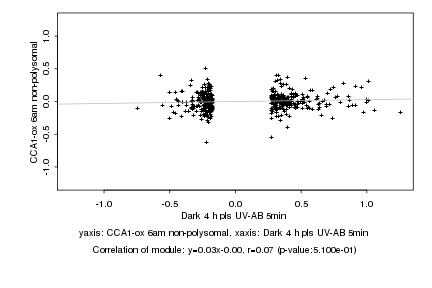
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Dark 4 h pls UV-AB 5min |
Log2 signal ratio
CCA1-ox 6am non-polysomal |
| 1 |
262517_at |
AT1G17180
|
ATGSTU25, glutathione S-transferase TAU 25, GSTU25, glutathione S-transferase TAU 25 |
1.253 |
-0.153 |
| 2 |
248434_at |
AT5G51440
|
[HSP20-like chaperones superfamily protein] |
1.052 |
-0.135 |
| 3 |
263231_at |
AT1G05680
|
UGT74E2, Uridine diphosphate glycosyltransferase 74E2 |
1.008 |
0.316 |
| 4 |
252123_at |
AT3G51240
|
F3'H, F3H, flavanone 3-hydroxylase, TT6, TRANSPARENT TESTA 6 |
1.007 |
0.023 |
| 5 |
264042_at |
AT2G03760
|
AtSOT1, AtSOT12, sulphotransferase 12, ATST1, ARABIDOPSIS THALIANA SULFOTRANSFERASE 1, AtSULT202A1, RAR047, SOT12, sulphotransferase 12, ST, ST1, SULFOTRANSFERASE 1, SULT202A1, sulfotransferase 202A1 |
0.990 |
0.033 |
| 6 |
249769_at |
AT5G24120
|
ATSIG5, SIGMA FACTOR 5, SIG5, SIGMA FACTOR 5, SIGE, sigma factor E |
0.990 |
-0.000 |
| 7 |
250207_at |
AT5G13930
|
ATCHS, CHS, CHALCONE SYNTHASE, TT4, TRANSPARENT TESTA 4 |
0.972 |
-0.158 |
| 8 |
260522_x_at |
AT2G41730
|
unknown |
0.954 |
0.225 |
| 9 |
258349_at |
AT3G17609
|
HYH, HY5-homolog |
0.913 |
-0.053 |
| 10 |
248683_at |
AT5G48490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.912 |
0.236 |
| 11 |
253943_at |
AT4G27030
|
FAD4, FATTY ACID DESATURASE 4, FADA, fatty acid desaturase A |
0.888 |
-0.057 |
| 12 |
253416_at |
AT4G33070
|
AtPDC1, PDC1, pyruvate decarboxylase 1 |
0.867 |
0.028 |
| 13 |
250533_at |
AT5G08640
|
ATFLS1, FLS, FLAVONOL SYNTHASE, FLS1, flavonol synthase 1 |
0.859 |
0.090 |
| 14 |
249125_at |
AT5G43450
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.857 |
-0.072 |
| 15 |
253046_at |
AT4G37370
|
CYP81D8, cytochrome P450, family 81, subfamily D, polypeptide 8 |
0.820 |
0.285 |
| 16 |
255298_at |
AT4G04840
|
ATMSRB6, methionine sulfoxide reductase B6, MSRB6, methionine sulfoxide reductase B6 |
0.797 |
-0.005 |
| 17 |
263402_at |
AT2G04050
|
[MATE efflux family protein] |
0.770 |
0.091 |
| 18 |
256766_at |
AT3G22231
|
PCC1, PATHOGEN AND CIRCADIAN CONTROLLED 1 |
0.758 |
0.069 |
| 19 |
251658_at |
AT3G57020
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.742 |
0.218 |
| 20 |
247780_at |
AT5G58770
|
AtcPT4, cPT4, cis-prenyltransferase 4 |
0.737 |
-0.248 |
| 21 |
263005_at |
AT1G54540
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.723 |
0.184 |
| 22 |
265668_at |
AT2G32020
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
0.711 |
-0.035 |
| 23 |
250794_at |
AT5G05270
|
AtCHIL, CHIL, chalcone isomerase like |
0.697 |
0.132 |
| 24 |
263515_at |
AT2G21640
|
unknown |
0.690 |
-0.067 |
| 25 |
250541_at |
AT5G09520
|
PELPK2, Pro-Glu-Leu|Ile|Val-Pro-Lys 2 |
0.687 |
-0.043 |
| 26 |
249063_at |
AT5G44110
|
ABCI21, ATP-binding cassette A21, ATNAP2, ARABIDOPSIS THALIANA NON-INTRINSIC ABC PROTEIN, ATPOP1, POP1 |
0.673 |
0.022 |
| 27 |
245560_at |
AT4G15480
|
UGT84A1 |
0.656 |
0.008 |
| 28 |
266578_at |
AT2G23910
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.652 |
-0.212 |
| 29 |
266097_at |
AT2G37970
|
AtHBP2, HBP2, haem-binding protein 2, SOUL-1 |
0.634 |
-0.084 |
| 30 |
251895_at |
AT3G54420
|
ATCHITIV, CHITINASE CLASS IV, ATEP3, homolog of carrot EP3-3 chitinase, CHIV, EP3, homolog of carrot EP3-3 chitinase |
0.633 |
-0.021 |
| 31 |
248049_at |
AT5G56090
|
COX15, cytochrome c oxidase 15 |
0.627 |
-0.041 |
| 32 |
258167_at |
AT3G21560
|
BRT1, Bright Trichomes 1, UGT84A2, UDP-glucosyl transferase 84A2 |
0.625 |
-0.114 |
| 33 |
251020_at |
AT5G02270
|
ABCI20, ATP-binding cassette I20, ATNAP9, ARABIDOPSIS THALIANA NON-INTRINSIC ABC PROTEIN 9, NAP9, non-intrinsic ABC protein 9 |
0.622 |
0.084 |
| 34 |
260072_at |
AT1G73650
|
[FUNCTIONS IN: oxidoreductase activity, acting on the CH-CH group of donors] |
0.620 |
0.057 |
| 35 |
246468_at |
AT5G17050
|
UGT78D2, UDP-glucosyl transferase 78D2 |
0.614 |
0.042 |
| 36 |
261192_at |
AT1G32870
|
ANAC013, Arabidopsis NAC domain containing protein 13, ANAC13, NAC domain protein 13, NAC13, NAC domain protein 13 |
0.586 |
0.179 |
| 37 |
248684_at |
AT5G48485
|
DIR1, DEFECTIVE IN INDUCED RESISTANCE 1 |
0.581 |
-0.114 |
| 38 |
258321_at |
AT3G22840
|
ELIP, ELIP1, EARLY LIGHT-INDUCABLE PROTEIN |
0.570 |
0.183 |
| 39 |
245306_at |
AT4G14690
|
ELIP2, EARLY LIGHT-INDUCIBLE PROTEIN 2 |
0.559 |
-0.104 |
| 40 |
258609_at |
AT3G02910
|
[AIG2-like (avirulence induced gene) family protein] |
0.557 |
0.001 |
| 41 |
261958_at |
AT1G64500
|
[Glutaredoxin family protein] |
0.553 |
-0.082 |
| 42 |
248625_at |
AT5G48880
|
KAT5, 3-KETO-ACYL-COENZYME A THIOLASE 5, PKT1, PEROXISOMAL-3-KETO-ACYL-COA THIOLASE 1, PKT2, peroxisomal 3-keto-acyl-CoA thiolase 2 |
0.545 |
0.075 |
| 43 |
250230_at |
AT5G13900
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.542 |
-0.098 |
| 44 |
250429_at |
AT5G10470
|
KAC1, kinesin like protein for actin based chloroplast movement 1, KCA1, KINESIN CDKA;1 ASSOCIATED 1 |
0.541 |
0.092 |
| 45 |
260877_at |
AT1G21500
|
unknown |
0.537 |
-0.051 |
| 46 |
248028_at |
AT5G55620
|
unknown |
0.526 |
0.363 |
| 47 |
256751_at |
AT3G27170
|
ATCLC-B, CLC-B, chloride channel B |
0.521 |
0.017 |
| 48 |
265290_at |
AT2G22590
|
[UDP-Glycosyltransferase superfamily protein] |
0.521 |
0.024 |
| 49 |
264415_at |
AT1G43160
|
RAP2.6, related to AP2 6 |
0.519 |
-0.003 |
| 50 |
257033_at |
AT3G19170
|
ATPREP1, presequence protease 1, ATZNMP, PREP1, presequence protease 1 |
0.514 |
0.068 |
| 51 |
248686_at |
AT5G48540
|
[receptor-like protein kinase-related family protein] |
0.510 |
-0.029 |
| 52 |
266368_at |
AT2G41380
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.509 |
0.029 |
| 53 |
251727_at |
AT3G56290
|
unknown |
0.507 |
0.015 |
