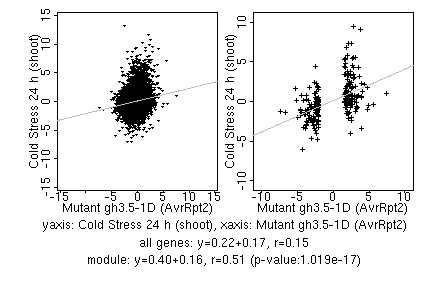
marker gene list
| Mutant gh3.5-1D (AvrRpt2) |
| Cold Stress 24 h (shoot) |
| probeID | AGICode | Annotation | gene expression |
|
|---|---|---|---|---|
| 1 | 259445_at | AT1G02400 | ATGA2OX6/DTA1 (GIBBERELLIN 2-OXIDASE 6); gibberellin 2-beta-dioxygenase | 2.82 |
| 2 | 260915_at | AT1G02660 | lipase class 3 family protein | 1.86 |
| 3 | 265066_at | AT1G03870 | FLA9 | -2.55 |
| 4 | 265042_at | AT1G04040 | acid phosphatase class B family protein | -1.93 |
| 5 | 264318_at | AT1G04220 | beta-ketoacyl-CoA synthase, putative | 2.14 |
| 6 | 263656_at | AT1G04240 | SHY2 (SHORT HYPOCOTYL 2); transcription factor | -2.24 |
| 7 | 263664_at | AT1G04250 | AXR3 (AUXIN RESISTANT 3); transcription factor | -2.56 |
| 8 | 264580_at | AT1G05340 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G32210.1) | 2.90 |
| 9 | 260831_at | AT1G06830 | glutaredoxin family protein | -2.61 |
| 10 | 261077_at | AT1G07430 | protein phosphatase 2C, putative / PP2C, putative | 2.96 |
| 11 | 264645_at | AT1G08940 | phosphoglycerate/bisphosphoglycerate mutase family protein | 2.53 |
| 12 | 264514_at | AT1G09500 | cinnamyl-alcohol dehydrogenase family / CAD family | 3.20 |
| 13 | 263236_at | AT1G10470 | ARR4 (RESPONSE REGULATOR 4); transcription regulator/ two-component response regulator | -2.31 |
| 14 | 261492_at | AT1G14290 | acid phosphatase, putative | -2.28 |
| 15 | 262603_at | AT1G15380 | lactoylglutathione lyase family protein / glyoxalase I family protein | 2.88 |
| 16 | 261763_at | AT1G15520 | ATPDR12/PDR12 (PLEIOTROPIC DRUG RESISTANCE 12); ATPase, coupled to transmembrane movement of substances | 2.17 |
| 17 | 259502_at | AT1G15670 | kelch repeat-containing F-box family protein | 2.30 |
| 18 | 262482_at | AT1G17020 | SRG1 (SENESCENCE-RELATED GENE 1); oxidoreductase, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors | 2.63 |
| 19 | 262539_at | AT1G17200 | integral membrane family protein | -2.06 |
| 20 | 259403_at | AT1G17745 | PGDH (3-PHOSPHOGLYCERATE DEHYDROGENASE); phosphoglycerate dehydrogenase | 2.57 |
| 21 | 259466_at | AT1G19050 | ARR7 (RESPONSE REGULATOR 7); transcription regulator/ two-component response regulator | -2.52 |
| 22 | 261226_at | AT1G20190 | ATEXPA11 (ARABIDOPSIS THALIANA EXPANSIN A11) | -3.78 |
| 23 | 261942_at | AT1G22590 | AGL87; transcription factor | -1.92 |
| 24 | 264195_at | AT1G22690 | gibberellin-responsive protein, putative | -2.05 |
| 25 | 261021_at | AT1G26380 | FAD-binding domain-containing protein | 1.72 |
| 26 | 261020_at | AT1G26390 | FAD-binding domain-containing protein | 2.66 |
| 27 | 261648_at | AT1G27730 | STZ (SALT TOLERANCE ZINC FINGER); nucleic acid binding / transcription factor/ zinc ion binding | 3.00 |
| 28 | 257506_at | AT1G29440 | auxin-responsive family protein | -4.42 |
| 29 | 259784_at | AT1G29450 | auxin-responsive protein, putative | -4.83 |
| 30 | 259787_at | AT1G29460 | auxin-responsive protein, putative | -3.59 |
| 31 | 259773_at | AT1G29500 | auxin-responsive protein, putative | -4.43 |
| 32 | 259783_at | AT1G29510 | SAUR68 (SMALL AUXIN UPREGULATED 68) | -7.30 |
| 33 | 259786_at | AT1G29660 | GDSL-motif lipase/hydrolase family protein | -3.29 |
| 34 | 263228_at | AT1G30700 | FAD-binding domain-containing protein | 3.35 |
| 35 | 262548_at | AT1G31280 | AGO2 (ARGONAUTE 2); nucleic acid binding | 2.93 |
