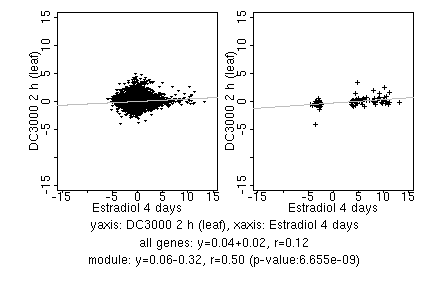
marker gene list
| Estradiol4 days |
| DC3000 2 h (leaf) |
| probeID | AGICode | Annotation | gene expression |
|
|---|---|---|---|---|
| 1 | 265095_at | AT1G03880 | CRU2 (CRUCIFERIN 2); nutrient reservoir | 11.37 |
| 2 | 264612_at | AT1G04560 | AWPM-19-like membrane family protein | 9.09 |
| 3 | 264606_at | AT1G04660 | glycine-rich protein | 7.31 |
| 4 | 263175_at | AT1G05510 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G31985.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO45247.1); contains InterPro domain Protein of unknown function DUF1264 (InterPro:IPR010686) | 10.02 |
| 5 | 264653_at | AT1G08980 | ATAMI1 (AMIDASE-LIKE PROTEIN 1); amidase | -2.56 |
| 6 | 261788_at | AT1G15980 | similar to unnamed protein product [Vitis vinifera] (GB:CAO49411.1); contains domain G3DSA:3.40.50.2000 (G3DSA:3.40.50.2000); contains domain SSF53756 (SSF53756) | -2.79 |
| 7 | 256115_at | AT1G16880 | uridylyltransferase-related | -2.50 |
| 8 | 255905_at | AT1G17810 | BETA-TIP (BETA-TONOPLAST INTRINSIC PROTEIN); water channel | 10.30 |
| 9 | 256073_at | AT1G18100 | E12A11; phosphatidylethanolamine binding | 3.83 |
| 10 | 260854_at | AT1G21970 | LEC1 (LEAFY COTYLEDON 1); transcription factor | 10.69 |
| 11 | 245634_at | AT1G25270 | similar to nodulin MtN21 family protein [Arabidopsis thaliana] (TAIR:AT1G68170.1); similar to Protein of unknown function DUF6, transmembrane [Medicago truncatula] (GB:ABD28596.1); contains InterPro domain Protein of unknown function DUF6, transmembrane; (InterPro:IPR000620) | 4.59 |
| 12 | 261005_at | AT1G26420 | FAD-binding domain-containing protein | 4.94 |
| 13 | 259580_at | AT1G28030 | oxidoreductase, 2OG-Fe(II) oxygenase family protein | 6.17 |
| 14 | 259782_at | AT1G29680 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G45690.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO47983.1); contains InterPro domain Protein of unknown function DUF1264 (InterPro:IPR010686) | 5.34 |
| 15 | 261190_at | AT1G32990 | PRPL11 (PLASTID RIBOSOMAL PROTEIN L11); structural constituent of ribosome | -2.41 |
| 16 | 262721_at | AT1G43560 | ATY2 (Arabidopsis thioredoxin y2); thiol-disulfide exchange intermediate | -3.42 |
| 17 | 260716_at | AT1G48130 | ATPER1 (Arabidopsis thaliana 1-cysteine peroxiredoxin 1); antioxidant | 11.33 |
| 18 | 261305_at | AT1G48470 | GLN1;5 (GLUTAMINE SYNTHETASE 1;5); glutamate-ammonia ligase | 4.57 |
| 19 | 264187_at | AT1G54860 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G19250.1); similar to GPI-anchored protein-like protein II [Cucumis melo] (GB:ABR67421.1) | 9.03 |
| 20 | 262086_at | AT1G56050 | GTP-binding protein-related | -2.59 |
| 21 | 264741_at | AT1G62290 | aspartyl protease family protein | 3.94 |
| 22 | 262878_at | AT1G64770 | carbohydrate binding / catalytic | -3.55 |
| 23 | 263138_at | AT1G65090 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G36100.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G36100.2); similar to hypothetical protein [Vitis vinifera] (GB:CAN81170.1) | 6.32 |
| 24 | 261503_at | AT1G71691 | GDSL-motif lipase/hydrolase family protein | 4.83 |
| 25 | 262175_at | AT1G74880 | NDH-O (NAD(P)H:PLASTOQUINONE DEHYDROGENASE COMPLEX SUBUNIT O) | -3.04 |
