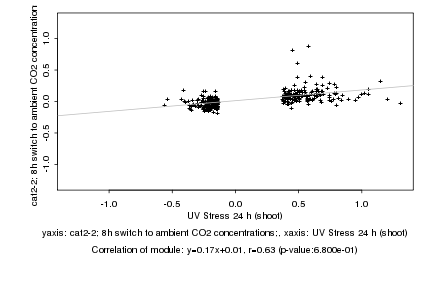
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
UV Stress 24 h (shoot) |
Log2 signal ratio
cat2-2; 8h switch to ambient CO2 concentrations; |
| 1 |
266070_at |
AT2G18660
|
AtPNP-A, PNP-A, plant natriuretic peptide A |
1.303 |
-0.030 |
| 2 |
248322_at |
AT5G52760
|
[Copper transport protein family] |
1.198 |
0.039 |
| 3 |
260116_at |
AT1G33960
|
AIG1, AVRRPT2-INDUCED GENE 1 |
1.146 |
0.328 |
| 4 |
251625_at |
AT3G57260
|
AtBG2, AtPR2, BG2, BETA-1,3-GLUCANASE 2, BGL2, beta-1,3-glucanase 2, PR-2, PATHOGENESIS-RELATED PROTEIN 2, PR2, PATHOGENESIS-RELATED PROTEIN 2 |
1.050 |
0.194 |
| 5 |
250445_at |
AT5G10760
|
[Eukaryotic aspartyl protease family protein] |
1.047 |
0.127 |
| 6 |
257623_at |
AT3G26210
|
CYP71B23, cytochrome P450, family 71, subfamily B, polypeptide 23 |
1.016 |
0.142 |
| 7 |
266385_at |
AT2G14610
|
ATPR1, PATHOGENESIS-RELATED GENE 1, PR 1, PATHOGENESIS-RELATED GENE 1, PR1, pathogenesis-related gene 1 |
0.996 |
0.118 |
| 8 |
259272_at |
AT3G01290
|
AtHIR2, HIR2, hypersensitive induced reaction 2 |
0.971 |
0.075 |
| 9 |
265837_at |
AT2G14560
|
LURP1, LATE UPREGULATED IN RESPONSE TO HYALOPERONOSPORA PARASITICA |
0.945 |
0.025 |
| 10 |
257365_x_at |
AT2G26020
|
PDF1.2b, plant defensin 1.2b |
0.891 |
0.043 |
| 11 |
263539_at |
AT2G24850
|
TAT, TYROSINE AMINOTRANSFERASE, TAT3, tyrosine aminotransferase 3 |
0.842 |
0.107 |
| 12 |
248327_at |
AT5G52750
|
[Heavy metal transport/detoxification superfamily protein ] |
0.839 |
0.030 |
| 13 |
249417_at |
AT5G39670
|
[Calcium-binding EF-hand family protein] |
0.815 |
0.056 |
| 14 |
260568_at |
AT2G43570
|
CHI, chitinase, putative |
0.797 |
0.137 |
| 15 |
256593_at |
AT3G28510
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.797 |
0.223 |
| 16 |
266578_at |
AT2G23910
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.796 |
-0.051 |
| 17 |
254869_at |
AT4G11890
|
ARCK1, ABA- AND OSMOTIC-STRESS-INDUCIBLE RECEPTOR-LIKE CYTOSOLIC KINASE1, CRK45, cysteine-rich receptor-like protein kinase 45 |
0.785 |
0.126 |
| 18 |
266292_at |
AT2G29350
|
SAG13, senescence-associated gene 13 |
0.779 |
0.278 |
| 19 |
252136_at |
AT3G50770
|
CML41, calmodulin-like 41 |
0.778 |
0.134 |
| 20 |
245329_at |
AT4G14365
|
XBAT34, XB3 ortholog 4 in Arabidopsis thaliana |
0.770 |
0.041 |
| 21 |
257382_at |
AT2G40750
|
ATWRKY54, WRKY DNA-BINDING PROTEIN 54, WRKY54, WRKY DNA-binding protein 54 |
0.756 |
0.002 |
| 22 |
263584_at |
AT2G17040
|
anac036, NAC domain containing protein 36, NAC036, NAC domain containing protein 36 |
0.744 |
0.029 |
| 23 |
257100_at |
AT3G25010
|
AtRLP41, receptor like protein 41, RLP41, receptor like protein 41 |
0.742 |
0.300 |
| 24 |
252345_at |
AT3G48640
|
unknown |
0.742 |
0.110 |
| 25 |
252417_at |
AT3G47480
|
[Calcium-binding EF-hand family protein] |
0.741 |
0.065 |
| 26 |
258203_at |
AT3G13950
|
unknown |
0.728 |
0.207 |
| 27 |
259550_at |
AT1G35230
|
AGP5, arabinogalactan protein 5 |
0.693 |
0.119 |
| 28 |
264434_at |
AT1G10340
|
[Ankyrin repeat family protein] |
0.687 |
0.163 |
| 29 |
254975_at |
AT4G10500
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.686 |
0.390 |
| 30 |
267546_at |
AT2G32680
|
AtRLP23, receptor like protein 23, RLP23, receptor like protein 23 |
0.686 |
0.259 |
| 31 |
259385_at |
AT1G13470
|
unknown |
0.679 |
0.166 |
| 32 |
246293_at |
AT3G56710
|
SIB1, sigma factor binding protein 1 |
0.676 |
-0.004 |
| 33 |
248062_at |
AT5G55450
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.668 |
0.024 |
| 34 |
246099_at |
AT5G20230
|
ATBCB, blue-copper-binding protein, BCB, BLUE COPPER BINDING PROTEIN, BCB, blue-copper-binding protein, SAG14, SENESCENCE ASSOCIATED GENE 14 |
0.667 |
0.098 |
| 35 |
265067_at |
AT1G03850
|
ATGRXS13, glutaredoxin 13, GRXS13, glutaredoxin 13 |
0.664 |
0.118 |
| 36 |
