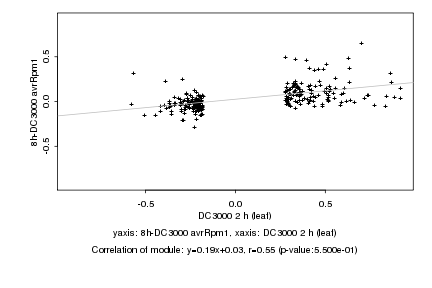
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
DC3000 2 h (leaf) |
Log2 signal ratio
8h-DC3000 avrRpm1 |
| 1 |
264648_at |
AT1G09080
|
BIP3, binding protein 3 |
0.916 |
0.037 |
| 2 |
264960_at |
AT1G76930
|
ATEXT1, EXTENSIN 1, ATEXT4, extensin 4, EXT1, extensin 1, EXT4, extensin 4, ORG5, OBP3-RESPONSIVE GENE 5 |
0.915 |
0.151 |
| 3 |
254819_at |
AT4G12500
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.883 |
0.047 |
| 4 |
262930_at |
AT1G65690
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.865 |
0.218 |
| 5 |
246368_at |
AT1G51890
|
[Leucine-rich repeat protein kinase family protein] |
0.857 |
0.314 |
| 6 |
266486_at |
AT2G47950
|
unknown |
0.839 |
0.057 |
| 7 |
248703_at |
AT5G48430
|
[Eukaryotic aspartyl protease family protein] |
0.831 |
-0.053 |
| 8 |
266975_at |
AT2G39380
|
ATEXO70H2, exocyst subunit exo70 family protein H2, EXO70H2, exocyst subunit exo70 family protein H2 |
0.772 |
-0.035 |
| 9 |
249045_at |
AT5G44380
|
[FAD-binding Berberine family protein] |
0.735 |
0.068 |
| 10 |
266967_at |
AT2G39530
|
[Uncharacterised protein family (UPF0497)] |
0.732 |
0.074 |
| 11 |
261005_at |
AT1G26420
|
[FAD-binding Berberine family protein] |
0.717 |
0.038 |
| 12 |
257592_at |
AT3G24982
|
ATRLP40, receptor like protein 40, RLP40, receptor like protein 40 |
0.696 |
0.650 |
| 13 |
264616_at |
AT2G17740
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.657 |
-0.002 |
| 14 |
251832_at |
AT3G55150
|
ATEXO70H1, exocyst subunit exo70 family protein H1, EXO70H1, exocyst subunit exo70 family protein H1 |
0.638 |
0.014 |
| 15 |
263948_at |
AT2G35980
|
ATNHL10, ARABIDOPSIS NDR1/HIN1-LIKE 10, NHL10, NDR1/HIN1-LIKE, YLS9, YELLOW-LEAF-SPECIFIC GENE 9 |
0.634 |
0.376 |
| 16 |
245148_at |
AT2G45220
|
PME17, pectin methylesterase 17 |
0.631 |
0.217 |
| 17 |
256252_at |
AT3G11340
|
UGT76B1, UDP-dependent glycosyltransferase 76B1 |
0.624 |
0.482 |
| 18 |
260335_at |
AT1G74000
|
SS3, strictosidine synthase 3 |
0.617 |
0.007 |
| 19 |
245089_at |
AT2G45290
|
TKL2, transketolase 2 |
0.604 |
0.149 |
| 20 |
248896_at |
AT5G46350
|
ATWRKY8, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 8, WRKY8, WRKY DNA-binding protein 8 |
0.599 |
0.091 |
| 21 |
251097_at |
AT5G01560
|
LecRK-VI.4, L-type lectin receptor kinase VI.4, LECRKA4.3, lectin receptor kinase a4.3 |
0.588 |
0.089 |
| 22 |
261474_at |
AT1G14540
|
PER4, peroxidase 4 |
0.587 |
-0.004 |
| 23 |
256370_at |
AT1G66780
|
[MATE efflux family protein] |
0.581 |
-0.037 |
| 24 |
262229_at |
AT1G68620
|
[alpha/beta-Hydrolases superfamily protein] |
0.554 |
0.259 |
| 25 |
267436_at |
AT2G19190
|
FRK1, FLG22-induced receptor-like kinase 1 |
0.553 |
0.146 |
| 26 |
266232_at |
AT2G02310
|
AtPP2-B6, phloem protein 2-B6, PP2-B6, phloem protein 2-B6 |
0.546 |
0.035 |
| 27 |
258349_at |
AT3G17609
|
HYH, HY5-homolog |
0.543 |
0.090 |
| 28 |
252098_at |
AT3G51330
|
[Eukaryotic aspartyl protease family protein] |
0.525 |
0.141 |
| 29 |
252984_at |
AT4G37990
|
ATCAD8, ARABIDOPSIS THALIANA CINNAMYL-ALCOHOL DEHYDROGENASE 8, CAD-B2, CINNAMYL-ALCOHOL DEHYDROGENASE B2, ELI3, ELICITOR-ACTIVATED GENE 3, ELI3-2, elicitor-activated gene 3-2 |
0.519 |
0.123 |
| 30 |
265359_at |
AT2G16720
|
ATMYB7, ARABIDOPSIS THALIANA MYB DOMAIN PROTEIN 7, ATY49, MYB7, myb domain protein 7 |
0.519 |
0.168 |
| 31 |
