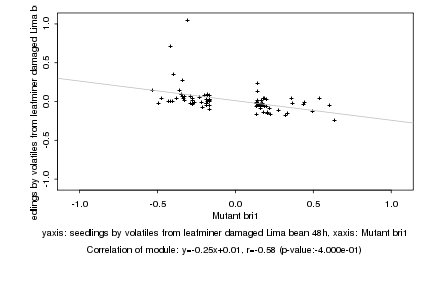
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Mutant bri1 |
Log2 signal ratio
seedlings by volatiles from leafminer damaged Lima bean 48h |
| 1 |
253305_at |
AT4G33666
|
unknown |
0.634 |
-0.244 |
| 2 |
258419_at |
AT3G16670
|
[Pollen Ole e 1 allergen and extensin family protein] |
0.598 |
-0.044 |
| 3 |
251072_at |
AT5G01740
|
[Nuclear transport factor 2 (NTF2) family protein] |
0.536 |
0.051 |
| 4 |
247880_at |
AT5G57780
|
P1R1, P1R1 |
0.491 |
-0.127 |
| 5 |
246122_at |
AT5G20380
|
PHT4;5, phosphate transporter 4;5 |
0.439 |
-0.003 |
| 6 |
256598_at |
AT3G30180
|
BR6OX2, brassinosteroid-6-oxidase 2, CYP85A2 |
0.434 |
-0.038 |
| 7 |
262135_at |
AT1G78080
|
AtWIND1, RAP2.4, related to AP2 4, WIND1, wound induced dedifferentiation 1 |
0.363 |
-0.020 |
| 8 |
249894_at |
AT5G22580
|
[Stress responsive A/B Barrel Domain] |
0.356 |
0.039 |
| 9 |
261292_at |
AT1G36940
|
unknown |
0.332 |
-0.152 |
| 10 |
249824_at |
AT5G23380
|
unknown |
0.320 |
-0.176 |
| 11 |
247882_at |
AT5G57785
|
unknown |
0.271 |
-0.105 |
| 12 |
264697_at |
AT1G70210
|
ATCYCD1;1, CYCD1;1, CYCLIN D1;1 |
0.219 |
-0.163 |
| 13 |
258456_at |
AT3G22420
|
ATWNK2, ARABIDOPSIS THALIANA WITH NO K 2, WNK2, with no lysine (K) kinase 2, ZIK3 |
0.214 |
-0.081 |
| 14 |
267010_at |
AT2G39250
|
SNZ, SCHNARCHZAPFEN |
0.205 |
-0.148 |
| 15 |
254638_at |
AT4G18740
|
[Rho termination factor] |
0.204 |
-0.145 |
| 16 |
252184_at |
AT3G50660
|
AtDWF4, CLM, CLOMAZONE-RESISTANT, CYP90B1, CYTOCHROME P450 90B1, DWF4, DWARF 4, PSC1, PARTIALLY SUPPRESSING COI1 INSENSITIVITY TO JA 1, SAV1, SHADE AVOIDANCE 1, SNP2, SUPPRESSOR OF NPH4 2 |
0.200 |
-0.133 |
| 17 |
255637_at |
AT4G00750
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.198 |
-0.059 |
| 18 |
256299_at |
AT1G69530
|
AT-EXP1, ATEXP1, ATEXPA1, expansin A1, ATHEXP ALPHA 1.2, EXP1, EXPANSIN 1, EXPA1, expansin A1 |
0.194 |
0.032 |
| 19 |
245523_at |
AT4G15910
|
ATDI21, drought-induced 21, DI21, drought-induced 21 |
0.186 |
0.047 |
| 20 |
251115_at |
AT5G01400
|
ESP4, ENHANCED SILENCING PHENOTYPE 4 |
0.184 |
-0.057 |
| 21 |
258939_at |
AT3G10020
|
unknown |
0.180 |
-0.137 |
| 22 |
245304_at |
AT4G15630
|
[Uncharacterised protein family (UPF0497)] |
0.177 |
-0.045 |
| 23 |
265359_at |
AT2G16720
|
ATMYB7, ARABIDOPSIS THALIANA MYB DOMAIN PROTEIN 7, ATY49, MYB7, myb domain protein 7 |
0.175 |
0.042 |
| 24 |
249164_at |
AT5G42820
|
ATU2AF35B, U2AF35B |
0.169 |
-0.013 |
| 25 |
264779_at |
AT1G08570
|
ACHT4, atypical CYS HIS rich thioredoxin 4 |
0.169 |
-0.048 |
| 26 |
246045_at |
AT5G19430
|
[RING/U-box superfamily protein] |
0.166 |
0.022 |
| 27 |
262496_at |
AT1G21790
|
[TRAM, LAG1 and CLN8 (TLC) lipid-sensing domain containing protein] |
