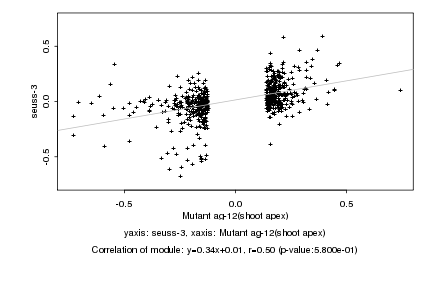
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Mutant ag-12(shoot apex) |
Log2 signal ratio
seuss-3 |
| 1 |
263497_at |
AT2G42540
|
COR15, COR15A, cold-regulated 15a |
0.742 |
0.100 |
| 2 |
247718_at |
AT5G59310
|
LTP4, lipid transfer protein 4 |
0.466 |
0.349 |
| 3 |
247717_at |
AT5G59320
|
LTP3, lipid transfer protein 3 |
0.458 |
0.327 |
| 4 |
260773_at |
AT1G78440
|
ATGA2OX1, Arabidopsis thaliana gibberellin 2-oxidase 1, GA2OX1, gibberellin 2-oxidase 1 |
0.446 |
0.100 |
| 5 |
249761_at |
AT5G23970
|
[HXXXD-type acyl-transferase family protein] |
0.445 |
0.117 |
| 6 |
245465_at |
AT4G16590
|
ATCSLA01, cellulose synthase-like A01, ATCSLA1, CELLULOSE SYNTHASE-LIKE A1, CSLA01, CSLA01, cellulose synthase-like A01 |
0.419 |
0.088 |
| 7 |
259161_at |
AT3G01500
|
ATBCA1, BETA CARBONIC ANHYDRASE 1, ATSABP3, ARABIDOPSIS THALIANA SALICYLIC ACID-BINDING PROTEIN 3, CA1, carbonic anhydrase 1, SABP3, SALICYLIC ACID-BINDING PROTEIN 3 |
0.413 |
-0.018 |
| 8 |
254189_at |
AT4G24000
|
ATCSLG2, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE LIKE G2, CSLG2, cellulose synthase like G2 |
0.408 |
0.191 |
| 9 |
248337_at |
AT5G52310
|
COR78, COLD REGULATED 78, LTI140, LTI78, LOW-TEMPERATURE-INDUCED 78, RD29A, RESPONSIVE TO DESSICATION 29A |
0.389 |
0.596 |
| 10 |
246380_at |
AT1G57750
|
CYP96A15, cytochrome P450, family 96, subfamily A, polypeptide 15, MAH1, MID-CHAIN ALKANE HYDROXYLASE 1 |
0.366 |
0.466 |
| 11 |
247760_at |
AT5G59130
|
[Subtilase family protein] |
0.361 |
0.020 |
| 12 |
261059_at |
AT1G01250
|
[Integrase-type DNA-binding superfamily protein] |
0.354 |
0.168 |
| 13 |
259789_at |
AT1G29395
|
COR413-TM1, COLD REGULATED 314 THYLAKOID MEMBRANE 1, COR413IM1, COLD REGULATED 314 INNER MEMBRANE 1, COR414-TM1, cold regulated 414 thylakoid membrane 1 |
0.345 |
0.382 |
| 14 |
265984_at |
AT2G24210
|
TPS10, terpene synthase 10 |
0.342 |
0.319 |
| 15 |
264606_at |
AT1G04660
|
[glycine-rich protein] |
0.338 |
0.216 |
| 16 |
256266_at |
AT3G12320
|
LNK3, night light-inducible and clock-regulated 3 |
0.330 |
-0.071 |
| 17 |
248496_at |
AT5G50790
|
AtSWEET10, SWEET10 |
0.320 |
0.284 |
| 18 |
262644_at |
AT1G62710
|
BETA-VPE, beta vacuolar processing enzyme, BETAVPE |
0.318 |
0.354 |
| 19 |
261285_at |
AT1G35720
|
ANN1, annexin 1, ANNAT1, annexin 1, AtANN1, ATOXY5, OXY5 |
0.318 |
0.220 |
| 20 |
263495_at |
AT2G42530
|
COR15B, cold regulated 15b |
0.316 |
0.113 |
| 21 |
248368_at |
AT5G51950
|
[Glucose-methanol-choline (GMC) oxidoreductase family protein] |
0.314 |
0.123 |
| 22 |
253736_at |
AT4G28780
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.307 |
0.117 |
| 23 |
259526_at |
AT1G12570
|
[Glucose-methanol-choline (GMC) oxidoreductase family protein] |
0.306 |
0.078 |
| 24 |
262612_at |
AT1G14150
|
PnsL2, Photosynthetic NDH subcomplex L 2, PQL1, PsbQ-like 1, PQL2, PsbQ-like 2 |
0.305 |
-0.004 |
| 25 |
251928_at |
AT3G53980
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.298 |
0.017 |
| 26 |
257628_at |
AT3G26290
|
CYP71B26, cytochrome P450, family 71, subfamily B, polypeptide 26 |
0.288 |
0.288 |
| 27 |
254066_at |
AT4G25480
|
ATCBF3, CBF3, C-REPEAT BINDING FACTOR 3, DREB1A, dehydration response element B1A |
0.287 |
-0.097 |
| 28 |
267528_at |
AT2G45650
|
AGL6, AGAMOUS-like 6, RSB1, REDUCED SHOOT BRANCHING 1 |
0.286 |
0.469 |
| 29 |
266617_at |
AT2G29670
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.284 |
0.316 |
| 30 |
258321_at |
AT3G22840
|
ELIP, ELIP1, EARLY LIGHT-INDUCABLE PROTEIN |
0.282 |
0.007 |
| 31 |
259737_at |
AT1G64400
|
LACS3, long-chain acyl-CoA synthetase 3 |
0.278 |
0.063 |
| 32 |
247447_at |
AT5G62730
|
[Major facilitator superfamily protein] |
0.275 |
0.079 |
| 33 |
258017_at |
AT3G19370
|
unknown |
0.265 |
0.321 |
| 34 |
251497_at |
AT3G59060
|
PIF5, PHYTOCHROME-INTERACTING FACTOR 5, PIL6, phytochrome interacting factor 3-like 6 |
0.264 |
0.048 |
| 35 |
259834_at |
AT1G69570
|
[Dof-type zinc finger DNA-binding family protein] |
0.262 |
0.088 |
| 36 |
259058_at |
AT3G03470
|
CYP89A9, cytochrome P450, family 87, subfamily A, polypeptide 9 |
0.260 |
0.159 |
| 37 |
257793_at |
AT3G26960
|
[Pollen Ole e 1 allergen and extensin family protein] |
0.257 |
0.217 |
| 38 |
251218_at |
AT3G62410
|
CP12, CP12 DOMAIN-CONTAINING PROTEIN 1, CP12-2, CP12 domain-containing protein 2 |
0.257 |
-0.132 |
| 39 |
254460_at |
AT4G21210
|
ATRP1, PPDK regulatory protein, RP1, PPDK regulatory protein |
0.254 |
0.038 |
| 40 |
250971_at |
AT5G02810
|
APRR7, PRR7, pseudo-response regulator 7 |
0.253 |
-0.024 |
| 41 |
264211_at |
AT1G22770
|
FB, GI, GIGANTEA |
0.252 |
0.004 |
| 42 |
258158_at |
AT3G17790
|
ATACP5, ATPAP17, PAP17, purple acid phosphatase 17 |
0.251 |
0.271 |
| 43 |
251885_at |
AT3G54050
|
HCEF1, high cyclic electron flow 1 |
0.249 |
-0.009 |
| 44 |
248540_at |
AT5G50160
|
ATFRO8, FERRIC REDUCTION OXIDASE 8, FRO8, ferric reduction oxidase 8 |
0.249 |
0.071 |
| 45 |
263350_at |
AT2G13360
|
AGT, alanine:glyoxylate aminotransferase, AGT1, ALANINE:GLYOXYLATE AMINOTRANSFERASE 1, SGAT, L-serine:glyoxylate aminotransferase |
0.245 |
0.074 |
| 46 |
249774_at |
AT5G24150
|
SQE5, SQUALENE MONOOXYGENASE 5, SQP1 |
0.244 |
0.077 |
| 47 |
252441_at |
AT3G46780
|
PTAC16, plastid transcriptionally active 16 |
0.241 |
0.110 |
| 48 |
253971_at |
AT4G26530
|
AtFBA5, FBA5, fructose-bisphosphate aldolase 5 |
0.240 |
-0.048 |
| 49 |
267595_at |
AT2G32990
|
AtGH9B8, glycosyl hydrolase 9B8, GH9B8, glycosyl hydrolase 9B8 |
0.238 |
0.204 |
| 50 |
262680_at |
AT1G75880
|
[SGNH hydrolase-type esterase superfamily protein] |
0.234 |
0.075 |
| 51 |
265698_at |
AT2G32160
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.231 |
0.015 |
| 52 |
256168_at |
AT1G51805
|
[Leucine-rich repeat protein kinase family protein] |
0.231 |
0.066 |
| 53 |
249688_at |
AT5G36160
|
[Tyrosine transaminase family protein] |
0.230 |
0.148 |
| 54 |
247024_at |
AT5G66985
|
unknown |
0.228 |
-0.129 |
| 55 |
254150_at |
AT4G24350
|
[Phosphorylase superfamily protein] |
0.227 |
-0.019 |
| 56 |
259429_at |
AT1G01600
|
CYP86A4, cytochrome P450, family 86, subfamily A, polypeptide 4 |
0.226 |
0.057 |
| 57 |
266279_at |
AT2G29290
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.225 |
-0.065 |
| 58 |
263384_at |
AT2G40130
|
SMXL8, SMAX1-like 8 |
0.224 |
-0.065 |
| 59 |
260241_at |
AT1G63710
|
CYP86A7, cytochrome P450, family 86, subfamily A, polypeptide 7 |
0.224 |
0.031 |
| 60 |
261819_at |
AT1G11410
|
[S-locus lectin protein kinase family protein] |
0.223 |
0.071 |
| 61 |
262745_at |
AT1G28600
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.223 |
0.047 |
| 62 |
261191_at |
AT1G32900
|
GBSS1, granule bound starch synthase 1 |
0.221 |
0.076 |
| 63 |
262050_at |
AT1G80130
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.220 |
0.261 |
| 64 |
266649_at |
AT2G25810
|
TIP4;1, tonoplast intrinsic protein 4;1 |
0.220 |
0.173 |
| 65 |
246844_at |
AT5G26660
|
ATMYB86, myb domain protein 86, MYB86, myb domain protein 86 |
0.219 |
0.058 |
| 66 |
262288_at |
AT1G70760
|
CRR23, CHLORORESPIRATORY REDUCTION 23, NdhL, NADH dehydrogenase-like complex L |
0.218 |
0.172 |
| 67 |
254416_at |
AT4G21380
|
ARK3, receptor kinase 3, RK3, receptor kinase 3 |
0.217 |
0.128 |
| 68 |
251155_at |
AT3G63160
|
OEP6, outer envelope protein 6 |
0.217 |
0.014 |
| 69 |
266353_at |
AT2G01520
|
MLP328, MLP-like protein 328, ZCE1, (Zusammen-CA)-enhanced 1 |
0.216 |
0.362 |
| 70 |
264400_at |
AT1G61800
|