267565_at |
AT2G30750
|
CYP71A12, cytochrome P450, family 71, subfamily A, polypeptide 12 |
0.517 |
0.070 |
| 32 |
247610_at |
AT5G60630
|
unknown |
0.516 |
0.010 |
| 33 |
254832_at |
AT4G12490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.515 |
0.018 |
| 34 |
262133_at |
AT1G78000
|
SEL1, SELENATE RESISTANT 1, SULTR1;2, sulfate transporter 1;2 |
0.513 |
0.052 |
| 35 |
260276_at |
AT1G80450
|
[VQ motif-containing protein] |
0.509 |
0.026 |
| 36 |
250287_at |
AT5G13330
|
Rap2.6L, related to AP2 6l |
0.504 |
0.415 |
| 37 |
264514_at |
AT1G09500
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.502 |
0.150 |
| 38 |
247215_at |
AT5G64905
|
PROPEP3, elicitor peptide 3 precursor |
0.501 |
0.095 |
| 39 |
266270_at |
AT2G29470
|
ATGSTU3, glutathione S-transferase tau 3, GST21, GLUTATHIONE S-TRANSFERASE 21, GSTU3, glutathione S-transferase tau 3 |
0.494 |
0.112 |
| 40 |
261240_at |
AT1G32940
|
ATSBT3.5, SBT3.5 |
0.485 |
0.364 |
| 41 |
258167_at |
AT3G21560
|
BRT1, Bright Trichomes 1, UGT84A2, UDP-glucosyl transferase 84A2 |
0.484 |
-0.048 |
| 42 |
250100_at |
AT5G16570
|
GLN1;4, glutamine synthetase 1;4 |
0.480 |
-0.033 |
| 43 |
259410_at |
AT1G13340
|
[Regulator of Vps4 activity in the MVB pathway protein] |
0.470 |
0.182 |
| 44 |
267238_at |
AT2G44130
|
KMD3, KISS ME DEADLY 3 |
0.467 |
0.226 |
| 45 |
261216_at |
AT1G33030
|
[O-methyltransferase family protein] |
0.461 |
0.359 |
| 46 |
257951_at |
AT3G21700
|
ATSGP2, SGP2 |
0.459 |
0.069 |
| 47 |
254652_at |
AT4G18170
|
ATWRKY28, WRKY28, WRKY DNA-binding protein 28 |
0.445 |
0.051 |
| 48 |
257654_at |
AT3G13310
|
DJC66, DNA J protein C66 |
0.440 |
0.172 |
| 49 |
252958_at |
AT4G38620
|
ATMYB4, myb domain protein 4, MYB4, myb domain protein 4 |
0.438 |
-0.046 |
| 50 |
261763_at |
AT1G15520
|
ABCG40, ATP-binding cassette G40, ATABCG40, Arabidopsis thaliana ATP-binding cassette G40, ATPDR12, PLEIOTROPIC DRUG RESISTANCE 12, PDR12, pleiotropic drug resistance 12 |
0.437 |
0.356 |
| 51 |
252364_at |
AT3G48450
|
[RPM1-interacting protein 4 (RIN4) family protein] |
0.434 |
0.005 |
| 52 |
253392_at |
AT4G32650
|
ATKC1, ARABIDOPSIS THALIANA K+ RECTIFYING CHANNEL 1, AtLKT1, A. thaliana low-K+-tolerant 1, KAT3, potassium channel in Arabidopsis thaliana 3, KC1 |
0.432 |
0.062 |
| 53 |
251739_at |
AT3G56170
|
CAN, Ca-2+ dependent nuclease, CAN1, calcium dependent nuclease 1 |
0.421 |
0.129 |
| 54 |
254543_at |
AT4G19810
|
ChiC, class V chitinase |
0.419 |
0.126 |
| 55 |
251096_at |
AT5G01550
|
LecRK-VI.3, L-type lectin receptor kinase VI.3, LECRKA4.2, lectin receptor kinase a4.1 |
0.419 |
0.096 |
| 56 |
260005_at |
AT1G67920
|
unknown |
0.417 |
0.190 |
| 57 |
249051_at |
AT5G44390
|
[FAD-binding Berberine family protein] |
0.416 |
0.014 |
| 58 |
259443_at |
AT1G02360
|
[Chitinase family protein] |
0.415 |
0.048 |
| 59 |
265208_at |
AT2G36690
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.409 |
0.007 |
| 60 |
259033_at |
AT3G09405
|
[Pectinacetylesterase family protein] |
0.408 |
0.145 |
| 61 |
267567_at |
AT2G30770
|
CYP71A13, cytochrome P450, family 71, subfamily A, polypeptide 13 |
0.407 |
0.379 |
| 62 |
264467_at |
AT1G10140
|
[Uncharacterised conserved protein UCP031279] |
0.406 |
0.177 |
| 63 |
267597_at |
AT2G33020
|
AtRLP24, receptor like protein 24, RLP24, receptor like protein 24 |
0.403 |
-0.014 |
| 64 |
254060_at |
