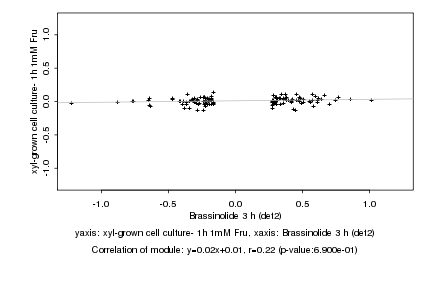
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Brassinolide 3 h (det2) |
Log2 signal ratio
xyl-grown cell culture- 1h 1mM Fru |
| 1 |
253103_at |
AT4G36110
|
SAUR9, SMALL AUXIN UPREGULATED RNA 9 |
1.011 |
0.024 |
| 2 |
247925_at |
AT5G57560
|
TCH4, Touch 4, XTH22, xyloglucan endotransglucosylase/hydrolase 22 |
0.858 |
0.031 |
| 3 |
247252_at |
AT5G64770
|
CLEL 9, CLE-like 9, GLV2, GOLVEN 2, RGF9, root meristem growth factor 9 |
0.765 |
0.073 |
| 4 |
245757_at |
AT1G35140
|
EXL1, EXORDIUM like 1, PHI-1, PHOSPHATE-INDUCED 1 |
0.745 |
0.028 |
| 5 |
267614_at |
AT2G26710
|
BAS1, PHYB ACTIVATION TAGGED SUPPRESSOR 1, CYP72B1, CYP734A1 |
0.699 |
-0.044 |
| 6 |
253608_at |
AT4G30290
|
ATXTH19, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 19, XTH19, xyloglucan endotransglucosylase/hydrolase 19 |
0.663 |
0.091 |
| 7 |
257506_at |
AT1G29440
|
SAUR63, SMALL AUXIN UP RNA 63 |
0.642 |
0.033 |
| 8 |
248282_at |
AT5G52900
|
MAKR6, MEMBRANE-ASSOCIATED KINASE REGULATOR 6 |
0.616 |
0.050 |
| 9 |
263207_at |
AT1G10550
|
XET, XYLOGLUCAN:XYLOGLUCOSYL TRANSFERASE 33, XTH33, xyloglucan:xyloglucosyl transferase 33 |
0.612 |
0.022 |
| 10 |
250292_at |
AT5G13220
|
JAS1, JASMONATE-ASSOCIATED 1, JAZ10, jasmonate-zim-domain protein 10, TIFY9, TIFY DOMAIN PROTEIN 9 |
0.611 |
-0.011 |
| 11 |
249071_at |
AT5G44050
|
[MATE efflux family protein] |
0.592 |
0.076 |
| 12 |
248912_at |
AT5G45670
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.580 |
-0.073 |
| 13 |
255028_at |
AT4G09890
|
unknown |
0.573 |
0.016 |
| 14 |
249011_at |
AT5G44670
|
GALS2, galactan synthase 2 |
0.570 |
0.115 |
| 15 |
252563_at |
AT3G45970
|
ATEXLA1, expansin-like A1, ATEXPL1, ATHEXP BETA 2.1, EXLA1, expansin-like A1, EXPL1, EXPANSIN L1 |
0.557 |
-0.009 |
| 16 |
255822_at |
AT2G40610
|
ATEXP8, ATEXPA8, expansin A8, ATHEXP ALPHA 1.11, EXP8, EXPA8, expansin A8 |
0.552 |
0.005 |
| 17 |
252624_at |
AT3G44735
|
ATPSK3, PHYTOSULFOKINE 3 PRECURSOR, PSK1, PSK3, PHYTOSULFOKINE 3 PRECURSOR |
0.504 |
0.036 |
| 18 |
258742_at |
AT3G05800
|
AIF1, AtBS1(activation-tagged BRI1 suppressor 1)-interacting factor 1 |
0.502 |
-0.003 |
| 19 |
252970_at |
AT4G38850
|
ATSAUR15, ARABIDOPSIS THALIANA SMALL AUXIN UPREGULATED 15, SAUR-AC1, ARABIDOPSIS COLUMBIA SAUR GENE 1, SAUR15, SMALL AUXIN UPREGULATED 15, SAUR_AC1, SMALL AUXIN UP RNA 1 FROM ARABIDOPSIS THALIANA ECOTYPE COLUMBIA |
0.486 |
-0.020 |
| 20 |
249306_at |
AT5G41400
|
[RING/U-box superfamily protein] |
0.485 |
0.052 |
| 21 |
246932_at |
AT5G25190
|
ESE3, ethylene and salt inducible 3 |
0.477 |
0.011 |
| 22 |
254606_at |
AT4G19030
|
AT-NLM1, ATNLM1, NOD26-LIKE MAJOR INTRINSIC PROTEIN 1, NIP1;1, NOD26-LIKE INTRINSIC PROTEIN 1;1, NLM1, NOD26-like major intrinsic protein 1 |