| 36 | 260693_at | AT1G32450 | proton-dependent oligopeptide transport (POT) family protein | 2.11 |
| 37 | 261242_at | AT1G32960 | ATSBT3.3; subtilase | 2.00 |
| 38 | 262010_at | AT1G35612 | transposable element gene | -2.06 |
| 39 | 256462_at | AT1G36230 | unknown protein | -3.60 |
| 40 | 264415_at | AT1G43160 | RAP2.6 (related to AP2 6); DNA binding / transcription factor | 4.88 |
| 41 | 259618_at | AT1G48000 | MYB112 (myb domain protein 112); DNA binding / transcription factor | 1.87 |
| 42 | 261629_at | AT1G49980 | similar to REV1 (Reversionless 1), damaged DNA binding / magnesium ion binding / nucleotidyltransferase [Arabidopsis thaliana] (TAIR:AT5G44750.2); similar to REV1 (Reversionless 1), damaged DNA binding / magnesium ion binding / nucleotidyltransferase [Arabidopsis thaliana] (TAIR:AT5G44750.1); similar to hypothetical protein OsJ_011272 [Oryza sativa (japonica cultivar-group)] (GB:EAZ27789.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO63978.1); similar to hypothetical protein OsI_012241 [Oryza sativa (indica cultivar-group)] (GB:EAY91008.1); contains InterPro domain UMUC-like DNA-repair protein; (InterPro:IPR001126) | -1.99 |
| 43 | 263157_at | AT1G54100 | ALDH7B4 (ALDEHYDE DEHYDROGENASE 7B4); 3-chloroallyl aldehyde dehydrogenase | 2.13 |
| 44 | 259660_at | AT1G55260 | lipid binding | -2.53 |
| 45 | 259664_at | AT1G55330 | AGP21 (ARABINOGALACTAN PROTEIN 21) | -3.88 |
| 46 | 245628_at | AT1G56650 | PAP1 (PRODUCTION OF ANTHOCYANIN PIGMENT 1); DNA binding / transcription factor | 1.96 |
| 47 | 264400_at | AT1G61800 | GPT2 (glucose-6-phosphate/phosphate translocator 2); antiporter/ glucose-6-phosphate transmembrane transporter | 1.80 |
| 48 | 264433_at | AT1G61810 | BGLU45; hydrolase, hydrolyzing O-glycosyl compounds | 2.06 |
| 49 | 262640_at | AT1G62760 | invertase/pectin methylesterase inhibitor family protein | 1.90 |
| 50 | 260243_at | AT1G63720 | similar to hydroxyproline-rich glycoprotein family protein [Arabidopsis thaliana] (TAIR:AT5G52430.1); similar to unknown [Thellungiella halophila] (GB:ABJ98061.1) | 1.73 |
| 51 | 260135_at | AT1G66400 | calmodulin-related protein, putative | 2.05 |
| 52 | 256378_at | AT1G66830 | leucine-rich repeat transmembrane protein kinase, putative | 2.55 |
| 53 | 260015_at | AT1G67980 | CCoAMT (caffeoyl-CoA 3-O-methyltransferase); caffeoyl-CoA O-methyltransferase | 1.79 |
| 54 | 259373_at | AT1G69160 | unknown protein | -2.10 |
| 55 | 256304_at | AT1G69523 | UbiE/COQ5 methyltransferase family protein | -1.91 |
| 56 | 256299_at | AT1G69530 | ATEXPA1 (ARABIDOPSIS THALIANA EXPANSIN A1) | -2.94 |
| 57 | 260410_at | AT1G69870 | proton-dependent oligopeptide transport (POT) family protein | 1.83 |
| 58 | 260405_at | AT1G69930 | ATGSTU11 (Arabidopsis thaliana Glutathione S-transferase (class tau) 11); glutathione transferase | 2.46 |
| 59 | 262304_at | AT1G70890 | MLP43 (MLP-LIKE PROTEIN 43) | -2.92 |
| 60 | 262263_at | AT1G70940 | PIN3 (PIN-FORMED 3); auxin:hydrogen symporter/ transporter | -2.29 |
| 61 | 262290_at | AT1G70985 | hydroxyproline-rich glycoprotein family protein | -3.62 |
| 62 | 260427_at | AT1G72430 | auxin-responsive protein-related | -2.16 |
| 63 | 262381_at | AT1G72900 | disease resistance protein (TIR-NBS class), putative | 2.08 |
| 64 | 260100_at | AT1G73177 | BNS (BONSAI) | -2.00 |
| 65 | 260070_at | AT1G73830 | BEE3 (BR ENHANCED EXPRESSION 3); DNA binding / transcription factor | -3.05 |