| 54 |
260955_at |
AT1G06000
|
[UDP-Glycosyltransferase superfamily protein] |
0.507 |
-0.017 |
| 55 |
261449_at |
AT1G21120
|
IGMT2, indole glucosinolate O-methyltransferase 2 |
0.504 |
-0.103 |
| 56 |
245126_at |
AT2G47460
|
ATMYB12, MYB DOMAIN PROTEIN 12, MYB12, myb domain protein 12, PFG1, PRODUCTION OF FLAVONOL GLYCOSIDES 1 |
0.504 |
0.043 |
| 57 |
254201_at |
AT4G24130
|
unknown |
0.503 |
0.120 |
| 58 |
257876_at |
AT3G17130
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.489 |
0.003 |
| 59 |
262473_at |
AT1G50250
|
FTSH1, FTSH protease 1 |
0.479 |
-0.030 |
| 60 |
265611_at |
AT2G25510
|
unknown |
0.470 |
0.007 |
| 61 |
259460_at |
AT1G44000
|
unknown |
0.468 |
-0.080 |
| 62 |
252131_at |
AT3G50930
|
BCS1, cytochrome BC1 synthesis |
0.467 |
-0.043 |
| 63 |
253039_at |
AT4G37760
|
SQE3, squalene epoxidase 3 |
0.466 |
0.105 |
| 64 |
262518_at |
AT1G17170
|
ATGSTU24, glutathione S-transferase TAU 24, GST, Arabidopsis thaliana Glutathione S-transferase (class tau) 24, GSTU24, glutathione S-transferase TAU 24 |
0.461 |
-0.087 |
| 65 |
251432_at |
AT3G59820
|
AtLETM1, LETM1, leucine zipper-EF-hand-containing transmembrane protein 1 |
0.458 |
0.105 |
| 66 |
263787_at |
AT2G46420
|
unknown |
0.456 |
-0.096 |
| 67 |
265846_at |
AT2G35770
|
scpl28, serine carboxypeptidase-like 28 |
0.456 |
0.092 |
| 68 |
248492_at |
AT5G51040
|
SDHAF2, Succinate dehydrogenase assembly factor 2 |
0.455 |
-0.044 |
| 69 |
252209_at |
AT3G50400
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.455 |
0.133 |
| 70 |
255764_at |
AT1G16720
|
HCF173, high chlorophyll fluorescence phenotype 173 |
0.455 |
-0.082 |
| 71 |
262626_at |
AT1G06430
|
FTSH8, FTSH protease 8 |
0.445 |
-0.088 |
| 72 |
262609_at |
AT1G13930
|
unknown |
0.443 |
-0.043 |
| 73 |
251814_at |
AT3G54890
|
LHCA1, photosystem I light harvesting complex gene 1 |
0.437 |
-0.016 |
| 74 |
259282_at |
AT3G11430
|
ATGPAT5, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 5, GPAT5, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 5 |
0.437 |
0.120 |
| 75 |
249798_at |
AT5G23730
|
EFO2, EARLY FLOWERING BY OVEREXPRESSION 2, RUP2, REPRESSOR OF UV-B PHOTOMORPHOGENESIS 2 |
0.435 |
0.069 |
| 76 |
250420_at |
AT5G11260
|
HY5, ELONGATED HYPOCOTYL 5, TED 5, REVERSAL OF THE DET PHENOTYPE 5 |
0.432 |
0.184 |
| 77 |
253790_at |
AT4G28660
|
PSB28, photosystem II reaction center PSB28 protein |
0.426 |
0.003 |
| 78 |
247252_at |
AT5G64770
|
CLEL 9, CLE-like 9, GLV2, GOLVEN 2, RGF9, root meristem growth factor 9 |
0.425 |
0.123 |
| 79 |
256497_at |
AT1G31580
|
CXC750, ECS1 |
0.424 |
-0.047 |
| 80 |
255793_at |
AT2G33250
|
unknown |
0.424 |
0.122 |
| 81 |
261488_at |
AT1G14345
|
[NAD(P)-linked oxidoreductase superfamily protein] |
0.424 |
0.032 |
| 82 |
251668_at |
AT3G57010
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.423 |
0.078 |
| 83 |
264452_at |
AT1G10270
|
GRP23, glutamine-rich protein 23 |
0.415 |
0.015 |
| 84 |
245113_at |
AT2G41660
|
MIZ1, mizu-kussei 1 |
0.408 |
-0.022 |
| 85 |
253247_at |
AT4G34610
|
BLH6, BEL1-like homeodomain 6 |
0.408 |
-0.221 |
| 86 |
259478_at |
AT1G18980
|
[RmlC-like cupins superfamily protein] |
0.406 |
-0.044 |
| 87 |
246584_at |
AT5G14730
|
unknown |
0.406 |
-0.078 |
| 88 |
266743_at |
AT2G02990
|
ATRNS1, RIBONUCLEASE 1, RNS1, ribonuclease 1 |
0.405 |
-0.038 |
| 89 |
263128_at |
AT1G78600
|
BBX22, B-box domain protein 22, DBB3, DOUBLE B-BOX 3, LZF1, light-regulated zinc finger protein 1, STH3, SALT TOLERANCE HOMOLOG 3 |
0.405 |
-0.085 |
| 90 |
256245_at |
AT3G12580
|
ATHSP70, ARABIDOPSIS HEAT SHOCK PROTEIN 70, HSP70, heat shock protein 70 |
0.404 |
-0.018 |
| 91 |
262645_at |
AT1G62750
|
ATSCO1, SNOWY COTYLEDON 1, ATSCO1/CPEF-G, SCO1, SNOWY COTYLEDON 1 |
0.404 |
0.200 |
| 92 |
246966_at |
AT5G24850
|
CRY3, cryptochrome 3 |
0.403 |
0.068 |
| 93 |
262945_at |
AT1G79510
|
[Uncharacterized conserved protein (DUF2358)] |
0.399 |
-0.039 |
| 94 |
263252_at |
AT2G31380
|
BBX25, B-box domain protein 25, STH, salt tolerance homologue |
0.398 |
0.022 |
| 95 |
249622_at |
AT5G37550
|
unknown |
0.394 |
-0.037 |
| 96 |
251824_at |
AT3G55090
|
ABCG16, ATP-binding cassette G16 |
0.394 |
-0.382 |
| 97 |
253013_at |
AT4G37910
|
mtHsc70-1, mitochondrial heat shock protein 70-1 |
0.394 |
0.008 |
| 98 |
256309_at |
AT1G30380
|
PSAK, photosystem I subunit K |
0.394 |
0.064 |
| 99 |
255248_at |
AT4G05180
|
PSBQ, PHOTOSYSTEM II SUBUNIT Q, PSBQ-2, photosystem II subunit Q-2, PSII-Q |
0.392 |
-0.006 |
| 100 |
260601_at |
AT1G55910
|
ZIP11, zinc transporter 11 precursor |
0.392 |
0.372 |
| 101 |
257058_at |
AT3G15352
|
ATCOX17, ARABIDOPSIS THALIANA CYTOCHROME C OXIDASE 17, COX17, cytochrome c oxidase 17 |
0.390 |
-0.048 |
| 102 |
263483_at |
AT2G04030
|
AtHsp90.5, HEAT SHOCK PROTEIN 90.5, AtHsp90C, CR88, EMB1956, EMBRYO DEFECTIVE 1956, Hsp88.1, HEAT SHOCK PROTEIN 88.1, HSP90.5, HEAT SHOCK PROTEIN 90.5 |
0.388 |
-0.011 |
| 103 |
264186_at |
AT1G54570
|
PES1, phytyl ester synthase 1 |
0.386 |
-0.033 |
| 104 |
256979_at |
AT3G21055
|
PSBTN, photosystem II subunit T |
0.385 |
-0.052 |
| 105 |
266237_at |
AT2G29540
|
ATRPAC14, ATRPC14, RNApolymerase 14 kDa subunit, RPC14, RNApolymerase 14 kDa subunit |
0.385 |
-0.109 |
| 106 |
259840_at |
AT1G52230
|
PSAH-2, PHOTOSYSTEM I SUBUNIT H-2, PSAH2, photosystem I subunit H2, PSI-H |
0.385 |
-0.046 |
| 107 |
265886_at |
AT2G25620
|
AtDBP1, DNA-binding protein phosphatase 1, DBP1, DNA-binding protein phosphatase 1 |
0.384 |
-0.185 |
| 108 |
258452_at |
AT3G22370
|
AOX1A, alternative oxidase 1A, ATAOX1A, AtHSR3, HSR3, hyper-sensitivity-related 3 |
0.383 |
0.051 |
| 109 |
264436_at |
AT1G10370
|
