2.60 |
| 102 | 251428_at | AT3G60140 | DIN2 (DARK INDUCIBLE 2); hydrolase, hydrolyzing O-glycosyl compounds | 4.88 |
| 103 | 251255_at | AT3G62280 | carboxylesterase | -2.24 |
| 104 | 251226_at | AT3G62680 | PRP3 (PROLINE-RICH PROTEIN 3); structural constituent of cell wall | -1.96 |
| 105 | 255695_at | AT4G00080 | UNE11 (unfertilized embryo sac 11); pectinesterase inhibitor | -2.29 |
| 106 | 255632_at | AT4G00680 | actin-depolymerizing factor, putative | -1.86 |
| 107 | 255543_at | AT4G01870 | tolB protein-related | 4.02 |
| 108 | 255005_at | AT4G09990 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G33800.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO16316.1); contains InterPro domain Protein of unknown function DUF579, plant (InterPro:IPR006514) | -1.73 |
| 109 | 254839_at | AT4G12400 | stress-inducible protein, putative | 2.96 |
| 110 | 254759_at | AT4G13180 | short-chain dehydrogenase/reductase (SDR) family protein | 2.71 |
| 111 | 245593_at | AT4G14550 | IAA14 (SOLITARY ROOT); transcription factor | -1.16 |
| 112 | 245561_at | AT4G15500 | UGT84A4; UDP-glycosyltransferase/ sinapate 1-glucosyltransferase/ transferase, transferring glycosyl groups / transferase, transferring hexosyl groups | -1.12 |
| 113 | 245317_at | AT4G15610 | integral membrane family protein | 2.33 |
| 114 | 254549_at | AT4G19880 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G45020.1); similar to Intracellular chloride channel [Medicago truncatula] (GB:ABC75353.2); contains InterPro domain Thioredoxin-like fold (InterPro:IPR012336); contains InterPro domain Glutathione S-transferase, C-terminal-like (InterPro:IPR010987); contains InterPro domain Glutathione S-transferase, C-terminal (InterPro:IPR004046); contains InterPro domain Glutathione S-transferase, predicted (InterPro:IPR016639) | 2.34 |
| 115 | 254447_at | AT4G20860 | FAD-binding domain-containing protein | 2.31 |
| 116 | 254461_at | AT4G21230 | protein kinase family protein | -1.10 |
| 117 | 254361_at | AT4G22212 | Encodes a defensin-like (DEFL) family protein. | -1.20 |
| 118 | 254318_at | AT4G22530 | embryo-abundant protein-related | 2.64 |
| 119 | 254326_at | AT4G22610 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | -2.52 |
| 120 | 254062_at | AT4G25380 | zinc finger (AN1-like) family protein | 5.10 |
| 121 | 253914_at | AT4G27400 | late embryogenesis abundant protein-related / LEA protein-related | -1.09 |
| 122 | 253763_at | AT4G28850 | xyloglucan:xyloglucosyl transferase, putative / xyloglucan endotransglycosylase, putative / endo-xyloglucan transferase, putative | -3.79 |
| 123 | 253608_at | AT4G30290 | ATXTH19 (XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 19); hydrolase, acting on glycosyl bonds | -2.23 |
| 124 | 253483_at | AT4G31910 | transferase family protein | -1.80 |
| 125 | 253309_at | AT4G33790 | acyl CoA reductase, putative | -1.13 |
| 126 | 246203_at | AT4G36610 | hydrolase, alpha/beta fold family protein | 3.32 |
| 127 | 253044_at | AT4G37290 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G23270.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN62855.1) | 2.52 |
| 128 | 253046_at | AT4G37370 | CYP81D8 (cytochrome P450, family 81, subfamily D, polypeptide 8); oxygen binding | 3.34 |
| 129 | 252984_at | AT4G37990 | ELI3-2 (ELICITOR-ACTIVATED GENE 3) | 2.89 |
| 130 | 252988_at | AT4G38410 | dehydrin, putative | -1.94 |
| 131 | 252833_at | AT4G40090 | AGP3 (ARABINOGALACTAN-PROTEIN 3) | -1.34 |
| 132 | 250983_at | AT5G02780 | In2-1 protein, putative | 2.56 |
| 133 | 250801_at | AT5G04960 | pectinesterase family protein | -2.08 |
| 134 | 250826_at | AT5G05220 | similar to hypothetical protein [Vitis vinifera] (GB:CAN82940.1) | 5.62 |
| 135 | 250781_at | AT5G05410 | DREB2A (DRE-BINDING PROTEIN 2A); DNA binding / transcription activator/ transcription factor | 4.48 |