| 66 | 260391_at | AT1G74020 | SS2 (STRICTOSIDINE SYNTHASE 2); strictosidine synthase | 1.89 |
| 67 | 260221_at | AT1G74670 | gibberellin-responsive protein, putative | -4.07 |
| 68 | 262211_at | AT1G74930 | ORA47; DNA binding / transcription factor | 2.77 |
| 69 | 256503_at | AT1G75250 | myb family transcription factor | -4.29 |
| 70 | 262947_at | AT1G75750 | GASA1 (GAST1 PROTEIN HOMOLOG 1) | -2.45 |
| 71 | 259884_at | AT1G76390 | armadillo/beta-catenin repeat family protein / U-box domain-containing protein | 2.09 |
| 72 | 262039_at | AT1G80050 | APT2 (ADENINE PHOSPHORIBOSYL TRANSFERASE 2); adenine phosphoribosyltransferase | -2.20 |
| 73 | 262047_at | AT1G80160 | lactoylglutathione lyase family protein / glyoxalase I family protein | 4.12 |
| 74 | 261881_at | AT1G80760 | NIP6;1 (NOD26-like intrinsic protein 6;1); water channel | -2.08 |
| 75 | 257474_at | AT1G80850 | methyladenine glycosylase family protein | -2.17 |
| 76 | 266329_at | AT2G01590 | similar to unnamed protein product [Vitis vinifera] (GB:CAO42320.1) | -2.06 |
| 77 | 263406_at | AT2G04160 | AIR3 (Auxin-Induced in Root cultures 3); subtilase | 2.03 |
| 78 | 265377_at | AT2G05790 | glycosyl hydrolase family 17 protein | -2.24 |
| 79 | 266215_at | AT2G06850 | EXGT-A1 (ENDOXYLOGLUCAN TRANSFERASE); hydrolase, acting on glycosyl bonds | -3.38 |
| 80 | 265400_at | AT2G10940 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | -3.19 |
| 81 | 265894_at | AT2G15050 | LTP; lipid binding | -2.05 |
| 82 | 264909_at | AT2G17300 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT4G35320.1) | -2.08 |
| 83 | 263073_at | AT2G17500 | auxin efflux carrier family protein | 2.45 |
| 84 | 264023_at | AT2G21195 | similar to unnamed protein product [Vitis vinifera] (GB:CAO44591.1) | -1.91 |
| 85 | 263549_at | AT2G21650 | MEE3 (maternal effect embryo arrest 3); DNA binding / transcription factor | -2.21 |
| 86 | 245076_at | AT2G23170 | GH3.3; indole-3-acetic acid amido synthetase | 2.54 |
| 87 | 266311_at | AT2G27130 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | -2.10 |
| 88 | 265276_at | AT2G28400 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G60680.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO21845.1); contains InterPro domain Protein of unknown function DUF584 (InterPro:IPR007608) | 2.03 |
| 89 | 266790_at | AT2G28950 | ATEXPA6 (ARABIDOPSIS THALIANA EXPANSIN A6) | -2.38 |
| 90 | 266285_at | AT2G29180 | similar to 80C09_19 [Brassica rapa subsp. pekinensis] (GB:AAZ41830.1) | -1.95 |
| 91 | 266267_at | AT2G29460 | ATGSTU4 (GLUTATHIONE S-TRANSFERASE 22); glutathione transferase | 1.74 |
| 92 | 263485_at | AT2G29890 | VLN1 (VILLIN 1); actin binding | 3.14 |
| 93 | 263475_at | AT2G31945 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G05575.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO22015.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN61524.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN71356.1) | 2.27 |
| 94 | 266995_at | AT2G34500 | CYP710A1 (cytochrome P450, family 710, subfamily A, polypeptide 1); C-22 sterol desaturase/ oxygen binding | 2.41 |
| 95 | 263951_at | AT2G35960 | NHL12 (NDR1/HIN1-like 12) | -1.92 |
| 96 | 267147_at | AT2G38240 | oxidoreductase, 2OG-Fe(II) oxygenase family protein | 3.37 |
| 97 | 267026_at | AT2G38340 | AP2 domain-containing transcription factor, putative (DRE2B) | 4.46 |