| 26 | 259956_at | AT1G75110 | RRA2 (REDUCED RESIDUAL ARABINOSE 2) | 4.92 |
| 27 | 261751_at | AT1G76080 | ATCDSP32/CDSP32 (CHLOROPLASTIC DROUGHT-INDUCED STRESS PROTEIN OF 32 KD); thiol-disulfide exchange intermediate | -3.19 |
| 28 | 261769_at | AT1G76100 | plastocyanin | -2.57 |
| 29 | 263131_at | AT1G78630 | EMB1473 (EMBRYO DEFECTIVE 1473); structural constituent of ribosome | -2.25 |
| 30 | 266034_at | AT2G06005 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G20580.1); similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G20580.2); similar to unknown [Populus trichocarpa] (GB:ABK93352.1) | 4.60 |
| 31 | 266570_at | AT2G24090 | ribosomal protein L35 family protein | -2.60 |
| 32 | 265644_at | AT2G27380 | ATEPR1 (Arabidopsis thaliana extensin proline-rich 1) | 5.06 |
| 33 | 265255_at | AT2G28420 | lactoylglutathione lyase family protein / glyoxalase I family protein | 4.23 |
| 34 | 255791_at | AT2G33430 | plastid developmental protein DAG, putative | -2.40 |
| 35 | 255840_at | AT2G33520 | similar to proline-rich family protein [Arabidopsis thaliana] (TAIR:AT1G12810.1) | 4.12 |
| 36 | 267317_at | AT2G34700 | pollen Ole e 1 allergen and extensin family protein | 8.36 |
| 37 | 267431_at | AT2G34870 | MEE26 (maternal effect embryo arrest 26) | 10.53 |
| 38 | 266608_at | AT2G35500 | shikimate kinase-related | -2.89 |
| 39 | 267088_at | AT2G38140 | PSRP4 (PLASTID-SPECIFIC RIBOSOMAL PROTEIN 4); structural constituent of ribosome | -2.39 |
| 40 | 267090_at | AT2G38270 | CXIP2 (CAX-INTERACTING PROTEIN 2); electron carrier/ protein disulfide oxidoreductase | -2.38 |
| 41 | 266421_at | AT2G38540 | LP1 (nonspecific lipid transfer protein 1) | -2.64 |
| 42 | 267075_at | AT2G41070 | EEL (ENHANCED EM LEVEL); DNA binding / transcription factor | 4.68 |
| 43 | 267586_at | AT2G41950 | similar to unnamed protein product [Vitis vinifera] (GB:CAO23423.1); contains domain S-adenosyl-L-methionine-dependent methyltransferases (SSF53335) | -2.46 |
| 44 | 266887_at | AT2G44650 | CHL-CPN10 (chloroplast chaperonin 10) | -2.40 |
| 45 | 266813_at | AT2G44920 | thylakoid lumenal 15 kDa protein, chloroplast | -2.87 |
| 46 | 266483_at | AT2G47910 | CRR6 (CHLORORESPIRATORY REDUCTION 6) | -4.30 |
| 47 | 265773_at | AT2G48070 | similar to unnamed protein product [Vitis vinifera] (GB:CAO22460.1) | -2.96 |
| 48 | 259167_at | AT3G01570 | glycine-rich protein / oleosin | 8.64 |
| 49 | 259098_at | AT3G04790 | ribose 5-phosphate isomerase-related | -2.80 |
| 50 | 259211_at | AT3G09020 | alpha 1,4-glycosyltransferase family protein / glycosyltransferase sugar-binding DXD motif-containing protein | 3.81 |
| 51 | 256290_at | AT3G12203 | SCPL17 (serine carboxypeptidase-like 17); serine carboxypeptidase | 9.73 |
| 52 | 256282_at | AT3G12550 | XH/XS domain-containing protein / XS zinc finger domain-containing protein | 3.85 |
| 53 | 257846_at | AT3G12910 | transcription factor | 4.30 |
| 54 | 257856_at | AT3G12930 | similar to unknown [Populus trichocarpa] (GB:ABK94112.1); contains InterPro domain Iojap-related protein (InterPro:IPR004394) | -2.42 |
| 55 | 256983_at | AT3G13470 | chaperonin, putative | -2.28 |
| 56 | 256780_at | AT3G13640 | ATRLI1 (Arabidopsis thaliana RNase L inhibitor protein 1) | 5.20 |
