| ID | C0186 |
| Compound name | NAD |
| Stereochemistry | - |
| Aracyc name | NAD+ |
| External link |
|
| Mass information |
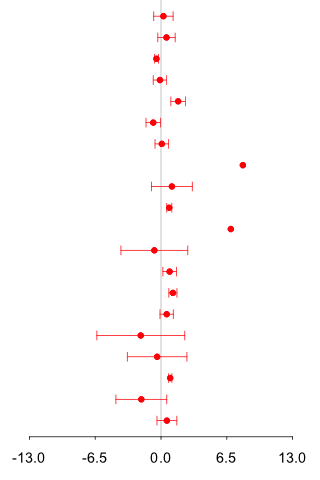
1st experiment : 2nd experiment : CE-TOF/MS (Anion analysis) |
| Molecular Formula | H26P2C21O14N7 |
| Canonical SMILES | C1(N=C(N)C2(N=CN(C(N=1)=2)C5(OC(COP(=O)([O-])OP(=O)([O-])OCC3(OC(C(O)C(O)3)[N+]4(C=CC=C(C=4)C(N)=O)))C(O)C(O)5))) |
| Molecular Weight | 664.438 |
| Pathway Information | ammonia assimilation cycle II, aspartate degradation II, adenosine nucleotides degradation I, plant sterol biosynthesis II, UDP-D-xylose and UDP-D-glucuronate biosynthesis, NAD biosynthesis I (from aspartate), ethanol degradation IV (peroxisomal), abscisic acid biosynthesis, palmitate biosynthesis II (bacteria and plants), zeaxanthin biosynthesis, sucrose degradation to ethanol and lactate (anaerobic), glutamate degradation IV, stearate biosynthesis II (plants), guanosine nucleotides degradation II, glutamate degradation I, mannitol degradation II, glycerol-3-phosphate shuttle, 2-ketoglutarate dehydrogenase complex, arginine degradation I (arginase pathway), aerobic respiration (alternative oxidase pathway), branched-chain alpha-keto acid dehydrogenase complex, 4-aminobutyrate degradation IV, glycolysis I, glycine cleavage complex, lysine degradation II, fatty acid beta-oxidation II (core pathway), nitrate reduction II (assimilatory), TCA cycle variation V (plant), leucine degradation I, thiamine biosynthesis II, acetaldehyde biosynthesis I, sorbitol degradation I, glutamine biosynthesis III, aerobic respiration (cytochrome c), ethanol degradation I, ascorbate biosynthesis I (L-galactose pathway), glycolysis IV (plant cytosol), benzoate biosynthesis II (CoA-independent, non-beta-oxidative), siroheme biosynthesis, TCA cycle variation III (eukaryotic), isoleucine degradation I, pyridine nucleotide cycling (plants), tyrosine biosynthesis I, gluconeogenesis I, glycine betaine biosynthesis III (plants), traumatin and (Z)-3-hexen-1-yl acetate biosynthesis, NAD/NADH phosphorylation and dephosphorylation, guanosine nucleotides degradation I, purine nucleotides de novo biosynthesis II, acetyl-CoA biosynthesis (from pyruvate), serine biosynthesis, photorespiration, histidine biosynthesis, glyoxylate cycle, jasmonic acid biosynthesis, alanine degradation II (to D-lactate), a ubiquinone + NADH + H+ -> a ubiquinol + NAD+, putrescine degradation IV, brassinosteroid biosynthesis III, fatty acid elongation -- saturated, pyruvate fermentation to ethanol II, beta-alanine biosynthesis II, 4-hydroxybenzoate biosynthesis V, plant sterol biosynthesis, urate biosynthesis/inosine 5'-phosphate degradation, phenylalanine degradation III, glutamate biosynthesis IV, leucine biosynthesis, fatty acid alpha-oxidation, a ubiquinone + NADH + H+ -> NAD+ + a ubiquinol, Fe(III)-reduction and Fe(II) transport, purine nucleotide metabolism (phosphotransfer and nucleotide modification), phenylethanol biosynthesis, pyruvate fermentation to lactate, superpathway of glyoxylate cycle and fatty acid degradation, fatty acid omega-oxidation, galactose degradation III, ethanol degradation II (cytosol), sucrose biosynthesis I, valine degradation I, folate transformations II |
| CAS ID | 53-84-9 |
| Synonym | β-nicotinamide adenine dinucleotide; coenzyme I; diphosphopyridine nucleotide; diphosphopyridine nucleotide oxidized; nicotinamide adenine dinucleotide; nicotinamide adenine dinucleotide oxidized; NAD-oxidized; NAD-ox; NAD+; DPN+; DPN-ox; DPN; β-nicotinamide adenine dinucleotide; NAD |
| External ID | CHEBI:57540, PUBCHEM:15938971, PUBCHEM:15938971, LIGAND-CPD:C00003 |


information