| 98 | 267008_at | AT2G39350 | ABC transporter family protein | 3.38 |
| 99 | 267364_at | AT2G40080 | ELF4 (EARLY FLOWERING 4) | 2.21 |
| 100 | 265722_at | AT2G40100 | LHCB4.3 (LIGHT HARVESTING COMPLEX PSII); chlorophyll binding | -2.18 |
| 101 | 255822_at | AT2G40610 | ATEXPA8 (ARABIDOPSIS THALIANA EXPANSIN A8) | -4.17 |
| 102 | 266368_at | AT2G41380 | embryo-abundant protein-related | 1.83 |
| 103 | 266873_at | AT2G44740 | CYCP4;1 (cyclin p4;1); cyclin-dependent protein kinase | -2.90 |
| 104 | 245148_at | AT2G45220 | pectinesterase family protein | 3.10 |
| 105 | 251395_at | AT2G45470 | FLA8 (Arabinogalactan protein 8) | -2.24 |
| 106 | 266327_at | AT2G46680 | ATHB-7 (ARABIDOPSIS THALIANA HOMEOBOX 7); transcription factor | 2.27 |
| 107 | 260581_at | AT2G47190 | MYB2 (myb domain protein 2); DNA binding / transcription factor | 1.91 |
| 108 | 258957_at | AT3G01420 | ALPHA-DOX1 (ALPHA-DIOXYGENASE 1) | 5.02 |
| 109 | 258976_at | AT3G01980 | short-chain dehydrogenase/reductase (SDR) family protein | -2.15 |
| 110 | 258610_at | AT3G02875 | ILR1 (IAA-LEUCINE RESISTANT 1); metallopeptidase | 2.00 |
| 111 | 259054_at | AT3G03480 | CHAT (ACETYL COA:(Z)-3-HEXEN-1-OL ACETYLTRANSFERASE); acetyl CoA:(Z)-3-hexen-1-ol acetyltransferase | 1.99 |
| 112 | 259331_at | AT3G03840 | auxin-responsive protein, putative | -6.57 |
| 113 | 258813_at | AT3G04060 | ANAC046 (Arabidopsis NAC domain containing protein 46); transcription factor | 2.23 |
| 114 | 258809_at | AT3G04070 | ANAC047 (Arabidopsis NAC domain containing protein 47); transcription factor | 2.43 |
| 115 | 258887_at | AT3G05630 | PLDP2 (PHOSPHOLIPASE D ZETA 2); phospholipase D | 3.73 |
| 116 | 258897_at | AT3G05730 | Encodes a defensin-like (DEFL) family protein. | -3.59 |
| 117 | 258468_at | AT3G06070 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G19190.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO71112.1) | -3.12 |
| 118 | 258529_at | AT3G06740 | zinc finger (GATA type) family protein | -2.03 |
| 119 | 258524_at | AT3G06810 | IBR3 (IBA-RESPONSE 3); acyl-CoA dehydrogenase/ oxidoreductase | 2.18 |
| 120 | 258983_at | AT3G08860 | alanine--glyoxylate aminotransferase, putative / beta-alanine-pyruvate aminotransferase, putative / AGT, putative | 2.17 |
| 121 | 259150_at | AT3G10320 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G41640.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO23616.1); contains InterPro domain Protein of unknown function DUF563 (InterPro:IPR007657) | 2.15 |
| 122 | 258920_at | AT3G10520 | AHB2 (NON-SYMBIOTIC HAEMOGLOBIN 2) | -2.30 |
| 123 | 256428_at | AT3G11080 | disease resistance family protein | 4.05 |
| 124 | 256237_at | AT3G12610 | DRT100 (DNA-DAMAGE REPAIR/TOLERATION 100); protein binding | -2.86 |
| 125 | 258207_at | AT3G14050 | RSH2 (RELA-SPOT HOMOLOG); catalytic | 2.31 |
| 126 | 258370_at | AT3G14395 | unknown protein | -3.20 |
| 127 | 257280_at | AT3G14440 | NCED3 (NINE-CIS-EPOXYCAROTENOID DIOXYGENASE3) | 1.73 |
| 128 | 258054_at | AT3G16240 | DELTA-TIP (delta tonoplast integral protein); water channel | -3.32 |
| 129 | 259375_at | AT3G16370 | GDSL-motif lipase/hydrolase family protein | -2.99 |
| 130 | 258462_at | AT3G17350 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G50290.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO70913.1); contains domain PROKAR_LIPOPROTEIN (PS51257) | -2.19 |