255406_at |
AT4G03450
|
[Ankyrin repeat family protein] |
0.657 |
0.174 |
| 37 |
261339_at |
AT1G35710
|
[Protein kinase family protein with leucine-rich repeat domain] |
0.646 |
0.197 |
| 38 |
263536_at |
AT2G25000
|
ATWRKY60, WRKY60, WRKY DNA-binding protein 60 |
0.644 |
0.080 |
| 39 |
246831_at |
AT5G26340
|
ATSTP13, SUGAR TRANSPORT PROTEIN 13, MSS1, STP13, SUGAR TRANSPORT PROTEIN 13 |
0.640 |
0.093 |
| 40 |
255341_at |
AT4G04500
|
CRK37, cysteine-rich RLK (RECEPTOR-like protein kinase) 37 |
0.640 |
0.173 |
| 41 |
258277_at |
AT3G26830
|
CYP71B15, PAD3, PHYTOALEXIN DEFICIENT 3 |
0.638 |
0.200 |
| 42 |
256596_at |
AT3G28540
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.638 |
0.272 |
| 43 |
252098_at |
AT3G51330
|
[Eukaryotic aspartyl protease family protein] |
0.638 |
0.142 |
| 44 |
245052_at |
AT2G26440
|
PME12, pectin methylesterase 12 |
0.623 |
0.054 |
| 45 |
259507_at |
AT1G43910
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.621 |
0.110 |
| 46 |
258791_at |
AT3G04720
|
AtPR4, HEL, HEVEIN-LIKE, PR-4, PR4, pathogenesis-related 4 |
0.609 |
0.028 |
| 47 |
247604_at |
AT5G60950
|
COBL5, COBRA-like protein 5 precursor |
0.606 |
0.047 |
| 48 |
254500_at |
AT4G20110
|
BP80-3;1, binding protein of 80 kDa 3;1, VSR3;1, VACUOLAR SORTING RECEPTOR 3;1, VSR7, VACUOLAR SORTING RECEPTOR 7 |
0.602 |
0.171 |
| 49 |
245038_at |
AT2G26560
|
PLA IIA, PHOSPHOLIPASE A 2A, PLA2A, phospholipase A 2A, PLAII alpha, PLP2, PATATIN-LIKE PROTEIN 2, PLP2, PHOSPHOLIPASE A 2A |
0.598 |
0.126 |
| 50 |
246405_at |
AT1G57630
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
0.595 |
0.151 |
| 51 |
252117_at |
AT3G51430
|
SSL5, STRICTOSIDINE SYNTHASE-LIKE 5, YLS2, YELLOW-LEAF-SPECIFIC GENE 2 |
0.590 |
0.039 |
| 52 |
256252_at |
AT3G11340
|
UGT76B1, UDP-dependent glycosyltransferase 76B1 |
0.590 |
0.413 |
| 53 |
264635_at |
AT1G65500
|
unknown |
0.579 |
0.048 |
| 54 |
253485_at |
AT4G31800
|
ATWRKY18, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 18, WRKY18, WRKY DNA-binding protein 18 |
0.579 |
0.110 |
| 55 |
254387_at |
AT4G21850
|
ATMSRB9, methionine sulfoxide reductase B9, MSRB9, methionine sulfoxide reductase B9 |
0.575 |
-0.045 |
| 56 |
250351_at |
AT5G12030
|
AT-HSP17.6A, heat shock protein 17.6A, HSP17.6, HEAT SHOCK PROTEIN 17.6, HSP17.6A, heat shock protein 17.6A |
0.572 |
0.887 |
| 57 |
250062_at |
AT5G17760
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.570 |
0.080 |
| 58 |
247602_at |
AT5G60900
|
RLK1, receptor-like protein kinase 1 |
0.569 |
0.010 |
| 59 |
266761_at |
AT2G47130
|
AtSDR3, SDR3, short-chain dehydrogenase/reductase 2 |
0.569 |
0.039 |
| 60 |
252060_at |
AT3G52430
|
ATPAD4, ARABIDOPSIS PHYTOALEXIN DEFICIENT 4, PAD4, PHYTOALEXIN DEFICIENT 4 |
0.566 |
0.052 |
| 61 |
252076_at |
AT3G51660
|
[Tautomerase/MIF superfamily protein] |
0.563 |
0.026 |
| 62 |
259561_at |
AT1G21250
|
AtWAK1, PRO25, WAK1, cell wall-associated kinase 1 |
0.560 |
0.109 |
| 63 |
267246_at |
AT2G30250
|
ATWRKY25, WRKY25, WRKY DNA-binding protein 25 |
0.555 |
0.089 |
| 64 |
262381_at |
AT1G72900
|
[Toll-Interleukin-Resistance (TIR) domain-containing protein] |
0.553 |
0.159 |
| 65 |
252403_at |
AT3G48080
|
[alpha/beta-Hydrolases superfamily protein] |
0.553 |
0.303 |
| 66 |
259559_at |
AT1G21240
|
WAK3, wall associated kinase 3 |
0.541 |
0.234 |
| 67 |
263783_at |
AT2G46400
|
ATWRKY46, WRKY DNA-BINDING PROTEIN 46, WRKY46, WRKY DNA-binding protein 46 |
0.541 |
0.109 |
| 68 |
248970_at |
AT5G45380
|
ATDUR3, DUR3, DEGRADATION OF UREA 3 |
0.540 |
0.186 |
| 69 |
266017_at |
AT2G18690
|
unknown |
0.530 |
0.110 |
| 70 |
256787_at |
AT3G13790
|
ATBFRUCT1, ATCWINV1, ARABIDOPSIS THALIANA CELL WALL INVERTASE 1 |
0.528 |
0.095 |
| 71 |
267385_at |
AT2G44380
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.525 |
0.144 |
| 72 |