AT4G25350
|
SHB1, SHORT HYPOCOTYL UNDER BLUE1 |
0.399 |
0.032 |
| 65 |
266267_at |
AT2G29460
|
ATGSTU4, glutathione S-transferase tau 4, GST22, GLUTATHIONE S-TRANSFERASE 22, GSTU4, glutathione S-transferase tau 4 |
0.393 |
0.462 |
| 66 |
245929_at |
AT5G24760
|
[GroES-like zinc-binding dehydrogenase family protein] |
0.384 |
0.044 |
| 67 |
250918_at |
AT5G03610
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.373 |
0.016 |
| 68 |
258201_at |
AT3G13910
|
unknown |
0.368 |
0.120 |
| 69 |
266752_at |
AT2G47000
|
ABCB4, ATP-binding cassette B4, AtABCB4, ATPGP4, ARABIDOPSIS P-GLYCOPROTEIN 4, MDR4, MULTIDRUG RESISTANCE 4, PGP4, P-GLYCOPROTEIN 4 |
0.368 |
0.112 |
| 70 |
246988_at |
AT5G67340
|
[ARM repeat superfamily protein] |
0.364 |
0.212 |
| 71 |
258209_at |
AT3G14060
|
unknown |
0.363 |
0.082 |
| 72 |
251790_at |
AT3G55470
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
0.359 |
0.110 |
| 73 |
256044_at |
AT1G07160
|
[Protein phosphatase 2C family protein] |
0.359 |
-0.028 |
| 74 |
264746_at |
AT1G62300
|
ATWRKY6, WRKY6 |
0.359 |
0.078 |
| 75 |
246370_at |
AT1G51920
|
unknown |
0.355 |
0.102 |
| 76 |
248298_at |
AT5G53110
|
[RING/U-box superfamily protein] |
0.353 |
0.062 |
| 77 |
259297_at |
AT3G05360
|
AtRLP30, receptor like protein 30, RLP30, receptor like protein 30 |
0.353 |
0.139 |
| 78 |
260362_at |
AT1G70530
|
CRK3, cysteine-rich RLK (RECEPTOR-like protein kinase) 3 |
0.352 |
0.082 |
| 79 |
252303_at |
AT3G49210
|
[O-acyltransferase (WSD1-like) family protein] |
0.352 |
0.018 |
| 80 |
250983_at |
AT5G02780
|
GSTL1, glutathione transferase lambda 1 |
0.351 |
0.141 |
| 81 |
254869_at |
AT4G11890
|
ARCK1, ABA- AND OSMOTIC-STRESS-INDUCIBLE RECEPTOR-LIKE CYTOSOLIC KINASE1, CRK45, cysteine-rich receptor-like protein kinase 45 |
0.349 |
0.156 |
| 82 |
246821_at |
AT5G26920
|
CBP60G, Cam-binding protein 60-like G |
0.347 |
0.157 |
| 83 |
254575_at |
AT4G19460
|
[UDP-Glycosyltransferase superfamily protein] |
0.345 |
-0.009 |
| 84 |
258203_at |
AT3G13950
|
unknown |
0.343 |
0.166 |
| 85 |
257690_at |
AT3G12830
|
SAUR72, SMALL AUXIN UPREGULATED 72 |
0.343 |
0.083 |
| 86 |
263179_at |
AT1G05710
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.342 |
0.177 |
| 87 |
246777_at |
AT5G27420
|
ATL31, Arabidopsis toxicos en levadura 31, CNI1, carbon/nitrogen insensitive 1 |
0.339 |
0.069 |
| 88 |
253401_at |
AT4G32870
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.337 |
0.164 |
| 89 |
267140_at |
AT2G38250
|
[Homeodomain-like superfamily protein] |
0.334 |
0.003 |
| 90 |
257571_at |
AT3G16870
|
GATA17, GATA transcription factor 17 |
0.333 |
-0.071 |
| 91 |
252908_at |
AT4G39670
|
[Glycolipid transfer protein (GLTP) family protein] |
0.332 |
0.199 |
| 92 |
260551_at |
AT2G43510
|
ATTI1, trypsin inhibitor protein 1, TI1, trypsin inhibitor protein 1 |
0.332 |
0.227 |
| 93 |
246401_at |
AT1G57560
|
AtMYB50, myb domain protein 50, MYB50, myb domain protein 50 |
0.331 |
0.210 |
| 94 |
252739_at |
AT3G43250
|
unknown |
0.330 |
0.006 |
| 95 |
257074_at |
AT3G19660
|
unknown |
0.330 |
0.126 |
| 96 |
261242_at |
AT1G32960
|
ATSBT3.3, SBT3.3 |
0.329 |
0.475 |
| 97 |
256969_at |
AT3G21080
|
[ABC transporter-related] |
0.328 |
0.114 |
| 98 |
254453_at |
AT4G21120
|
AAT1, amino acid transporter 1, CAT1, CATIONIC AMINO ACID TRANSPORTER 1 |
0.327 |
0.183 |
| 99 |