| 57 | 256855_at | AT3G15190 | chloroplast 30S ribosomal protein S20, putative | -2.41 |
| 58 | 258398_at | AT3G15360 | ATHM4 (Arabidopsis thioredoxin M-type 4); thiol-disulfide exchange intermediate | -2.58 |
| 59 | 257021_at | AT3G19710 | BCAT4 (BRANCHED-CHAIN AMINOTRANSFERASE4); catalytic/ methionine-oxo-acid transaminase | -2.62 |
| 60 | 258327_at | AT3G22640 | cupin family protein | 13.37 |
| 61 | 258258_at | AT3G26790 | FUS3 (FUSCA 3); DNA binding / transcription factor | 4.89 |
| 62 | 258240_at | AT3G27660 | OLEO4 (OLEOSIN4) | 10.74 |
| 63 | 252618_at | AT3G45140 | LOX2 (LIPOXYGENASE 2) | -3.44 |
| 64 | 252409_at | AT3G47650 | bundle-sheath defective protein 2 family / bsd2 family | -2.87 |
| 65 | 252181_at | AT3G50685 | similar to unnamed protein product [Vitis vinifera] (GB:CAO22868.1) | -2.53 |
| 66 | 251838_at | AT3G54940 | cysteine proteinase, putative | 5.98 |
| 67 | 246294_at | AT3G56910 | PSRP5 (PLASTID-SPECIFIC 50S RIBOSOMAL PROTEIN 5) | -2.88 |
| 68 | 251576_at | AT3G58200 | meprin and TRAF homology domain-containing protein / MATH domain-containing protein | 4.71 |
| 69 | 251442_at | AT3G59980 | tRNA-binding region domain-containing protein | -3.38 |
| 70 | 251155_at | AT3G63160 | unknown protein | -3.41 |
| 71 | 255471_at | AT4G03050 | AOP3 (2-oxoglutarate?dependent dioxygenase 3); oxidoreductase, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors | 10.23 |
| 72 | 245347_at | AT4G14890 | ferredoxin family protein | -2.53 |
| 73 | 245512_at | AT4G15770 | 60S ribosome subunit biogenesis protein, putative | -2.60 |
| 74 | 254398_at | AT4G21280 | PSBQ/PSBQ-1/PSBQA; calcium ion binding | -2.39 |
| 75 | 254095_at | AT4G25140 | OLEO1 (OLEOSIN1) | 8.95 |
| 76 | 254020_at | AT4G25700 | BETA-OHASE 1 (BETA-HYDROXYLASE 1); beta-carotene hydroxylase | -2.51 |
| 77 | 254036_at | AT4G25980 | cationic peroxidase, putative | 4.64 |
| 78 | 253972_at | AT4G26500 | ATSUFE/CPSUFE/EMB1374 (EMBRYO DEFECTIVE 1374); enzyme activator/ transcription regulator | -2.35 |
| 79 | 253904_at | AT4G27140 | 2S seed storage protein 1 / 2S albumin storage protein / NWMU1-2S albumin 1 | 8.40 |
| 80 | 253894_at | AT4G27150 | 2S seed storage protein 2 / 2S albumin storage protein / NWMU2-2S albumin 2 | 10.38 |
| 81 | 253895_at | AT4G27160 | AT2S3; lipid binding / nutrient reservoir | 9.32 |
| 82 | 253902_at | AT4G27170 | 2S seed storage protein 4 / 2S albumin storage protein / NWMU2-2S albumin 4 | 9.42 |
| 83 | 253767_at | AT4G28520 | CRU3 (CRUCIFERIN 3); nutrient reservoir | 7.92 |
| 84 | 253627_at | AT4G30650 | hydrophobic protein, putative / low temperature and salt responsive protein, putative | -2.53 |
| 85 | 253342_at | AT4G33520 | PAA1 (metal-transporting P-type ATPase 1); ATPase, coupled to transmembrane movement of ions, phosphorylative mechanism | -3.08 |
| 86 | 253272_at | AT4G34190 | SEP1 (STRESS ENHANCED PROTEIN 1) | -2.54 |
| 87 | 253241_at | AT4G34520 | FAE1 (FATTY ACID ELONGATION1); acyltransferase | 8.31 |
| 88 | 246273_at | AT4G36700 | cupin family protein | 11.15 |
| 89 | 250733_at | AT5G06290 | 2-cys peroxiredoxin, chloroplast, putative | -2.35 |
| 90 | 250620_at | AT5G07190 | ATS3 (ARABIDOPSIS THALIANA SEED GENE 3) | 6.74 |