0.471 |
0.063 |
| 23 |
253736_at |
AT4G28780
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.465 |
0.025 |
| 24 |
250012_x_at |
AT5G18060
|
SAUR23, SMALL AUXIN UP RNA 23 |
0.449 |
0.109 |
| 25 |
252965_at |
AT4G38860
|
SAUR16, SMALL AUXIN UPREGULATED RNA 16 |
0.449 |
0.024 |
| 26 |
266693_at |
AT2G19800
|
MIOX2, myo-inositol oxygenase 2 |
0.446 |
-0.123 |
| 27 |
247255_at |
AT5G64780
|
[Uncharacterised conserved protein UCP009193] |
0.430 |
-0.105 |
| 28 |
252997_at |
AT4G38400
|
ATEXLA2, expansin-like A2, ATEXPL2, ATHEXP BETA 2.2, EXLA2, expansin-like A2, EXPL2, EXPANSIN L2 |
0.423 |
0.034 |
| 29 |
254042_at |
AT4G25810
|
XTH23, xyloglucan endotransglucosylase/hydrolase 23, XTR6, xyloglucan endotransglycosylase 6 |
0.418 |
0.022 |
| 30 |
253628_at |
AT4G30280
|
ATXTH18, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 18, XTH18, xyloglucan endotransglucosylase/hydrolase 18 |
0.417 |
-0.002 |
| 31 |
245262_at |
AT4G16563
|
[Eukaryotic aspartyl protease family protein] |
0.417 |
0.011 |
| 32 |
263467_at |
AT2G31730
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.389 |
0.018 |
| 33 |
266873_at |
AT2G44740
|
CYCP4;1, cyclin p4;1 |
0.378 |
0.030 |
| 34 |
248921_at |
AT5G45950
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.374 |
0.065 |
| 35 |
256627_at |
AT3G19970
|
[alpha/beta-Hydrolases superfamily protein] |
0.368 |
0.118 |
| 36 |
258468_at |
AT3G06070
|
unknown |
0.367 |
0.046 |
| 37 |
251922_at |
AT3G54030
|
BSK6, brassinosteroid-signaling kinase 6 |
0.358 |
0.039 |
| 38 |
264016_at |
AT2G21220
|
SAUR12, SMALL AUXIN UPREGULATED RNA 12 |
0.358 |
0.050 |
| 39 |
249750_at |
AT5G24570
|
unknown |
0.356 |
-0.022 |
| 40 |
249337_at |
AT5G41080
|
AtGDPD2, GDPD2, glycerophosphodiester phosphodiesterase 2 |
0.341 |
0.113 |
| 41 |
245574_at |
AT4G14750
|
IQD19, IQ-domain 19 |
0.336 |
-0.031 |
| 42 |
251861_at |
AT3G54810
|
BME3, BLUE MICROPYLAR END 3, BME3-ZF, BLUE MICROPYLAR END 3-ZINC FINGER, GATA8, GATA TRANSCRIPTION FACTOR 8 |
0.332 |
0.053 |
| 43 |
248819_at |
AT5G47050
|
[SBP (S-ribonuclease binding protein) family protein] |
0.328 |
0.047 |
| 44 |
258528_at |
AT3G06770
|
[Pectin lyase-like superfamily protein] |
0.308 |
0.028 |
| 45 |
264783_at |
AT1G08650
|
ATPPCK1, PHOSPHOENOLPYRUVATE CARBOXYLASE KINASE 1, PPCK1, phosphoenolpyruvate carboxylase kinase 1 |
0.305 |
0.071 |
| 46 |
266956_at |
AT2G34510
|
unknown |
0.305 |
0.025 |
| 47 |
259331_at |
AT3G03840
|
SAUR27, SMALL AUXIN UP RNA 27 |
0.305 |
-0.037 |
| 48 |
256633_at |
AT3G28340
|
GATL10, galacturonosyltransferase-like 10, GolS8, galactinol synthase 8 |
0.303 |
0.055 |
| 49 |
266749_at |
