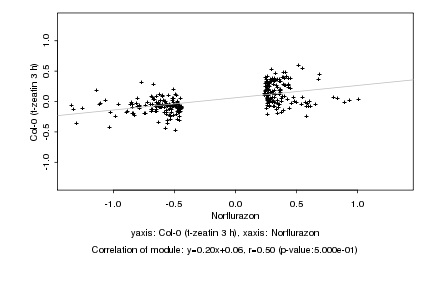
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Norflurazon |
Log2 signal ratio
Col-0 (t-zeatin 3 h) |
| 1 |
263402_at |
AT2G04050
|
[MATE efflux family protein] |
1.007 |
0.034 |
| 2 |
256459_at |
AT1G36180
|
ACC2, acetyl-CoA carboxylase 2 |
0.929 |
0.021 |
| 3 |
260522_x_at |
AT2G41730
|
unknown |
0.893 |
-0.012 |
| 4 |
263515_at |
AT2G21640
|
unknown |
0.830 |
0.065 |
| 5 |
263403_at |
AT2G04040
|
ATDTX1, DTX1, detoxification 1, TX1 |
0.796 |
0.078 |
| 6 |
254079_at |
AT4G25730
|
[FtsJ-like methyltransferase family protein] |
0.686 |
0.456 |
| 7 |
245392_at |
AT4G15680
|
[Thioredoxin superfamily protein] |
0.675 |
0.365 |
| 8 |
265588_at |
AT2G19970
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
0.651 |
-0.044 |
| 9 |
261449_at |
AT1G21120
|
IGMT2, indole glucosinolate O-methyltransferase 2 |
0.609 |
-0.080 |
| 10 |
247615_at |
AT5G60250
|
[zinc finger (C3HC4-type RING finger) family protein] |
0.606 |
0.011 |
| 11 |
264346_at |
AT1G12010
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.593 |
-0.074 |
| 12 |
261135_at |
AT1G19610
|
LCR78, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 78, PDF1.4 |
0.589 |
-0.017 |
| 13 |
267472_at |
AT2G02850
|
ARPN, plantacyanin |
0.578 |
-0.066 |
| 14 |
251248_at |
AT3G62150
|
ABCB21, ATP-binding cassette B21, PGP21, P-glycoprotein 21 |
0.577 |
-0.236 |
| 15 |
257947_at |
AT3G21720
|
ICL, isocitrate lyase |
0.572 |
-0.013 |
| 16 |
255500_at |
AT4G02390
|
APP, poly(ADP-ribose) polymerase, PARP2, poly(ADP-ribose) polymerase 2, PP, poly(ADP-ribose) polymerase |
0.544 |
0.081 |
| 17 |
260824_at |
AT1G06720
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.542 |
0.551 |
| 18 |
259511_at |
AT1G12520
|
ATCCS, copper chaperone for SOD1, CCS, copper chaperone for SOD1 |
0.541 |
-0.036 |
| 19 |
252305_at |
AT3G49240
|
emb1796, embryo defective 1796 |
0.510 |
0.608 |
| 20 |
249867_at |
AT5G23020
|
IMS2, 2-isopropylmalate synthase 2, MAM-L, METHYLTHIOALKYMALATE SYNTHASE-LIKE, MAM3 |
0.499 |
0.000 |
| 21 |
266165_at |
AT2G28190
|
CSD2, copper/zinc superoxide dismutase 2, CZSOD2, COPPER/ZINC SUPEROXIDE DISMUTASE 2 |
0.482 |
0.015 |
| 22 |
251281_at |
AT3G61640
|
AGP20, arabinogalactan protein 20, AtAGP20, Arabinogalactan protein 20 |
0.470 |
0.076 |
| 23 |
264460_at |
AT1G10170
|
ATNFXL1, NF-X-like 1, NFXL1, NF-X-like 1 |
0.453 |
-0.018 |
| 24 |
259579_at |
AT1G28010
|
ABCB14, ATP-binding cassette B14, ATABCB14, Arabidopsis thaliana ATP-binding cassette B14, MDR12, multi-drug resistance 12, PGP14, P-glycoprotein 14 |
0.445 |
0.221 |
| 25 |
256230_at |
AT3G12340
|
[FKBP-like peptidyl-prolyl cis-trans isomerase family protein] |
0.445 |
0.389 |
| 26 |
255110_at |
AT4G08770
|
Prx37, peroxidase 37 |
0.442 |
-0.105 |
| 27 |
260605_at |
AT2G43780
|
unknown |
0.434 |
0.274 |
| 28 |
245063_at |
AT2G39795
|
[Mitochondrial glycoprotein family protein] |
0.434 |
0.380 |
| 29 |
266532_at |
AT2G16890
|
[UDP-Glycosyltransferase superfamily protein] |
0.432 |
0.234 |
| 30 |
265061_at |
AT1G61640
|
[Protein kinase superfamily protein] |
0.429 |
0.292 |
| 31 |
261192_at |
AT1G32870
|
ANAC013, Arabidopsis NAC domain containing protein 13, ANAC13, NAC domain protein 13, NAC13, NAC domain protein 13 |
0.425 |
0.045 |
| 32 |
247497_at |
AT5G61770
|
PPAN, PETER PAN-like protein |
0.414 |
0.424 |
| 33 |
250825_at |
AT5G05210
|
[Surfeit locus protein 6] |
0.405 |
0.274 |
| 34 |
267371_at |