ATGPT2, ARABIDOPSIS GLUCOSE-6-PHOSPHATE/PHOSPHATE TRANSLOCATOR 2, GPT2, glucose-6-phosphate/phosphate translocator 2 |
0.215 |
0.586 |
| 71 |
250286_at |
AT5G13320
|
AtGH3.12, GDG1, GH3-LIKE DEFENSE GENE 1, GH3.12, GRETCHEN HAGEN 3.12, PBS3, AVRPPHB SUSCEPTIBLE 3, WIN3, HOPW1-1-INTERACTING 3 |
0.215 |
0.067 |
| 72 |
254202_at |
AT4G24140
|
[alpha/beta-Hydrolases superfamily protein] |
0.214 |
0.015 |
| 73 |
257300_at |
AT3G28080
|
UMAMIT47, Usually multiple acids move in and out Transporters 47 |
0.213 |
0.134 |
| 74 |
250432_at |
AT5G10420
|
[MATE efflux family protein] |
0.212 |
0.063 |
| 75 |
262811_at |
AT1G11700
|
unknown |
0.211 |
0.269 |
| 76 |
247323_at |
AT5G64170
|
LNK1, night light-inducible and clock-regulated 1 |
0.210 |
-0.048 |
| 77 |
266965_at |
AT2G39510
|
UMAMIT14, Usually multiple acids move in and out Transporters 14 |
0.210 |
0.305 |
| 78 |
258719_at |
AT3G09540
|
[Pectin lyase-like superfamily protein] |
0.209 |
-0.019 |
| 79 |
256736_at |
AT3G29410
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
0.209 |
0.147 |
| 80 |
254971_at |
AT4G10380
|
AtNIP5;1, NIP5;1, NOD26-like intrinsic protein 5;1, NLM6, NOD26-LIKE MIP 6, NLM8, NOD26-LIKE MIP 8 |
0.209 |
0.336 |
| 81 |
250633_at |
AT5G07460
|
ATMSRA2, ARABIDOPSIS THALIANA METHIONINE SULFOXIDE REDUCTASE 2, PMSR2, peptidemethionine sulfoxide reductase 2 |
0.208 |
0.221 |
| 82 |
257262_at |
AT3G21890
|
BBX31, B-box domain protein 31 |
0.207 |
-0.056 |
| 83 |
258956_at |
AT3G01440
|
PnsL3, Photosynthetic NDH subcomplex L 3, PQL1, PsbQ-like 1, PQL2, PsbQ-like 2 |
0.206 |
0.062 |
| 84 |
259442_at |
AT1G02310
|
MAN1, endo-beta-mannanase 1 |
0.206 |
0.056 |
| 85 |
266672_at |
AT2G29650
|
ANTR1, anion transporter 1, PHT4;1, phosphate transporter 4;1 |
0.204 |
-0.053 |
| 86 |
252429_at |
AT3G47500
|
CDF3, cycling DOF factor 3 |
0.204 |
-0.083 |
| 87 |
250054_at |
AT5G17860
|
CAX7, calcium exchanger 7 |
0.203 |
0.235 |
| 88 |
262748_at |
AT1G28610
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.203 |
0.079 |
| 89 |
262629_at |
AT1G06460
|
ACD31.2, ALPHA-CRYSTALLIN DOMAIN 31.2, ACD32.1, alpha-crystallin domain 32.1 |
0.202 |
0.120 |
| 90 |
255604_at |
AT4G01080
|
TBL26, TRICHOME BIREFRINGENCE-LIKE 26 |
0.201 |
0.150 |
| 91 |
255694_at |
AT4G00050
|
UNE10, unfertilized embryo sac 10 |
0.200 |
0.130 |
| 92 |
254465_at |
AT4G20420
|
[Tapetum specific protein TAP35/TAP44] |
0.200 |
-0.014 |
| 93 |
250688_at |
AT5G06510
|
NF-YA10, nuclear factor Y, subunit A10 |
0.200 |
-0.007 |
| 94 |
259783_at |
AT1G29510
|
SAUR67, SMALL AUXIN UPREGULATED RNA 67 |
0.200 |
0.215 |
| 95 |
266293_at |
AT2G29360
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.199 |
-0.034 |
| 96 |
250741_at |
AT5G05790
|
[Duplicated homeodomain-like superfamily protein] |
0.198 |
-0.026 |
| 97 |
258239_at |
AT3G27690
|
LHCB2, LIGHT-HARVESTING CHLOROPHYLL B-BINDING 2, LHCB2.3, photosystem II light harvesting complex gene 2.3, LHCB2.4 |
0.198 |
0.017 |
| 98 |
252462_at |
AT3G47250
|
unknown |
0.198 |
-0.068 |
| 99 |
258223_at |
AT3G15840
|
PIFI, post-illumination chlorophyll fluorescence increase |
0.197 |
0.140 |
| 100 |
259188_at |
AT3G01510
|
LSF1, like SEX4 1 |
0.197 |
0.007 |
| 101 |
248353_at |
AT5G52320
|
CYP96A4, cytochrome P450, family 96, subfamily A, polypeptide 4 |
0.197 |
0.189 |
| 102 |
262448_at |
AT1G49450
|
[Transducin/WD40 repeat-like superfamily protein] |
0.197 |
0.269 |
| 103 |
252092_at |
AT3G51420
|
ATSSL4, STRICTOSIDINE SYNTHASE-LIKE 4, SSL4, strictosidine synthase-like 4 |
0.197 |
0.145 |
| 104 |
253298_at |
AT4G33560
|
[Wound-responsive family protein] |
0.197 |
-0.207 |
| 105 |
259373_at |
AT1G69160
|
unknown |
0.196 |
0.240 |
| 106 |
246997_at |
AT5G67390
|
unknown |
0.195 |
-0.061 |
| 107 |
264287_at |
AT1G61930
|
unknown |
0.195 |
0.057 |
| 108 |
257057_at |
AT3G15310
|
unknown |
0.194 |
-0.009 |
| 109 |
260007_at |
AT1G67870
|
[glycine-rich protein] |
0.194 |
0.079 |
| 110 |
260141_at |
AT1G66350
|
RGL, RGL1, RGA-like 1 |
0.193 |
0.131 |
| 111 |
246939_at |
AT5G25390
|
SHN3, shine3 |
0.193 |
0.088 |
| 112 |
260618_at |
AT1G53230
|
TCP3, TEOSINTE BRANCHED 1, cycloidea and PCF transcription factor 3 |
0.193 |
0.229 |
| 113 |
250665_at |
AT5G06980
|
LNK4, night light-inducible and clock-regulated 4 |
0.193 |
-0.047 |
| 114 |
263320_at |
AT2G47180
|
AtGolS1, galactinol synthase 1, GolS1, galactinol synthase 1 |
0.192 |
0.273 |
| 115 |
266820_at |
AT2G44940
|
[Integrase-type DNA-binding superfamily protein] |
0.192 |
0.032 |
| 116 |
255716_at |
AT4G00330
|
CRCK2, calmodulin-binding receptor-like cytoplasmic kinase 2 |
0.192 |
0.220 |
| 117 |
260412_at |
AT1G69830
|
AMY3, alpha-amylase-like 3, ATAMY3, ALPHA-AMYLASE-LIKE 3 |
0.191 |
0.034 |
| 118 |
252130_at |
AT3G50820
|
OEC33, OXYGEN EVOLVING COMPLEX SUBUNIT 33 KDA, PSBO-2, PHOTOSYSTEM II SUBUNIT O-2, PSBO2, photosystem II subunit O-2 |
0.191 |
-0.052 |
| 119 |
259371_at |
AT1G69080
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
0.191 |
0.001 |
| 120 |
263319_at |
AT2G47160
|
AtBOR1, BOR1, REQUIRES HIGH BORON 1 |
0.191 |
0.149 |
| 121 |
246861_at |
AT5G25890
|
IAA28, indole-3-acetic acid inducible 28, IAR2, IAA-ALANINE RESISTANT 2 |
0.191 |
0.076 |
| 122 |
248153_at |
AT5G54250
|
ATCNGC4, cyclic nucleotide-gated cation channel 4, CNGC4, cyclic nucleotide-gated cation channel 4, DND2, DEFENSE, NO DEATH 2, HLM1 |
0.191 |
0.241 |
| 123 |
250892_at |
AT5G03760
|
ATCSLA09, ATCSLA9, CSLA09, CSLA9, CELLULOSE SYNTHASE LIKE A9, RAT4, RESISTANT TO AGROBACTERIUM TRANSFORMATION 4 |
0.190 |
-0.082 |
| 124 |
258287_at |
AT3G15990
|
SULTR3;4, sulfate transporter 3;4 |
0.190 |
0.092 |
| 125 |
255719_at |
AT1G32080
|
AtLrgB, LrgB |
0.190 |
0.115 |
| 126 |
251157_at |
AT3G63140
|
CSP41A, chloroplast stem-loop binding protein of 41 kDa |
0.188 |
-0.002 |
| 127 |
259185_at |
AT3G01550
|
ATPPT2, PHOSPHOENOLPYRUVATE (PEP)/PHOSPHATE TRANSLOCATOR 2, PPT2, phosphoenolpyruvate (pep)/phosphate translocator 2 |
0.187 |
-0.031 |
| 128 |
248333_at |
AT5G52390
|
[PAR1 protein] |
0.187 |
0.277 |
| 129 |
264343_at |
AT1G11850
|
unknown |
0.187 |
0.077 |
| 130 |
255768_at |
AT1G16705
|
[p300/CBP acetyltransferase-related protein-related] |
0.186 |
0.241 |
| 131 |
262309_at |
AT1G70820
|
[phosphoglucomutase, putative / glucose phosphomutase, putative] |
0.185 |
0.009 |
| 132 |
266572_at |
AT2G23840
|
[HNH endonuclease] |
0.185 |
0.132 |
| 133 |
254065_at |
AT4G25420
|
AT2301, ATGA20OX1, ARABIDOPSIS THALIANA GIBBERELLIN 20-OXIDASE 1, GA20OX1, GA5, GA REQUIRING 5 |
0.184 |
0.199 |
| 134 |
251753_at |
AT3G55760
|
unknown |
0.182 |
0.114 |
| 135 |
256799_at |
AT3G18560
|
unknown |
0.181 |
0.048 |
| 136 |
251169_at |
AT3G63210
|
MARD1, MEDIATOR OF ABA-REGULATED DORMANCY 1 |
0.180 |
0.208 |
| 137 |
258562_at |
AT3G05980
|
unknown |
0.179 |
0.052 |
| 138 |
260419_at |
AT1G69730
|
[Wall-associated kinase family protein] |
0.178 |
0.174 |
| 139 |
261023_at |
AT1G12200
|
FMO, flavin monooxygenase |
0.177 |
0.102 |
| 140 |
248160_at |
AT5G54470
|
BBX29, B-box domain protein 29 |
0.177 |
-0.059 |
| 141 |
259275_at |
AT3G01060
|
unknown |
0.177 |
-0.017 |
| 142 |
249927_at |
AT5G19220
|
ADG2, ADP GLUCOSE PYROPHOSPHORYLASE 2, APL1, ADP glucose pyrophosphorylase large subunit 1 |
0.177 |
0.128 |
| 143 |
263715_at |
AT2G20570
|
GBF's pro-rich region-interacting factor 1 |
0.177 |
0.166 |
| 144 |
248073_at |
AT5G55720
|
[Pectin lyase-like superfamily protein] |
0.177 |
0.028 |
| 145 |
258472_at |
AT3G06080
|
TBL10, TRICHOME BIREFRINGENCE-LIKE 10 |
0.176 |
-0.001 |