| 91 | 246054_at | AT5G08360 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G23380.1); similar to Os04g0528000 [Oryza sativa (japonica cultivar-group)] (GB:NP_001053372.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO68101.1); similar to Os10g0494000 [Oryza sativa (japonica cultivar-group)] (GB:NP_001064942.1); contains InterPro domain Protein of unknown function DUF789 (InterPro:IPR008507) | 4.03 |
| 92 | 250501_at | AT5G09640 | SNG2 (SINAPOYLGLUCOSE ACCUMULATOR 2); serine carboxypeptidase | 8.38 |
| 93 | 250389_at | AT5G11320 | YUC4 (YUCCA4); monooxygenase | 3.84 |
| 94 | 250371_at | AT5G11450 | oxygen-evolving complex-related | -2.47 |
| 95 | 250255_at | AT5G13730 | SIG4 (SIGMA FACTOR 4); DNA binding / DNA-directed RNA polymerase/ transcription factor | -2.90 |
| 96 | 250258_at | AT5G13790 | AGL15 (AGAMOUS-LIKE 15); DNA binding / transcription factor | 5.90 |
| 97 | 250058_at | AT5G17870 | PSRP6 (PLASTID-SPECIFIC 50S RIBOSOMAL PROTEIN 6) | -2.89 |
| 98 | 246004_at | AT5G20630 | GLP3 (GERMIN-LIKE PROTEIN 3); manganese ion binding / metal ion binding / nutrient reservoir | -3.97 |
| 99 | 246747_at | AT5G27700 | 40S ribosomal protein S21 (RPS21C) | -2.32 |
| 100 | 249353_at | AT5G40420 | OLEO2 (OLEOSIN 2) | 8.06 |
| 101 | 249162_at | AT5G42765 | similar to unnamed protein product [Vitis vinifera] (GB:CAO61157.1); contains InterPro domain Twin-arginine translocation pathway signal (InterPro:IPR006311) | -2.29 |
| 102 | 249082_at | AT5G44120 | CRA1 (CRUCIFERINA); nutrient reservoir | 8.24 |
| 103 | 249043_at | AT5G44360 | FAD-binding domain-containing protein | 4.70 |
| 104 | 248962_at | AT5G45680 | FK506-binding protein 1 (FKBP13) | -2.74 |
| 105 | 248754_at | AT5G47670 | CCAAT-box binding transcription factor family protein / leafy cotyledon 1-related (L1L) | 6.12 |
| 106 | 248647_at | AT5G49190 | SUS2 (SUCROSE SYNTHASE 2); UDP-glycosyltransferase/ sucrose synthase/ transferase, transferring glycosyl groups | 8.28 |
| 107 | 248467_at | AT5G50800 | nodulin MtN3 family protein | -2.52 |
| 108 | 248435_at | AT5G51210 | OLEO3 (OLEOSIN3) | 6.80 |
| 109 | 248348_at | AT5G52190 | sugar isomerase (SIS) domain-containing protein | -2.85 |
| 110 | 248125_at | AT5G54740 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | 8.16 |
| 111 | 248094_at | AT5G55220 | trigger factor type chaperone family protein | -2.74 |
| 112 | 248062_at | AT5G55450 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | -3.44 |
| 113 | 247943_at | AT5G57170 | macrophage migration inhibitory factor family protein / MIF family protein | -3.20 |
| 114 | 247816_at | AT5G58260 | Encodes subunit NDH-N of NAD(P)H:plastoquinone dehydrogenase complex (Ndh complex) present in the thylakoid membrane of chloroplasts. This subunit is thought to be required for Ndh complex assembly. | -3.28 |
| 115 | 247762_at | AT5G59170 | proline-rich family protein | 5.81 |
| 116 | 247702_at | AT5G59500 | protein-S-isoprenylcysteine O-methyltransferase | -3.24 |
| 117 | 247421_at | AT5G62800 | seven in absentia (SINA) family protein | 6.29 |
| 118 | 247400_at | AT5G62840 | phosphoglycerate/bisphosphoglycerate mutase family protein | -2.25 |
| 119 | 247376_at | AT5G63310 | NDPK2 (NUCLEOSIDE DIPHOSPHATE KINASE 2); ATP binding / nucleoside diphosphate kinase | -2.84 |
| 120 | 244997_at | ATCG00170 | RNA polymerase beta' subunit-2 | -2.69 |