259489_at |
AT1G15790
|
unknown |
0.522 |
0.094 |
| 73 |
248932_at |
AT5G46050
|
AtNPF5.2, ATPTR3, ARABIDOPSIS THALIANA PEPTIDE TRANSPORTER 3, NPF5.2, NRT1/ PTR family 5.2, PTR3, peptide transporter 3 |
0.517 |
0.188 |
| 74 |
249983_at |
AT5G18470
|
[Curculin-like (mannose-binding) lectin family protein] |
0.506 |
0.122 |
| 75 |
249719_at |
AT5G35735
|
[Auxin-responsive family protein] |
0.506 |
0.002 |
| 76 |
249346_at |
AT5G40780
|
LHT1, lysine histidine transporter 1 |
0.505 |
0.092 |
| 77 |
246821_at |
AT5G26920
|
CBP60G, Cam-binding protein 60-like G |
0.505 |
0.158 |
| 78 |
259410_at |
AT1G13340
|
[Regulator of Vps4 activity in the MVB pathway protein] |
0.503 |
0.056 |
| 79 |
267083_at |
AT2G41100
|
ATCAL4, ARABIDOPSIS THALIANA CALMODULIN LIKE 4, TCH3, TOUCH 3 |
0.502 |
0.046 |
| 80 |
249481_at |
AT5G38900
|
[Thioredoxin superfamily protein] |
0.502 |
0.161 |
| 81 |
267567_at |
AT2G30770
|
CYP71A13, cytochrome P450, family 71, subfamily A, polypeptide 13 |
0.499 |
0.161 |
| 82 |
254255_at |
AT4G23220
|
CRK14, cysteine-rich RLK (RECEPTOR-like protein kinase) 14 |
0.497 |
0.057 |
| 83 |
248330_at |
AT5G52810
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.491 |
0.108 |
| 84 |
252515_at |
AT3G46230
|
ATHSP17.4, ARABIDOPSIS THALIANA HEAT SHOCK PROTEIN 17.4, HSP17.4, heat shock protein 17.4 |
0.490 |
0.616 |
| 85 |
252131_at |
AT3G50930
|
BCS1, cytochrome BC1 synthesis |
0.487 |
0.188 |
| 86 |
256100_at |
AT1G13750
|
[Purple acid phosphatases superfamily protein] |
0.486 |
0.090 |
| 87 |
246368_at |
AT1G51890
|
[Leucine-rich repeat protein kinase family protein] |
0.486 |
0.384 |
| 88 |
254093_at |
AT4G25110
|
AtMC2, metacaspase 2, AtMCP1c, metacaspase 1c, MC2, metacaspase 2, MCP1c, metacaspase 1c |
0.485 |
0.113 |
| 89 |
248551_at |
AT5G50200
|
ATNRT3.1, NRT3.1, NITRATE TRANSPORTER 3.1, WR3, WOUND-RESPONSIVE 3 |
0.484 |
0.105 |
| 90 |
265917_at |
AT2G15080
|
AtRLP19, receptor like protein 19, RLP19, receptor like protein 19 |
0.484 |
0.154 |
| 91 |
245035_at |
AT2G26400
|
ARD, ACIREDUCTONE DIOXYGENASE, ARD3, acireductone dioxygenase 3, ATARD3, acireductone dioxygenase 3 |
0.481 |
0.018 |
| 92 |
259992_at |
AT1G67970
|
AT-HSFA8, HSFA8, heat shock transcription factor A8 |
0.479 |
0.004 |
| 93 |
259841_at |
AT1G52200
|
[PLAC8 family protein] |
0.475 |
0.109 |
| 94 |
257061_at |
AT3G18250
|
[Putative membrane lipoprotein] |
0.474 |
0.106 |
| 95 |
256877_at |
AT3G26470
|
[Powdery mildew resistance protein, RPW8 domain] |
0.472 |
0.059 |
| 96 |
251054_at |
AT5G01540
|
LecRK-VI.2, L-type lectin receptor kinase-VI.2, LECRKA4.1, lectin receptor kinase a4.1 |
0.468 |
0.143 |
| 97 |
266993_at |
AT2G39210
|
[Major facilitator superfamily protein] |
0.467 |
0.070 |
| 98 |
254271_at |
AT4G23150
|
CRK7, cysteine-rich RLK (RECEPTOR-like protein kinase) 7 |
0.467 |
0.260 |
| 99 |
265260_at |
AT2G43000
|
ANAC042, NAC domain containing protein 42, JUB1, JUNGBRUNNEN 1, NAC042, NAC domain containing protein 42 |
0.463 |
0.089 |
| 100 |
255148_at |
AT4G08470
|
MAPKKK10, MEKK3, MAPK/ERK kinase kinase 3 |
0.461 |
0.030 |
| 101 |
265161_at |
AT1G30900
|
BP80-3;3, binding protein of 80 kDa 3;3, VSR3;3, VACUOLAR SORTING RECEPTOR 3;3, VSR6, VACUOLAR SORTING RECEPTOR 6 |
0.460 |
0.186 |
| 102 |
259479_at |
AT1G19020
|
unknown |
0.459 |
0.119 |
| 103 |
256576_at |
AT3G28210
|
PMZ, SAP12, STRESS-ASSOCIATED PROTEIN 12 |
0.450 |
0.826 |
| 104 |
248092_at |
AT5G55170
|
ATSUMO3, SUM3, SMALL UBIQUITIN-LIKE MODIFIER 3, SUMO 3, SMALL UBIQUITIN-LIKE MODIFIER 3, SUMO3, small ubiquitin-like modifier 3 |
0.449 |
-0.001 |
| 105 |
252387_at |
AT3G47800
|
[Galactose mutarotase-like superfamily protein] |
0.449 |
0.041 |
| 106 |
256933_at |
AT3G22600
|
LTPG5, glycosylphosphatidylinositol-anchored lipid protein transfer 5 |
0.443 |
0.191 |
| 107 |
255595_at |