257271_at |
AT3G28007
|
AtSWEET4, SWEET4 |
0.325 |
0.101 |
| 100 |
249481_at |
AT5G38900
|
[Thioredoxin superfamily protein] |
0.324 |
0.195 |
| 101 |
252940_at |
AT4G39270
|
[Leucine-rich repeat protein kinase family protein] |
0.322 |
0.174 |
| 102 |
258975_at |
AT3G01970
|
ATWRKY45, WRKY DNA-BINDING PROTEIN 45, WRKY45, WRKY DNA-binding protein 45 |
0.320 |
0.207 |
| 103 |
262925_at |
AT1G65730
|
YSL7, YELLOW STRIPE like 7 |
0.319 |
0.073 |
| 104 |
261899_at |
AT1G80820
|
ATCCR2, CCR2, cinnamoyl coa reductase |
0.314 |
0.160 |
| 105 |
260405_at |
AT1G69930
|
ATGSTU11, glutathione S-transferase TAU 11, GSTU11, glutathione S-transferase TAU 11 |
0.313 |
0.161 |
| 106 |
260391_at |
AT1G74020
|
SS2, strictosidine synthase 2 |
0.311 |
0.043 |
| 107 |
264574_at |
AT1G05300
|
ZIP5, zinc transporter 5 precursor |
0.306 |
-0.048 |
| 108 |
254673_at |
AT4G18430
|
AtRABA1e, RAB GTPase homolog A1E, RABA1e, RAB GTPase homolog A1E |
0.304 |
0.086 |
| 109 |
262619_at |
AT1G06550
|
[ATP-dependent caseinolytic (Clp) protease/crotonase family protein] |
0.303 |
-0.055 |
| 110 |
264756_at |
AT1G61370
|
[S-locus lectin protein kinase family protein] |
0.301 |
0.136 |
| 111 |
247205_at |
AT5G64890
|
PROPEP2, elicitor peptide 2 precursor |
0.301 |
0.032 |
| 112 |
260648_at |
AT1G08050
|
[Zinc finger (C3HC4-type RING finger) family protein] |
0.300 |
0.135 |
| 113 |
253281_at |
AT4G34138
|
UGT73B1, UDP-glucosyl transferase 73B1 |
0.299 |
-0.044 |
| 114 |
255595_at |
AT4G01700
|
[Chitinase family protein] |
0.297 |
0.099 |
| 115 |
261564_at |
AT1G01720
|
ANAC002, Arabidopsis NAC domain containing protein 2, ATAF1 |
0.296 |
0.088 |
| 116 |
259721_at |
AT1G60890
|
[Phosphatidylinositol-4-phosphate 5-kinase family protein] |
0.292 |
0.055 |
| 117 |
254413_at |
AT4G21440
|
ATM4, A. THALIANA MYB 4, ATMYB102, MYB-like 102, MYB102, MYB102, MYB-like 102 |
0.292 |
-0.016 |
| 118 |
263919_at |
AT2G36470
|
unknown |
0.290 |
0.026 |
| 119 |
250153_at |
AT5G15130
|
ATWRKY72, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 72, WRKY72, WRKY DNA-binding protein 72 |
0.289 |
-0.001 |
| 120 |
249467_at |
AT5G39610
|
ANAC092, Arabidopsis NAC domain containing protein 92, ATNAC2, NAC domain containing protein 2, ATNAC6, NAC domain containing protein 6, NAC2, NAC domain containing protein 2, NAC6, NAC domain containing protein 6, ORE1, ORESARA 1 |
0.288 |
0.134 |
| 121 |
264953_at |
AT1G77120
|
ADH, ALCOHOL DEHYDROGENASE, ADH1, alcohol dehydrogenase 1, ATADH, ARABIDOPSIS THALIANA ALCOHOL DEHYDROGENASE, ATADH1 |
0.288 |
0.025 |
| 122 |
258277_at |
AT3G26830
|
CYP71B15, PAD3, PHYTOALEXIN DEFICIENT 3 |
0.286 |
0.202 |
| 123 |
263184_at |
AT1G05560
|
UGT1, UDP-GLUCOSE TRANSFERASE 1, UGT75B1, UDP-glucosyltransferase 75B1 |
0.285 |
0.005 |
| 124 |
250803_at |
AT5G04980
|
[DNAse I-like superfamily protein] |
0.284 |
0.042 |
| 125 |
249730_at |
AT5G24430
|
[Calcium-dependent protein kinase (CDPK) family protein] |
0.284 |
0.066 |
| 126 |
264893_at |
AT1G23140
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
0.284 |
-0.029 |
| 127 |
247655_at |
AT5G59820
|
AtZAT12, RHL41, RESPONSIVE TO HIGH LIGHT 41, ZAT12 |
0.283 |
0.161 |
| 128 |
257058_at |
AT3G15352
|
ATCOX17, ARABIDOPSIS THALIANA CYTOCHROME C OXIDASE 17, COX17, cytochrome c oxidase 17 |
0.280 |
0.117 |
| 129 |
250234_at |
AT5G13420
|
TRA2, transaldolase 2 |