AT2G47060
|
PTI1-4, Pto-interacting 1-4 |
0.299 |
0.074 |
| 50 |
251584_at |
AT3G58620
|
TTL4, tetratricopetide-repeat thioredoxin-like 4 |
0.296 |
-0.001 |
| 51 |
260840_at |
AT1G29050
|
TBL38, TRICHOME BIREFRINGENCE-LIKE 38 |
0.293 |
0.074 |
| 52 |
254608_at |
AT4G18910
|
ATNLM2, NOD26-LIKE INTRINSIC PROTEIN 2, NIP1;2, NOD26-like intrinsic protein 1;2, NLM2, NOD26-LIKE INTRINSIC PROTEIN 2 |
0.287 |
-0.012 |
| 53 |
256755_at |
AT3G25600
|
[Calcium-binding EF-hand family protein] |
0.286 |
-0.020 |
| 54 |
248179_at |
AT5G54380
|
THE1, THESEUS1 |
0.280 |
-0.036 |
| 55 |
260494_at |
AT2G41820
|
PXC3, PXY/TDR-correlated 3 |
0.280 |
0.091 |
| 56 |
263800_at |
AT2G24600
|
[Ankyrin repeat family protein] |
0.279 |
0.023 |
| 57 |
254110_at |
AT4G25260
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.279 |
-0.043 |
| 58 |
250875_at |
AT5G04020
|
[calmodulin binding] |
0.276 |
-0.092 |
| 59 |
245244_at |
AT1G44350
|
ILL6, IAA-leucine resistant (ILR)-like gene 6 |
0.276 |
-0.051 |
| 60 |
250437_at |
AT5G10430
|
AGP4, arabinogalactan protein 4, ATAGP4 |
0.275 |
-0.003 |
| 61 |
252711_at |
AT3G43720
|
LTPG2, glycosylphosphatidylinositol-anchored lipid protein transfer 2 |
0.273 |
0.004 |
| 62 |
266805_at |
AT2G30010
|
TBL45, TRICHOME BIREFRINGENCE-LIKE 45 |
0.271 |
0.020 |
| 63 |
267592_at |
AT2G39710
|
[Eukaryotic aspartyl protease family protein] |
0.271 |
-0.001 |
| 64 |
256598_at |
AT3G30180
|
BR6OX2, brassinosteroid-6-oxidase 2, CYP85A2 |
-1.229 |
-0.025 |
| 65 |
251072_at |
AT5G01740
|
[Nuclear transport factor 2 (NTF2) family protein] |
-0.886 |
-0.004 |
| 66 |
250752_at |
AT5G05690
|
CBB3, CABBAGE 3, CPD, CONSTITUTIVE PHOTOMORPHOGENIC DWARF, CYP90, CYP90A, CYP90A1, CYTOCHROME P450 90A1, DWF3, DWARF 3 |
-0.774 |
0.005 |
| 67 |
267305_at |
AT2G30070
|
ATKT1, potassium transporter 1, ATKT1P, ATKUP1, KT1, potassium transporter 1, KUP1, POTASSIUM UPTAKE TRANSPORTER 1 |
-0.761 |
0.003 |
| 68 |
255538_at |
AT4G01680
|
AtMYB55, myb domain protein 55, MYB55, myb domain protein 55 |
-0.655 |
0.018 |
| 69 |
262635_at |
AT1G06570
|
HPD, 4-hydroxyphenylpyruvate dioxygenase, HPPD, 4-hydroxyphenylpyruvate dioxygenase, PDS1, phytoene desaturation 1 |
-0.644 |
-0.047 |
| 70 |
246284_at |
AT4G36780
|
BEH2, BES1/BZR1 homolog 2 |
-0.644 |
0.050 |
| 71 |
249467_at |
AT5G39610
|
ANAC092, Arabidopsis NAC domain containing protein 92, ATNAC2, NAC domain containing protein 2, ATNAC6, NAC domain containing protein 6, NAC2, NAC domain containing protein 2, NAC6, NAC domain containing protein 6, ORE1, ORESARA 1 |
-0.638 |
-0.063 |
| 72 |
251785_at |
AT3G55130
|
ABCG19, ATP-binding cassette G19, ATWBC19, white-brown complex homolog 19, WBC19, white-brown complex homolog 19 |