0.163 |
-0.027 |
| 28 |
257766_at |
AT3G23030
|
IAA2, indole-3-acetic acid inducible 2 |
0.163 |
-0.027 |
| 29 |
259166_at |
AT3G01670
|
AtSEOR2, Arabidopsis thaliana sieve element occlusion-related 2, SEOa, sieve element occlusion a, SEOR2, sieve element occlusion-related 2 |
0.161 |
-0.050 |
| 30 |
256353_at |
AT1G55000
|
[peptidoglycan-binding LysM domain-containing protein] |
0.160 |
-0.035 |
| 31 |
260983_at |
AT1G53560
|
[Ribosomal protein L18ae family] |
0.157 |
-0.058 |
| 32 |
246236_at |
AT4G36470
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.157 |
-0.087 |
| 33 |
266793_at |
AT2G03060
|
AGL30, AGAMOUS-like 30 |
0.146 |
-0.039 |
| 34 |
258746_at |
AT3G05950
|
[RmlC-like cupins superfamily protein] |
0.142 |
-0.025 |
| 35 |
265592_at |
AT2G20110
|
[Tesmin/TSO1-like CXC domain-containing protein] |
0.141 |
0.022 |
| 36 |
250109_at |
AT5G15230
|
GASA4, GAST1 protein homolog 4 |
0.141 |
-0.047 |
| 37 |
258604_at |
AT3G02980
|
MCC1, MEIOTIC CONTROL OF CROSSOVERS1 |
0.138 |
0.137 |
| 38 |
266098_at |
AT2G37870
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.138 |
0.237 |
| 39 |
254006_at |
AT4G26340
|
[F-box/RNI-like/FBD-like domains-containing protein] |
0.136 |
-0.051 |
| 40 |
260495_at |
AT2G41810
|
unknown |
0.132 |
-0.002 |
| 41 |
254175_at |
AT4G24050
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.131 |
-0.158 |
| 42 |
245733_at |
AT1G73380
|
unknown |
0.129 |
-0.021 |
| 43 |
265495_at |
AT2G15695
|
unknown |
0.129 |
-0.055 |
| 44 |
267121_at |
AT2G23540
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.533 |
0.149 |
| 45 |
256497_at |
AT1G31580
|
CXC750, ECS1 |
-0.496 |
-0.024 |
| 46 |
245181_at |
AT5G12420
|
[O-acyltransferase (WSD1-like) family protein] |
-0.476 |
0.041 |
| 47 |
262657_at |
AT1G14210
|
[Ribonuclease T2 family protein] |
-0.431 |
0.010 |
| 48 |
266625_at |
AT2G35380
|
[Peroxidase superfamily protein] |
-0.417 |
0.001 |
| 49 |
249215_at |
AT5G42800
|
DFR, dihydroflavonol 4-reductase, M318, TT3 |
-0.417 |
0.722 |
| 50 |
256811_at |
AT3G21340
|
[Leucine-rich repeat protein kinase family protein] |
-0.410 |
0.011 |
| 51 |
252888_at |
AT4G39210
|
APL3 |
-0.403 |
0.350 |
| 52 |
247765_at |
AT5G58860
|
CYP86, CYP86A1, cytochrome P450, family 86, subfamily A, polypeptide 1, HORST, hydroxylase of root suberized tissue |
-0.382 |
0.040 |
| 53 |
263544_at |
AT2G21590
|
APL4 |
-0.360 |
0.144 |
| 54 |
250541_at |
AT5G09520
|
PELPK2, Pro-Glu-Leu|Ile|Val-Pro-Lys 2 |
-0.347 |
0.100 |
| 55 |
250500_at |
AT5G09530
|
PELPK1, Pro-Glu-Leu|Ile|Val-Pro-Lys 1, PRP10, proline-rich protein 10 |
-0.341 |
0.280 |
| 56 |
260234_at |
AT1G74460
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.340 |
0.076 |
| 57 |
251531_at |