| 131 | 258162_at | AT3G17810 | dihydroorotate dehydrogenase family protein / dihydroorotate oxidase family protein | 1.91 |
| 132 | 258160_at | AT3G17820 | ATGSKB6 (Arabidopsis thaliana glutamine synthase clone KB6); glutamate-ammonia ligase | 1.99 |
| 133 | 257085_at | AT3G20630 | UBP14 (UBIQUITIN-SPECIFIC PROTEASE 14); ubiquitin-specific protease | 1.75 |
| 134 | 258038_at | AT3G21260 | GLTP3 (GLYCOLIPID TRANSFER PROTEIN 3); glycolipid binding / glycolipid transporter | -2.20 |
| 135 | 258182_at | AT3G21500 | DXPS1; 1-deoxy-D-xylulose-5-phosphate synthase | 2.23 |
| 136 | 256796_at | AT3G22210 | unknown protein | -1.92 |
| 137 | 257769_at | AT3G23050 | IAA7 (AUXIN RESISTANT 2); transcription factor | -3.28 |
| 138 | 257793_at | AT3G26960 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G41050.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO49052.1) | -4.43 |
| 139 | 252711_at | AT3G43720 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | -2.14 |
| 140 | 252679_at | AT3G44260 | CCR4-NOT transcription complex protein, putative | 2.64 |
| 141 | 252624_at | AT3G44735 | PSK1 (PHYTOSULFOKINE 3 PRECURSOR); growth factor | -1.90 |
| 142 | 252549_at | AT3G45860 | receptor-like protein kinase, putative | -1.95 |
| 143 | 252501_at | AT3G46880 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G59080.1) | -2.35 |
| 144 | 252415_at | AT3G47340 | ASN1 (DARK INDUCIBLE 6) | 1.99 |
| 145 | 252368_at | AT3G48520 | CYP94B3 (cytochrome P450, family 94, subfamily B, polypeptide 3); oxygen binding | 3.83 |
| 146 | 252317_at | AT3G48720 | transferase family protein | -2.73 |
| 147 | 252303_at | AT3G49210 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G49200.1); similar to condensation domain-containing protein [Arabidopsis thaliana] (TAIR:AT3G49190.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO48523.1); contains InterPro domain Protein of unknown function UPF0089 (InterPro:IPR004255); contains InterPro domain Protein of unknown function DUF1298 (InterPro:IPR009721) | 3.05 |
| 148 | 252220_at | AT3G49940 | LBD38 (LOB DOMAIN-CONTAINING PROTEIN 38) | -2.32 |
| 149 | 252167_at | AT3G50560 | short-chain dehydrogenase/reductase (SDR) family protein | -2.40 |
| 150 | 252102_at | AT3G50970 | LTI30 (LOW TEMPERATURE-INDUCED 30) | 2.97 |
| 151 | 256674_at | AT3G52360 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT2G35850.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO39911.1) | -2.04 |
| 152 | 252011_at | AT3G52720 | carbonic anhydrase family protein | -2.00 |
| 153 | 251766_at | AT3G55910 | unknown protein | 3.73 |
| 154 | 251665_at | AT3G57040 | ARR9 (RESPONSE REACTOR 4); transcription regulator | -2.37 |
| 155 | 251635_at | AT3G57510 | ADPG1 (endo-polygalacturonase 1); polygalacturonase | 4.04 |
| 156 | 251428_at | AT3G60140 | DIN2 (DARK INDUCIBLE 2); hydrolase, hydrolyzing O-glycosyl compounds | 5.74 |
| 157 | 251191_at | AT3G62590 | lipase class 3 family protein | 2.07 |
| 158 | 255692_at | AT4G00400 | GPAT8 (GLYCEROL-3-PHOSPHATE ACYLTRANSFERASE 8); acyltransferase/ glycerol-3-phosphate O-acyltransferase | -1.95 |
| 159 | 255302_at | AT4G04830 | methionine sulfoxide reductase domain-containing protein / SeIR domain-containing protein | -2.01 |
| 160 | 255298_at | AT4G04840 | methionine sulfoxide reductase domain-containing protein / SeIR domain-containing protein | -3.32 |
| 161 | 255104_at | AT4G08685 | SAH7 | -2.88 |