0.278 |
0.109 |
| 130 |
254759_at |
AT4G13180
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.277 |
0.117 |
| 131 |
266536_at |
AT2G16900
|
[Arabidopsis phospholipase-like protein (PEARLI 4) family] |
0.277 |
0.017 |
| 132 |
247327_at |
AT5G64120
|
AtPRX71, PRX71, peroxidase 71 |
0.277 |
0.494 |
| 133 |
250936_at |
AT5G03120
|
unknown |
-0.579 |
-0.027 |
| 134 |
247866_at |
AT5G57550
|
XTH25, xyloglucan endotransglucosylase/hydrolase 25, XTR3, xyloglucan endotransglycosylase 3 |
-0.568 |
0.316 |
| 135 |
251402_at |
AT3G60290
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.511 |
-0.151 |
| 136 |
251575_at |
AT3G58120
|
ATBZIP61, BZIP61 |
-0.450 |
-0.149 |
| 137 |
255774_at |
AT1G18620
|
TRM3, TON1 Recruiting Motif 3 |
-0.421 |
-0.104 |
| 138 |
246011_at |
AT5G08330
|
AtTCP11, TCP11, TCP domain protein 11 |
-0.417 |
-0.050 |
| 139 |
261873_at |
AT1G11350
|
CBRLK1, CALMODULIN-BINDING RECEPTOR-LIKE PROTEIN KINASE, RKS2, SD1-13, S-domain-1 13 |
-0.396 |
-0.037 |
| 140 |
245262_at |
AT4G16563
|
[Eukaryotic aspartyl protease family protein] |
-0.390 |
0.229 |
| 141 |
265066_at |
AT1G03870
|
FLA9, FASCICLIN-like arabinoogalactan 9 |
-0.384 |
-0.077 |
| 142 |
258736_at |
AT3G05900
|
[neurofilament protein-related] |
-0.372 |
-0.021 |
| 143 |
252433_at |
AT3G47560
|
[alpha/beta-Hydrolases superfamily protein] |
-0.367 |
0.001 |
| 144 |
249008_at |
AT5G44680
|
[DNA glycosylase superfamily protein] |
-0.367 |
-0.057 |
| 145 |
266956_at |
AT2G34510
|
unknown |
-0.362 |
-0.053 |
| 146 |
247845_at |
AT5G58090
|
[O-Glycosyl hydrolases family 17 protein] |
-0.362 |
-0.044 |
| 147 |
265042_at |
AT1G04040
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
-0.357 |
-0.108 |
| 148 |
253278_at |
AT4G34220
|
[Leucine-rich repeat protein kinase family protein] |
-0.356 |
-0.142 |
| 149 |
265342_at |
AT2G18300
|
HBI1, HOMOLOG OF BEE2 INTERACTING WITH IBH 1 |
-0.342 |
-0.040 |
| 150 |
248419_at |
AT5G51550
|
EXL3, EXORDIUM like 3 |
-0.341 |
-0.021 |
| 151 |
259915_at |
AT1G72790
|
[hydroxyproline-rich glycoprotein family protein] |
-0.341 |
0.053 |
| 152 |
265724_at |
AT2G32100
|
ATOFP16, RABIDOPSIS THALIANA OVATE FAMILY PROTEIN 16, OFP16, ovate family protein 16 |
-0.336 |
-0.019 |
| 153 |
264704_at |
AT1G70090
|
GATL9, GALACTURONOSYLTRANSFERASE-LIKE 9, LGT8, glucosyl transferase family 8 |
-0.318 |
0.037 |
| 154 |
264209_at |
AT1G22740
|
ATRABG3B, RAB7, RAB75, RABG3B, RAB GTPase homolog G3B |
-0.308 |
0.028 |
| 155 |
255818_at |
AT2G33570
|
GALS1, galactan synthase 1 |
-0.307 |
-0.034 |
| 156 |
263876_at |
AT2G21880
|
ATRAB7A, RAB GTPase homolog 7A, ATRABG2, ARABIDOPSIS RAB GTPASE HOMOLOG G2, RAB7A, RAB GTPase homolog 7A |
-0.306 |
-0.005 |
| 157 |
255433_at |
AT4G03210
|
XTH9, xyloglucan endotransglucosylase/hydrolase 9 |
-0.305 |
-0.030 |
| 158 |
257964_at |
AT3G19850
|
[Phototropic-responsive NPH3 family protein] |
-0.305 |
-0.011 |
| 159 |
255856_at |
AT1G66940
|
[protein kinase-related] |
-0.304 |
-0.087 |
| 160 |
248095_at |
AT5G55230
|
ATMAP65-1, microtubule-associated proteins 65-1, MAP65-1, MAP65-1, microtubule-associated proteins 65-1 |
-0.304 |
-0.031 |
| 161 |
246019_at |
AT5G10690
|
[pentatricopeptide (PPR) repeat-containing protein / CBS domain-containing protein] |
-0.302 |
-0.106 |
| 162 |
252563_at |
AT3G45970
|
