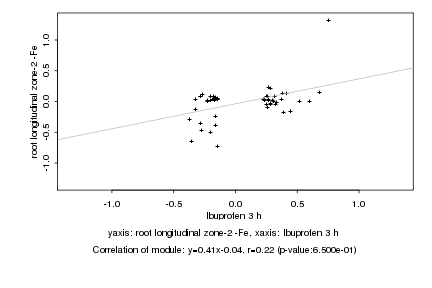
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Ibuprofen 3 h |
Log2 signal ratio
root longitudinal zone-2 -Fe |
| 1 |
251677_at |
AT3G56980
|
BHLH039, bHLH39, basic helix-loop-helix 39, ORG3, OBP3-RESPONSIVE GENE 3 |
0.748 |
1.333 |
| 2 |
260803_at |
AT1G78340
|
ATGSTU22, glutathione S-transferase TAU 22, GSTU22, glutathione S-transferase TAU 22 |
0.680 |
0.157 |
| 3 |
256589_at |
AT3G28740
|
CYP81D11, cytochrome P450, family 81, subfamily D, polypeptide 11 |
0.595 |
0.012 |
| 4 |
259705_at |
AT1G77450
|
anac032, NAC domain containing protein 32, NAC032, NAC domain containing protein 32 |
0.511 |
0.012 |
| 5 |
263184_at |
AT1G05560
|
UGT1, UDP-GLUCOSE TRANSFERASE 1, UGT75B1, UDP-glucosyltransferase 75B1 |
0.439 |
-0.153 |
| 6 |
254056_at |
AT4G25250
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.407 |
0.132 |
| 7 |
257823_at |
AT3G25190
|
[Vacuolar iron transporter (VIT) family protein] |
0.389 |
-0.177 |
| 8 |
264042_at |
AT2G03760
|
AtSOT1, AtSOT12, sulphotransferase 12, ATST1, ARABIDOPSIS THALIANA SULFOTRANSFERASE 1, AtSULT202A1, RAR047, SOT12, sulphotransferase 12, ST, ST1, SULFOTRANSFERASE 1, SULT202A1, sulfotransferase 202A1 |
0.380 |
0.136 |
| 9 |
254759_at |
AT4G13180
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.367 |
0.043 |
| 10 |
256877_at |
AT3G26470
|
[Powdery mildew resistance protein, RPW8 domain] |
0.326 |
-0.009 |
| 11 |
255543_at |
AT4G01870
|
[tolB protein-related] |
0.318 |
-0.035 |
| 12 |
246273_at |
AT4G36700
|
[RmlC-like cupins superfamily protein] |
0.312 |
0.093 |
| 13 |
266296_at |
AT2G29420
|
ATGSTU7, glutathione S-transferase tau 7, GST25, GLUTATHIONE S-TRANSFERASE 25, GSTU7, glutathione S-transferase tau 7 |
0.302 |
0.007 |
| 14 |
262124_at |
AT1G59660
|
[Nucleoporin autopeptidase] |
0.292 |
0.027 |
| 15 |
266294_at |
AT2G29500
|
[HSP20-like chaperones superfamily protein] |
0.283 |
-0.041 |
| 16 |
256252_at |
AT3G11340
|
UGT76B1, UDP-dependent glycosyltransferase 76B1 |
0.280 |
-0.017 |
| 17 |
250248_at |
AT5G13740
|
ZIF1, zinc induced facilitator 1 |
0.278 |
0.223 |
| 18 |
255591_at |
AT4G01630
|
ATEXP17, EXPANSIN 17, ATEXPA17, expansin A17, ATHEXP ALPHA 1.13, EXPA17, expansin A17 |
0.264 |
0.018 |
| 19 |
259228_at |
AT3G07720
|
[Galactose oxidase/kelch repeat superfamily protein] |
0.263 |
0.229 |
| 20 |
251293_at |
AT3G61930
|
unknown |
0.262 |
0.034 |
| 21 |
251942_at |
AT3G53480
|
ABCG37, ATP-binding cassette G37, ATPDR9, PLEIOTROPIC DRUG RESISTANCE 9, PDR9, pleiotropic drug resistance 9, PIS1, polar auxin transport inhibitor sensitive 1 |
0.255 |
0.092 |
| 22 |
247488_at |
AT5G61820
|
unknown |
0.253 |
-0.095 |
| 23 |
257217_at |
AT3G14940
|
ATPPC3, phosphoenolpyruvate carboxylase 3, PPC3, phosphoenolpyruvate carboxylase 3 |
0.250 |
0.089 |
| 24 |
258646_at |
AT3G08040