ATGSTU17, GLUTATHIONE S-TRANSFERASE TAU 17, ERD9, EARLY-RESPONSIVE TO DEHYDRATION 9, GST30, GLUTATHIONE S-TRANSFERASE 30, GST30B, GLUTATHIONE S-TRANSFERASE 30B, GSTU17, GLUTATHIONE S-TRANSFERASE U17 |
0.382 |
0.274 |
| 110 |
266514_at |
AT2G47890
|
[B-box type zinc finger protein with CCT domain] |
0.377 |
0.017 |
| 111 |
254241_at |
AT4G23190
|
AT-RLK3, RECEPTOR LIKE PROTEIN KINASE 3, CRK11, cysteine-rich RLK (RECEPTOR-like protein kinase) 11 |
0.376 |
0.045 |
| 112 |
253009_at |
AT4G37930
|
SHM1, serine transhydroxymethyltransferase 1, SHMT1, SERINE HYDROXYMETHYLTRANSFERASE 1, STM, SERINE TRANSHYDROXYMETHYLTRANSFERASE |
0.374 |
-0.003 |
| 113 |
262970_at |
AT1G75690
|
LQY1, LOW QUANTUM YIELD OF PHOTOSYSTEM II 1 |
0.372 |
0.007 |
| 114 |
261907_at |
AT1G65060
|
4CL3, 4-coumarate:CoA ligase 3 |
0.372 |
-0.144 |
| 115 |
261746_at |
AT1G08380
|
PSAO, photosystem I subunit O |
0.367 |
-0.021 |
| 116 |
249645_at |
AT5G36910
|
THI2.2, thionin 2.2 |
0.365 |
-0.112 |
| 117 |
250563_at |
AT5G08050
|
unknown |
0.365 |
0.093 |
| 118 |
250239_at |
AT5G13580
|
ABCG6, ATP-binding cassette G6 |
0.364 |
0.098 |
| 119 |
249289_at |
AT5G41040
|
ASFT, aliphatic suberin feruloyl-transferase, HHT, hydroxycinnamoyl- CoA:ω-hydroxyacid O-hydroxycinnamoyltransferase, RWP1, REDUCED LEVELS OF WALL-BOUND PHENOLICS 1 |
0.362 |
0.273 |
| 120 |
250502_at |
AT5G09590
|
HSC70-5, HEAT SHOCK COGNATE, MTHSC70-2, mitochondrial HSO70 2 |
0.361 |
0.066 |
| 121 |
255594_at |
AT4G01660
|
ABC1, ABC transporter 1, ATABC1, ABC transporter 1, ATATH10 |
0.361 |
0.047 |
| 122 |
263841_at |
AT2G36870
|
AtXTH32, XTH32, xyloglucan endotransglucosylase/hydrolase 32 |
0.360 |
0.061 |
| 123 |
257149_at |
AT3G27280
|
ATPHB4, prohibitin 4, PHB4, prohibitin 4 |
0.360 |
0.089 |
| 124 |
249677_at |
AT5G35970
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.358 |
0.018 |
| 125 |
249244_at |
AT5G42270
|
FTSH5, VAR1, VARIEGATED 1 |
0.358 |
-0.034 |
| 126 |
267364_at |
AT2G40080
|
ELF4, EARLY FLOWERING 4 |
0.358 |
-0.005 |
| 127 |
255457_at |
AT4G02770
|
PSAD-1, photosystem I subunit D-1 |
0.355 |
-0.068 |
| 128 |
246203_at |
AT4G36610
|
[alpha/beta-Hydrolases superfamily protein] |
0.354 |
-0.060 |
| 129 |
253387_at |
AT4G33010
|
AtGLDP1, glycine decarboxylase P-protein 1, GLDP1, glycine decarboxylase P-protein 1 |
0.353 |
-0.012 |
| 130 |
255046_at |
AT4G09650
|
ATPD, ATP synthase delta-subunit gene, PDE332, PIGMENT DEFECTIVE 332 |
0.351 |
-0.101 |
| 131 |
257252_at |
AT3G24170
|
ATGR1, glutathione-disulfide reductase, GR1, glutathione-disulfide reductase |
0.351 |
-0.007 |
| 132 |
251309_at |
AT3G61220
|
SDR1, short-chain dehydrogenase/reductase 1 |
0.350 |
0.230 |
| 133 |
258607_at |
AT3G02730
|
ATF1, TRXF1, thioredoxin F-type 1 |
0.350 |
-0.200 |
| 134 |
251827_at |
AT3G55120
|
A11, AtCHI, CFI, CHALCONE FLAVANONE ISOMERASE, CHI, chalcone isomerase, TT5, TRANSPARENT TESTA 5 |
0.349 |
-0.032 |
| 135 |
263403_at |
AT2G04040
|
ATDTX1, DTX1, detoxification 1, TX1 |
0.348 |
-0.032 |
| 136 |
267076_at |
AT2G41090
|
[Calcium-binding EF-hand family protein] |
0.347 |
0.002 |
| 137 |
248151_at |
AT5G54270
|
LHCB3, light-harvesting chlorophyll B-binding protein 3, LHCB3*1 |
0.347 |
-0.039 |
| 138 |
253197_at |
AT4G35250
|
HCF244, high chlorophyll fluorescence phenotype 244 |
0.344 |
0.122 |
| 139 |
252365_at |
AT3G48350
|
CEP3, cysteine endopeptidase 3 |
0.344 |
0.092 |
| 140 |
248138_at |
AT5G54960
|
PDC2, pyruvate decarboxylase-2 |
0.343 |
0.113 |
| 141 |
267070_at |
AT2G41000
|
[Chaperone DnaJ-domain superfamily protein] |
0.342 |
-0.093 |
| 142 |
254447_at |
AT4G20860
|
[FAD-binding Berberine family protein] |
0.341 |
-0.106 |
| 143 |
253738_at |
AT4G28750
|
PSAE-1, PSA E1 KNOCKOUT |
0.341 |
0.285 |
| 144 |
255440_at |
AT4G02530
|
[chloroplast thylakoid lumen protein] |
0.338 |
0.072 |
| 145 |
258094_at |
AT3G14690
|
CYP72A15, cytochrome P450, family 72, subfamily A, polypeptide 15 |
0.338 |
-0.287 |
| 146 |
245445_at |
AT4G16750
|
[Integrase-type DNA-binding superfamily protein] |
0.337 |
0.024 |
| 147 |
248101_at |
AT5G55200
|
MGE1, mitochondrial GrpE 1 |
0.337 |
0.348 |
| 148 |
255877_at |
AT2G40460
|
[Major facilitator superfamily protein] |
0.336 |
-0.019 |
| 149 |
253777_at |
AT4G28450
|
[nucleotide binding] |
0.333 |
-0.106 |
| 150 |
253841_at |
AT4G27830
|
AtBGLU10, BGLU10, beta glucosidase 10 |
0.332 |
0.060 |
| 151 |
259507_at |
AT1G43910
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.331 |
0.065 |
| 152 |
255259_at |
AT4G05020
|
NDB2, NAD(P)H dehydrogenase B2 |
0.331 |
-0.083 |
| 153 |
257932_at |
AT3G17040
|
HCF107, high chlorophyll fluorescent 107 |
0.331 |
-0.078 |
| 154 |
265442_at |
AT2G20940
|
unknown |
0.330 |
0.004 |
| 155 |
249918_at |
AT5G19240
|
[Glycoprotein membrane precursor GPI-anchored] |
0.330 |
-0.005 |
| 156 |
261618_at |
AT1G33110
|
[MATE efflux family protein] |
0.328 |
0.046 |
| 157 |
262533_at |
AT1G17090
|
unknown |
0.327 |
0.106 |
| 158 |
264832_at |
AT1G03660
|
[Ankyrin-repeat containing protein] |
0.324 |
0.409 |
| 159 |
246401_at |
AT1G57560
|
AtMYB50, myb domain protein 50, MYB50, myb domain protein 50 |
0.324 |
0.072 |
| 160 |
266958_at |
AT2G34630
|
GPPS, GERANYLPYROPHOSPHATE SYNTHASE, GPS1, geranyl diphosphate synthase 1 |
0.324 |
0.013 |
| 161 |
246488_at |
AT5G16010
|
[3-oxo-5-alpha-steroid 4-dehydrogenase family protein] |
0.321 |
-0.214 |
| 162 |
252661_at |
AT3G44450
|
unknown |
0.319 |
-0.013 |
| 163 |
261240_at |
AT1G32940
|
ATSBT3.5, SBT3.5 |
0.318 |
0.071 |
| 164 |
255982_at |
AT1G34000
|
OHP2, one-helix protein 2 |
0.317 |
0.029 |
| 165 |
261065_at |
AT1G07500
|
SMR5, SIAMESE-RELATED 5 |
0.316 |
-0.013 |
| 166 |
246954_at |
AT5G04830
|
[Nuclear transport factor 2 (NTF2) family protein] |
0.316 |
0.053 |
| 167 |
262736_at |
AT1G28570
|