ATEXLA1, expansin-like A1, ATEXPL1, ATHEXP BETA 2.1, EXLA1, expansin-like A1, EXPL1, EXPANSIN L1 |
-0.298 |
0.253 |
| 163 |
245743_at |
AT1G51080
|
unknown |
-0.297 |
-0.210 |
| 164 |
254609_at |
AT4G18970
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.292 |
-0.201 |
| 165 |
259671_at |
AT1G52290
|
AtPERK15, proline-rich extensin-like receptor kinase 15, PERK15, proline-rich extensin-like receptor kinase 15 |
-0.291 |
0.005 |
| 166 |
259658_at |
AT1G55370
|
NDF5, NDH-dependent cyclic electron flow 5 |
-0.286 |
-0.080 |
| 167 |
258537_at |
AT3G04210
|
[Disease resistance protein (TIR-NBS class)] |
-0.285 |
0.015 |
| 168 |
251669_at |
AT3G57180
|
BPG2, BRASSINAZOLE(BRZ) INSENSITIVE PALE GREEN 2 |
-0.284 |
-0.129 |
| 169 |
264037_at |
AT2G03750
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.283 |
-0.058 |
| 170 |
264698_at |
AT1G70200
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.282 |
-0.089 |
| 171 |
254553_at |
AT4G19530
|
[disease resistance protein (TIR-NBS-LRR class) family] |
-0.278 |
-0.043 |
| 172 |
260840_at |
AT1G29050
|
TBL38, TRICHOME BIREFRINGENCE-LIKE 38 |
-0.276 |
0.098 |
| 173 |
250366_at |
AT5G11420
|
unknown |
-0.276 |
-0.054 |
| 174 |
250910_at |
AT5G03720
|
AT-HSFA3, ARABIDOPSIS THALIANA HEAT SHOCK TRANSCRIPTION FACTOR A3, HSFA3, heat shock transcription factor A3 |
-0.274 |
0.087 |
| 175 |
259958_at |
AT1G53730
|
SRF6, STRUBBELIG-receptor family 6 |
-0.271 |
0.001 |
| 176 |
266745_at |
AT2G02950
|
PKS1, phytochrome kinase substrate 1 |
-0.270 |
0.035 |
| 177 |
247540_at |
AT5G61590
|
[Integrase-type DNA-binding superfamily protein] |
-0.269 |
0.044 |
| 178 |
254032_at |
AT4G25940
|
[ENTH/ANTH/VHS superfamily protein] |
-0.266 |
0.017 |
| 179 |
258708_at |
AT3G09580
|
[FAD/NAD(P)-binding oxidoreductase family protein] |
-0.264 |
-0.022 |
| 180 |
266215_at |
AT2G06850
|
EXGT-A1, endoxyloglucan transferase A1, EXT, ENDOXYLOGLUCAN TRANSFERASE, XTH4, xyloglucan endotransglucosylase/hydrolase 4 |
-0.262 |
-0.066 |
| 181 |
262569_at |
AT1G15180
|
[MATE efflux family protein] |
-0.261 |
-0.077 |
| 182 |
252485_at |
AT3G46530
|
RPP13, RECOGNITION OF PERONOSPORA PARASITICA 13 |
-0.259 |
-0.031 |
| 183 |
246487_at |
AT5G16030
|
unknown |
-0.259 |
-0.059 |
| 184 |
263319_at |
AT2G47160
|
AtBOR1, BOR1, REQUIRES HIGH BORON 1 |
-0.258 |
-0.024 |
| 185 |
263656_at |
AT1G04240
|
IAA3, indole-3-acetic acid inducible 3, SHY2, SHORT HYPOCOTYL 2 |
-0.255 |
-0.004 |
| 186 |
256525_at |
AT1G66180
|
[Eukaryotic aspartyl protease family protein] |
-0.255 |
0.077 |
| 187 |
247596_at |
AT5G60840
|
unknown |
-0.245 |
0.011 |
| 188 |
252950_at |
AT4G38690
|
[PLC-like phosphodiesterases superfamily protein] |
-0.244 |
-0.009 |
| 189 |
260427_at |
AT1G72430
|
SAUR78, SMALL AUXIN UPREGULATED RNA 78 |
-0.243 |
0.056 |
| 190 |
246371_at |
AT1G51940
|
LYK3, LysM-containing receptor-like kinase 3 |
-0.243 |
0.031 |
| 191 |
261975_at |
AT1G64640
|
AtENODL8, ENODL8, early nodulin-like protein 8 |
-0.239 |
-0.072 |
| 192 |
261851_at |
AT1G50460
|
ATHKL1, HKL1, hexokinase-like 1 |
-0.238 |
-0.005 |
| 193 |
246313_at |
AT1G31920
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.236 |
-0.101 |
| 194 |
263920_at |
AT2G36410
|
unknown |
-0.234 |
-0.055 |
| 195 |
262245_at |
AT1G48240
|
ATNPSN12, NPSN12, novel plant snare 12 |
-0.234 |
-0.070 |
| 196 |
265771_at |
AT2G48030
|
[DNAse I-like superfamily protein] |