| 146 |
263951_at |
AT2G35960
|
NHL12, NDR1/HIN1-like 12 |
0.175 |
0.146 |
| 147 |
249783_at |
AT5G24270
|
ATSOS3, SALT OVERLY SENSITIVE 3, CBL4, CALCINEURIN B-LIKE PROTEIN 4, SOS3, SALT OVERLY SENSITIVE 3 |
0.175 |
0.223 |
| 148 |
260083_at |
AT1G63220
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
0.175 |
0.258 |
| 149 |
265261_at |
AT2G42990
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.174 |
0.144 |
| 150 |
257008_at |
AT3G14210
|
ESM1, epithiospecifier modifier 1 |
0.174 |
0.048 |
| 151 |
248793_at |
AT5G47240
|
atnudt8, nudix hydrolase homolog 8, NUDT8, nudix hydrolase homolog 8 |
0.174 |
-0.109 |
| 152 |
245449_at |
AT4G16870
|
[copia-like retrotransposon family, has a 0. P-value blast match to dbj|BAA78426.1| polyprotein (AtRE2-1) (Arabidopsis thaliana) (Ty1_Copia-element)] |
0.174 |
0.205 |
| 153 |
267127_at |
AT2G23610
|
ATMES3, ARABIDOPSIS THALIANA METHYL ESTERASE 3, MES3, methyl esterase 3 |
0.174 |
0.209 |
| 154 |
263073_at |
AT2G17500
|
[Auxin efflux carrier family protein] |
0.173 |
0.014 |
| 155 |
250435_at |
AT5G10380
|
ATRING1, RING1 |
0.173 |
0.235 |
| 156 |
247906_at |
AT5G57420
|
IAA33, indole-3-acetic acid inducible 33 |
0.173 |
0.088 |
| 157 |
258114_at |
AT3G14660
|
CYP72A13, cytochrome P450, family 72, subfamily A, polypeptide 13 |
0.173 |
0.034 |
| 158 |
261118_at |
AT1G75460
|
[ATP-dependent protease La (LON) domain protein] |
0.173 |
-0.096 |
| 159 |
253416_at |
AT4G33070
|
AtPDC1, PDC1, pyruvate decarboxylase 1 |
0.172 |
-0.018 |
| 160 |
253238_at |
AT4G34480
|
[O-Glycosyl hydrolases family 17 protein] |
0.172 |
0.108 |
| 161 |
250201_at |
AT5G14230
|
unknown |
0.172 |
0.109 |
| 162 |
245302_at |
AT4G17695
|
KAN3, KANADI 3 |
0.171 |
-0.059 |
| 163 |
247356_at |
AT5G63800
|
BGAL6, beta-galactosidase 6, MUM2, MUCILAGE-MODIFIED 2 |
0.171 |
0.022 |
| 164 |
261068_at |
AT1G07450
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.171 |
0.120 |
| 165 |
245306_at |
AT4G14690
|
ELIP2, EARLY LIGHT-INDUCIBLE PROTEIN 2 |
0.171 |
0.003 |
| 166 |
257294_at |
AT3G15570
|
[Phototropic-responsive NPH3 family protein] |
0.170 |
0.005 |
| 167 |
248619_at |
AT5G49630
|
AAP6, amino acid permease 6 |
0.169 |
0.279 |
| 168 |
248062_at |
AT5G55450
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.169 |
0.274 |
| 169 |
261845_at |
AT1G15960
|
ATNRAMP6, NRAMP6, NRAMP metal ion transporter 6 |
0.169 |
0.065 |
| 170 |
249128_at |
AT5G43440
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.169 |
-0.031 |
| 171 |
260266_at |
AT1G68520
|
BBX14, B-box domain protein 14 |
0.169 |
0.034 |
| 172 |
254305_at |
AT4G22200
|
AKT2, AKT2/3, potassium transport 2/3, AKT3, KT2/3, potassium transport 2/3 |
0.168 |
0.236 |
| 173 |
254962_at |
AT4G11050
|
AtGH9C3, glycosyl hydrolase 9C3, GH9C3, glycosyl hydrolase 9C3 |
0.168 |
0.016 |
| 174 |
248377_at |
AT5G51720
|
At-NEET, NEET, NEET group protein |
0.168 |
0.111 |
| 175 |
256655_at |
AT3G18890
|
AtTic62, translocon at the inner envelope membrane of chloroplasts 62, Tic62, translocon at the inner envelope membrane of chloroplasts 62 |
0.167 |
0.060 |
| 176 |
262175_at |
AT1G74880
|
NDH-O, NAD(P)H:plastoquinone dehydrogenase complex subunit O, NdhO, NADH dehydrogenase-like complex ) |
0.167 |
0.096 |
| 177 |
263931_at |
AT2G36220
|
unknown |
0.167 |
0.153 |
| 178 |
248773_at |
AT5G47820
|
FRA1, FRAGILE FIBER 1 |
0.166 |
0.093 |
| 179 |
254794_at |
AT4G12970
|
EPFL9, STOMAGEN, STOMAGEN |
0.166 |
0.033 |
| 180 |
263410_at |
AT2G04039
|
unknown |
0.166 |
0.046 |
| 181 |
263330_at |
AT2G15320
|
[Leucine-rich repeat (LRR) family protein] |
0.165 |
0.122 |
| 182 |
262736_at |
AT1G28570
|
[SGNH hydrolase-type esterase superfamily protein] |
0.165 |
0.029 |
| 183 |
245861_at |
AT5G28300
|
AtGT2L, GT2L, GT-2Like protein |
0.165 |
0.206 |
| 184 |
261788_at |
AT1G15980
|
NDF1, NDH-dependent cyclic electron flow 1, NDH48, NAD(P)H DEHYDROGENASE SUBUNIT 48, PnsB1, Photosynthetic NDH subcomplex B 1 |
0.164 |
-0.105 |
| 185 |
265722_at |
AT2G40100
|
LHCB4.3, light harvesting complex photosystem II |
0.164 |
0.120 |
| 186 |
254687_at |
AT4G13770
|
CYP83A1, cytochrome P450, family 83, subfamily A, polypeptide 1, REF2, REDUCED EPIDERMAL FLUORESCENCE 2 |
0.164 |
0.023 |
| 187 |
263574_at |
AT2G16990
|
[Major facilitator superfamily protein] |
0.164 |
0.104 |
| 188 |
248094_at |
AT5G55220
|
[trigger factor type chaperone family protein] |
0.164 |
-0.002 |
| 189 |
258409_at |
AT3G17640
|
[Leucine-rich repeat (LRR) family protein] |
0.163 |
0.182 |
| 190 |
263676_at |
AT1G09340
|
CRB, chloroplast RNA binding, CSP41B, CHLOROPLAST STEM-LOOP BINDING PROTEIN OF 41 KDA, HIP1.3, heteroglycan-interacting protein 1.3 |
0.163 |
-0.025 |
| 191 |
253886_at |
AT4G27710
|
CYP709B3, cytochrome P450, family 709, subfamily B, polypeptide 3 |
0.163 |
0.076 |
| 192 |
265646_at |
AT2G27360
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.162 |
0.069 |
| 193 |
257748_at |
AT3G18710
|
ATPUB29, ARABIDOPSIS THALIANA PLANT U-BOX 29, PUB29, plant U-box 29 |
0.162 |
0.056 |
| 194 |
245024_at |
ATCG00120
|
ATPA, ATP synthase subunit alpha |
0.162 |
0.055 |
| 195 |
256620_at |
AT3G24480
|
[Leucine-rich repeat (LRR) family protein] |
0.162 |
0.008 |
| 196 |
248247_at |
AT5G53210
|
SPCH, SPEECHLESS |
0.162 |
0.230 |
| 197 |
263287_at |
AT2G36145
|
unknown |
0.161 |
0.025 |
| 198 |
255852_at |
AT1G66970
|
GDPDL1, Glycerophosphodiester phosphodiesterase (GDPD) like 1, SVL2, SHV3-like 2 |
0.161 |
0.189 |
| 199 |
257835_at |
AT3G25180
|
CYP82G1, cytochrome P450, family 82, subfamily G, polypeptide 1 |
0.161 |
0.016 |
| 200 |
248772_at |
AT5G47800
|
[Phototropic-responsive NPH3 family protein] |
0.161 |
0.193 |
| 201 |
262543_at |
AT1G34245
|
EPF2, EPIDERMAL PATTERNING FACTOR 2 |
0.161 |
0.072 |
| 202 |
248167_at |
AT5G54530
|
unknown |
0.160 |
0.047 |
| 203 |
263318_at |
AT2G24762
|
AtGDU4, glutamine dumper 4, GDU4, glutamine dumper 4 |
0.160 |
0.173 |
| 204 |
247691_at |
AT5G59720
|
HSP18.2, heat shock protein 18.2 |
0.160 |
0.120 |
| 205 |
259658_at |
AT1G55370
|
NDF5, NDH-dependent cyclic electron flow 5 |
0.160 |
0.064 |
| 206 |
255437_at |
AT4G03060
|
AOP2, alkenyl hydroxalkyl producing 2 |
0.159 |
0.102 |
| 207 |
251968_at |
AT3G53100
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.159 |
0.088 |
| 208 |
259516_at |
AT1G20450
|
ERD10, EARLY RESPONSIVE TO DEHYDRATION 10, LTI29, LOW TEMPERATURE INDUCED 29, LTI45, LOW TEMPERATURE INDUCED 45 |
0.158 |
0.325 |
| 209 |
246036_at |
AT5G08370
|
AGAL2, alpha-galactosidase 2, AtAGAL2, alpha-galactosidase 2 |
0.158 |
0.283 |
| 210 |
267592_at |
AT2G39710
|
[Eukaryotic aspartyl protease family protein] |
0.158 |
-0.004 |
| 211 |
267164_at |
AT2G37700
|
[Fatty acid hydroxylase superfamily] |
0.158 |
0.045 |
| 212 |
244937_at |
ATCG01110
|
NDHH, NAD(P)H dehydrogenase subunit H |
0.158 |
0.100 |
| 213 |
248371_at |
AT5G51810
|
AT2353, ATGA20OX2, GA20OX2, gibberellin 20 oxidase 2 |
0.157 |
0.102 |
| 214 |
246488_at |
AT5G16010
|
[3-oxo-5-alpha-steroid 4-dehydrogenase family protein] |
0.157 |
0.025 |
| 215 |
245842_at |
AT1G58430
|
RXF26 |
0.157 |
0.180 |
| 216 |
245346_at |
AT4G17090
|
AtBAM3, BAM3, BETA-AMYLASE 3, BMY8, BETA-AMYLASE 8, CT-BMY, chloroplast beta-amylase |
0.156 |
0.345 |
| 217 |
262693_at |
AT1G62780
|
unknown |
0.156 |
0.034 |
| 218 |
245393_at |
AT4G16260
|
[Glycosyl hydrolase superfamily protein] |
0.156 |
-0.013 |
| 219 |
266990_at |
AT2G39190
|
ATATH8 |
0.156 |
-0.026 |
| 220 |
257709_at |
AT3G27325
|
[hydrolases, acting on ester bonds] |