-0.473 |
0.055 |
| 73 |
247880_at |
AT5G57780
|
P1R1, P1R1 |
-0.471 |
0.043 |
| 74 |
245784_at |
AT1G32190
|
[alpha/beta-Hydrolases superfamily protein] |
-0.422 |
0.007 |
| 75 |
246389_at |
AT1G77380
|
AAP3, amino acid permease 3, ATAAP3 |
-0.414 |
0.005 |
| 76 |
247351_at |
AT5G63790
|
ANAC102, NAC domain containing protein 102, NAC102, NAC domain containing protein 102 |
-0.396 |
-0.043 |
| 77 |
259681_at |
AT1G77760
|
GNR1, NIA1, nitrate reductase 1, NR1, NITRATE REDUCTASE 1 |
-0.391 |
0.011 |
| 78 |
248895_at |
AT5G46330
|
FLS2, FLAGELLIN-SENSITIVE 2 |
-0.381 |
-0.096 |
| 79 |
265208_at |
AT2G36690
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.371 |
-0.004 |
| 80 |
245319_at |
AT4G16146
|
[cAMP-regulated phosphoprotein 19-related protein] |
-0.367 |
-0.032 |
| 81 |
261191_at |
AT1G32900
|
GBSS1, granule bound starch synthase 1 |
-0.364 |
0.111 |
| 82 |
266983_at |
AT2G39400
|
[alpha/beta-Hydrolases superfamily protein] |
-0.350 |
-0.092 |
| 83 |
255177_at |
AT4G08040
|
ACS11, 1-aminocyclopropane-1-carboxylate synthase 11 |
-0.348 |
0.009 |
| 84 |
256062_at |
AT1G07090
|
LSH6, LIGHT SENSITIVE HYPOCOTYLS 6 |
-0.332 |
0.022 |
| 85 |
248381_at |
AT5G51830
|
[pfkB-like carbohydrate kinase family protein] |
-0.326 |
0.035 |
| 86 |
251181_at |
AT3G62820
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.315 |
0.008 |
| 87 |
250907_at |
AT5G03670
|
TRM28, TON1 Recruiting Motif 28 |
-0.310 |
0.055 |
| 88 |
253043_at |
AT4G37540
|
LBD39, LOB domain-containing protein 39 |
-0.309 |
-0.001 |
| 89 |
253042_at |
AT4G37550
|
[Acetamidase/Formamidase family protein] |
-0.294 |
-0.024 |
| 90 |
257232_at |
AT3G16500
|
IAA26, indole-3-acetic acid inducible 26, PAP1, phytochrome-associated protein 1 |
-0.290 |
0.018 |
| 91 |
253999_at |
AT4G26200
|
ACCS7, 1-amino-cyclopropane-1-carboxylate synthase 7, ACS7, 1-amino-cyclopropane-1-carboxylate synthase 7, ATACS7 |
-0.289 |
0.023 |
| 92 |
261292_at |
AT1G36940
|
unknown |
-0.286 |
-0.125 |
| 93 |
260230_at |
AT1G74500
|
ATBS1, activation-tagged BRI1(brassinosteroid-insensitive 1)-suppressor 1, bHLH135, basic helix-loop-helix protein 135, BS1, activation-tagged BRI1(brassinosteroid-insensitive 1)-suppressor 1, PRE3, PACLOBUTRAZOL RESISTANCE 3, TMO7, TARGET OF MONOPTEROS 7 |
-0.286 |
0.036 |
| 94 |
260979_at |
AT1G53510
|
ATMPK18, ARABIDOPSIS THALIANA MAP KINASE 18, MPK18, mitogen-activated protein kinase 18 |
-0.281 |
-0.042 |
| 95 |
264510_at |
AT1G09530
|
PAP3, PHYTOCHROME-ASSOCIATED PROTEIN 3, PIF3, phytochrome interacting factor 3, POC1, PHOTOCURRENT 1 |
-0.269 |
-0.025 |
| 96 |
261456_at |
AT1G21050
|
unknown |
-0.266 |
0.069 |
| 97 |
264037_at |
AT2G03750