AT4G01700
|
[Chitinase family protein] |
0.443 |
0.185 |
| 108 |
256366_at |
AT1G66880
|
[Protein kinase superfamily protein] |
0.442 |
0.113 |
| 109 |
257101_at |
AT3G25020
|
AtRLP42, receptor like protein 42, RBPG1, response to Botrytis polygalacturonases 1, RLP42, receptor like protein 42 |
0.441 |
0.104 |
| 110 |
260783_at |
AT1G06160
|
ORA59, octadecanoid-responsive Arabidopsis AP2/ERF 59 |
0.440 |
-0.110 |
| 111 |
252976_s_at |
AT4G38550
|
[Arabidopsis phospholipase-like protein (PEARLI 4) family] |
0.440 |
-0.026 |
| 112 |
263402_at |
AT2G04050
|
[MATE efflux family protein] |
0.434 |
0.093 |
| 113 |
251336_at |
AT3G61190
|
BAP1, BON association protein 1 |
0.433 |
0.062 |
| 114 |
246302_at |
AT3G51860
|
ATCAX3, ATHCX1, CAX1-LIKE, CAX3, cation exchanger 3 |
0.432 |
0.123 |
| 115 |
256969_at |
AT3G21080
|
[ABC transporter-related] |
0.431 |
0.090 |
| 116 |
263963_at |
AT2G36080
|
ABS2, ABNORMAL SHOOT 2, NGAL1, NGATHA-Like 1 |
0.429 |
0.034 |
| 117 |
265597_at |
AT2G20142
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
0.429 |
0.056 |
| 118 |
259211_at |
AT3G09020
|
[alpha 1,4-glycosyltransferase family protein] |
0.426 |
0.109 |
| 119 |
263515_at |
AT2G21640
|
unknown |
0.424 |
0.171 |
| 120 |
264867_at |
AT1G24150
|
ATFH4, FORMIN HOMOLOGUE 4, FH4, formin homologue 4 |
0.423 |
0.051 |
| 121 |
260943_at |
AT1G45145
|
ATH5, THIOREDOXIN H-TYPE 5, ATTRX5, thioredoxin H-type 5, LIV1, LOCUS OF INSENSITIVITY TO VICTORIN 1, TRX-h5, THIOREDOXIN H-TYPE 5, TRX5, thioredoxin H-type 5 |
0.421 |
0.051 |
| 122 |
252373_at |
AT3G48090
|
ATEDS1, EDS1, enhanced disease susceptibility 1 |
0.421 |
0.132 |
| 123 |
263478_at |
AT2G31880
|
EVR, EVERSHED, SOBIR1, SUPPRESSOR OF BIR1 1 |
0.419 |
0.081 |
| 124 |
256050_at |
AT1G07000
|
ATEXO70B2, exocyst subunit exo70 family protein B2, EXO70B2, exocyst subunit exo70 family protein B2 |
0.417 |
-0.013 |
| 125 |
256883_at |
AT3G26440
|
unknown |
0.416 |
0.166 |
| 126 |
252698_at |
AT3G43670
|
[Copper amine oxidase family protein] |
0.416 |
-0.032 |
| 127 |
249850_at |
AT5G23240
|
DJC76, DNA J protein C76 |
0.416 |
0.058 |
| 128 |
249188_at |
AT5G42830
|
[HXXXD-type acyl-transferase family protein] |
0.410 |
0.112 |
| 129 |
254266_at |
AT4G23130
|
CRK5, cysteine-rich RLK (RECEPTOR-like protein kinase) 5, RLK6, RECEPTOR-LIKE PROTEIN KINASE 6 |
0.409 |
-0.045 |
| 130 |
259560_at |
AT1G21270
|
WAK2, wall-associated kinase 2 |
0.409 |
0.049 |
| 131 |
260656_at |
AT1G19380
|
unknown |
0.407 |
-0.042 |
| 132 |
248169_at |
AT5G54610
|
ANK, ankyrin, BDA1, bian da 2 (becoming big in Chinese) |
0.406 |
0.115 |
| 133 |
264809_at |
AT1G08830
|
CSD1, copper/zinc superoxide dismutase 1 |
0.405 |
0.002 |
| 134 |
259925_at |
AT1G75040
|
PR-5, PR5, pathogenesis-related gene 5 |
0.405 |
0.045 |
| 135 |
255319_at |
AT4G04220
|
AtRLP46, receptor like protein 46, RLP46, receptor like protein 46 |
0.400 |
0.106 |
| 136 |
249754_at |
AT5G24530
|
DMR6, DOWNY MILDEW RESISTANT 6 |
0.397 |
0.120 |
| 137 |
251422_at |
AT3G60540
|
[Preprotein translocase Sec, Sec61-beta subunit protein] |
0.393 |
0.052 |
| 138 |
252346_at |
AT3G48650
|
[pseudogene] |
0.390 |
0.218 |
| 139 |
257535_at |
AT3G09490
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.390 |
0.083 |
| 140 |
251705_at |
AT3G56400
|
ATWRKY70, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 70, WRKY70, WRKY DNA-binding protein 70 |
0.389 |
0.075 |
| 141 |
247685_at |
AT5G59680
|
[Leucine-rich repeat protein kinase family protein] |
0.389 |
0.139 |
| 142 |
258100_at |
AT3G23550
|
[MATE efflux family protein] |
0.389 |
0.004 |
| 143 |
257591_at |
AT3G24900
|
AtRLP39, receptor like protein 39, RLP39, receptor like protein 39 |
0.386 |
0.174 |
| 144 |
248668_at |
AT5G48720
|
XRI, X-RAY INDUCED TRANSCRIPT, XRI1, X-RAY INDUCED TRANSCRIPT 1 |
0.384 |