-0.233 |
0.126 |
| 197 |
253736_at |
AT4G28780
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.230 |
-0.282 |
| 198 |
254221_at |
AT4G23820
|
[Pectin lyase-like superfamily protein] |
-0.229 |
-0.027 |
| 199 |
245383_at |
AT4G17810
|
[C2H2 and C2HC zinc fingers superfamily protein] |
-0.229 |
-0.003 |
| 200 |
266279_at |
AT2G29290
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.228 |
-0.075 |
| 201 |
251413_at |
AT3G60320
|
unknown |
-0.228 |
-0.003 |
| 202 |
245176_at |
AT2G47440
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.226 |
-0.015 |
| 203 |
258432_at |
AT3G16570
|
ATRALF23, ARABIDOPSIS RAPID ALKALINIZATION FACTOR 23, RALF23, rapid alkalinization factor 23 |
-0.225 |
0.027 |
| 204 |
261594_at |
AT1G33240
|
AT-GTL1, GT2-LIKE 1, ATGTL1, GTL1, GT-2-like 1 |
-0.225 |
0.008 |
| 205 |
261611_at |
AT1G49730
|
[Protein kinase superfamily protein] |
-0.224 |
-0.052 |
| 206 |
266957_at |
AT2G34640
|
HMR, HEMERA, PTAC12, plastid transcriptionally active 12, TAC12 |
-0.224 |
-0.105 |
| 207 |
262376_at |
AT1G72970
|
EDA17, embryo sac development arrest 17, HTH, HOTHEAD |
-0.223 |
-0.140 |
| 208 |
246200_at |
AT4G37240
|
unknown |
-0.223 |
-0.031 |
| 209 |
256516_at |
AT1G66150
|
TMK1, transmembrane kinase 1 |
-0.222 |
-0.022 |
| 210 |
246492_at |
AT5G16140
|
[Peptidyl-tRNA hydrolase family protein] |
-0.221 |
-0.109 |
| 211 |
251072_at |
AT5G01740
|
[Nuclear transport factor 2 (NTF2) family protein] |
-0.220 |
0.103 |
| 212 |
263606_at |
AT2G16280
|
KCS9, 3-ketoacyl-CoA synthase 9 |
-0.220 |
-0.031 |
| 213 |
262871_at |
AT1G65010
|
[INVOLVED IN: flower development] |
-0.219 |
-0.056 |
| 214 |
262891_at |
AT1G79460
|
ATKS, ARABIDOPSIS THALIANA ENT-KAURENE SYNTHASE, ATKS1, ARABIDOPSIS THALIANA ENT-KAURENE SYNTHASE 1, GA2, GA REQUIRING 2, KS, KS1, ENT-KAURENE SYNTHASE 1 |
-0.219 |
-0.191 |
| 215 |
261420_at |
AT1G07720
|
KCS3, 3-ketoacyl-CoA synthase 3 |
-0.217 |
0.038 |
| 216 |
264521_at |
AT1G10020
|
unknown |
-0.217 |
0.021 |
| 217 |
251142_at |
AT5G01015
|
unknown |
-0.216 |
-0.099 |
| 218 |
261822_at |
AT1G11380
|
[PLAC8 family protein] |
-0.216 |
-0.019 |
| 219 |
245242_at |
AT1G44446
|
ATCAO, ARABIDOPSIS THALIANA CHLOROPHYLL A OXYGENASE, CAO, CHLOROPHYLL A OXYGENASE, CH1, CHLORINA 1 |
-0.215 |
-0.027 |
| 220 |
255506_at |
AT4G02130
|
GATL6, galacturonosyltransferase-like 6, LGT10 |
-0.214 |
0.012 |
| 221 |
246275_at |
AT4G36540
|
BEE2, BR enhanced expression 2 |
-0.214 |
-0.060 |
| 222 |
257912_at |
AT3G25500
|
AFH1, formin homology 1, AHF1, ATFH1, ARABIDOPSIS THALIANA FORMIN HOMOLOGY 1, FH1, FORMIN HOMOLOGY 1 |
-0.213 |
0.003 |
| 223 |
250975_at |
AT5G03050
|
unknown |
-0.211 |
-0.032 |
| 224 |
261769_at |
AT1G76100
|
PETE1, plastocyanin 1 |
-0.211 |
-0.004 |
| 225 |
261825_at |
AT1G11545
|
XTH8, xyloglucan endotransglucosylase/hydrolase 8 |
-0.211 |
-0.101 |
| 226 |
249493_at |
AT5G39080
|
[HXXXD-type acyl-transferase family protein] |
-0.210 |
0.029 |
| 227 |
246253_at |
AT4G37260
|
ATMYB73, MYB73, myb domain protein 73 |
-0.208 |
0.068 |
| 228 |
253283_at |
AT4G34090
|
unknown |
-0.207 |
-0.079 |
| 229 |
264339_at |
AT1G70290
|
ATTPS8, ATTPSC, TPS8, trehalose-6-phosphatase synthase S8 |
-0.206 |
0.069 |
| 230 |
252317_at |
AT3G48720
|
DCF, DEFICIENT IN CUTIN FERULATE |
-0.206 |
-0.046 |
| 231 |
259786_at |
AT1G29660