0.156 |
-0.048 |
| 221 |
254232_at |
AT4G23600
|
CORI3, CORONATINE INDUCED 1, JR2, JASMONIC ACID RESPONSIVE 2 |
0.156 |
-0.383 |
| 222 |
247759_at |
AT5G59040
|
COPT3, copper transporter 3 |
0.155 |
0.011 |
| 223 |
252543_at |
AT3G45780
|
JK224, NPH1, NONPHOTOTROPIC HYPOCOTYL 1, PHOT1, phototropin 1, RPT1, ROOT PHOTOTROPISM 1 |
0.155 |
0.330 |
| 224 |
259797_at |
AT1G64410
|
unknown |
0.155 |
-0.137 |
| 225 |
255294_at |
AT4G04750
|
[Major facilitator superfamily protein] |
0.155 |
0.045 |
| 226 |
263120_at |
AT1G78490
|
CYP708A3, cytochrome P450, family 708, subfamily A, polypeptide 3 |
0.155 |
0.440 |
| 227 |
264319_at |
AT1G04110
|
AtSDD1, SDD1, STOMATAL DENSITY AND DISTRIBUTION 1 |
0.154 |
-0.055 |
| 228 |
254605_at |
AT4G18950
|
[Integrin-linked protein kinase family] |
0.154 |
0.055 |
| 229 |
262577_at |
AT1G15290
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.153 |
-0.063 |
| 230 |
260140_at |
AT1G66390
|
ATMYB90, MYB90, myb domain protein 90, PAP2, PRODUCTION OF ANTHOCYANIN PIGMENT 2 |
0.153 |
0.157 |
| 231 |
254572_at |
AT4G19380
|
[Long-chain fatty alcohol dehydrogenase family protein] |
0.153 |
0.119 |
| 232 |
258894_at |
AT3G05650
|
AtRLP32, receptor like protein 32, RLP32, receptor like protein 32 |
0.152 |
0.172 |
| 233 |
247853_at |
AT5G58140
|
NPL1, NON PHOTOTROPIC HYPOCOTYL 1-LIKE, PHOT2, phototropin 2 |
0.152 |
-0.048 |
| 234 |
255877_at |
AT2G40460
|
[Major facilitator superfamily protein] |
0.151 |
0.163 |
| 235 |
260262_at |
AT1G68470
|
[Exostosin family protein] |
0.151 |
0.295 |
| 236 |
264348_at |
AT1G12110
|
AtNPF6.3, ATNRT1, ARABIDOPSIS THALIANA NITRATE TRANSPORTER 1, B-1, CHL1, CHLORINA 1, CHL1-1, NPF6.3, NRT1/ PTR family 6.3, NRT1, NITRATE TRANSPORTER 1, NRT1.1, nitrate transporter 1.1 |
0.151 |
-0.138 |
| 237 |
251448_at |
AT3G59845
|
[Zinc-binding dehydrogenase family protein] |
0.150 |
0.083 |
| 238 |
262231_at |
AT1G68740
|
PHO1;H1 |
0.150 |
0.017 |
| 239 |
254090_at |
AT4G25010
|
AtSWEET14, SWEET14 |
0.150 |
0.034 |
| 240 |
263177_at |
AT1G05540
|
unknown |
0.150 |
0.052 |
| 241 |
261663_at |
AT1G18330
|
EPR1, EARLY-PHYTOCHROME-RESPONSIVE1, RVE7, REVEILLE 7 |
0.150 |
-0.018 |
| 242 |
255907_at |
AT1G17920
|
HDG12, homeodomain GLABROUS 12 |
0.150 |
0.064 |
| 243 |
252661_at |
AT3G44450
|
unknown |
0.150 |
-0.034 |
| 244 |
256514_at |
AT1G66130
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.149 |
-0.043 |
| 245 |
250445_at |
AT5G10760
|
[Eukaryotic aspartyl protease family protein] |
0.149 |
0.016 |
| 246 |
262826_at |
AT1G13080
|
CYP71B2, cytochrome P450, family 71, subfamily B, polypeptide 2 |
0.149 |
0.137 |
| 247 |
263761_at |
AT2G21330
|
AtFBA1, FBA1, fructose-bisphosphate aldolase 1 |
0.149 |
-0.079 |
| 248 |
257066_at |
AT3G18280
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.149 |
0.312 |
| 249 |
262436_at |
AT1G47610
|
[Transducin/WD40 repeat-like superfamily protein] |
0.149 |
0.128 |
| 250 |
264846_at |
AT2G17850
|
[Rhodanese/Cell cycle control phosphatase superfamily protein] |
0.148 |
-0.015 |
| 251 |
267503_at |
AT2G45600
|
[alpha/beta-Hydrolases superfamily protein] |
0.148 |
0.040 |
| 252 |
257081_at |
AT3G30460
|
[RING/U-box superfamily protein] |
0.148 |
-0.004 |
| 253 |
247252_at |
AT5G64770
|
CLEL 9, CLE-like 9, GLV2, GOLVEN 2, RGF9, root meristem growth factor 9 |
0.148 |
-0.057 |
| 254 |
266291_at |
AT2G29320
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.147 |
0.031 |
| 255 |
249167_at |
AT5G42860
|
unknown |
0.146 |
0.197 |
| 256 |
263252_at |
AT2G31380
|
BBX25, B-box domain protein 25, STH, salt tolerance homologue |
0.146 |
-0.058 |
| 257 |
250045_at |
AT5G17700
|
[MATE efflux family protein] |
0.146 |
-0.054 |
| 258 |
264137_at |
AT1G78960
|
ATLUP2, lupeol synthase 2, LUP2, lupeol synthase 2 |
0.146 |
-0.010 |
| 259 |
258326_at |
AT3G22760
|
SOL1 |
0.146 |
-0.025 |
| 260 |
261772_at |
AT1G76240
|
unknown |
0.145 |
-0.068 |
| 261 |
247455_at |
AT5G62470
|
ATMYB96, MYB DOMAIN PROTEIN 96, MYB96, myb domain protein 96, MYBCOV1 |
0.145 |
0.036 |
| 262 |
266514_at |
AT2G47890
|
[B-box type zinc finger protein with CCT domain] |
0.145 |
0.040 |
| 263 |
245819_at |
AT1G26310
|
AGL10, AGAMOUS-like 10, CAL, CAULIFLOWER, CAL1 |
0.145 |
-0.010 |
| 264 |
263947_at |
AT2G35820
|
[ureidoglycolate hydrolases] |
0.145 |
0.137 |
| 265 |
248329_at |
AT5G52780
|
unknown |
0.144 |
0.151 |
| 266 |
263796_at |
AT2G24540
|
AFR, ATTENUATED FAR-RED RESPONSE |
0.144 |
-0.032 |
| 267 |
255088_at |
AT4G09350
|
CRRJ, CHLORORESPIRATORY REDUCTION J, DJC75, DNA J protein C75, NdhT, NADH dehydrogenase-like complex T |
0.144 |
0.059 |
| 268 |
254174_at |
AT4G24120
|
ATYSL1, YELLOW STRIPE LIKE 1, YSL1, YELLOW STRIPE like 1 |
0.143 |
0.012 |
| 269 |
264900_at |
AT1G23080
|
ATPIN7, ARABIDOPSIS PIN-FORMED 7, PIN7, PIN-FORMED 7 |
0.143 |
-0.001 |
| 270 |
245712_at |
AT5G04360
|
ATLDA, limit dextrinase, ATPU1, PULLULANASE 1, LDA, limit dextrinase, PU1, PULLULANASE 1 |
0.143 |
-0.029 |
| 271 |
249876_at |
AT5G23060
|
CaS, calcium sensing receptor |
0.143 |
0.024 |
| 272 |
252414_at |
AT3G47420
|
AtG3Pp1, Glycerol-3-phosphate permease 1, ATPS3, phosphate starvation-induced gene 3, G3Pp1, Glycerol-3-phosphate permease 1, PS3, phosphate starvation-induced gene 3 |
0.143 |
0.280 |
| 273 |
263406_at |
AT2G04160
|
AIR3, AUXIN-INDUCED IN ROOT CULTURES 3 |
0.143 |
0.071 |
| 274 |
247295_at |
AT5G64180
|
unknown |
0.142 |
-0.025 |
| 275 |
245226_at |
AT3G29970
|
[B12D protein] |
0.142 |
-0.032 |
| 276 |
258984_at |
AT3G08970
|
ATERDJ3A, TMS1, THERMOSENSITIVE MALE STERILE 1 |
0.142 |
0.047 |
| 277 |
256999_at |
AT3G14200
|
[Chaperone DnaJ-domain superfamily protein] |
0.142 |
0.036 |
| 278 |
246595_at |
AT5G14780
|
FDH, formate dehydrogenase |
0.142 |
0.116 |
| 279 |
256524_at |
AT1G66200
|
ATGSR2, glutamine synthase clone F11, GLN1;2, glutamine synthetase 1;2, GSR2, glutamine synthase clone F11 |
0.142 |
0.034 |
| 280 |
258232_at |
AT3G27750
|
EMB3123, EMBRYO DEFECTIVE 3123, THA8, Thylakoid Assembly 8 |
0.142 |
-0.016 |
| 281 |
246302_at |
AT3G51860
|
ATCAX3, ATHCX1, CAX1-LIKE, CAX3, cation exchanger 3 |
0.142 |
0.183 |
| 282 |
253849_at |
AT4G28080
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.141 |
-0.082 |
| 283 |
264953_at |
AT1G77120
|
ADH, ALCOHOL DEHYDROGENASE, ADH1, alcohol dehydrogenase 1, ATADH, ARABIDOPSIS THALIANA ALCOHOL DEHYDROGENASE, ATADH1 |
0.141 |
0.027 |
| 284 |
253309_at |
AT4G33790
|
CER4, ECERIFERUM 4, FAR3, FATTY ACID REDUCTASE 3, G7 |
0.141 |
0.016 |
| 285 |
260521_at |
AT2G41740
|
ATVLN2, VLN2, villin 2 |
0.141 |
-0.002 |
| 286 |
260371_at |
AT1G69690
|
AtTCP15, TCP15, TEOSINTE BRANCHED1/CYCLOIDEA/PCF 15 |
0.141 |
0.041 |
| 287 |
262700_at |
AT1G76020
|
[Thioredoxin superfamily protein] |
0.141 |
0.070 |
| 288 |
254255_at |
AT4G23220
|
CRK14, cysteine-rich RLK (RECEPTOR-like protein kinase) 14 |
0.141 |
-0.009 |
| 289 |
258299_at |
AT3G23410
|
ATFAO3, ARABIDOPSIS FATTY ALCOHOL OXIDASE 3, FAO3, fatty alcohol oxidase 3 |
0.140 |
0.234 |
| 290 |
266175_at |
AT2G02450
|
ANAC034, Arabidopsis NAC domain containing protein 34, ANAC035, NAC domain containing protein 35, AtLOV1, LOV1, LONG VEGETATIVE PHASE 1, NAC035, NAC domain containing protein 35 |
0.140 |
0.084 |
| 291 |
255644_at |
AT4G00870
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.139 |
0.181 |
| 292 |
263028_at |
AT1G24030
|
[Protein kinase superfamily protein] |
0.139 |
0.085 |
| 293 |
253411_at |
AT4G32980
|
ATH1, homeobox gene 1 |
0.139 |
0.069 |
| 294 |
262882_at |