0.008 |
| 145 |
261161_at |
AT1G34420
|
[leucine-rich repeat transmembrane protein kinase family protein] |
0.383 |
0.080 |
| 146 |
250942_at |
AT5G03350
|
[Legume lectin family protein] |
0.378 |
0.026 |
| 147 |
261167_at |
AT1G04980
|
ATPDI10, ARABIDOPSIS THALIANA PROTEIN DISULFIDE ISOMERASE 10, ATPDIL2-2, PDI-like 2-2, PDI10, PROTEIN DISULFIDE ISOMERASE, PDIL2-2, PDI-like 2-2 |
0.378 |
0.095 |
| 148 |
249494_at |
AT5G39050
|
PMAT1, phenolic glucoside malonyltransferase 1 |
0.378 |
0.195 |
| 149 |
260904_at |
AT1G02450
|
NIMIN-1, NIMIN1, NIM1-interacting 1 |
0.378 |
-0.017 |
| 150 |
267288_at |
AT2G23680
|
[Cold acclimation protein WCOR413 family] |
0.376 |
0.023 |
| 151 |
258537_at |
AT3G04210
|
[Disease resistance protein (TIR-NBS class)] |
0.373 |
0.006 |
| 152 |
253414_at |
AT4G33050
|
AtIQM1, EDA39, embryo sac development arrest 39, IQM1, IQ-motif protein 1 |
0.370 |
0.080 |
| 153 |
245265_at |
AT4G14400
|
ACD6, ACCELERATED CELL DEATH 6 |
0.369 |
0.036 |
| 154 |
263497_at |
AT2G42540
|
COR15, COR15A, cold-regulated 15a |
-0.566 |
-0.048 |
| 155 |
245982_at |
AT5G13170
|
AtSWEET15, SAG29, senescence-associated gene 29, SWEET15 |
-0.539 |
0.037 |
| 156 |
257017_at |
AT3G19620
|
[Glycosyl hydrolase family protein] |
-0.430 |
0.039 |
| 157 |
252269_at |
AT3G49580
|
LSU1, RESPONSE TO LOW SULFUR 1 |
-0.413 |
0.181 |
| 158 |
248467_at |
AT5G50800
|
AtSWEET13, SWEET13 |
-0.410 |
0.003 |
| 159 |
262737_at |
AT1G28560
|
SRD2, SHOOT REDIFFERENTIATION DEFECTIVE 2 |
-0.402 |
-0.009 |
| 160 |
247760_at |
AT5G59130
|
[Subtilase family protein] |
-0.376 |
0.005 |
| 161 |
250478_at |
AT5G10250
|
DOT3, DEFECTIVELY ORGANIZED TRIBUTARIES 3 |
-0.371 |
-0.061 |
| 162 |
249645_at |
AT5G36910
|
THI2.2, thionin 2.2 |
-0.365 |
-0.103 |
| 163 |
256304_at |
AT1G69523
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.362 |
-0.118 |
| 164 |
247055_at |
AT5G66740
|
unknown |
-0.353 |
-0.067 |
| 165 |
252607_at |
AT3G44990
|
AtXTH31, ATXTR8, XTH31, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 31, XTR8, xyloglucan endo-transglycosylase-related 8 |
-0.348 |
-0.130 |
| 166 |
264100_at |
AT1G78970
|
ATLUP1, ARABIDOPSIS THALIANA LUPEOL SYNTHASE 1, LUP1, lupeol synthase 1 |
-0.346 |
0.019 |
| 167 |
249588_at |
AT5G37790
|
[Protein kinase superfamily protein] |
-0.344 |
-0.049 |
| 168 |
253254_at |
AT4G34650
|
SQS2, squalene synthase 2 |
-0.337 |
-0.038 |
| 169 |
253946_at |
AT4G26790
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.334 |
-0.057 |
| 170 |
249071_at |
AT5G44050
|
[MATE efflux family protein] |
-0.320 |
-0.064 |
| 171 |
266320_at |
AT2G46640
|
TAC1, Tiller Angle Control 1 |
-0.307 |
-0.016 |
| 172 |
254633_at |
AT4G18640
|
MRH1, morphogenesis of root hair 1 |
-0.306 |
0.029 |
| 173 |
252143_at |
AT3G51150
|
[ATP binding microtubule motor family protein] |
-0.305 |
0.024 |
| 174 |
259760_at |
AT1G77580
|
unknown |
-0.298 |
0.048 |
| 175 |
256926_at |
AT3G22540
|
unknown |
-0.296 |
-0.050 |
| 176 |
256302_at |
AT1G69526
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.294 |
-0.090 |
| 177 |
253344_at |
AT4G33550
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.281 |
-0.078 |
| 178 |
264157_at |
AT1G65310
|
ATXTH17, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 17, XTH17, xyloglucan endotransglucosylase/hydrolase 17 |
-0.274 |
-0.051 |
| 179 |
262232_at |
AT1G68600
|
[Aluminium activated malate transporter family protein] |
-0.273 |
-0.003 |
| 180 |
261399_at |
AT1G79620
|
[Leucine-rich repeat protein kinase family protein] |
-0.270 |
0.108 |
| 181 |
250820_at |
AT5G05160
|
RUL1, REDUCED IN LATERAL GROWTH1 |
-0.269 |
-0.012 |
| 182 |
252365_at |
AT3G48350
|
CEP3, cysteine endopeptidase 3 |
-0.261 |
0.018 |
| 183 |
259001_at |
AT3G01960
|
unknown |
-0.259 |
-0.096 |
| 184 |
260363_at |
AT1G70550
|
unknown |