| 162 | 254889_at | AT4G11650 | ATOSM34 (OSMOTIN 34) | 4.14 |
| 163 | 254789_at | AT4G12880 | plastocyanin-like domain-containing protein | -2.17 |
| 164 | 254746_at | AT4G12980 | auxin-responsive protein, putative | -2.00 |
| 165 | 254685_at | AT4G13790 | auxin-responsive protein, putative | 4.90 |
| 166 | 245624_at | AT4G14090 | UDP-glucoronosyl/UDP-glucosyl transferase family protein | 2.65 |
| 167 | 245262_at | AT4G16563 | aspartyl protease family protein | 3.34 |
| 168 | 245444_at | AT4G16740 | ATTPS03 (Arabidopsis thaliana terpene synthase 03) | 2.15 |
| 169 | 245318_at | AT4G16980 | arabinogalactan-protein family | -2.10 |
| 170 | 245383_at | AT4G17810 | nucleic acid binding / transcription factor/ zinc ion binding | -2.09 |
| 171 | 254598_at | AT4G18990 | xyloglucan:xyloglucosyl transferase, putative / xyloglucan endotransglycosylase, putative / endo-xyloglucan transferase, putative | 2.79 |
| 172 | 254562_at | AT4G19230 | CYP707A1 (cytochrome P450, family 707, subfamily A, polypeptide 1); oxygen binding | 2.12 |
| 173 | 254543_at | AT4G19810 | glycosyl hydrolase family 18 protein | 2.68 |
| 174 | 254447_at | AT4G20860 | FAD-binding domain-containing protein | 2.04 |
| 175 | 254396_at | AT4G21680 | proton-dependent oligopeptide transport (POT) family protein | 4.26 |
| 176 | 254314_at | AT4G22470 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | 3.09 |
| 177 | 254215_at | AT4G23700 | ATCHX17 (CATION/H+ EXCHANGER 17); monovalent cation:proton antiporter | 2.12 |
| 178 | 254189_at | AT4G24000 | ATCSLG2 (Cellulose synthase-like G2); transferase/ transferase, transferring glycosyl groups | 3.49 |
| 179 | 254102_at | AT4G25050 | ACP4 (ACYL CARRIER PROTEIN 4) | -2.00 |
| 180 | 254066_at | AT4G25480 | DREB1A (DEHYDRATION RESPONSE ELEMENT B1A); DNA binding / transcription activator/ transcription factor | 2.17 |
| 181 | 254030_at | AT4G25890 | 60S acidic ribosomal protein P3 (RPP3A) | -2.14 |
| 182 | 253908_at | AT4G27260 | GH3.5/WES1; indole-3-acetic acid amido synthetase | 7.51 |
| 183 | 253843_at | AT4G27910 | PHD finger protein-related / SET domain-containing protein (TX4) | 2.21 |
| 184 | 253749_at | AT4G29080 | PAP2 (PHYTOCHROME-ASSOCIATED PROTEIN 2); transcription factor | -2.28 |
| 185 | 253608_at | AT4G30290 | ATXTH19 (XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 19); hydrolase, acting on glycosyl bonds | -2.34 |
| 186 | 253373_at | AT4G33150 | LKR (SACCHAROPINE DEHYDROGENASE) | 3.53 |
| 187 | 253343_at | AT4G33540 | metallo-beta-lactamase family protein | 1.83 |
| 188 | 253288_at | AT4G34310 | binding | 3.13 |
| 189 | 253046_at | AT4G37370 | CYP81D8 (cytochrome P450, family 81, subfamily D, polypeptide 8); oxygen binding | 1.98 |
| 190 | 253101_at | AT4G37430 | CYP91A2 (CYTOCHROME P450 MONOOXYGENASE 91A2); oxygen binding | 2.47 |
| 191 | 253040_at | AT4G37800 | xyloglucan:xyloglucosyl transferase, putative / xyloglucan endotransglycosylase, putative / endo-xyloglucan transferase, putative | -3.48 |
| 192 | 252984_at | AT4G37990 | ELI3-2 (ELICITOR-ACTIVATED GENE 3) | 2.88 |
| 193 | 252972_at | AT4G38840 | auxin-responsive protein, putative | -2.44 |
| 194 | 252970_at | AT4G38850 | SAUR_AC1 (SMALL AUXIN UP RNA 1 FROM ARABIDOPSIS THALIANA ECOTYPE COLUMBIA) | -3.65 |
| 195 | 252965_at | AT4G38860 | auxin-responsive protein, putative | -3.83 |