AT1G64900
|
CYP89, CYP89A2, cytochrome P450, family 89, subfamily A, polypeptide 2 |
0.139 |
0.104 |
| 295 |
247902_at |
AT5G57350
|
AHA3, H(+)-ATPase 3, ATAHA3, ARABIDOPSIS THALIANA ARABIDOPSIS H(+)-ATPASE, HA3, H(+)-ATPase 3 |
0.139 |
0.276 |
| 296 |
261751_at |
AT1G76080
|
ATCDSP32, ARABIDOPSIS THALIANA CHLOROPLASTIC DROUGHT-INDUCED STRESS PROTEIN OF 32 KD, CDSP32, chloroplastic drought-induced stress protein of 32 kD |
0.139 |
-0.058 |
| 297 |
264314_at |
AT1G70420
|
unknown |
0.138 |
0.036 |
| 298 |
263114_at |
AT1G03130
|
PSAD-2, photosystem I subunit D-2 |
0.138 |
0.011 |
| 299 |
254746_at |
AT4G12980
|
[Auxin-responsive family protein] |
0.138 |
0.114 |
| 300 |
263709_at |
AT1G09310
|
unknown |
0.138 |
0.305 |
| 301 |
260941_at |
AT1G44970
|
[Peroxidase superfamily protein] |
-0.733 |
-0.134 |
| 302 |
257930_at |
AT3G17010
|
REM22, REPRODUCTIVE MERISTEM 22 |
-0.731 |
-0.304 |
| 303 |
246888_at |
AT5G26270
|
unknown |
-0.709 |
-0.008 |
| 304 |
253161_at |
AT4G35770
|
ATSEN1, ARABIDOPSIS THALIANA SENESCENCE 1, DIN1, DARK INDUCIBLE 1, SEN1, SENESCENCE 1, SEN1, SENESCENCE ASSOCIATED GENE 1 |
-0.649 |
-0.015 |
| 305 |
254595_at |
AT4G18960
|
AG, AGAMOUS |
-0.615 |
0.048 |
| 306 |
250491_at |
AT5G09780
|
[Transcriptional factor B3 family protein] |
-0.599 |
-0.119 |
| 307 |
246025_at |
AT5G21150
|
AGO9, ARGONAUTE 9 |
-0.593 |
-0.404 |
| 308 |
264433_at |
AT1G61810
|
BGLU45, beta-glucosidase 45 |
-0.564 |
0.159 |
| 309 |
256940_at |
AT3G30720
|
QQS, QUA-QUINE STARCH |
-0.551 |
-0.062 |
| 310 |
252415_at |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
-0.548 |
0.340 |
| 311 |
263092_at |
AT2G16210
|
[Transcriptional factor B3 family protein] |
-0.506 |
-0.062 |
| 312 |
261815_at |
AT1G08320
|
bZIP21, TGA9, TGACG (TGA) motif-binding protein 9 |
-0.480 |
-0.124 |
| 313 |
256987_at |
AT3G28560
|
[BCS1 AAA-type ATPase] |
-0.479 |
-0.360 |
| 314 |
244994_at |
ATCG01010
|
NDHF |
-0.478 |
-0.013 |
| 315 |
261960_at |
AT1G36550
|
[similar to retrotransposon protein, putative, Ty1-copia subclass [Oryza sativa (japonica cultivar-group)] (GB:ABA98367.2)] |
-0.461 |
-0.095 |
| 316 |
260287_at |
AT1G80440
|
KFB20, Kelch repeat F-box 20, KMD1, KISS ME DEADLY 1 |
-0.449 |
-0.054 |
| 317 |
260883_at |
AT1G29270
|
unknown |
-0.431 |
0.002 |
| 318 |
254777_at |
AT4G12960
|
AtGILT, GILT, gamma-interferon-responsive lysosomal thiol reductase |
-0.415 |
0.008 |
| 319 |
254665_at |
AT4G18340
|
[Glycosyl hydrolase superfamily protein] |
-0.412 |
0.000 |
| 320 |
257330_at |
ATMG01290
|
ORF111C |
-0.406 |
0.026 |
| 321 |
247925_at |
AT5G57560
|
TCH4, Touch 4, XTH22, xyloglucan endotransglucosylase/hydrolase 22 |
-0.390 |
0.037 |
| 322 |
253862_at |
AT4G27330
|
NZZ, NOZZLE, SPL, SPOROCYTELESS |
-0.390 |
-0.073 |
| 323 |
260355_at |
AT1G69180
|
CRC, CRABS CLAW |
-0.389 |
-0.083 |
| 324 |
264066_at |
AT2G27880
|
AGO5, ARGONAUTE 5, AtAGO5 |
-0.385 |
-0.038 |
| 325 |
248752_at |
AT5G47600
|
[HSP20-like chaperones superfamily protein] |
-0.378 |
-0.026 |
| 326 |
248332_at |
AT5G52640
|
ATHS83, AtHsp90-1, HEAT SHOCK PROTEIN 90-1, ATHSP90.1, heat shock protein 90.1, HSP81-1, HEAT SHOCK PROTEIN 81-1, HSP81.1, HSP83, HEAT SHOCK PROTEIN 83, HSP90.1, heat shock protein 90.1 |
-0.358 |
-0.231 |
| 327 |
253259_at |
AT4G34410
|
RRTF1, redox responsive transcription factor 1 |
-0.349 |
0.014 |
| 328 |
267461_at |
AT2G33830
|
AtDRM2, DRM2, dormancy associated gene 2 |
-0.347 |
0.018 |
| 329 |
245005_at |
ATCG00330
|
RPS14, chloroplast ribosomal protein S14 |
-0.336 |
-0.030 |
| 330 |
253689_at |
AT4G29770
|
unknown |
-0.334 |
-0.511 |
| 331 |
264788_at |
AT2G17880
|
DJC24, DNA J protein C24 |
-0.330 |
-0.094 |
| 332 |
252367_at |
AT3G48360
|
ATBT2, BT2, BTB and TAZ domain protein 2 |
-0.316 |
-0.004 |
| 333 |
250223_at |
AT5G14070
|
ROXY2 |
-0.316 |
-0.088 |
| 334 |
265234_at |
AT2G07721
|
unknown |
-0.313 |
0.018 |
| 335 |
248926_at |
AT5G45880
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.307 |
-0.470 |
| 336 |
264339_at |
AT1G70290
|
ATTPS8, ATTPSC, TPS8, trehalose-6-phosphatase synthase S8 |
-0.306 |
-0.189 |
| 337 |
253061_at |
AT4G37610
|
BT5, BTB and TAZ domain protein 5 |
-0.306 |
-0.037 |
| 338 |
262443_at |
AT1G47655
|
[Dof-type zinc finger DNA-binding family protein] |
-0.302 |
-0.154 |
| 339 |
257798_at |
AT3G15950
|
NAI2 |
-0.300 |
0.143 |
| 340 |
256966_at |
AT3G13400
|
sks13, SKU5 similar 13 |
-0.298 |
-0.610 |
| 341 |
254001_at |
AT4G26260
|
MIOX4, myo-inositol oxygenase 4 |
-0.292 |
-0.271 |
| 342 |
256459_at |
AT1G36180
|
ACC2, acetyl-CoA carboxylase 2 |
-0.285 |
-0.063 |
| 343 |
256245_at |
AT3G12580
|
ATHSP70, ARABIDOPSIS HEAT SHOCK PROTEIN 70, HSP70, heat shock protein 70 |
-0.283 |
-0.421 |
| 344 |
261410_at |
AT1G07610
|
MT1C, metallothionein 1C |
-0.279 |
-0.068 |
| 345 |
265511_at |
AT2G05540
|
[Glycine-rich protein family] |
-0.279 |
-0.013 |
| 346 |
244977_at |
ATCG00730
|
PETD, photosynthetic electron transfer D |
-0.275 |
0.012 |
| 347 |
255568_at |
AT4G01250
|
AtWRKY22, WRKY22 |
-0.273 |
-0.031 |
| 348 |
265387_at |
AT2G20670
|
unknown |
-0.273 |
0.061 |
| 349 |
251868_at |
AT3G54490
|
RPB5E, RNA polymerase II fifth largest subunit, E |
-0.273 |
0.018 |
| 350 |
257026_at |
AT3G19200
|
unknown |
-0.271 |
0.013 |
| 351 |
258225_at |
AT3G15630
|
unknown |
-0.270 |
-0.043 |
| 352 |
256021_at |
AT1G58270
|
ZW9 |
-0.270 |
-0.474 |
| 353 |
260744_at |
AT1G15010
|
unknown |
-0.268 |
0.033 |
| 354 |
256017_at |
AT1G19180
|
AtJAZ1, JAZ1, jasmonate-zim-domain protein 1, TIFY10A |
-0.265 |
0.228 |
| 355 |
262603_at |
AT1G15380
|
GLYI4, glyoxylase I 4 |
-0.263 |
-0.023 |
| 356 |
259831_at |
AT1G69600
|
ATHB29, ARABIDOPSIS THALIANA HOMEOBOX PROTEIN 29, ZFHD1, zinc finger homeodomain 1, ZHD11, ZINC FINGER HOMEODOMAIN 11 |
-0.263 |
-0.037 |
| 357 |
247875_at |
AT5G57720
|
[AP2/B3-like transcriptional factor family protein] |
-0.261 |
-0.101 |
| 358 |
259802_at |
AT1G72260
|
THI2.1, thionin 2.1, THI2.1.1 |
-0.261 |
-0.066 |
| 359 |
256175_at |
AT1G51670
|
unknown |
-0.260 |
-0.119 |
| 360 |
248240_at |
AT5G53950
|
ANAC098, Arabidopsis NAC domain containing protein 98, ATCUC2, CUP-SHAPED COTYLEDON 2, CUC2, CUP-SHAPED COTYLEDON 2 |
-0.258 |
-0.122 |
| 361 |
259760_at |
AT1G77580
|
unknown |
-0.253 |
-0.008 |
| 362 |
258639_at |
AT3G07820
|
[Pectin lyase-like superfamily protein] |
-0.252 |
-0.671 |
| 363 |
265483_at |
AT2G15790
|
CYP40, CYCLOPHILIN 40, SQN, SQUINT |
-0.251 |
-0.052 |
| 364 |
264280_at |
AT1G61820
|
BGLU46, beta glucosidase 46 |
-0.250 |
0.127 |
| 365 |
253737_at |
AT4G28703
|
[RmlC-like cupins superfamily protein] |
-0.249 |
-0.109 |
| 366 |
245000_at |
ATCG00210
|
YCF6 |
-0.248 |
-0.003 |
| 367 |
254203_at |
AT4G24150
|
AtGRF8, growth-regulating factor 8, GRF8, growth-regulating factor 8 |
-0.248 |
-0.041 |
| 368 |
248467_at |
AT5G50800
|
AtSWEET13, SWEET13 |
-0.248 |
-0.264 |
| 369 |
262760_at |
AT1G10770
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.246 |
-0.594 |
| 370 |
255937_at |
AT1G12610
|
DDF1, DWARF AND DELAYED FLOWERING 1 |
-0.246 |
-0.000 |
| 371 |
253769_at |
AT4G28560
|
RIC7, ROP-interactive CRIB motif-containing protein 7 |
-0.245 |
-0.027 |
| 372 |
263680_at |
AT1G26930
|
[Galactose oxidase/kelch repeat superfamily protein] |
-0.244 |
-0.197 |
| 373 |
262001_at |
AT1G33790
|
[jacalin lectin family protein] |
-0.243 |
-0.239 |
| 374 |
249817_at |
AT5G23820
|
ML3, MD2-related lipid recognition 3 |
-0.240 |
-0.054 |
| 375 |
261395_at |
AT1G79700
|
WRI4, WRINKLED 4 |
-0.239 |
-0.051 |