-0.258 |
-0.088 |
| 185 |
262113_at |
AT1G02820
|
AtLEA3, LEA3, late embryogenesis abundant 3 |
-0.256 |
0.088 |
| 186 |
256860_at |
AT3G23840
|
CER26-LIKE, CER26-LIKE |
-0.256 |
-0.012 |
| 187 |
256599_at |
AT3G14760
|
unknown |
-0.255 |
-0.113 |
| 188 |
248888_at |
AT5G46240
|
AtKAT1, KAT1, potassium channel in Arabidopsis thaliana 1 |
-0.255 |
0.170 |
| 189 |
255127_at |
AT4G08300
|
UMAMIT17, Usually multiple acids move in and out Transporters 17 |
-0.255 |
0.050 |
| 190 |
258139_at |
AT3G24520
|
AT-HSFC1, HSFC1, heat shock transcription factor C1 |
-0.253 |
-0.124 |
| 191 |
251141_at |
AT5G01075
|
[Glycosyl hydrolase family 35 protein] |
-0.251 |
-0.153 |
| 192 |
261226_at |
AT1G20190
|
ATEXP11, ATEXPA11, expansin 11, ATHEXP ALPHA 1.14, EXP11, EXPANSIN 11, EXPA11, expansin 11 |
-0.249 |
-0.123 |
| 193 |
245417_at |
AT4G17360
|
[Formyl transferase] |
-0.245 |
-0.064 |
| 194 |
250968_at |
AT5G02890
|
[HXXXD-type acyl-transferase family protein] |
-0.243 |
-0.052 |
| 195 |
258017_at |
AT3G19370
|
unknown |
-0.242 |
0.173 |
| 196 |
253285_at |
AT4G34250
|
KCS16, 3-ketoacyl-CoA synthase 16 |
-0.239 |
0.007 |
| 197 |
261957_at |
AT1G64660
|
ATMGL, methionine gamma-lyase, MGL, methionine gamma-lyase |
-0.238 |
-0.031 |
| 198 |
261068_at |
AT1G07450
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.236 |
-0.078 |
| 199 |
259081_at |
AT3G05030
|
ATNHX2, NHX2, sodium hydrogen exchanger 2 |
-0.236 |
-0.088 |
| 200 |
253723_at |
AT4G29240
|
[Leucine-rich repeat (LRR) family protein] |
-0.235 |
-0.135 |
| 201 |
263136_at |
AT1G78580
|
ATTPS1, trehalose-6-phosphate synthase, TPS1, trehalose-6-phosphate synthase, TPS1, TREHALOSE-6-PHOSPHATE SYNTHASE 1 |
-0.229 |
0.063 |
| 202 |
247415_at |
AT5G63060
|
[Sec14p-like phosphatidylinositol transfer family protein] |
-0.224 |
-0.095 |
| 203 |
266802_at |
AT2G22900
|
[Galactosyl transferase GMA12/MNN10 family protein] |
-0.224 |
-0.026 |
| 204 |
245748_at |
AT1G51140
|
AKS1, ABA-responsive kinase substrate 1, FBH3, FLOWERING BHLH 3 |
-0.222 |
-0.037 |
| 205 |
264444_at |
AT1G27360
|
SPL11, squamosa promoter-like 11 |
-0.221 |
-0.052 |
| 206 |
246827_at |
AT5G26330
|
[Cupredoxin superfamily protein] |
-0.221 |
-0.026 |
| 207 |
255604_at |
AT4G01080
|
TBL26, TRICHOME BIREFRINGENCE-LIKE 26 |
-0.220 |
-0.097 |
| 208 |
260727_at |
AT1G48100
|
[Pectin lyase-like superfamily protein] |
-0.218 |
-0.087 |
| 209 |
260667_at |
AT1G19440
|
KCS4, 3-ketoacyl-CoA synthase 4 |
-0.217 |
0.002 |
| 210 |
254125_at |
AT4G24670
|
TAR2, tryptophan aminotransferase related 2 |
-0.215 |
-0.081 |
| 211 |
264324_at |
AT1G04160
|
ATXIB, ARABIDOPSIS THALIANA MYOSIN XI B, XI-8, MYOSIN XI-8, XI-B, MYOSIN XI B, XIB, myosin XI B |
-0.215 |
0.028 |
| 212 |
263486_at |
AT2G22200
|
[Integrase-type DNA-binding superfamily protein] |
-0.215 |
-0.144 |
| 213 |
262748_at |
AT1G28610
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.214 |
-0.005 |
| 214 |
261421_at |
AT1G18840
|
IQD30, IQ-domain 30 |
-0.214 |
0.071 |
| 215 |
258932_at |
AT3G10150
|
ATPAP16, PAP16, purple acid phosphatase 16 |
-0.214 |
-0.010 |
| 216 |
260034_at |
AT1G68810
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.213 |
-0.059 |
| 217 |
247085_at |
AT5G66310
|
[ATP binding microtubule motor family protein] |
-0.212 |
-0.005 |
| 218 |
252014_at |
AT3G52870
|
[IQ calmodulin-binding motif family protein] |
-0.211 |
-0.030 |
| 219 |
245676_at |
AT1G56670
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.211 |
-0.049 |
| 220 |
261522_at |
AT1G71710
|
[DNAse I-like superfamily protein] |
-0.208 |
0.052 |
| 221 |
253736_at |
AT4G28780
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.205 |
-0.078 |
| 222 |
266663_at |
AT2G25790
|
SKM1, STERILITY-REGULATING KINASE MEMBER 1 |
-0.205 |
0.004 |
| 223 |
248232_at |
AT5G53760
|