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.205 |
-0.089 |
| 232 |
245877_at |
AT1G26220
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
-0.205 |
0.018 |
| 233 |
248656_at |
AT5G48460
|
[Actin binding Calponin homology (CH) domain-containing protein] |
-0.203 |
-0.080 |
| 234 |
250102_at |
AT5G16590
|
LRR1, Leucine rich repeat protein 1 |
-0.202 |
-0.084 |
| 235 |
259848_at |
AT1G72180
|
[Leucine-rich receptor-like protein kinase family protein] |
-0.201 |
0.057 |
| 236 |
258920_at |
AT3G10520
|
AHB2, haemoglobin 2, ARATH GLB2, ATGLB2, ARABIDOPSIS HEMOGLOBIN 2, GLB2, HEMOGLOBIN 2, HB2, haemoglobin 2, NSHB2, NON-SYMBIOTIC HAEMOGLOBIN 2 |
-0.200 |
-0.054 |
| 237 |
250189_at |
AT5G14410
|
unknown |
-0.199 |
0.006 |
| 238 |
249783_at |
AT5G24270
|
ATSOS3, SALT OVERLY SENSITIVE 3, CBL4, CALCINEURIN B-LIKE PROTEIN 4, SOS3, SALT OVERLY SENSITIVE 3 |
-0.199 |
-0.047 |
| 239 |
253597_at |
AT4G30690
|
[Translation initiation factor 3 protein] |
-0.199 |
-0.015 |
| 240 |
252972_at |
AT4G38840
|
SAUR14, SMALL AUXIN UPREGULATED RNA 14 |
-0.198 |
-0.065 |
| 241 |
250884_at |
AT5G03940
|
54CP, 54 CHLOROPLAST PROTEIN, CPSRP54, chloroplast signal recognition particle 54 kDa subunit, FFC, FIFTY-FOUR CHLOROPLAST HOMOLOGUE, SRP54CP, SIGNAL RECOGNITION PARTICLE 54 KDA SUBUNIT CHLOROPLAST PROTEIN |
-0.198 |
-0.048 |
| 242 |
254239_at |
AT4G23400
|
PIP1;5, plasma membrane intrinsic protein 1;5, PIP1D |
-0.197 |
-0.042 |
| 243 |
267532_at |
AT2G42040
|
unknown |
-0.196 |
0.057 |
| 244 |
260790_at |
AT1G06240
|
unknown |
-0.196 |
-0.019 |
| 245 |
258333_at |
AT3G16000
|
MFP1, MAR binding filament-like protein 1 |
-0.195 |
-0.034 |
| 246 |
255817_at |
AT2G33330
|
PDLP3, plasmodesmata-located protein 3 |
-0.195 |
-0.156 |
| 247 |
246483_at |
AT5G16000
|
AtNIK1, NIK1, NSP-interacting kinase 1 |
-0.192 |
-0.093 |
| 248 |
249378_at |
AT5G40450
|
unknown |
-0.192 |
0.038 |
| 249 |
258332_at |
AT3G16180
|
NRT1.12, nitrate transporter 1.12 |
-0.191 |
-0.074 |
| 250 |
253092_at |
AT4G37380
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.191 |
-0.156 |
| 251 |
252178_at |
AT3G50750
|
BEH1, BES1/BZR1 homolog 1 |
-0.191 |
0.001 |
| 252 |
265823_at |
AT2G35760
|
[Uncharacterised protein family (UPF0497)] |
-0.191 |
0.014 |
| 253 |
259853_at |
AT1G72300
|
PSY1R, PSY1 receptor |
-0.190 |
0.017 |
| 254 |
259763_at |
AT1G77630
|
LYM3, lysin-motif (LysM) domain protein 3, LYP3, LysM-containing receptor protein 3 |
-0.189 |
-0.143 |
| 255 |
266254_at |
AT2G27810
|
ATNAT12, ARABIDOPSIS NUCLEOBASE-ASCORBATE TRANSPORTER 12, NAT12, nucleobase-ascorbate transporter 12 |
-0.189 |
-0.068 |
| 256 |
257516_at |
AT1G69040
|
ACR4, ACT domain repeat 4 |
-0.188 |
-0.134 |
| 257 |
252175_at |
AT3G50700
|
AtIDD2, indeterminate(ID)-domain 2, IDD2, indeterminate(ID)-domain 2 |
-0.187 |
-0.058 |
| 258 |
245141_at |
AT2G45400
|
BEN1, BRI1-5 ENHANCED 1 |
-0.187 |
-0.085 |
| 259 |
262612_at |
AT1G14150
|
PnsL2, Photosynthetic NDH subcomplex L 2, PQL1, PsbQ-like 1, PQL2, PsbQ-like 2 |
-0.186 |
-0.035 |
| 260 |
266889_at |
AT2G44640
|
unknown |
-0.186 |
-0.086 |
| 261 |
260686_at |
AT1G17620
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
-0.185 |
0.068 |
| 262 |
255652_at |
AT4G00950
|
MEE47, maternal effect embryo arrest 47 |
-0.185 |
-0.046 |
| 263 |
258653_at |
AT3G09870
|
SAUR48, SMALL AUXIN UPREGULATED RNA 48 |
-0.183 |
0.067 |