| 196 | 252908_at | AT4G39670 | glycolipid binding / glycolipid transporter | 3.17 |
| 197 | 251142_at | AT5G01015 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT1G65295.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN82885.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO45347.1) | -2.70 |
| 198 | 251031_at | AT5G02120 | OHP (ONE HELIX PROTEIN) | -1.91 |
| 199 | 250936_at | AT5G03120 | similar to hypothetical protein [Vitis vinifera] (GB:CAN68657.1) | -2.98 |
| 200 | 250895_at | AT5G03850 | 40S ribosomal protein S28 (RPS28B) | -1.93 |
| 201 | 245711_at | AT5G04340 | C2H2 (ZINC FINGER OF ARABIDOPSIS THALIANA 6); nucleic acid binding / transcription factor/ zinc ion binding | 1.81 |
| 202 | 250798_at | AT5G05340 | peroxidase, putative | 2.20 |
| 203 | 250781_at | AT5G05410 | DREB2A (DRE-BINDING PROTEIN 2A); DNA binding / transcription activator/ transcription factor | 2.07 |
| 204 | 250764_at | AT5G05960 | protease inhibitor/seed storage/lipid transfer protein (LTP) family protein | -4.43 |
| 205 | 250662_at | AT5G07010 | sulfotransferase family protein | 2.14 |
| 206 | 250580_at | AT5G07440 | GDH2 (GLUTAMATE DEHYDROGENASE 2); oxidoreductase | 2.38 |
| 207 | 250366_at | AT5G11420 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G25460.1); similar to unknown [Ricinus communis] (GB:CAB02653.1); contains InterPro domain Protein of unknown function DUF642 (InterPro:IPR006946); contains InterPro domain Galactose-binding like (InterPro:IPR008979) | -2.09 |
| 208 | 250327_at | AT5G12050 | similar to unnamed protein product [Vitis vinifera] (GB:CAO45643.1) | -3.23 |
| 209 | 250155_at | AT5G15160 | bHLH family protein | -2.25 |
| 210 | 250158_at | AT5G15190 | unknown protein | 1.80 |
| 211 | 246522_at | AT5G15830 | ATBZIP3 (ARABIDOPSIS THALIANA BASIC LEUCINE-ZIPPER 3); DNA binding / transcription factor | -2.67 |
| 212 | 250058_at | AT5G17870 | PSRP6 (PLASTID-SPECIFIC 50S RIBOSOMAL PROTEIN 6) | -1.92 |
| 213 | 249996_at | AT5G18600 | glutaredoxin family protein | -2.24 |
| 214 | 245998_at | AT5G20830 | SUS1 (SUCROSE SYNTHASE 1); UDP-glycosyltransferase/ sucrose synthase | 2.79 |
| 215 | 249894_at | AT5G22580 | Identical to Uncharacterized protein At5g22580 [Arabidopsis Thaliana] (GB:Q9FK81); similar to stable protein 1-related [Arabidopsis thaliana] (TAIR:AT3G17210.1); similar to hypothetical protein [Vitis vinifera] (GB:CAN68720.1); contains InterPro domain Dimeric alpha-beta barrel (InterPro:IPR011008); contains InterPro domain Stress responsive alpha-beta barrel (InterPro:IPR013097) | -5.05 |
| 216 | 249850_at | AT5G23240 | DNAJ heat shock N-terminal domain-containing protein | 2.49 |
| 217 | 246922_at | AT5G25110 | CIPK25 (CBL-INTERACTING PROTEIN KINASE 25); kinase | 3.97 |
| 218 | 246932_at | AT5G25190 | ethylene-responsive element-binding protein, putative | -1.94 |
| 219 | 246919_at | AT5G25460 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G11420.1); similar to unknown [Ricinus communis] (GB:CAB02653.1); contains InterPro domain Protein of unknown function DUF642 (InterPro:IPR006946) | -4.06 |
| 220 | 246909_at | AT5G25770 | similar to unnamed protein product [Vitis vinifera] (GB:CAO44054.1); contains domain PTHR10992 (PTHR10992); contains domain SSF53474 (SSF53474); contains domain G3DSA:3.40.50.1820 (G3DSA:3.40.50.1820) | 2.21 |
| 221 | 249527_at | AT5G38710 | proline oxidase, putative / osmotic stress-responsive proline dehydrogenase, putative | 2.66 |