ATMLO11, MILDEW RESISTANCE LOCUS O 11, MLO11, MILDEW RESISTANCE LOCUS O 11 |
-0.203 |
0.095 |
| 224 |
260041_at |
AT1G68780
|
[RNI-like superfamily protein] |
-0.203 |
-0.114 |
| 225 |
248696_at |
AT5G48360
|
[Actin-binding FH2 (formin homology 2) family protein] |
-0.202 |
-0.099 |
| 226 |
251142_at |
AT5G01015
|
unknown |
-0.198 |
-0.136 |
| 227 |
245364_at |
AT4G15790
|
unknown |
-0.197 |
-0.105 |
| 228 |
261382_at |
AT1G05470
|
CVP2, COTYLEDON VASCULAR PATTERN 2 |
-0.197 |
-0.085 |
| 229 |
245088_at |
AT2G39850
|
[Subtilisin-like serine endopeptidase family protein] |
-0.193 |
0.039 |
| 230 |
252412_at |
AT3G47295
|
unknown |
-0.193 |
-0.097 |
| 231 |
253440_at |
AT4G32570
|
TIFY8, TIFY domain protein 8 |
-0.192 |
-0.054 |
| 232 |
261247_at |
AT1G20070
|
unknown |
-0.191 |
-0.024 |
| 233 |
248160_at |
AT5G54470
|
BBX29, B-box domain protein 29 |
-0.190 |
0.018 |
| 234 |
251058_at |
AT5G01790
|
unknown |
-0.189 |
-0.063 |
| 235 |
254118_at |
AT4G24790
|
[AAA-type ATPase family protein] |
-0.188 |
-0.025 |
| 236 |
250233_at |
AT5G13460
|
IQD11, IQ-domain 11 |
-0.186 |
0.075 |
| 237 |
265870_at |
AT2G01660
|
PDLP6, plasmodesmata-located protein 6 |
-0.186 |
-0.039 |
| 238 |
250561_at |
AT5G08030
|
AtGDPD6, GDPD6, glycerophosphodiester phosphodiesterase 6 |
-0.185 |
-0.054 |
| 239 |
249090_at |
AT5G43745
|
unknown |
-0.185 |
0.030 |
| 240 |
258938_at |
AT3G10080
|
[RmlC-like cupins superfamily protein] |
-0.184 |
-0.067 |
| 241 |
256674_at |
AT3G52360
|
unknown |
-0.182 |
-0.055 |
| 242 |
256527_at |
AT1G66100
|
[Plant thionin] |
-0.182 |
-0.001 |
| 243 |
261605_at |
AT1G49580
|
[Calcium-dependent protein kinase (CDPK) family protein] |
-0.181 |
-0.071 |
| 244 |
248839_at |
AT5G46690
|
bHLH071, beta HLH protein 71 |
-0.181 |
-0.161 |
| 245 |
264250_at |
AT1G78680
|
ATGGH2, gamma-glutamyl hydrolase 2, GGH2, gamma-glutamyl hydrolase 2 |
-0.180 |
-0.025 |
| 246 |
259832_at |
AT1G69580
|
[Homeodomain-like superfamily protein] |
-0.179 |
-0.035 |
| 247 |
265880_at |
AT2G42300
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.179 |
0.006 |
| 248 |
262414_at |
AT1G49430
|
LACS2, long-chain acyl-CoA synthetase 2, LRD2, LATERAL ROOT DEVELOPMENT 2 |
-0.178 |
0.021 |
| 249 |
263793_at |
AT2G24630
|
ATCSLC08, ATCSLC8, CELLULOSE-SYNTHASE LIKE C8, CSLC08 |
-0.177 |
-0.070 |
| 250 |
259528_at |
AT1G12330
|
unknown |
-0.176 |
-0.043 |
| 251 |
254789_at |
AT4G12880
|
AtENODL19, ENODL19, early nodulin-like protein 19 |
-0.174 |
-0.043 |
| 252 |
249059_at |
AT5G44530
|
[Subtilase family protein] |
-0.174 |
0.022 |
| 253 |
247170_at |
AT5G65530
|
[Protein kinase superfamily protein] |
-0.169 |
-0.060 |
| 254 |
267141_at |
AT2G38090
|
[Duplicated homeodomain-like superfamily protein] |
-0.169 |
-0.108 |
| 255 |
247175_at |
AT5G65280
|
GCL1, GCR2-like 1 |
-0.168 |
-0.047 |
| 256 |
257740_at |
AT3G27330
|
[zinc finger (C3HC4-type RING finger) family protein] |
-0.167 |
-0.040 |
| 257 |
266866_at |
AT2G29940
|
ABCG31, ATP-binding cassette G31, ATPDR3, PLEIOTROPIC DRUG RESISTANCE 3, PDR3, pleiotropic drug resistance 3 |
-0.166 |
0.026 |
| 258 |
247427_at |
AT5G62580
|
[ARM repeat superfamily protein] |
-0.166 |
0.011 |
| 259 |
256237_at |
AT3G12610
|
DRT100, DNA-DAMAGE REPAIR/TOLERATION 100 |
-0.166 |
-0.075 |
| 260 |
248028_at |
AT5G55620
|
unknown |
-0.165 |
-0.068 |
| 261 |
253876_at |
AT4G27430
|
CIP7, COP1-interacting protein 7 |
-0.163 |
0.065 |
| 262 |
260857_at |
AT1G21880
|
LYM1, lysm domain GPI-anchored protein 1 precursor, LYP2, LysM-containing receptor protein 2 |
-0.162 |
-0.042 |
| 263 |
251546_at |
AT3G58830
|
[haloacid dehalogenase (HAD) superfamily protein] |
-0.162 |
-0.072 |
| 264 |
254545_at |
AT4G19830
|
[FKBP-like peptidyl-prolyl cis-trans isomerase family protein] |
-0.162 |
-0.104 |
| 265 |
257650_at |
AT3G16800
|