| 376 |
246114_at |
AT5G20250
|
DIN10, DARK INDUCIBLE 10, RS6, raffinose synthase 6 |
-0.239 |
0.047 |
| 377 |
261340_at |
AT1G35730
|
APUM9, pumilio 9, PUM9, pumilio 9 |
-0.232 |
-0.184 |
| 378 |
255733_at |
AT1G25400
|
unknown |
-0.228 |
0.033 |
| 379 |
246526_at |
AT5G15720
|
GLIP7, GDSL-motif lipase 7 |
-0.225 |
-0.067 |
| 380 |
261892_at |
AT1G80840
|
ATWRKY40, WRKY40, WRKY DNA-binding protein 40 |
-0.224 |
0.090 |
| 381 |
251195_at |
AT3G62930
|
[Thioredoxin superfamily protein] |
-0.223 |
-0.069 |
| 382 |
255734_at |
AT1G25550
|
[myb-like transcription factor family protein] |
-0.222 |
0.039 |
| 383 |
249862_at |
AT5G22920
|
[CHY-type/CTCHY-type/RING-type Zinc finger protein] |
-0.222 |
-0.056 |
| 384 |
256766_at |
AT3G22231
|
PCC1, PATHOGEN AND CIRCADIAN CONTROLLED 1 |
-0.221 |
-0.115 |
| 385 |
258707_at |
AT3G09480
|
[Histone superfamily protein] |
-0.220 |
-0.008 |
| 386 |
266141_at |
AT2G02120
|
LCR70, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 70, PDF2.1 |
-0.219 |
0.013 |
| 387 |
250174_at |
AT5G14380
|
AGP6, arabinogalactan protein 6 |
-0.218 |
-0.425 |
| 388 |
261901_at |
AT1G80920
|
AtJ8, AtToc12, translocon at the outer envelope membrane of chloroplasts 12, DJC22, DNA J protein C22, J8, Toc12, translocon at the outer envelope membrane of chloroplasts 12 |
-0.218 |
-0.069 |
| 389 |
267075_at |
AT2G41070
|
ATBZIP12, DPBF4, EEL, ENHANCED EM LEVEL |
-0.218 |
0.033 |
| 390 |
263237_at |
AT1G10610
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.218 |
-0.162 |
| 391 |
256584_at |
AT3G28750
|
unknown |
-0.218 |
-0.528 |
| 392 |
261938_at |
AT1G22510
|
[FUNCTIONS IN: zinc ion binding] |
-0.218 |
0.008 |
| 393 |
249401_at |
AT5G40260
|
AtSWEET8, RPG1, RUPTURED POLLEN GRAIN1, SWEET8 |
-0.217 |
-0.016 |
| 394 |
257777_x_at |
AT3G29210
|
[similar to Ulp1 protease family protein [Arabidopsis thaliana] (TAIR:AT2G14130.1)] |
-0.217 |
0.006 |
| 395 |
248855_at |
AT5G46590
|
anac096, NAC domain containing protein 96, NAC096, NAC domain containing protein 96 |
-0.216 |
0.194 |
| 396 |
267488_at |
AT2G19110
|
ATHMA4, ARABIDOPSIS HEAVY METAL ATPASE 4, HMA4, heavy metal atpase 4 |
-0.215 |
0.044 |
| 397 |
245422_at |
AT4G17470
|
[alpha/beta-Hydrolases superfamily protein] |
-0.215 |
0.025 |
| 398 |
263016_at |
AT1G23410
|
[Ribosomal protein S27a / Ubiquitin family protein] |
-0.214 |
0.054 |
| 399 |
266415_at |
AT2G38530
|
cdf3, cell growth defect factor-3, LP2, LTP2, lipid transfer protein 2 |
-0.213 |
-0.045 |
| 400 |
256977_at |
AT3G21040
|
[copia-like retrotransposon family, has a 5.7e-20 P-value blast match to gb|AAO73527.1| gag-pol polyprotein (Glycine max) (SIRE1) (Ty1_Copia-family)] |
-0.212 |
-0.200 |
| 401 |
248550_at |
AT5G50210
|
OLD5, ONSET OF LEAF DEATH 5, QS, quinolinate synthase, SUFE3, SULFUR E 3 |
-0.212 |
-0.003 |
| 402 |
259793_at |
AT1G64380
|
[Integrase-type DNA-binding superfamily protein] |
-0.210 |
0.116 |
| 403 |
264783_at |
AT1G08650
|
ATPPCK1, PHOSPHOENOLPYRUVATE CARBOXYLASE KINASE 1, PPCK1, phosphoenolpyruvate carboxylase kinase 1 |
-0.208 |
-0.018 |
| 404 |
253488_at |
AT4G31610
|
ATREM1, REPRODUCTIVE MERISTEM 1, REM1, REPRODUCTIVE MERISTEM 1 |
-0.208 |
-0.136 |
| 405 |
250258_at |
AT5G13790
|
AGL15, AGAMOUS-like 15 |
-0.208 |
-0.088 |
| 406 |
250869_at |
AT5G03840
|
TFL-1, TERMINAL FLOWER 1, TFL1, TERMINAL FLOWER 1 |
-0.206 |
-0.009 |
| 407 |
250701_at |
AT5G06839
|
bZIP65, TGA10, TGACG (TGA) motif-binding protein 10 |
-0.206 |
-0.116 |
| 408 |
262212_at |
AT1G74890
|
ARR15, response regulator 15 |
-0.205 |
-0.165 |
| 409 |
258764_at |
AT3G10720
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.203 |
0.171 |
| 410 |
263819_x_at |
AT2G10140
|
[CACTA-like transposase family (Tnp2/En/Spm), has a 1.7e-140 P-value blast match to GB:CAA40555 TNP2 (CACTA-element) (Antirrhinum majus)] |
-0.203 |
-0.050 |
| 411 |
251986_at |
AT3G53310
|
[AP2/B3-like transcriptional factor family protein] |
-0.202 |
-0.087 |
| 412 |
245668_at |
AT1G28330
|
AtDRM1, DRM1, DORMANCY-ASSOCIATED PROTEIN 1, DYL1, dormancy-associated protein-like 1 |
-0.202 |
-0.224 |
| 413 |
261211_at |
AT1G12780
|
ATUGE1, A. THALIANA UDP-GLC 4-EPIMERASE 1, UGE1, UDP-D-glucose/UDP-D-galactose 4-epimerase 1 |
-0.201 |
-0.014 |
| 414 |
254197_at |
AT4G24040
|
ATTRE1, TRE1, trehalase 1 |
-0.199 |
-0.008 |
| 415 |
257229_at |
AT3G16490
|
IQD26, IQ-domain 26 |
-0.198 |
0.005 |
| 416 |
267083_at |
AT2G41100
|
ATCAL4, ARABIDOPSIS THALIANA CALMODULIN LIKE 4, TCH3, TOUCH 3 |
-0.198 |
0.106 |
| 417 |
265475_at |
AT2G15620
|
ATHNIR, ARABIDOPSIS THALIANA NITRITE REDUCTASE, NIR, NITRITE REDUCTASE, NIR1, nitrite reductase 1 |
-0.197 |
0.016 |
| 418 |
256976_at |
AT3G21020
|
[copia-like retrotransposon family, has a 5.4e-14 P-value blast match to gb|AAO73529.1| gag-pol polyprotein (Glycine max) (SIRE1) (Ty1_Copia-family)] |
-0.196 |
-0.052 |
| 419 |
265481_at |
AT2G15960
|
unknown |
-0.196 |
0.221 |
| 420 |
262122_at |
AT1G02790
|
PGA4, polygalacturonase 4 |
-0.195 |
-0.565 |
| 421 |
252691_at |
AT3G44050
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.195 |
0.038 |
| 422 |
256538_x_at |
AT3G32900
|
[similar to Ulp1 protease family protein [Arabidopsis thaliana] (TAIR:AT3G47260.1)] |
-0.194 |
-0.037 |
| 423 |
258108_at |
AT3G23570
|
[alpha/beta-Hydrolases superfamily protein] |
-0.193 |
-0.097 |
| 424 |
246099_at |
AT5G20230
|
ATBCB, blue-copper-binding protein, BCB, BLUE COPPER BINDING PROTEIN, BCB, blue-copper-binding protein, SAG14, SENESCENCE ASSOCIATED GENE 14 |
-0.193 |
-0.393 |
| 425 |
244931_at |
ATMG00630
|
ORF110B |
-0.193 |
0.062 |
| 426 |
245191_at |
AT1G67770
|
TEL2, terminal EAR1-like 2 |
-0.191 |
-0.135 |
| 427 |
259694_at |
AT1G63180
|
UGE3, UDP-D-glucose/UDP-D-galactose 4-epimerase 3 |
-0.190 |
-0.051 |
| 428 |
251196_at |
AT3G62950
|
[Thioredoxin superfamily protein] |
-0.190 |
-0.071 |
| 429 |
253064_at |
AT4G37730
|
AtbZIP7, basic leucine-zipper 7, bZIP7, basic leucine-zipper 7 |
-0.190 |
-0.102 |
| 430 |
251469_at |
AT3G59530
|
LAP3, LESS ADHERENT POLLEN 3 |
-0.187 |
0.167 |
| 431 |
247228_at |
AT5G65140
|
TPPJ, trehalose-6-phosphate phosphatase J |
-0.187 |
-0.015 |
| 432 |
249337_at |
AT5G41080
|
AtGDPD2, GDPD2, glycerophosphodiester phosphodiesterase 2 |
-0.184 |
0.132 |
| 433 |
260551_at |
AT2G43510
|
ATTI1, trypsin inhibitor protein 1, TI1, trypsin inhibitor protein 1 |
-0.183 |
-0.086 |
| 434 |
260461_at |
AT1G10980
|
[Lung seven transmembrane receptor family protein] |
-0.182 |
-0.008 |
| 435 |
247562_at |
AT5G61120
|
unknown |
-0.181 |
0.025 |
| 436 |
246034_at |
AT5G08350
|
[GRAM domain-containing protein / ABA-responsive protein-related] |
-0.179 |
0.001 |
| 437 |
253811_at |
AT4G28190
|
ULT, ULTRAPETALA, ULT1, ULTRAPETALA1 |
-0.179 |
-0.002 |
| 438 |
266637_at |
AT2G35600
|
ATBRXL1, ARABIDOPSIS THALIANA BREVIS RADIX LIKE 1, BRXL1, BREVIS RADIX-like 1 |
-0.178 |
-0.234 |
| 439 |
249920_at |
AT5G19260
|
FAF3, FANTASTIC FOUR 3 |
-0.178 |
-0.119 |
| 440 |
265680_at |
AT2G32150
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.177 |
-0.006 |
| 441 |
251617_at |
AT3G58000
|
[VQ motif-containing protein] |
-0.177 |
-0.085 |
| 442 |
249587_at |
AT5G37860
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.177 |
-0.085 |
| 443 |
262986_at |
AT1G23390
|
[Kelch repeat-containing F-box family protein] |
-0.176 |
0.096 |
| 444 |
266072_at |
AT2G18700
|
ATTPS11, trehalose phosphatase/synthase 11, ATTPSB, TPS11, trehalose phosphatase/synthase 11, TPS11, TREHALOSE-6-PHOSPHATE SYNTHASE 11 |
