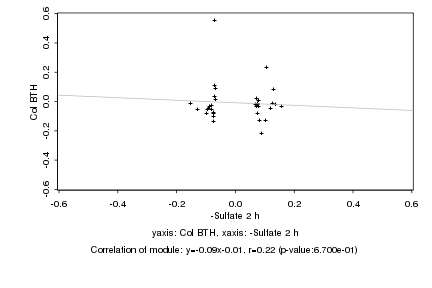
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
-Sulfate 2 h |
Log2 signal ratio
Col BTH |
| 1 |
247070_at |
AT5G66815
|
unknown |
0.154 |
-0.031 |
| 2 |
247215_at |
AT5G64905
|
PROPEP3, elicitor peptide 3 precursor |
0.135 |
-0.016 |
| 3 |
259325_at |
AT3G05320
|
[O-fucosyltransferase family protein] |
0.128 |
0.084 |
| 4 |
260584_at |
AT2G43660
|
[Carbohydrate-binding X8 domain superfamily protein] |
0.124 |
-0.008 |
| 5 |
266443_at |
AT2G43270
|
[F-box and associated interaction domains-containing protein] |
0.119 |
-0.047 |
| 6 |
261443_at |
AT1G28480
|
GRX480, roxy19 |
0.103 |
0.236 |
| 7 |
267265_at |
AT2G22980
|
SCPL13, serine carboxypeptidase-like 13 |
0.101 |
-0.126 |
| 8 |
245041_at |
AT2G26530
|
AR781 |
0.087 |
-0.213 |
| 9 |
267038_at |
AT2G38480
|
[Uncharacterised protein family (UPF0497)] |
0.079 |
-0.128 |
| 10 |
248686_at |
AT5G48540
|
[receptor-like protein kinase-related family protein] |
0.076 |
0.007 |
| 11 |
245855_at |
AT5G13550
|
SULTR4;1, sulfate transporter 4.1 |
0.076 |
-0.028 |
| 12 |
245188_at |
AT1G67660
|
[Restriction endonuclease, type II-like superfamily protein] |
0.075 |
-0.081 |
| 13 |
258420_at |
AT3G16680
|
[DNA binding] |
0.074 |
-0.009 |
| 14 |
247393_at |
AT5G63130
|
[Octicosapeptide/Phox/Bem1p family protein] |
0.072 |
-0.017 |
| 15 |
250213_at |
AT5G13820
|
ATBP-1, ATBP1, ATTBP1, HPPBF-1, H-PROTEIN PROMOTE, TBP1, telomeric DNA binding protein 1 |
0.069 |
0.021 |
| 16 |
259035_at |
AT3G09430
|
unknown |
0.069 |
-0.033 |
| 17 |
265809_at |
AT2G18100
|
unknown |
0.068 |
-0.014 |
| 18 |
255186_at |
AT4G07750
|
[CACTA-like transposase family (Tnp2/En/Spm), has a 2.9e-168 P-value blast match to gb|AAG52024.1|AC022456_5 Tam1-homologous transposon protein TNP2, putative] |
-0.155 |
-0.012 |
| 19 |
252130_at |
AT3G50820
|
OEC33, OXYGEN EVOLVING COMPLEX SUBUNIT 33 KDA, PSBO-2, PHOTOSYSTEM II SUBUNIT O-2, PSBO2, photosystem II subunit O-2 |
-0.131 |
-0.048 |
| 20 |
265620_at |
AT2G27310
|
[F-box family protein] |
-0.100 |
-0.081 |
| 21 |
260633_at |
AT1G62400
|
HT1, high leaf temperature 1 |
-0.096 |
-0.050 |
| 22 |
258333_at |
AT3G16000
|
MFP1, MAR binding filament-like protein 1 |
-0.095 |
-0.042 |
| 23 |
258541_at |
AT3G07000
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.089 |
-0.031 |
| 24 |
245171_at |
AT2G47560
|
[RING/U-box superfamily protein] |
-0.082 |
-0.050 |
| 25 |
254729_at |
AT4G13700
|
ATPAP23, PAP23, purple acid phosphatase 23 |
-0.082 |
-0.027 |
| 26 |
266110_at |
AT2G02080
|
AtIDD4, indeterminate(ID)-domain 4, IDD4, indeterminate(ID)-domain 4 |
-0.078 |
-0.077 |
| 27 |
265602_at |
AT2G14380
|
[gypsy-like retrotransposon family, has a 1.6e-123 P-value blast match to GB:AAD19359 polyprotein (gypsy_Ty3-element) (Sorghum bicolor)] |
-0.077 |
-0.069 |
| 28 |
245946_at |
AT5G19580
|
[glyoxal oxidase-related protein] |
-0.076 |
-0.134 |
| 29 |
262040_at |
AT1G80080
|
AtRLP17, Receptor Like Protein 17, TMM, TOO MANY MOUTHS |
-0.075 |
-0.097 |
| 30 |
266142_at |
AT2G39030
|
NATA1, N-acetyltransferase activity 1 |
-0.073 |
0.112 |
| 31 |
255340_at |
AT4G04490
|
CRK36, cysteine-rich RLK (RECEPTOR-like protein kinase) 36 |
-0.073 |
0.560 |
| 32 |
265741_at |
AT2G01320
|
ABCG7, ATP-binding cassette G7 |
-0.072 |
0.039 |
| 33 |
252087_at |
AT3G52080
|
chx28, cation/hydrogen exchanger 28 |
-0.071 |
0.089 |
| 34 |
264472_at |
AT1G67140
|
SWEETIE, SWEETIE |
-0.070 |
0.014 |