[SGNH hydrolase-type esterase superfamily protein] |
0.315 |
0.114 |
| 168 |
252911_at |
AT4G39510
|
CYP96A12, cytochrome P450, family 96, subfamily A, polypeptide 12 |
0.314 |
-0.000 |
| 169 |
255886_at |
AT1G20340
|
DRT112, DNA-DAMAGE-REPAIR/TOLERATION PROTEIN 112, PETE2, PLASTOCYANIN 2 |
0.314 |
0.330 |
| 170 |
258179_at |
AT3G21690
|
[MATE efflux family protein] |
0.314 |
-0.121 |
| 171 |
252010_at |
AT3G52740
|
unknown |
0.312 |
-0.166 |
| 172 |
260035_at |
AT1G68850
|
[Peroxidase superfamily protein] |
0.311 |
-0.094 |
| 173 |
258696_at |
AT3G09650
|
CRM3, HCF152, HIGH CHLOROPHYLL FLUORESCENCE 152 |
0.311 |
-0.117 |
| 174 |
248607_at |
AT5G49480
|
ATCP1, Ca2+-binding protein 1, CP1, Ca2+-binding protein 1 |
0.311 |
-0.219 |
| 175 |
265569_at |
AT2G05620
|
AtPGR5, PGR5, proton gradient regulation 5 |
0.310 |
-0.004 |
| 176 |
261417_at |
AT1G07700
|
[Thioredoxin superfamily protein] |
0.307 |
0.009 |
| 177 |
262168_at |
AT1G74730
|
unknown |
0.306 |
-0.157 |
| 178 |
247779_at |
AT5G58760
|
DDB2, damaged DNA binding 2 |
0.306 |
-0.052 |
| 179 |
251713_at |
AT3G56080
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.306 |
0.001 |
| 180 |
247435_at |
AT5G62480
|
ATGSTU9, glutathione S-transferase tau 9, GST14, GLUTATHIONE S-TRANSFERASE 14, GST14B, GLUTATHIONE S-TRANSFERASE 14B, GSTU9, glutathione S-transferase tau 9 |
0.305 |
0.409 |
| 181 |
247162_at |
AT5G65730
|
XTH6, xyloglucan endotransglucosylase/hydrolase 6 |
0.304 |
-0.040 |
| 182 |
253949_at |
AT4G26780
|
AR192, MGE2, mitochondrial GrpE 2 |
0.302 |
0.046 |
| 183 |
259892_at |
AT1G72610
|
ATGER1, A. THALIANA GERMIN-LIKE PROTEIN 1, GER1, germin-like protein 1, GLP1, GERMIN-LIKE PROTEIN 1 |
0.302 |
0.065 |
| 184 |
255787_at |
AT2G33590
|
AtCRL1, CRL1, CCR(Cinnamoyl coA:NADP oxidoreductase)-like 1 |
0.302 |
-0.061 |
| 185 |
261751_at |
AT1G76080
|
ATCDSP32, ARABIDOPSIS THALIANA CHLOROPLASTIC DROUGHT-INDUCED STRESS PROTEIN OF 32 KD, CDSP32, chloroplastic drought-induced stress protein of 32 kD |
0.301 |
0.154 |
| 186 |
249770_at |
AT5G24110
|
ATWRKY30, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 30, WRKY30, WRKY DNA-binding protein 30 |
0.301 |
-0.119 |
| 187 |
267005_at |
AT2G34460
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.300 |
-0.026 |
| 188 |
258295_at |
AT3G23400
|
FIB4, fibrillin 4 |
0.300 |
-0.115 |
| 189 |
256076_at |
AT1G18060
|
unknown |
0.300 |
0.308 |
| 190 |
264207_at |
AT1G22750
|
unknown |
0.300 |
0.065 |
| 191 |
253688_at |
AT4G29590
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.299 |
0.069 |
| 192 |
253251_at |
AT4G34730
|
RBF1, RbfA domain-containing protein 1 |
0.298 |
0.097 |
| 193 |
252340_at |
AT3G48920
|
AtMYB45, myb domain protein 45, MYB45, myb domain protein 45 |
0.298 |
0.024 |
| 194 |
263401_at |
AT2G04070
|
[MATE efflux family protein] |
0.297 |
-0.076 |
| 195 |
262349_at |
AT2G48130
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.295 |
-0.082 |
| 196 |
266267_at |
AT2G29460
|
ATGSTU4, glutathione S-transferase tau 4, GST22, GLUTATHIONE S-TRANSFERASE 22, GSTU4, glutathione S-transferase tau 4 |
0.292 |
-0.064 |
| 197 |
246597_at |
AT5G14760
|
AO, L-aspartate oxidase, FIN4, FLAGELLIN-INSENSITIVE 4 |
0.291 |
0.109 |
| 198 |
246911_at |
AT5G25810
|
tny, TINY |
0.291 |
0.054 |
| 199 |
267425_at |
AT2G34810
|
[FAD-binding Berberine family protein] |
0.290 |
0.099 |
| 200 |
261794_at |
AT1G16060
|
ADAP, ARIA-interacting double AP2 domain protein, WRI3, WRINKLED 3 |
0.288 |
0.074 |
| 201 |
258589_at |
AT3G04290
|
ATLTL1, LTL1, Li-tolerant lipase 1 |
0.287 |
0.212 |
| 202 |
266510_at |
AT2G47990
|
EDA13, EMBRYO SAC DEVELOPMENT ARREST 13, EDA19, EMBRYO SAC DEVELOPMENT ARREST 19, SWA1, SLOW WALKER1 |
0.285 |
-0.044 |
| 203 |
261296_at |
AT1G48460
|
unknown |
0.283 |
0.019 |
| 204 |
264460_at |
AT1G10170
|
ATNFXL1, NF-X-like 1, NFXL1, NF-X-like 1 |
0.283 |
0.161 |
| 205 |
254398_at |
AT4G21280
|
PSBQ, PHOTOSYSTEM II SUBUNIT Q, PSBQ-1, PHOTOSYSTEM II SUBUNIT Q-1, PSBQA, photosystem II subunit QA |
0.282 |
0.027 |
| 206 |
265852_at |
AT2G42350
|
[RING/U-box superfamily protein] |
0.280 |
-0.147 |
| 207 |
256762_at |
AT3G25655
|
IDL1, inflorescence deficient in abscission (IDA)-like 1 |
0.279 |
0.031 |
| 208 |
252102_at |
AT3G50970
|
LTI30, LOW TEMPERATURE-INDUCED 30, XERO2 |
0.278 |
0.018 |
| 209 |
263533_at |
AT2G24820
|
AtTic55, translocon at the inner envelope membrane of chloroplasts 55, Tic55, translocon at the inner envelope membrane of chloroplasts 55, TIC55-II, translocon at the inner envelope membrane of chloroplasts 55-II |
0.277 |
-0.004 |
| 210 |
245501_at |
AT4G15620
|
[Uncharacterised protein family (UPF0497)] |
0.277 |
0.035 |
| 211 |
259297_at |
AT3G05360
|
AtRLP30, receptor like protein 30, RLP30, receptor like protein 30 |
0.277 |
0.156 |
| 212 |
262916_at |
AT1G59700
|
ATGSTU16, glutathione S-transferase TAU 16, GSTU16, glutathione S-transferase TAU 16 |
0.277 |
0.092 |
| 213 |
251476_at |
AT3G59670
|
unknown |
0.276 |
0.071 |
| 214 |
253922_at |
AT4G26850
|
VTC2, vitamin c defective 2 |
0.276 |
0.195 |
| 215 |
261376_at |
AT1G18660
|
[zinc finger (C3HC4-type RING finger) family protein] |
0.275 |
0.046 |
| 216 |
251029_at |
AT5G02050
|
[Mitochondrial glycoprotein family protein] |
0.275 |
-0.111 |
| 217 |
260036_at |
AT1G68830
|
STN7, STT7 homolog STN7 |
0.273 |
0.048 |
| 218 |
251091_at |
AT5G01410
|
ATPDX1, ATPDX1.3, ARABIDOPSIS THALIANA PYRIDOXINE BIOSYNTHESIS 1.3, PDX1, PDX1.3, PYRIDOXINE BIOSYNTHESIS 1.3, RSR4, REDUCED SUGAR RESPONSE 4 |
0.273 |
0.071 |
| 219 |
259040_at |
AT3G09270
|
ATGSTU8, glutathione S-transferase TAU 8, GSTU8, glutathione S-transferase TAU 8 |
0.272 |
-0.175 |
| 220 |
251282_at |
AT3G61630
|
CRF6, cytokinin response factor 6 |
0.271 |