AT3G58550
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.336 |
0.044 |
| 58 |
264100_at |
AT1G78970
|
ATLUP1, ARABIDOPSIS THALIANA LUPEOL SYNTHASE 1, LUP1, lupeol synthase 1 |
-0.333 |
0.017 |
| 59 |
245889_at |
AT5G09480
|
[hydroxyproline-rich glycoprotein family protein] |
-0.329 |
0.065 |
| 60 |
245275_at |
AT4G15210
|
AT-BETA-AMY, ATBETA-AMY, ARABIDOPSIS THALIANA BETA-AMYLASE, BAM5, beta-amylase 5, BMY1, RAM1, REDUCED BETA AMYLASE 1 |
-0.311 |
1.057 |
| 61 |
256675_at |
AT3G52170
|
[DNA binding] |
-0.290 |
0.072 |
| 62 |
249061_at |
AT5G44550
|
[Uncharacterised protein family (UPF0497)] |
-0.289 |
-0.014 |
| 63 |
266157_at |
AT2G28100
|
ATFUC1, alpha-L-fucosidase 1, FUC1, alpha-L-fucosidase 1 |
-0.282 |
-0.035 |
| 64 |
260824_at |
AT1G06720
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.281 |
0.043 |
| 65 |
264470_at |
AT1G67110
|
CYP735A2, cytochrome P450, family 735, subfamily A, polypeptide 2 |
-0.280 |
-0.023 |
| 66 |
249955_at |
AT5G18840
|
[Major facilitator superfamily protein] |
-0.278 |
0.023 |
| 67 |
262961_at |
AT1G54490
|
AIN1, ACC INSENSITIVE 1, ATXRN4, EXORIBONUCLEASE 4, EIN5, ETHYLENE INSENSITIVE 5, XRN4, exoribonuclease 4 |
-0.263 |
-0.005 |
| 68 |
265326_at |
AT2G18220
|
[Noc2p family] |
-0.234 |
0.056 |
| 69 |
260035_at |
AT1G68850
|
[Peroxidase superfamily protein] |
-0.221 |
-0.005 |
| 70 |
256460_at |
AT1G36240
|
[Ribosomal protein L7Ae/L30e/S12e/Gadd45 family protein] |
-0.213 |
-0.069 |
| 71 |
258474_at |
AT3G02650
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.205 |
0.089 |
| 72 |
262317_at |
AT2G48140
|
EDA4, embryo sac development arrest 4 |
-0.192 |
-0.020 |
| 73 |
266617_at |
AT2G29670
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.189 |
-0.019 |
| 74 |
253605_at |
AT4G30990
|
[ARM repeat superfamily protein] |
-0.186 |
-0.039 |
| 75 |
256978_at |
AT3G21110
|
ATPURC, PUR7, purin 7, PURC, PURIN C |
-0.186 |
0.038 |
| 76 |
252638_at |
AT3G44540
|
FAR4, fatty acid reductase 4 |
-0.181 |
0.006 |
| 77 |
267309_at |
AT2G19385
|
[zinc ion binding] |
-0.181 |
0.101 |
| 78 |
258965_at |
AT3G10530
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.180 |
0.085 |
| 79 |
264987_at |
AT1G27030
|
unknown |
-0.172 |
0.086 |
| 80 |
267013_at |
AT2G39180
|
ATCRR2, CCR2, CRINKLY4 related 2 |
-0.172 |
-0.044 |
| 81 |
247183_at |
AT5G65440
|
unknown |
-0.171 |
-0.097 |
| 82 |
258589_at |
AT3G04290
|
ATLTL1, LTL1, Li-tolerant lipase 1 |
-0.170 |
0.021 |
| 83 |
253566_at |
AT4G31210
|
[DNA topoisomerase, type IA, core] |
-0.169 |
0.028 |
| 84 |
254202_at |
AT4G24140
|
[alpha/beta-Hydrolases superfamily protein] |
-0.168 |
0.008 |
| 85 |
263766_at |
AT2G21440
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.168 |
0.028 |