[Protein phosphatase 2C family protein] |
-0.161 |
0.010 |
| 266 |
267066_at |
AT2G41040
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.159 |
0.086 |
| 267 |
249860_at |
AT5G22860
|
[Serine carboxypeptidase S28 family protein] |
-0.158 |
0.162 |
| 268 |
261624_at |
AT1G02000
|
GAE2, UDP-D-glucuronate 4-epimerase 2 |
-0.157 |
-0.059 |
| 269 |
254209_at |
AT4G23490
|
unknown |
-0.157 |
-0.040 |
| 270 |
255488_at |
AT4G02630
|
[Protein kinase superfamily protein] |
-0.156 |
-0.066 |
| 271 |
245671_at |
AT1G28230
|
ATPUP1, PUP1, purine permease 1 |
-0.155 |
-0.013 |
| 272 |
245190_at |
AT1G67690
|
[Zincin-like metalloproteases family protein] |
-0.155 |
0.016 |
| 273 |
266277_at |
AT2G29310
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.154 |
-0.087 |
| 274 |
253579_at |
AT4G30610
|
BRS1, BRI1 SUPPRESSOR 1, SCPL24, SERINE CARBOXYPEPTIDASE 24 PRECURSOR |
-0.154 |
-0.065 |
| 275 |
266079_at |
AT2G37860
|
LCD1, LOWER CELL DENSITY 1 |
-0.154 |
-0.055 |
| 276 |
256220_at |
AT1G56230
|
unknown |
-0.154 |
0.035 |
| 277 |
248291_at |
AT5G53020
|
[Ribonuclease P protein subunit P38-related] |
-0.153 |
0.056 |
| 278 |
260388_at |
AT1G74070
|
[Cyclophilin-like peptidyl-prolyl cis-trans isomerase family protein] |
-0.151 |
-0.115 |
| 279 |
248104_at |
AT5G55250
|
AtIAMT1, IAMT1, IAA carboxylmethyltransferase 1 |
-0.151 |
0.009 |
| 280 |
263677_at |
AT1G04520
|
PDLP2, plasmodesmata-located protein 2 |
-0.151 |
-0.021 |
| 281 |
264738_at |
AT1G62250
|
unknown |
-0.150 |
-0.069 |
| 282 |
252716_at |
AT3G43920
|
ATDCL3, DICER-LIKE 3, DCL3, dicer-like 3 |
-0.150 |
0.042 |
| 283 |
265377_at |
AT2G05790
|
[O-Glycosyl hydrolases family 17 protein] |
-0.149 |
-0.068 |
| 284 |
249208_at |
AT5G42650
|
AOS, allene oxide synthase, CYP74A, CYTOCHROME P450 74A, DDE2, DELAYED DEHISCENCE 2 |
-0.149 |
-0.020 |
| 285 |
249134_at |
AT5G43150
|
unknown |
-0.148 |
-0.089 |
| 286 |
263437_at |
AT2G28670
|
ESB1, ENHANCED SUBERIN 1 |
-0.148 |
-0.036 |
| 287 |
265665_at |
AT2G27420
|
[Cysteine proteinases superfamily protein] |
-0.148 |
-0.107 |
| 288 |
265312_at |
AT2G20240
|
TRM17, TON1 Recruiting Motif 17 |
-0.148 |
0.060 |
| 289 |
257663_at |
AT3G20260
|
[FUNCTIONS IN: structural constituent of ribosome] |
-0.147 |
-0.074 |
| 290 |
257396_at |
AT2G20875
|
EPF1, EPIDERMAL PATTERNING FACTOR 1 |
-0.147 |
-0.035 |
| 291 |
245449_at |
AT4G16870
|
[copia-like retrotransposon family, has a 0. P-value blast match to dbj|BAA78426.1| polyprotein (AtRE2-1) (Arabidopsis thaliana) (Ty1_Copia-element)] |
-0.146 |
0.043 |
| 292 |
260629_at |
AT1G62330
|
[O-fucosyltransferase family protein] |
-0.146 |
0.010 |
| 293 |
259021_at |
AT3G07540
|
[Actin-binding FH2 (formin homology 2) family protein] |
-0.145 |
-0.089 |
| 294 |
267359_at |
AT2G40020
|
[Nucleolar histone methyltransferase-related protein] |
-0.145 |
-0.066 |
| 295 |
258633_at |
AT3G07990
|
SCPL27, serine carboxypeptidase-like 27 |
-0.145 |
-0.027 |
| 296 |
247672_at |
AT5G60220
|
TET4, tetraspanin4 |
-0.144 |
0.001 |
| 297 |
266503_at |
AT2G47780
|
[Rubber elongation factor protein (REF)] |
-0.144 |
-0.125 |
| 298 |
254746_at |
AT4G12980
|
[Auxin-responsive family protein] |
-0.143 |
-0.044 |
| 299 |
266760_at |
AT2G46870
|
NGA1, NGATHA1 |
-0.143 |
-0.183 |
| 300 |
250960_at |
AT5G02940
|
unknown |
-0.143 |
0.038 |
| 301 |
265694_at |
AT2G24440
|
[selenium binding] |
-0.142 |
-0.041 |
| 302 |
256075_at |
AT1G18150
|
ATMPK8, MPK8 |
-0.142 |
-0.009 |
| 303 |
263841_at |
AT2G36870
|
AtXTH32, XTH32, xyloglucan endotransglucosylase/hydrolase 32 |
-0.142 |
-0.067 |
| 304 |
250471_at |
AT5G10170
|
ATMIPS3, MYO-INOSITOL-1-PHOSTPATE SYNTHASE 3, MIPS3, myo-inositol-1-phosphate synthase 3 |
-0.142 |
0.018 |
| 305 |
253394_at |
AT4G32770
|
ATSDX1, SUCROSE EXPORT DEFECTIVE 1, VTE1, VITAMIN E DEFICIENT 1 |
-0.141 |
-0.005 |