-0.175 |
-0.115 |
| 445 |
253122_at |
AT4G35987
|
CaM KMT, calmodulin N-methyltransferase |
-0.175 |
-0.035 |
| 446 |
251535_at |
AT3G58540
|
unknown |
-0.173 |
0.042 |
| 447 |
253908_at |
AT4G27260
|
GH3.5, WES1 |
-0.173 |
-0.291 |
| 448 |
260973_at |
AT1G53490
|
HEI10, homolog of human HEI10 ( Enhancer of cell Invasion No.10) |
-0.172 |
-0.065 |
| 449 |
261004_at |
AT1G26450
|
[Carbohydrate-binding X8 domain superfamily protein] |
-0.171 |
-0.046 |
| 450 |
265821_at |
AT2G17950
|
PGA6, WUS, WUSCHEL, WUS1, WUSCHEL 1 |
-0.171 |
0.039 |
| 451 |
244903_at |
ATMG00660
|
ORF149 |
-0.170 |
0.059 |
| 452 |
251200_at |
AT3G63010
|
ATGID1B, GID1B, GA INSENSITIVE DWARF1B |
-0.169 |
-0.154 |
| 453 |
246962_s_at |
AT5G24800
|
ATBZIP9, ARABIDOPSIS THALIANA BASIC LEUCINE ZIPPER 9, BZIP9, basic leucine zipper 9, BZO2H2, BASIC LEUCINE ZIPPER O2 HOMOLOG 2 |
-0.169 |
0.127 |
| 454 |
255080_at |
AT4G09030
|
AGP10, arabinogalactan protein 10, ATAGP10 |
-0.169 |
0.199 |
| 455 |
264342_at |
AT1G12080
|
[Vacuolar calcium-binding protein-related] |
-0.168 |
0.258 |
| 456 |
267477_at |
AT2G02710
|
PLP, PAS/LOV PROTEIN, PLPA, PAS/LOV PROTEIN A, PLPB, PAS/LOV protein B, PLPC, PAS/LOV PROTEIN C |
-0.168 |
0.008 |
| 457 |
253883_at |
AT4G27660
|
unknown |
-0.167 |
-0.001 |
| 458 |
263963_at |
AT2G36080
|
ABS2, ABNORMAL SHOOT 2, NGAL1, NGATHA-Like 1 |
-0.167 |
-0.102 |
| 459 |
251964_at |
AT3G53370
|
[S1FA-like DNA-binding protein] |
-0.167 |
-0.019 |
| 460 |
261979_at |
AT1G37130
|
ATNR2, ARABIDOPSIS NITRATE REDUCTASE 2, B29, CHL3, CHLORATE RESISTANT 3, NIA2, nitrate reductase 2, NIA2-1, NR, NITRATE REDUCTASE, NR2, NITRATE REDUCTASE 2 |
-0.167 |
0.029 |
| 461 |
254057_at |
AT4G25170
|
[Uncharacterised conserved protein (UCP012943)] |
-0.166 |
-0.328 |
| 462 |
265345_at |
AT2G22680
|
WAVH1, WAV3 homolog 1 |
-0.165 |
-0.038 |
| 463 |
266901_at |
AT2G34600
|
JAZ7, jasmonate-zim-domain protein 7, TIFY5B |
-0.165 |
0.044 |
| 464 |
266106_at |
AT2G45170
|
ATATG8E, AUTOPHAGY 8E, ATG8E, AUTOPHAGY 8E |
-0.165 |
-0.075 |
| 465 |
263681_at |
AT1G26840
|
ATORC6, ARABIDOPSIS THALIANA ORIGIN RECOGNITION COMPLEX PROTEIN 6, ORC6, origin recognition complex protein 6 |
-0.165 |
-0.011 |
| 466 |
257110_x_at |
AT3G30440
|
[similar to Ulp1 protease family protein [Arabidopsis thaliana] (TAIR:AT2G14130.1)] |
-0.164 |
-0.061 |
| 467 |
248385_at |
AT5G51910
|
[TCP family transcription factor ] |
-0.164 |
-0.101 |
| 468 |
247337_at |
AT5G63660
|
LCR74, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 74, PDF2.5 |
-0.163 |
-0.223 |
| 469 |
256577_at |
AT3G28220
|
[TRAF-like family protein] |
-0.163 |
-0.119 |
| 470 |
263482_at |
AT2G03980
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.163 |
-0.172 |
| 471 |
266032_x_at |
AT2G05890
|
unknown |
-0.162 |
0.056 |
| 472 |
247739_at |
AT5G59240
|
[Ribosomal protein S8e family protein] |
-0.162 |
0.117 |
| 473 |
246396_at |
AT1G58180
|
ATBCA6, A. THALIANA BETA CARBONIC ANHYDRASE 6, BCA6, beta carbonic anhydrase 6 |
-0.161 |
0.038 |
| 474 |
259293_at |
AT3G11580
|
[AP2/B3-like transcriptional factor family protein] |
-0.161 |
-0.072 |
| 475 |
250471_at |
AT5G10170
|
ATMIPS3, MYO-INOSITOL-1-PHOSTPATE SYNTHASE 3, MIPS3, myo-inositol-1-phosphate synthase 3 |
-0.161 |
-0.029 |
| 476 |
244957_at |
ATMG00400
|
ORF157 |
-0.160 |
0.024 |
| 477 |
247023_at |
AT5G67060
|
HEC1, HECATE 1 |
-0.160 |
-0.227 |
| 478 |
256481_at |
AT1G33430
|
[Galactosyltransferase family protein] |
-0.160 |
-0.134 |
| 479 |
245757_at |
AT1G35140
|
EXL1, EXORDIUM like 1, PHI-1, PHOSPHATE-INDUCED 1 |
-0.160 |
-0.006 |
| 480 |
260389_at |
AT1G74055
|
unknown |
-0.159 |
0.033 |
| 481 |
262129_at |
AT1G52500
|
ATFPG-1, A. THALIANA FORMAMIDOPYRIMIDINE-DNA GLYCOSYLASE 1, ATFPG-2, FORMAMIDOPYRIMIDINE-DNA GLYCOSYLASE 2, ATMMH-1, MUTM homolog-1, ATMMH-2, FPG-1, FORMAMIDOPYRIMIDINE-DNA GLYCOSYLASE 1, FPG-2, FORMAMIDOPYRIMIDINE-DNA GLYCOSYLASE 2, MMH, MUTM homolog, MMH-1, MUTM homolog-1, MMH-2 |
-0.159 |
-0.036 |
| 482 |
261727_at |
AT1G76090
|
SMT3, sterol methyltransferase 3 |
-0.159 |
0.027 |
| 483 |
259269_at |
AT3G01270
|
[Pectate lyase family protein] |
-0.158 |
-0.496 |
| 484 |
256787_at |
AT3G13790
|
ATBFRUCT1, ATCWINV1, ARABIDOPSIS THALIANA CELL WALL INVERTASE 1 |
-0.157 |
-0.040 |
| 485 |
257336_at |
ATMG01410
|
ORF204, open reading frame 204 |
-0.157 |
0.008 |
| 486 |
253842_at |
AT4G27860
|
MEB1, MEMBRANE OF ER BODY 1 |
-0.157 |
-0.066 |
| 487 |
261111_at |
AT1G75430
|
BLH11, BEL1-like homeodomain 11 |
-0.157 |
-0.510 |
| 488 |
267008_at |
AT2G39350
|
ABCG1, ATP-binding cassette G1 |
-0.156 |
-0.004 |
| 489 |
262616_at |
AT1G06620
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.156 |
0.016 |
| 490 |
246841_at |
AT5G26700
|
[RmlC-like cupins superfamily protein] |
-0.156 |
-0.540 |
| 491 |
259966_at |
AT1G76500
|
AHL29, AT-hook motif nuclear-localized protein 29, SOB3, SUPPRESSOR OF PHYB-4#3 |
-0.156 |
-0.103 |
| 492 |
260012_at |
AT1G67865
|
unknown |
-0.155 |
-0.101 |
| 493 |
250070_at |
AT5G17980
|
[C2 calcium/lipid-binding plant phosphoribosyltransferase family protein] |
-0.155 |
0.108 |
| 494 |
251252_at |
AT3G62230
|
DAF1, DUO1-activated F-box 1 |
-0.155 |
-0.524 |
| 495 |
266197_at |
AT2G39120
|
WTF9, what's this factor 9 |
-0.155 |
0.006 |
| 496 |
252057_at |
AT3G52480
|
unknown |
-0.154 |
-0.049 |
| 497 |
254175_at |
AT4G24050
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.154 |
-0.081 |
| 498 |
258402_at |
AT3G15450
|
[Aluminium induced protein with YGL and LRDR motifs] |
-0.154 |
-0.138 |
| 499 |
259502_at |
AT1G15670
|
KFB01, Kelch repeat F-box 1, KMD2, KISS ME DEADLY 2 |
-0.154 |
0.078 |
| 500 |
253117_at |
AT4G35905
|
unknown |
-0.153 |
-0.001 |
| 501 |
254120_at |
AT4G24570
|
DIC2, dicarboxylate carrier 2 |
-0.153 |
-0.125 |
| 502 |
251010_at |
AT5G02550
|
unknown |
-0.153 |
-0.114 |
| 503 |
261820_at |
AT1G11420
|
unknown |
-0.153 |
-0.012 |
| 504 |
262010_at |
AT1G35612
|
[expressed protein] |
-0.153 |
-0.038 |
| 505 |
253574_at |
AT4G31030
|
[Putative membrane lipoprotein] |
-0.153 |
0.034 |
| 506 |
263498_at |
AT2G42610
|
LSH10, LIGHT SENSITIVE HYPOCOTYLS 10 |
-0.152 |
-0.182 |
| 507 |
258167_at |
AT3G21560
|
BRT1, Bright Trichomes 1, UGT84A2, UDP-glucosyl transferase 84A2 |
-0.151 |
-0.180 |
| 508 |
248430_at |
AT5G51800
|
[Protein kinase superfamily protein] |
-0.151 |
-0.081 |
| 509 |
251035_at |
AT5G02220
|
SMR4, SIAMESE-RELATED 4 |
-0.150 |
-0.017 |
| 510 |
247176_at |
AT5G65110
|
ACX2, acyl-CoA oxidase 2, ATACX2 |
-0.150 |
-0.094 |
| 511 |
245906_at |
AT5G11070
|
unknown |
-0.150 |
0.016 |
| 512 |
245051_at |
AT2G23320
|
AtWRKY15, WRKY15, WRKY DNA-binding protein 15 |
-0.149 |
-0.037 |
| 513 |
253322_at |
AT4G33980
|
unknown |
-0.148 |
-0.002 |
| 514 |
260010_at |
AT1G68020
|
ATTPS6, TPS6, TREHALOSE -6-PHOSPHATASE SYNTHASE S6 |
-0.148 |
-0.089 |
| 515 |
257536_at |
AT3G02800
|
AtPFA-DSP3, PFA-DSP3, plant and fungi atypical dual-specificity phosphatase 3 |
-0.148 |
-0.078 |
| 516 |
250881_at |
AT5G04080
|
unknown |
-0.148 |
-0.034 |
| 517 |
262399_at |
AT1G49500
|
unknown |
-0.148 |
0.020 |
| 518 |
256159_at |
AT1G30135
|
JAZ8, jasmonate-zim-domain protein 8, TIFY5A |
-0.146 |
0.038 |
| 519 |
266693_at |
AT2G19800
|
MIOX2, myo-inositol oxygenase 2 |
-0.146 |
0.171 |
| 520 |
257380_at |
AT2G28090
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.146 |
-0.014 |
| 521 |
261265_at |
AT1G26800
|
[RING/U-box superfamily protein] |
-0.146 |
-0.105 |
| 522 |
262033_at |
AT1G37140
|
MCT1, MEI2 C-terminal RRM only like 1 |