|
ATFRD3, FRD3, FERRIC REDUCTASE DEFECTIVE 3, MAN1, MANGANESE ACCUMULATOR 1 |
0.248 |
-0.047 |
| 25 |
246858_at |
AT5G25930
|
[Protein kinase family protein with leucine-rich repeat domain] |
0.231 |
0.044 |
| 26 |
258287_at |
AT3G15990
|
SULTR3;4, sulfate transporter 3;4 |
0.228 |
0.017 |
| 27 |
246098_at |
AT5G20400
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.223 |
0.034 |
| 28 |
250801_at |
AT5G04960
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.379 |
-0.289 |
| 29 |
252698_at |
AT3G43670
|
[Copper amine oxidase family protein] |
-0.359 |
-0.643 |
| 30 |
250942_at |
AT5G03350
|
[Legume lectin family protein] |
-0.331 |
0.047 |
| 31 |
245966_at |
AT5G19790
|
RAP2.11, related to AP2 11 |
-0.328 |
-0.115 |
| 32 |
259195_at |
AT3G01730
|
unknown |
-0.285 |
0.087 |
| 33 |
247871_at |
AT5G57530
|
AtXTH12, XTH12, xyloglucan endotransglucosylase/hydrolase 12 |
-0.283 |
-0.345 |
| 34 |
258035_at |
AT3G21180
|
ACA9, autoinhibited Ca(2+)-ATPase 9, ATACA9 |
-0.283 |
0.087 |
| 35 |
252238_at |
AT3G49960
|
[Peroxidase superfamily protein] |
-0.281 |
-0.459 |
| 36 |
245979_at |
AT5G13150
|
ATEXO70C1, exocyst subunit exo70 family protein C1, EXO70C1, exocyst subunit exo70 family protein C1 |
-0.273 |
0.125 |
| 37 |
265837_at |
AT2G14560
|
LURP1, LATE UPREGULATED IN RESPONSE TO HYALOPERONOSPORA PARASITICA |
-0.232 |
0.028 |
| 38 |
267037_at |
AT2G38320
|
TBL34, TRICHOME BIREFRINGENCE-LIKE 34 |
-0.229 |
0.010 |
| 39 |
245636_at |
AT1G25240
|
[ENTH/VHS/GAT family protein] |
-0.209 |
0.093 |
| 40 |
259071_at |
AT3G11650
|
NHL2, NDR1/HIN1-like 2 |
-0.208 |
0.028 |
| 41 |
253998_at |
AT4G26010
|
[Peroxidase superfamily protein] |
-0.206 |
-0.496 |
| 42 |
264680_at |
AT1G65510
|
unknown |
-0.187 |
0.041 |
| 43 |
260628_at |
AT1G62320
|
[ERD (early-responsive to dehydration stress) family protein] |
-0.181 |
0.041 |
| 44 |
261870_at |
AT1G11570
|
NTL, NTF2-like |
-0.179 |
0.097 |
| 45 |
247199_at |
AT5G65210
|
TGA1, TGACG sequence-specific binding protein 1 |
-0.173 |
0.025 |
| 46 |
256283_at |
AT3G12540
|
unknown |
-0.168 |
-0.229 |
| 47 |
265114_at |
AT1G62440
|
LRX2, leucine-rich repeat/extensin 2 |
-0.168 |
0.074 |
| 48 |
256020_at |
AT1G58290
|
AtHEMA1, Arabidopsis thaliana hemA 1, HEMA1 |
-0.163 |
-0.378 |
| 49 |
259180_at |
AT3G01680
|
AtSEOR1, Arabidopsis thaliana sieve element occlusion-related 1, SEOb, sieve element occlusion b, SEOR1, Sieve-Element-Occlusion-Related 1 |
-0.162 |
0.057 |
| 50 |
253216_at |
AT4G34960
|
[Cyclophilin-like peptidyl-prolyl cis-trans isomerase family protein] |
-0.149 |
0.038 |
| 51 |
251109_at |
AT5G01600
|
ATFER1, ARABIDOPSIS THALIANA FERRETIN 1, FER1, ferretin 1 |
-0.148 |
-0.717 |
| 52 |
258476_at |
AT3G02400
|
[SMAD/FHA domain-containing protein ] |
-0.148 |
0.057 |
| 53 |
252836_at |
AT3G42070
|
unknown |
-0.146 |
0.041 |