-0.018 |
| 221 |
257311_at |
AT3G26570
|
ORF02, PHT2;1, phosphate transporter 2;1 |
0.271 |
-0.130 |
| 222 |
257262_at |
AT3G21890
|
BBX31, B-box domain protein 31 |
0.269 |
-0.015 |
| 223 |
254790_at |
AT4G12800
|
PSAL, photosystem I subunit l |
0.269 |
-0.253 |
| 224 |
259392_at |
AT1G06380
|
[Ribosomal protein L1p/L10e family] |
0.269 |
0.078 |
| 225 |
263553_at |
AT2G16430
|
ATPAP10, PAP10, purple acid phosphatase 10 |
0.269 |
-0.545 |
| 226 |
262557_at |
AT1G31330
|
PSAF, photosystem I subunit F |
0.268 |
0.193 |
| 227 |
261981_at |
AT1G33811
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.268 |
0.095 |
| 228 |
258679_at |
AT3G08590
|
iPGAM2, 2,3-biphosphoglycerate-independent phosphoglycerate mutase 2 |
0.267 |
-0.104 |
| 229 |
251974_at |
AT3G53200
|
AtMYB27, myb domain protein 27, MYB27, myb domain protein 27 |
-0.750 |
-0.106 |
| 230 |
259799_at |
AT1G72290
|
[Kunitz family trypsin and protease inhibitor protein] |
-0.575 |
0.398 |
| 231 |
249174_at |
AT5G42900
|
COR27, cold regulated gene 27 |
-0.559 |
-0.049 |
| 232 |
256266_at |
AT3G12320
|
LNK3, night light-inducible and clock-regulated 3 |
-0.507 |
0.147 |
| 233 |
248819_at |
AT5G47050
|
[SBP (S-ribonuclease binding protein) family protein] |
-0.502 |
-0.251 |
| 234 |
254644_at |
AT4G18510
|
CLE2, CLAVATA3/ESR-related 2 |
-0.493 |
-0.066 |
| 235 |
257670_at |
AT3G20340
|
unknown |
-0.474 |
-0.153 |
| 236 |
255905_at |
AT1G17810
|
BETA-TIP, beta-tonoplast intrinsic protein |
-0.461 |
0.139 |
| 237 |
262198_at |
AT1G53830
|
ATPME2, pectin methylesterase 2, PME2, pectin methylesterase 2 |
-0.457 |
-0.178 |
| 238 |
251012_at |
AT5G02580
|
unknown |
-0.449 |
0.035 |
| 239 |
266393_at |
AT2G41260
|
ATM17, M17 |
-0.445 |
0.017 |
| 240 |
250907_at |
AT5G03670
|
TRM28, TON1 Recruiting Motif 28 |
-0.436 |
-0.059 |
| 241 |
248395_at |
AT5G52120
|
AtPP2-A14, phloem protein 2-A14, PP2-A14, phloem protein 2-A14 |
-0.434 |
0.008 |
| 242 |
253322_at |
AT4G33980
|
unknown |
-0.418 |
0.159 |
| 243 |
245233_at |
AT4G25580
|
[CAP160 protein] |
-0.412 |
-0.216 |
| 244 |
250214_at |
AT5G13870
|
EXGT-A4, endoxyloglucan transferase A4, XTH5, xyloglucan endotransglucosylase/hydrolase 5 |
-0.406 |
-0.004 |
| 245 |
250158_at |
AT5G15190
|
unknown |
-0.405 |
-0.002 |
| 246 |
252054_at |
AT3G52540
|
ATOFP18, ARABIDOPSIS THALIANA OVATE FAMILY PROTEIN 18, OFP18, ovate family protein 18 |
-0.403 |
0.160 |
| 247 |
266974_at |
AT2G39370
|
MAKR4, MEMBRANE-ASSOCIATED KINASE REGULATOR 4 |
-0.387 |
-0.151 |
| 248 |
258338_at |
AT3G16150
|
ASPGB1, asparaginase B1 |
-0.379 |
-0.012 |
| 249 |
262105_at |
AT1G02810
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.376 |
-0.056 |
| 250 |
248435_at |
AT5G51210
|
OLEO3, oleosin3 |
-0.364 |
-0.144 |
| 251 |
264403_at |
AT2G25150
|
[HXXXD-type acyl-transferase family protein] |
-0.360 |
-0.146 |
| 252 |
266526_at |
AT2G16980
|
[Major facilitator superfamily protein] |
-0.343 |
0.254 |
| 253 |
260668_at |
AT1G19530
|
unknown |
-0.341 |
0.007 |
| 254 |
264527_at |
AT1G30760
|
[FAD-binding Berberine family protein] |
-0.340 |
-0.079 |
| 255 |
254300_at |
AT4G22780
|
ACR7, ACT domain repeat 7 |
-0.339 |
-0.094 |
| 256 |
264598_at |
AT1G04610
|
YUC3, YUCCA 3 |
-0.338 |
0.323 |
| 257 |
264524_at |
AT1G10070
|
ATBCAT-2, branched-chain amino acid transaminase 2, BCAT-2, branched-chain amino acid transaminase 2, BCAT2, branched-chain amino acid transferase 2 |
-0.333 |
0.011 |
| 258 |
258395_at |
AT3G15500
|
ANAC055, NAC domain containing protein 55, ATNAC3, NAC domain containing protein 3, NAC055, NAC domain containing protein 55, NAC3, NAC domain containing protein 3 |
-0.330 |
0.028 |
| 259 |
261423_at |
AT1G18750
|
AGL65, AGAMOUS-like 65 |
-0.328 |
0.015 |
| 260 |
259616_at |
AT1G47960
|
ATC/VIF1, CELL WALL / VACUOLAR INHIBITOR OF FRUCTOSIDASE 1, C/VIF1, cell wall / vacuolar inhibitor of fructosidase 1 |
-0.328 |
-0.044 |
| 261 |
253667_at |
AT4G30170
|
[Peroxidase family protein] |
-0.327 |
-0.121 |
| 262 |
249824_at |
AT5G23380
|
unknown |
-0.327 |
0.014 |
| 263 |
261778_at |
AT1G76220
|
unknown |
-0.327 |
0.069 |
| 264 |
245970_at |
AT5G20710
|
BGAL7, beta-galactosidase 7 |
-0.324 |
-0.213 |
| 265 |
253522_at |
AT4G31290
|
AtGGCT2;2, GGCT2;2, gamma-glutamyl cyclotransferase 2;2 |
-0.323 |
-0.062 |
| 266 |
245513_at |
AT4G15780
|
ATVAMP724, vesicle-associated membrane protein 724, VAMP724, vesicle-associated membrane protein 724 |
-0.322 |
-0.093 |
| 267 |
266901_at |
AT2G34600
|
JAZ7, jasmonate-zim-domain protein 7, TIFY5B |
-0.313 |
-0.070 |
| 268 |
261558_at |
AT1G01770
|
unknown |
-0.300 |
-0.148 |
| 269 |
266546_at |
AT2G35270
|
AHL21, AT-hook motif nuclear localized protein 21, GIK, GIANT KILLER |
-0.299 |
0.140 |
| 270 |
260774_at |
AT1G78290
|
SNRK2-8, SNF1-RELATED PROTEIN KINASE 2-8, SNRK2.8, SNF1-RELATED PROTEIN KINASE 2.8, SRK2C, SNF1-RELATED PROTEIN KINASE 2C |
-0.297 |
-0.067 |
| 271 |
248969_at |
AT5G45310
|
unknown |
-0.294 |
0.019 |
| 272 |
261814_at |
AT1G08310
|
[alpha/beta-Hydrolases superfamily protein] |
-0.293 |
0.149 |
| 273 |
247047_at |
AT5G66650
|
unknown |
-0.292 |
-0.073 |
| 274 |
260741_at |
AT1G15040
|
GAT, glutamine amidotransferase, GAT1_2.1, glutamine amidotransferase 1_2.1 |
-0.287 |
0.131 |
| 275 |
246978_at |
AT5G24910
|
CYP714A1, cytochrome P450, family 714, subfamily A, polypeptide 1, ELA1, EUI-like p450 A1 |
-0.285 |
-0.049 |
| 276 |
267035_at |
AT2G38400
|
AGT3, alanine:glyoxylate aminotransferase 3 |
-0.283 |
0.072 |
| 277 |
249454_at |
AT5G39520
|
unknown |
-0.281 |
0.035 |
| 278 |
253827_at |
AT4G28085
|
unknown |
-0.279 |
-0.071 |
| 279 |
258080_at |
AT3G25930
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
-0.272 |
-0.012 |
| 280 |
254648_at |