| 136 | 250778_at | AT5G05500 | pollen Ole e 1 allergen and extensin family protein | -1.93 |
| 137 | 250670_at | AT5G06860 | PGIP1 (POLYGALACTURONASE INHIBITING PROTEIN 1); protein binding | 2.31 |
| 138 | 246018_at | AT5G10695 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G57123.1); similar to unknown [Picea sitchensis] (GB:ABK22689.1) | 3.82 |
| 139 | 250351_at | AT5G12030 | AT-HSP17.6A (Arabidopsis thaliana heat shock protein 17.6A) | 5.00 |
| 140 | 245976_at | AT5G13080 | WRKY75 (WRKY DNA-BINDING PROTEIN 75); transcription factor | 3.31 |
| 141 | 250252_at | AT5G13750 | ZIFL1 (ZINC INDUCED FACILITATOR-LIKE 1); tetracycline:hydrogen antiporter/ transporter | 2.69 |
| 142 | 250142_at | AT5G14650 | polygalacturonase, putative / pectinase, putative | -2.16 |
| 143 | 246540_at | AT5G15600 | SP1L4 (SPIRAL1-LIKE4) | -1.54 |
| 144 | 246464_at | AT5G16980 | NADP-dependent oxidoreductase, putative | 2.36 |
| 145 | 246149_at | AT5G19890 | peroxidase, putative | -3.12 |
| 146 | 246093_at | AT5G20550 | oxidoreductase, 2OG-Fe(II) oxygenase family protein | -1.30 |
| 147 | 249942_at | AT5G22300 | NIT4 (NITRILASE 4) | 2.80 |
| 148 | 249934_at | AT5G22410 | peroxidase, putative | -2.55 |
| 149 | 246922_at | AT5G25110 | CIPK25 (CBL-INTERACTING PROTEIN KINASE 25); kinase | -1.12 |
| 150 | 246858_at | AT5G25930 | leucine-rich repeat family protein / protein kinase family protein | 2.46 |
| 151 | 246825_at | AT5G26260 | meprin and TRAF homology domain-containing protein / MATH domain-containing protein | -1.25 |
| 152 | 246807_at | AT5G27100 | ATGLR2.1 (Arabidopsis thaliana glutamate receptor 2.1) | -1.14 |
| 153 | 249522_at | AT5G38700 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G02170.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO40081.1) | -1.47 |
| 154 | 249494_at | AT5G39050 | transferase family protein | 2.84 |
| 155 | 249227_at | AT5G42180 | peroxidase 64 (PER64) (P64) (PRXR4) | -1.44 |
| 156 | 249097_at | AT5G43520 | DC1 domain-containing protein | -2.30 |
| 157 | 248725_at | AT5G47980 | transferase family protein | -1.30 |
| 158 | 248728_at | AT5G48000 | CYP708A2 (cytochrome P450, family 708, subfamily A, polypeptide 2); oxygen binding | -1.73 |
| 159 | 248657_at | AT5G48570 | peptidyl-prolyl cis-trans isomerase, putative / FK506-binding protein, putative | 2.79 |
| 160 | 248381_at | AT5G51830 | pfkB-type carbohydrate kinase family protein | 2.37 |
| 161 | 248332_at | AT5G52640 | HSP81-1 (HEAT SHOCK PROTEIN 81-1); ATP binding / unfolded protein binding | 3.58 |
| 162 | 248183_at | AT5G54040 | DC1 domain-containing protein | -1.85 |
| 163 | 248178_at | AT5G54370 | late embryogenesis abundant protein-related / LEA protein-related | -1.59 |
| 164 | 247871_at | AT5G57530 | xyloglucan:xyloglucosyl transferase, putative / xyloglucan endotransglycosylase, putative / endo-xyloglucan transferase, putative | -2.60 |
| 165 | 247914_at | AT5G57540 | xyloglucan:xyloglucosyl transferase, putative / xyloglucan endotransglycosylase, putative / endo-xyloglucan transferase, putative | -4.24 |
| 166 | 247655_at | AT5G59820 | RHL41 (RESPONSIVE TO HIGH LIGHT 41); nucleic acid binding / transcription factor/ zinc ion binding | 3.25 |
| 167 | 247645_at | AT5G60530 | late embryogenesis abundant protein-related / LEA protein-related | -1.28 |
| 168 | 247488_at | AT5G61820 | similar to MtN19-like protein [Pisum sativum] (GB:AAU14999.2); contains InterPro domain Stress up-regulated Nod 19 (InterPro:IPR011692) | 2.53 |
| 169 | 247287_at | AT5G64230 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G19920.1); similar to hypothetical protein [Trifolium pratense] (GB:BAE71289.1) | 2.42 |
| 170 | 247283_at | AT5G64250 | 2-nitropropane dioxygenase family / NPD family | 3.06 |
| 171 | 247074_at | AT5G66590 | allergen V5/Tpx-1-related family protein | -1.21 |
| 172 | 247069_at | AT5G66920 | SKS17 (SKU5 Similar 17); copper ion binding / oxidoreductase | -1.09 |