-0.146 |
-0.162 |
| 523 |
246183_at |
AT5G20940
|
[Glycosyl hydrolase family protein] |
-0.145 |
0.067 |
| 524 |
258383_at |
AT3G15440
|
unknown |
-0.145 |
-0.025 |
| 525 |
254110_at |
AT4G25260
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.145 |
-0.207 |
| 526 |
260100_at |
AT1G73177
|
APC13, anaphase-promoting complex 13, BNS, BONSAI |
-0.145 |
-0.010 |
| 527 |
258476_at |
AT3G02400
|
[SMAD/FHA domain-containing protein ] |
-0.145 |
-0.004 |
| 528 |
248657_at |
AT5G48570
|
ATFKBP65, FKBP65, ROF2 |
-0.144 |
-0.162 |
| 529 |
263019_at |
AT1G23870
|
ATTPS9, trehalose-phosphatase/synthase 9, TPS9, TREHALOSE -6-PHOSPHATASE SYNTHASE S9, TPS9, trehalose-phosphatase/synthase 9 |
-0.143 |
-0.075 |
| 530 |
255172_x_at |
AT4G08000
|
[CACTA-like transposase family (Tnp2/En/Spm), has a 1.0e-136 P-value blast match to GB:CAA40555 TNP2 (CACTA-element) (Antirrhinum majus)] |
-0.143 |
-0.013 |
| 531 |
250941_at |
AT5G03320
|
[Protein kinase superfamily protein] |
-0.143 |
0.011 |
| 532 |
245244_at |
AT1G44350
|
ILL6, IAA-leucine resistant (ILR)-like gene 6 |
-0.143 |
0.005 |
| 533 |
253011_at |
AT4G37890
|
EDA40, embryo sac development arrest 40 |
-0.142 |
-0.232 |
| 534 |
259110_at |
AT3G05570
|
unknown |
-0.142 |
0.054 |
| 535 |
244933_at |
ATCG01070
|
NDHE |
-0.142 |
0.084 |
| 536 |
253540_at |
AT4G31615
|
[Transcriptional factor B3 family protein] |
-0.142 |
-0.168 |
| 537 |
247835_at |
AT5G57910
|
unknown |
-0.142 |
-0.239 |
| 538 |
261782_at |
AT1G76110
|
[HMG (high mobility group) box protein with ARID/BRIGHT DNA-binding domain] |
-0.142 |
-0.236 |
| 539 |
251650_at |
AT3G57360
|
unknown |
-0.141 |
0.098 |
| 540 |
250616_at |
AT5G07280
|
EMS1, EXCESS MICROSPOROCYTES1, EXS, EXTRA SPOROGENOUS CELLS |
-0.141 |
-0.072 |
| 541 |
262041_at |
AT1G80100
|
AHP6, histidine phosphotransfer protein 6, HP6, histidine phosphotransfer protein 6 |
-0.141 |
0.067 |
| 542 |
263549_at |
AT2G21650
|
ATRL2, ARABIDOPSIS RAD-LIKE 2, MEE3, MATERNAL EFFECT EMBRYO ARREST 3, RSM1, RADIALIS-LIKE SANT/MYB 1 |
-0.140 |
0.125 |
| 543 |
261109_at |
AT1G75450
|
ATCKX5, ARABIDOPSIS THALIANA CYTOKININ OXIDASE 5, ATCKX6, CYTOKININ OXIDASE 6, CKX5, cytokinin oxidase 5 |
-0.140 |
-0.185 |
| 544 |
259002_at |
AT3G02000
|
ROXY1 |
-0.139 |
-0.150 |
| 545 |
251640_at |
AT3G57450
|
unknown |
-0.139 |
-0.013 |
| 546 |
253864_at |
AT4G27460
|
CBSX5, CBS domain containing protein 5 |
-0.139 |
-0.063 |
| 547 |
266011_at |
AT2G37440
|
[DNAse I-like superfamily protein] |
-0.139 |
-0.081 |
| 548 |
252128_at |
AT3G50870
|
GATA18, GATA TRANSCRIPTION FACTOR 18, HAN, HANABA TANARU, MNP, MONOPOLE |
-0.138 |
-0.144 |
| 549 |
253243_at |
AT4G34560
|
unknown |
-0.138 |
0.126 |
| 550 |
259730_at |
AT1G77660
|
[Histone H3 K4-specific methyltransferase SET7/9 family protein] |
-0.138 |
0.086 |
| 551 |
250509_at |
AT5G09970
|
CYP78A7, cytochrome P450, family 78, subfamily A, polypeptide 7 |
-0.138 |
-0.238 |
| 552 |
259976_at |
AT1G76560
|
CP12-3, CP12 domain-containing protein 3 |
-0.137 |
-0.078 |
| 553 |
254933_at |
AT4G11130
|
RDR2, RNA-dependent RNA polymerase 2, SMD1, SILENCING MOVEMENT DEFICIENT 1 |
-0.137 |
-0.057 |
| 554 |
249332_at |
AT5G40980
|
unknown |
-0.136 |
0.199 |
| 555 |
251590_at |
AT3G57690
|
AGP23, arabinogalactan protein 23, ATAGP23, ARABINOGALACTAN-PROTEIN 23 |
-0.136 |
-0.390 |
| 556 |
259998_at |
AT1G68120
|
ATBPC3, BPC3, basic pentacysteine 3 |
-0.136 |
-0.106 |
| 557 |
252563_at |
AT3G45970
|
ATEXLA1, expansin-like A1, ATEXPL1, ATHEXP BETA 2.1, EXLA1, expansin-like A1, EXPL1, EXPANSIN L1 |
-0.136 |
0.135 |
| 558 |
249941_at |
AT5G22270
|
unknown |
-0.136 |
-0.164 |
| 559 |
249113_at |
AT5G43790
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.136 |
-0.021 |
| 560 |
247893_at |
AT5G57980
|
RPB5C, RNA polymerase II fifth largest subunit, C |
-0.136 |
-0.010 |
| 561 |
256528_at |
AT1G66140
|
ZFP4, zinc finger protein 4 |
-0.135 |
-0.110 |
| 562 |
249188_at |
AT5G42830
|
[HXXXD-type acyl-transferase family protein] |
-0.135 |
-0.002 |
| 563 |
253720_at |
AT4G29270
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
-0.135 |
0.018 |
| 564 |
259189_at |
AT3G01700
|
AGP11, arabinogalactan protein 11, ATAGP11, ARABINOGALACTAN PROTEIN 11 |
-0.135 |
-0.520 |
| 565 |
252991_at |
AT4G38470
|
STY46, serine/threonine/tyrosine kinase 46 |
-0.135 |
-0.030 |
| 566 |
259856_at |
AT1G68440
|
unknown |
-0.134 |
-0.019 |
| 567 |
247954_at |
AT5G56870
|
BGAL4, beta-galactosidase 4 |
-0.134 |
0.025 |
| 568 |
259225_at |
AT3G07590
|
[Small nuclear ribonucleoprotein family protein] |
-0.134 |
0.015 |
| 569 |
251099_at |
AT5G01660
|
unknown |
-0.133 |
-0.036 |
| 570 |
247610_at |
AT5G60630
|
unknown |
-0.133 |
-0.176 |
| 571 |
253880_at |
AT4G27590
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.133 |
0.043 |
| 572 |
260301_at |
AT1G80290
|
[Nucleotide-diphospho-sugar transferases superfamily protein] |
-0.133 |
-0.027 |
| 573 |
249606_at |
AT5G37260
|
CIR1, CIRCADIAN 1, RVE2, REVEILLE 2 |
-0.132 |
-0.211 |
| 574 |
262704_at |
AT1G16530
|
ASL9, ASYMMETRIC LEAVES 2-like 9, LBD3, LOB DOMAIN-CONTAINING PROTEIN 3 |
-0.132 |
-0.057 |
| 575 |
256230_at |
AT3G12340
|
[FKBP-like peptidyl-prolyl cis-trans isomerase family protein] |
-0.132 |
-0.006 |
| 576 |
253168_at |
AT4G35070
|
[SBP (S-ribonuclease binding protein) family protein] |
-0.132 |
-0.482 |
| 577 |
261317_at |
AT1G53030
|
[Cytochrome C oxidase copper chaperone (COX17)] |
-0.132 |
-0.012 |
| 578 |
259076_at |
AT3G02140
|
AFP4, ABI FIVE BINDING PROTEIN 4, TMAC2, TWO OR MORE ABRES-CONTAINING GENE 2 |
-0.132 |
0.032 |
| 579 |
244934_at |
ATCG01080
|
NDHG |
-0.132 |
0.002 |
| 580 |
247903_at |
AT5G57340
|
unknown |
-0.131 |
0.110 |
| 581 |
245076_at |
AT2G23170
|
GH3.3 |
-0.130 |
-0.020 |
| 582 |
258792_at |
AT3G04640
|
[glycine-rich protein] |
-0.130 |
-0.038 |
| 583 |
249732_at |
AT5G24420
|
PGL5, 6-phosphogluconolactonase 5 |
-0.130 |
-0.051 |
| 584 |
249923_at |
AT5G19120
|
[Eukaryotic aspartyl protease family protein] |
-0.130 |
0.049 |
| 585 |
254907_at |
AT4G11190
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.129 |
0.001 |
| 586 |
251745_at |
AT3G55980
|
ATSZF1, SZF1, salt-inducible zinc finger 1 |
-0.129 |
-0.039 |
| 587 |
265414_at |
AT2G16660
|
[Major facilitator superfamily protein] |
-0.129 |
0.099 |
| 588 |
259982_at |
AT1G76410
|
ATL8 |
-0.129 |
-0.068 |
| 589 |
258201_at |
AT3G13910
|
unknown |
-0.128 |
-0.081 |
| 590 |
250572_at |
AT5G08210
|
MIR834A, microRNA834A |
-0.128 |
-0.078 |
| 591 |
253762_at |
AT4G28830
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.128 |
0.017 |
| 592 |
250860_at |
AT5G04770
|
ATCAT6, ARABIDOPSIS THALIANA CATIONIC AMINO ACID TRANSPORTER 6, CAT6, cationic amino acid transporter 6 |
-0.128 |
-0.095 |
| 593 |
244909_at |
ATMG00740
|
ORF100A |
-0.128 |
-0.024 |
| 594 |
264514_at |
AT1G09500
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.127 |
-0.028 |
| 595 |
265429_at |
AT2G20710
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.127 |
0.009 |
| 596 |
255742_at |
AT1G25560
|
AtTEM1, EDF1, ETHYLENE RESPONSE DNA BINDING FACTOR 1, TEM1, TEMPRANILLO 1 |
-0.126 |
-0.026 |
| 597 |
263046_at |
AT2G05380
|
GRP3S, glycine-rich protein 3 short isoform |
-0.126 |
0.070 |
| 598 |
250064_at |
AT5G17890
|
CHS3, CHILLING SENSITIVE 3, DAR4, DA1-related protein 4 |
-0.126 |
-0.004 |
| 599 |
247772_at |
AT5G58610
|
[PHD finger transcription factor, putative] |
-0.126 |
-0.238 |
| 600 |
250327_at |
AT5G12050
|
unknown |
-0.125 |
-0.060 |