AT4G18550
|
AtDSEL, Arabidopsis thaliana DAD1-like seeding establishment-related lipase, DSEL, DAD1-like seeding establishment-related lipase |
-0.271 |
0.101 |
| 281 |
248486_at |
AT5G51060
|
ATRBOHC, A. THALIANA RESPIRATORY BURST OXIDASE HOMOLOG C, RBOHC, RESPIRATORY BURST OXIDASE HOMOLOG C, RHD2, ROOT HAIR DEFECTIVE 2 |
-0.269 |
0.136 |
| 282 |
267265_at |
AT2G22980
|
SCPL13, serine carboxypeptidase-like 13 |
-0.268 |
0.001 |
| 283 |
248793_at |
AT5G47240
|
atnudt8, nudix hydrolase homolog 8, NUDT8, nudix hydrolase homolog 8 |
-0.268 |
-0.201 |
| 284 |
261272_at |
AT1G26665
|
[Mediator complex, subunit Med10] |
-0.267 |
0.144 |
| 285 |
250905_at |
AT5G03640
|
[Protein kinase superfamily protein] |
-0.265 |
-0.266 |
| 286 |
250032_at |
AT5G18170
|
GDH1, glutamate dehydrogenase 1 |
-0.265 |
-0.115 |
| 287 |
248869_at |
AT5G46840
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.263 |
-0.008 |
| 288 |
262971_at |
AT1G75640
|
[Leucine-rich receptor-like protein kinase family protein] |
-0.262 |
0.122 |
| 289 |
260662_at |
AT1G19540
|
[NmrA-like negative transcriptional regulator family protein] |
-0.261 |
0.147 |
| 290 |
247878_at |
AT5G57760
|
unknown |
-0.259 |
-0.138 |
| 291 |
246390_at |
AT1G77330
|
ACO5, ACC oxidase 5 |
-0.258 |
0.227 |
| 292 |
256712_at |
AT2G34020
|
[Calcium-binding EF-hand family protein] |
-0.253 |
0.128 |
| 293 |
256965_at |
AT3G13450
|
DIN4, DARK INDUCIBLE 4 |
-0.253 |
0.155 |
| 294 |
263118_at |
AT1G03090
|
MCCA |
-0.253 |
0.117 |
| 295 |
250665_at |
AT5G06980
|
LNK4, night light-inducible and clock-regulated 4 |
-0.252 |
0.067 |
| 296 |
246952_at |
AT5G04820
|
ATOFP13, ARABIDOPSIS THALIANA OVATE FAMILY PROTEIN 13, OFP13, ovate family protein 13 |
-0.251 |
0.058 |
| 297 |
249768_at |
AT5G24100
|
[Leucine-rich repeat protein kinase family protein] |
-0.249 |
-0.112 |
| 298 |
255007_at |
AT4G10020
|
AtHSD5, hydroxysteroid dehydrogenase 5, HSD5, hydroxysteroid dehydrogenase 5 |
-0.248 |
-0.106 |
| 299 |
260693_at |
AT1G32450
|
AtNPF7.3, NPF7.3, NRT1/ PTR family 7.3, NRT1.5, nitrate transporter 1.5 |
-0.248 |
-0.188 |
| 300 |
261032_at |
AT1G17430
|
[alpha/beta-Hydrolases superfamily protein] |
-0.247 |
0.059 |
| 301 |
266581_at |
AT2G46140
|
[Late embryogenesis abundant protein] |
-0.247 |
-0.132 |
| 302 |
265276_at |
AT2G28400
|
unknown |
-0.246 |
0.140 |
| 303 |
253279_at |
AT4G34030
|
MCCB, 3-methylcrotonyl-CoA carboxylase |
-0.246 |
-0.093 |
| 304 |
247324_at |
AT5G64190
|
unknown |
-0.245 |
0.078 |
| 305 |
263985_at |
AT2G42750
|
DJC77, DNA J protein C77 |
-0.244 |
-0.059 |
| 306 |
262667_at |
AT1G62810
|
CuAO1, COPPER AMINE OXIDASE1 |
-0.241 |
0.005 |
| 307 |
248167_at |
AT5G54530
|
unknown |
-0.240 |
-0.214 |
| 308 |
246506_at |
AT5G16110
|
unknown |
-0.239 |
0.108 |
| 309 |
252991_at |
AT4G38470
|
STY46, serine/threonine/tyrosine kinase 46 |
-0.239 |
-0.042 |
| 310 |
254580_at |
AT4G19390
|
[Uncharacterised protein family (UPF0114)] |
-0.238 |
-0.072 |
| 311 |
247425_at |
AT5G62550
|
unknown |
-0.238 |
0.022 |
| 312 |
248193_at |
AT5G54080
|
AtHGO, HGO, homogentisate 1,2-dioxygenase |
-0.237 |
0.074 |
| 313 |
264647_at |
AT1G09090
|
ATRBOHB, respiratory burst oxidase homolog B, ATRBOHB-BETA, RBOHB, respiratory burst oxidase homolog B |
-0.237 |
-0.036 |
| 314 |
262505_at |
AT1G21680
|
[DPP6 N-terminal domain-like protein] |
-0.237 |
0.263 |
| 315 |
264902_at |
AT1G23060
|
MDP40, MICROTUBULE DESTABILIZING PROTEIN 40 |
-0.237 |
-0.092 |
| 316 |
261658_at |
AT1G50040
|
unknown |
-0.235 |
-0.000 |
| 317 |
246913_at |
AT5G25830
|
GATA12, GATA transcription factor 12 |
-0.235 |
0.164 |
| 318 |
253804_at |
AT4G28230
|
unknown |
-0.234 |
0.155 |
| 319 |
249626_at |
AT5G37540
|
[Eukaryotic aspartyl protease family protein] |
-0.234 |
0.047 |
| 320 |
251013_at |
AT5G02540
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.234 |
0.510 |
| 321 |
266656_at |
AT2G25900
|
ATCTH, ATTZF1, A. THALIANA TANDEM ZINC FINGER PROTEIN 1 |
-0.233 |
-0.013 |
| 322 |
264658_at |
AT1G09910
|
[Rhamnogalacturonate lyase family protein] |
-0.233 |
0.040 |
| 323 |
259962_at |
AT1G53690
|
[DNA directed RNA polymerase, 7 kDa subunit] |
-0.232 |
-0.213 |
| 324 |
261382_at |
AT1G05470
|
CVP2, COTYLEDON VASCULAR PATTERN 2 |
-0.232 |
-0.143 |
| 325 |
258527_at |
AT3G06850
|
BCE2, DIN3, DARK INDUCIBLE 3, LTA1 |
-0.232 |
-0.065 |
| 326 |
257975_at |
AT3G20830
|
AGC2-4, AGC2 kinase 4, UCNL, UNICORN-like |
-0.232 |
0.008 |
| 327 |
267388_at |
AT2G44450
|
BGLU15, beta glucosidase 15 |
-0.230 |
-0.178 |
| 328 |
266783_at |
AT2G29130
|
ATLAC2, LAC2, laccase 2 |
-0.230 |
-0.093 |
| 329 |
248744_at |
AT5G48250
|
BBX8, B-box domain protein 8 |
-0.229 |
-0.060 |
| 330 |
253434_at |
AT4G32500
|
AKT5, K+ transporter 5, KT5, K+ transporter 5 |
-0.228 |
0.220 |
| 331 |
245258_at |
AT4G15340
|
04C11, ATPEN1, pentacyclic triterpene synthase 1, PEN1, pentacyclic triterpene synthase 1 |
-0.228 |
-0.132 |
| 332 |
264933_at |
AT1G61160
|
unknown |
-0.228 |
-0.091 |
| 333 |
252740_at |
AT3G43270
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.227 |
-0.141 |
| 334 |
264697_at |
AT1G70210
|
ATCYCD1;1, CYCD1;1, CYCLIN D1;1 |
-0.227 |
0.033 |
| 335 |
253958_at |
AT4G26400
|
[RING/U-box superfamily protein] |
-0.226 |
0.013 |
| 336 |
261224_at |
AT1G20160
|
ATSBT5.2 |
-0.224 |
-0.031 |
| 337 |
254571_at |
AT4G19370
|
unknown |
-0.223 |
-0.033 |
| 338 |
255381_at |
AT4G03510
|
ATRMA1, RMA1, RING membrane-anchor 1 |
-0.222 |
0.118 |
| 339 |
245528_at |
AT4G15530
|
PPDK, pyruvate orthophosphate dikinase |
-0.222 |
0.131 |
| 340 |
267337_at |
AT2G39980
|
[HXXXD-type acyl-transferase family protein] |
-0.222 |
0.023 |
| 341 |
262452_at |
AT1G11210
|
unknown |
-0.222 |
-0.029 |
| 342 |
262512_at |