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.245 |
-0.125 |
| 98 |
261224_at |
AT1G20160
|
ATSBT5.2 |
-0.244 |
0.015 |
| 99 |
252661_at |
AT3G44450
|
unknown |
-0.244 |
0.068 |
| 100 |
247191_at |
AT5G65310
|
ATHB-5, ATHB5, homeobox protein 5, HB5, homeobox protein 5 |
-0.236 |
0.065 |
| 101 |
264400_at |
AT1G61800
|
ATGPT2, ARABIDOPSIS GLUCOSE-6-PHOSPHATE/PHOSPHATE TRANSLOCATOR 2, GPT2, glucose-6-phosphate/phosphate translocator 2 |
-0.236 |
0.064 |
| 102 |
261070_at |
AT1G07390
|
AtRLP1, receptor like protein 1, RLP1, receptor like protein 1 |
-0.232 |
-0.037 |
| 103 |
253425_at |
AT4G32190
|
[Myosin heavy chain-related protein] |
-0.229 |
-0.066 |
| 104 |
265245_at |
AT2G43060
|
AtIBH1, IBH1, ILI1 binding bHLH 1 |
-0.227 |
0.017 |
| 105 |
255829_at |
AT2G40540
|
ATKT2, ATKUP2, KT2, potassium transporter 2, KUP2, SHY3, TRK2 |
-0.226 |
-0.018 |
| 106 |
264859_at |
AT1G24280
|
G6PD3, glucose-6-phosphate dehydrogenase 3 |
-0.226 |
0.046 |
| 107 |
260073_at |
AT1G73660
|
SIS8, SUGAR INSENSITIVE 8 |
-0.225 |
-0.020 |
| 108 |
262505_at |
AT1G21680
|
[DPP6 N-terminal domain-like protein] |
-0.225 |
-0.017 |
| 109 |
257964_at |
AT3G19850
|
[Phototropic-responsive NPH3 family protein] |
-0.217 |
-0.029 |
| 110 |
264200_at |
AT1G22650
|
A/N-InvD, alkaline/neutral invertase D |
-0.215 |
0.057 |
| 111 |
267337_at |
AT2G39980
|
[HXXXD-type acyl-transferase family protein] |
-0.213 |
-0.033 |
| 112 |
266095_at |
AT2G38050
|
ATDET2, DET2, DE-ETIOLATED 2, DWF6, DWARF 6 |
-0.204 |
0.010 |
| 113 |
249325_at |
AT5G40850
|
UPM1, urophorphyrin methylase 1 |
-0.204 |
0.034 |
| 114 |
259723_at |
AT1G60960
|
ATIRT3, IRON REGULATED TRANSPORTER 3, IRT3, iron regulated transporter 3 |
-0.200 |
0.042 |
| 115 |
251459_at |
AT3G60200
|
unknown |
-0.195 |
-0.034 |
| 116 |
247136_at |
AT5G66170
|
STR18, sulfurtransferase 18 |
-0.188 |
0.028 |
| 117 |
264246_at |
AT1G60140
|
ATTPS10, trehalose phosphate synthase, TPS10, trehalose phosphate synthase, TPS10, TREHALOSE PHOSPHATE SYNTHASE 10 |
-0.185 |
0.037 |
| 118 |
247407_at |
AT5G62900
|
unknown |
-0.185 |
0.047 |
| 119 |
253812_at |
AT4G28240
|
[Wound-responsive family protein] |
-0.184 |
0.030 |
| 120 |
257460_at |
AT1G75580
|
SAUR51, SMALL AUXIN UPREGULATED RNA 51 |
-0.182 |
0.077 |
| 121 |
262388_at |
AT1G49320
|
unknown |
-0.181 |
-0.030 |
| 122 |
250911_at |
AT5G03730
|
AtCTR1, CTR1, CONSTITUTIVE TRIPLE RESPONSE 1, SIS1, SUGAR-INSENSITIVE 1 |
-0.171 |
-0.012 |
| 123 |
259643_at |
AT1G68890
|
PHYLLO, PHYLLO |
-0.167 |
0.140 |
| 124 |
263866_at |
AT2G36950
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.167 |
-0.038 |
| 125 |
249124_at |
AT5G43740
|
[Disease resistance protein (CC-NBS-LRR class) family] |
-0.166 |
-0.016 |