| 222 | 249454_at | AT5G39520 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT5G39530.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO15021.1) | 3.12 |
| 223 | 249383_at | AT5G39860 | PRE1 (PACLOBUTRAZOL RESISTANCE1); DNA binding / transcription factor | -3.44 |
| 224 | 249230_at | AT5G42070 | similar to unnamed protein product [Vitis vinifera] (GB:CAO69519.1) | -2.02 |
| 225 | 249215_at | AT5G42800 | DFR (DIHYDROFLAVONOL 4-REDUCTASE); dihydrokaempferol 4-reductase | 2.35 |
| 226 | 249073_at | AT5G44020 | acid phosphatase class B family protein | -3.54 |
| 227 | 249008_at | AT5G44680 | methyladenine glycosylase family protein | -2.68 |
| 228 | 248975_at | AT5G45040 | cytochrome c6 (ATC6) | -1.99 |
| 229 | 248962_at | AT5G45680 | FK506-binding protein 1 (FKBP13) | -2.06 |
| 230 | 248713_at | AT5G48180 | kelch repeat-containing protein | 2.05 |
| 231 | 248528_at | AT5G50760 | auxin-responsive family protein | 2.62 |
| 232 | 248460_at | AT5G50915 | basic helix-loop-helix (bHLH) family protein | -3.73 |
| 233 | 248487_at | AT5G51070 | ERD1 (EARLY RESPONSIVE TO DEHYDRATION 1); ATP binding / ATPase | 2.20 |
| 234 | 248377_at | AT5G51720 | similar to Os07g0467200 [Oryza sativa (japonica cultivar-group)] (GB:NP_001059590.1); similar to hypothetical protein OsI_025030 [Oryza sativa (indica cultivar-group)] (GB:EAZ03798.1); contains domain PTHR13680 (PTHR13680); contains domain PTHR13680:SF1 (PTHR13680:SF1) | -1.95 |
| 235 | 248337_at | AT5G52310 | COR78 (COLD REGULATED 78) | 2.70 |
| 236 | 248246_at | AT5G53200 | TRY (TRIPTYCHON); DNA binding / transcription factor | -2.26 |
| 237 | 248185_at | AT5G54060 | UF3GT (UDP-GLUCOSE:FLAVONOID 3-O-GLUCOSYLTRANSFERASE); transferase, transferring glycosyl groups | 1.90 |
| 238 | 248163_at | AT5G54510 | DFL1/GH3.6 (DWARF IN LIGHT 1); indole-3-acetic acid amido synthetase | 3.41 |
| 239 | 248028_at | AT5G55620 | similar to unknown protein [Arabidopsis thaliana] (TAIR:AT3G09950.1); similar to unnamed protein product [Vitis vinifera] (GB:CAO39940.1) | -2.10 |
| 240 | 247880_at | AT5G57780 | transcription regulator | -2.65 |
| 241 | 247814_at | AT5G58310 | hydrolase, alpha/beta fold family protein | -2.29 |
| 242 | 247723_at | AT5G59220 | protein phosphatase 2C, putative / PP2C, putative | 3.31 |
| 243 | 247668_at | AT5G60100 | APRR3 (PSEUDO-RESPONSE REGULATOR 3); transcription regulator | 2.97 |
| 244 | 247406_at | AT5G62920 | ARR6 (RESPONSE REGULATOR 6); transcription regulator/ two-component response regulator | -5.12 |
| 245 | 247279_at | AT5G64310 | AGP1 (ARABINOGALACTAN-PROTEIN 1) | 1.73 |
| 246 | 247252_at | AT5G64770 | similar to 80C09_10 [Brassica rapa subsp. pekinensis] (GB:AAZ41821.1) | -4.27 |
| 247 | 247176_at | AT5G65110 | ACX2 (ACYL-COA OXIDASE 2); acyl-CoA oxidase | 2.09 |
| 248 | 247162_at | AT5G65730 | xyloglucan:xyloglucosyl transferase, putative / xyloglucan endotransglycosylase, putative / endo-xyloglucan transferase, putative | -3.62 |
| 249 | 247074_at | AT5G66590 | allergen V5/Tpx-1-related family protein | -2.50 |
| 250 | 247037_at | AT5G67070 | RALFL34 (RALF-LIKE 34) | -2.31 |
| 251 | 246984_at | AT5G67310 | CYP81G1 (cytochrome P450, family 81, subfamily G, polypeptide 1); oxygen binding | 2.81 |
| 252 | 247013_at | AT5G67480 | BT4 (BTB AND TAZ DOMAIN PROTEIN 4); protein binding / transcription regulator | 2.35 |