AT1G17145
|
[RING/U-box superfamily protein] |
-0.221 |
-0.617 |
| 343 |
251289_at |
AT3G61830
|
ARF18, auxin response factor 18 |
-0.221 |
-0.027 |
| 344 |
249606_at |
AT5G37260
|
CIR1, CIRCADIAN 1, RVE2, REVEILLE 2 |
-0.221 |
-0.209 |
| 345 |
264561_at |
AT1G55810
|
UKL3, uridine kinase-like 3 |
-0.220 |
0.015 |
| 346 |
263894_at |
AT2G21910
|
CYP96A5, cytochrome P450, family 96, subfamily A, polypeptide 5 |
-0.220 |
0.038 |
| 347 |
250649_at |
AT5G06690
|
WCRKC1, WCRKC thioredoxin 1 |
-0.220 |
-0.199 |
| 348 |
265817_at |
AT2G18050
|
HIS1-3, histone H1-3 |
-0.219 |
0.087 |
| 349 |
260483_at |
AT1G11000
|
ATMLO4, MILDEW RESISTANCE LOCUS O 4, MLO4, MILDEW RESISTANCE LOCUS O 4 |
-0.219 |
-0.022 |
| 350 |
263738_at |
AT1G60060
|
[Serine/threonine-protein kinase WNK (With No Lysine)-related] |
-0.219 |
0.339 |
| 351 |
262635_at |
AT1G06570
|
HPD, 4-hydroxyphenylpyruvate dioxygenase, HPPD, 4-hydroxyphenylpyruvate dioxygenase, PDS1, phytoene desaturation 1 |
-0.218 |
-0.005 |
| 352 |
255625_at |
AT4G01120
|
ATBZIP54, BASIC REGION/LEUCINE ZIPPER MOTIF 5, GBF2, G-box binding factor 2 |
-0.218 |
-0.064 |
| 353 |
264597_at |
AT1G04620
|
HCAR, 7-hydroxymethyl chlorophyll a (HMChl) reductase |
-0.218 |
-0.039 |
| 354 |
252855_at |
AT4G39660
|
AGT2, alanine:glyoxylate aminotransferase 2 |
-0.217 |
-0.057 |
| 355 |
261211_at |
AT1G12780
|
ATUGE1, A. THALIANA UDP-GLC 4-EPIMERASE 1, UGE1, UDP-D-glucose/UDP-D-galactose 4-epimerase 1 |
-0.217 |
0.121 |
| 356 |
259076_at |
AT3G02140
|
AFP4, ABI FIVE BINDING PROTEIN 4, TMAC2, TWO OR MORE ABRES-CONTAINING GENE 2 |
-0.217 |
0.215 |
| 357 |
261567_at |
AT1G33055
|
unknown |
-0.216 |
-0.054 |
| 358 |
248050_at |
AT5G56100
|
[glycine-rich protein / oleosin] |
-0.216 |
-0.041 |
| 359 |
245463_at |
AT4G17030
|
AT-EXPR, ATEXLB1, expansin-like B1, ATEXPR1, ATHEXP BETA 3.1, EXLB1, expansin-like B1, EXPR |
-0.216 |
-0.158 |
| 360 |
257163_at |
AT3G24310
|
ATMYB71, MYB DOMAIN PROTEIN 71, MYB305, myb domain protein 305 |
-0.215 |
0.069 |
| 361 |
246436_at |
AT5G17440
|
[LUC7 related protein] |
-0.214 |
-0.064 |
| 362 |
263373_at |
AT2G20515
|
unknown |
-0.214 |
0.034 |
| 363 |
254233_at |
AT4G23800
|
3xHMG-box2, 3xHigh Mobility Group-box2 |
-0.214 |
-0.061 |
| 364 |
245382_at |
AT4G17800
|
AHL23, AT-hook motif nuclear localized protein 23 |
-0.214 |
0.075 |
| 365 |
261757_at |
AT1G08210
|
[Eukaryotic aspartyl protease family protein] |
-0.213 |
-0.284 |
| 366 |
258610_at |
AT3G02875
|
ILR1, IAA-LEUCINE RESISTANT 1 |
-0.213 |
-0.043 |
| 367 |
252833_at |
AT4G40090
|
AGP3, arabinogalactan protein 3 |
-0.212 |
-0.080 |
| 368 |
251272_at |
AT3G61890
|
ATHB-12, homeobox 12, ATHB12, ARABIDOPSIS THALIANA HOMEOBOX 12, HB-12, homeobox 12 |
-0.212 |
-0.068 |
| 369 |
250992_at |
AT5G02260
|
ATEXP9, ATEXPA9, expansin A9, ATHEXP ALPHA 1.10, EXP9, EXPA9, expansin A9 |
-0.211 |
0.127 |
| 370 |
261826_at |
AT1G11580
|
ATPMEPCRA, PMEPCRA, methylesterase PCR A |
-0.211 |
-0.237 |
| 371 |
253373_at |
AT4G33150
|
LKR, LKR/SDH, LYSINE-KETOGLUTARATE REDUCTASE/SACCHAROPINE DEHYDROGENASE, SDH, SACCHAROPINE DEHYDROGENASE |
-0.210 |
-0.122 |
| 372 |
263732_at |
AT1G59980
|
ARL2, ARG1-like 2, ATDJC39, GPS4, gravity persistence signal 4 |
-0.210 |
0.278 |
| 373 |
264405_at |
AT1G10210
|
ATMPK1, mitogen-activated protein kinase 1, MPK1, mitogen-activated protein kinase 1 |
-0.210 |
0.033 |
| 374 |
262275_at |
AT1G68710
|
[ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein] |
-0.210 |
-0.047 |
| 375 |
262264_at |
AT1G42470
|
[Patched family protein] |
-0.210 |
0.019 |
| 376 |
261559_at |
AT1G01780
|
PLIM2b, PLIM2b |
-0.209 |
0.164 |
| 377 |
266309_at |
AT2G27140
|
[HSP20-like chaperones superfamily protein] |
-0.209 |
-0.089 |
| 378 |
253684_at |
AT4G29690
|
[Alkaline-phosphatase-like family protein] |
-0.208 |
0.023 |
| 379 |
267101_at |
AT2G41480
|
AtPRX25, PRX25, peroxidase 25 |
-0.208 |
0.187 |
| 380 |
267077_at |
AT2G40970
|
MYBC1 |
-0.208 |
-0.086 |
| 381 |
261250_at |
AT1G05890
|
ARI5, ARIADNE 5, ATARI5, ARABIDOPSIS ARIADNE 5 |
-0.207 |
-0.315 |
| 382 |
260411_at |
AT1G69890
|
unknown |
-0.207 |
0.071 |
| 383 |
254431_at |
AT4G20840
|
[FAD-binding Berberine family protein] |
-0.207 |
0.064 |
| 384 |
253519_at |
AT4G31240
|
AtNRX2, NRX2, nucleoredoxin 2 |
-0.207 |
0.180 |
| 385 |
263275_at |
AT2G14170
|
ALDH6B2, aldehyde dehydrogenase 6B2 |
-0.206 |
-0.030 |
| 386 |
252077_at |
AT3G51720
|
unknown |
-0.205 |
0.251 |
| 387 |
259834_at |
AT1G69570
|
[Dof-type zinc finger DNA-binding family protein] |
-0.205 |
0.018 |
| 388 |
259977_at |
AT1G76590
|
[PLATZ transcription factor family protein] |
-0.204 |
-0.144 |
| 389 |
265070_at |
AT1G55510
|
BCDH BETA1, branched-chain alpha-keto acid decarboxylase E1 beta subunit |
-0.204 |
0.231 |
| 390 |
258795_at |
AT3G04570
|
AHL19, AT-hook motif nuclear-localized protein 19 |
-0.204 |
0.035 |
| 391 |
258648_at |
AT3G07900
|
[O-fucosyltransferase family protein] |
-0.204 |
-0.129 |
| 392 |
252236_at |
AT3G49930
|
[C2H2 and C2HC zinc fingers superfamily protein] |
-0.203 |
-0.077 |
| 393 |
248160_at |
AT5G54470
|
BBX29, B-box domain protein 29 |
-0.203 |
-0.129 |
| 394 |
246951_at |
AT5G04880
|
[expressed protein] |
-0.202 |
0.271 |
| 395 |
262003_at |
AT1G64460
|
[Protein kinase superfamily protein] |
-0.202 |
-0.039 |
| 396 |
260284_at |
AT1G80380
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.202 |
-0.090 |
| 397 |
260361_at |
AT1G69360
|
unknown |
-0.201 |
0.082 |
| 398 |
254754_at |
AT4G13210
|
[Pectin lyase-like superfamily protein] |
-0.200 |
-0.156 |
| 399 |
254949_at |
AT4G11020
|
unknown |
-0.200 |
0.077 |
| 400 |
247979_at |
AT5G56750
|
NDL1, N-MYC downregulated-like 1 |
-0.200 |
-0.123 |
| 401 |
258445_at |
AT3G22400
|
ATLOX5, Arabidopsis thaliana lipoxygenase 5, LOX5 |
-0.199 |
0.047 |
| 402 |
264045_at |
AT2G22450
|
AtRIBA2, RIBA2, homolog of ribA 2 |
-0.199 |
0.039 |
| 403 |
248257_at |
AT5G53410
|
unknown |
-0.199 |
-0.094 |
| 404 |
258141_at |
AT3G18035
|
HON4 |
-0.199 |
-0.138 |
| 405 |
252570_at |
AT3G45300
|
ATIVD, IVD, isovaleryl-CoA-dehydrogenase, IVDH, ISOVALERYL-COA-DEHYDROGENASE |
-0.199 |
0.206 |
| 406 |
252937_at |
AT4G39180
|
ATSEC14, ARABIDOPSIS THALIANA SECRETION 14, SEC14, SECRETION 14 |
-0.199 |
-0.002 |
| 407 |
246311_at |
AT3G51880
|
AtHMGB1, HMGB1, high mobility group B1, NFD1, NUCLEOSOME/CHROMATIN ASSEMBLY FACTOR D1 |
-0.198 |
0.004 |
| 408 |
259058_at |
AT3G03470
|
CYP89A9, cytochrome P450, family 87, subfamily A, polypeptide 9 |
-0.197 |
-0.034 |
| 409 |
252415_at |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
-0.197 |
-0.011 |
| 410 |
253874_at |
AT4G27450
|
[Aluminium induced protein with YGL and LRDR motifs] |
-0.196 |
0.211 |
| 411 |
267075_at |
AT2G41070
|
ATBZIP12, DPBF4, EEL, ENHANCED EM LEVEL |
-0.196 |
-0.020 |
| 412 |
260991_at |
AT1G12160
|
[Flavin-binding monooxygenase family protein] |
-0.196 |
-0.025 |
| 413 |
255265_at |
AT4G05190
|
ATK5, kinesin 5 |
-0.196 |
-0.253 |
| 414 |
253161_at |
AT4G35770
|
ATSEN1, ARABIDOPSIS THALIANA SENESCENCE 1, DIN1, DARK INDUCIBLE 1, SEN1, SENESCENCE 1, SEN1, SENESCENCE ASSOCIATED GENE 1 |
-0.195 |
0.178 |
| 415 |
247924_at |
AT5G57655
|
[xylose isomerase family protein] |
-0.194 |
-0.090 |
| 416 |
262204_at |
AT2G01100
|
unknown |
-0.194 |
-0.001 |
| 417 |
263295_at |
AT2G14210
|
AGL44, AGAMOUS-like 44, ANR1, ARABIDOPSIS NITRATE REGULATED 1 |
-0.194 |
-0.134 |
| 418 |
251100_at |
AT5G01670
|
[NAD(P)-linked oxidoreductase superfamily protein] |
-0.193 |
0.001 |
| 419 |
252872_at |
AT4G40010
|
SNRK2-7, SUCROSE NONFERMENTING 1-RELATED PROTEIN KINASE 2-7, SNRK2.7, SNF1-related protein kinase 2.7, SRK2F |
-0.193 |
0.085 |
| 420 |
247348_at |
AT5G63810
|
AtBGAL10, Arabidopsis thaliana beta-galactosidase 10, BGAL10, beta-galactosidase 10 |
-0.193 |
-0.164 |
| 421 |
261253_at |
AT1G05840
|
[Eukaryotic aspartyl protease family protein] |
-0.193 |
-0.113 |
| 422 |
261205_at |
AT1G12790
|
unknown |
-0.192 |
-0.051 |
| 423 |
247356_at |
AT5G63800
|
BGAL6, beta-galactosidase 6, MUM2, MUCILAGE-MODIFIED 2 |
-0.191 |
-0.186 |
| 424 |
257197_at |
AT3G23800
|
SBP3, selenium-binding protein 3 |
-0.191 |
0.121 |
| 425 |
259169_at |
AT3G03520
|
NPC3, non-specific phospholipase C3 |
-0.191 |
-0.110 |
| 426 |
260317_at |
AT1G63800
|
UBC5, ubiquitin-conjugating enzyme 5 |
-0.190 |
-0.113 |
| 427 |
250769_at |
AT5G05680
|
EMB2789, EMBRYO DEFECTIVE 2789, MOS7, MODIFIER OF SNC1,7 |
-0.190 |
0.122 |
| 428 |
253882_at |
AT4G27650
|
PEL1, PELOTA |
-0.189 |
-0.124 |
| 429 |
251026_at |
AT5G02200
|
FHL, far-red-elongated hypocotyl1-like |
-0.189 |
-0.076 |
| 430 |
253980_at |
AT4G26620
|
[Sucrase/ferredoxin-like family protein] |
-0.188 |
0.057 |
| 431 |
260097_at |
AT1G73220
|
AtOCT1, organic cation/carnitine transporter1, OCT1, organic cation/carnitine transporter1 |
-0.188 |
-0.140 |
| 432 |
260332_at |
AT1G70470
|
unknown |
-0.188 |
-0.078 |
| 433 |
249944_at |
AT5G22290
|
anac089, NAC domain containing protein 89, FSQ6, fructose-sensing quantitative trait locus 6, NAC089, NAC domain containing protein 89 |
-0.187 |
-0.122 |
| 434 |
267386_at |
AT2G44430
|
[DNA-binding bromodomain-containing protein] |
-0.187 |
0.042 |
| 435 |
261581_at |
AT1G01140
|
CIPK9, CBL-interacting protein kinase 9, PKS6, PROTEIN KINASE 6, SnRK3.12, SNF1-RELATED PROTEIN KINASE 3.12 |
-0.186 |
-0.030 |
| 436 |
246396_at |
AT1G58180
|
ATBCA6, A. THALIANA BETA CARBONIC ANHYDRASE 6, BCA6, beta carbonic anhydrase 6 |
-0.186 |
0.114 |
| 437 |
266428_at |
AT2G07180
|
[Protein kinase superfamily protein] |
-0.186 |
0.238 |
| 438 |
251403_at |
AT3G60300
|
[RWD domain-containing protein] |
-0.185 |
0.116 |
| 439 |
245668_at |
AT1G28330
|
AtDRM1, DRM1, DORMANCY-ASSOCIATED PROTEIN 1, DYL1, dormancy-associated protein-like 1 |
-0.185 |
-0.015 |
| 440 |
247603_at |
AT5G60930
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.184 |
-0.058 |
| 441 |
251870_at |
AT3G54510
|
[Early-responsive to dehydration stress protein (ERD4)] |
-0.184 |
-0.059 |
| 442 |
258347_at |
AT3G17520
|
[Late embryogenesis abundant protein (LEA) family protein] |
-0.184 |
-0.117 |
| 443 |
250228_at |
AT5G13840
|
CCS52B, cell cycle switch protein 52 B, FZR3, FIZZY-related 3 |
-0.183 |
-0.217 |
| 444 |
253864_at |
AT4G27460
|
CBSX5, CBS domain containing protein 5 |
-0.183 |
-0.099 |
| 445 |
260635_at |
AT1G62422
|
unknown |
-0.183 |
0.001 |
| 446 |
245174_at |
AT2G47500
|
[P-loop nucleoside triphosphate hydrolases superfamily protein with CH (Calponin Homology) domain] |
-0.182 |
0.100 |
| 447 |
265680_at |
AT2G32150
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.181 |
-0.081 |
| 448 |
264466_at |
AT1G10380
|
[Putative membrane lipoprotein] |
-0.181 |
-0.008 |
| 449 |
247696_at |
AT5G59780
|
ATMYB59, MYB DOMAIN PROTEIN 59, ATMYB59-1, ATMYB59-2, ATMYB59-3, MYB59, myb domain protein 59 |
-0.181 |
-0.070 |
| 450 |
259978_at |
AT1G76540
|
CDKB2;1, cyclin-dependent kinase B2;1 |
-0.181 |
0.058 |
| 451 |
265573_at |
AT2G28200
|
[C2H2-type zinc finger family protein] |
-0.180 |
-0.009 |
| 452 |
245645_at |
AT1G24764
|
ATMAP70-2, microtubule-associated proteins 70-2, MAP70-2, microtubule-associated proteins 70-2 |
-0.180 |
-0.092 |
| 453 |
261780_at |
AT1G76310
|
CYCB2;4, CYCLIN B2;4 |
-0.180 |
0.025 |
| 454 |
253483_at |
AT4G31910
|
BAT1, BR-related AcylTransferase1, PIZ, PIZZA |
-0.180 |
-0.112 |
| 455 |
266828_at |
AT2G22930
|
[UDP-Glycosyltransferase superfamily protein] |
-0.179 |
0.032 |