AT2G44510
|
[CDK inhibitor P21 binding protein] |
0.404 |
0.491 |
| 35 |
252281_at |
AT3G49320
|
[Metal-dependent protein hydrolase] |
0.400 |
0.386 |
| 36 |
250680_at |
AT5G06570
|
[alpha/beta-Hydrolases superfamily protein] |
0.397 |
0.269 |
| 37 |
256285_at |
AT3G12510
|
[MADS-box family protein] |
0.395 |
0.093 |
| 38 |
253777_at |
AT4G28450
|
[nucleotide binding] |
0.391 |
0.398 |
| 39 |
261317_at |
AT1G53030
|
[Cytochrome C oxidase copper chaperone (COX17)] |
0.390 |
-0.133 |
| 40 |
264895_at |
AT1G23100
|
[GroES-like family protein] |
0.387 |
0.487 |
| 41 |
247491_at |
AT5G61880
|
[Protein Transporter, Pam16] |
0.386 |
0.283 |
| 42 |
259444_at |
AT1G02370
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.380 |
0.407 |
| 43 |
258019_at |
AT3G19470
|
[F-box and associated interaction domains-containing protein] |
0.380 |
0.092 |
| 44 |
255011_at |
AT4G10040
|
CYTC-2, cytochrome c-2 |
0.376 |
0.009 |
| 45 |
248228_at |
AT5G53800
|
unknown |
0.374 |
0.066 |
| 46 |
250152_at |
AT5G15120
|
unknown |
0.374 |
-0.177 |
| 47 |
264723_at |
AT1G22960
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.367 |
0.155 |
| 48 |
258545_at |
AT3G07050
|
NSN1, nucleostemin-like 1 |
0.364 |
0.342 |
| 49 |
263161_at |
AT1G54020
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.363 |
0.199 |
| 50 |
247855_at |
AT5G58210
|
[hydroxyproline-rich glycoprotein family protein] |
0.362 |
0.187 |
| 51 |
256998_at |
AT3G14180
|
ASIL2, Arabidopsis 6B-interacting protein 1-like 2 |
0.358 |
-0.046 |
| 52 |
261524_at |
AT1G14300
|
[ARM repeat superfamily protein] |
0.358 |
0.186 |
| 53 |
249253_at |
AT5G42060
|
[DEK, chromatin associated protein] |
0.358 |
0.119 |
| 54 |
263766_at |
AT2G21440
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
0.357 |
0.335 |
| 55 |
254778_at |
AT4G12750
|
[Homeodomain-like transcriptional regulator] |
0.356 |
0.367 |
| 56 |
265999_at |
AT2G24100
|
ASG1, ALTERED SEED GERMINATION 1 |
0.352 |
-0.033 |
| 57 |
261222_at |
AT1G20120
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.350 |
-0.097 |
| 58 |
257058_at |
AT3G15352
|
ATCOX17, ARABIDOPSIS THALIANA CYTOCHROME C OXIDASE 17, COX17, cytochrome c oxidase 17 |
0.342 |
0.243 |
| 59 |
250196_at |
AT5G14580
|
[polyribonucleotide nucleotidyltransferase, putative] |
0.341 |
0.371 |
| 60 |
259110_at |
AT3G05570
|
unknown |
0.341 |
0.032 |
| 61 |
248611_at |
AT5G49520
|
ATWRKY48, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 48, WRKY48, WRKY DNA-binding protein 48 |
0.340 |
-0.195 |
| 62 |
253175_at |
AT4G35050
|
MSI3, MULTICOPY SUPPRESSOR OF IRA1 3, NFC3, NUCLEOSOME/CHROMATIN ASSEMBLY FACTOR GROUP C 3 |
0.339 |
-0.031 |
| 63 |
265067_at |
AT1G03850
|
ATGRXS13, glutaredoxin 13, GRXS13, glutaredoxin 13 |
0.337 |
-0.030 |
| 64 |
264809_at |
AT1G08830
|
CSD1, copper/zinc superoxide dismutase 1 |
0.335 |
-0.121 |
| 65 |
266264_at |
AT2G27775
|
unknown |
0.335 |
0.266 |
| 66 |
258910_at |
AT3G06480
|
[DEAD box RNA helicase family protein] |
0.327 |
0.122 |
| 67 |
259899_at |
AT1G71210
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.327 |
0.360 |
| 68 |
263039_at |
AT1G23280
|
[MAK16 protein-related] |
0.325 |
0.475 |
| 69 |
258747_at |
AT3G05810
|
unknown |
0.318 |
0.190 |
| 70 |
250506_at |
AT5G09930
|
ABCF2, ATP-binding cassette F2 |
0.318 |
-0.007 |
| 71 |
253566_at |
AT4G31210
|
[DNA topoisomerase, type IA, core] |
0.315 |
0.330 |
| 72 |
261027_at |
AT1G01340
|
ACBK1, ATCNGC10, cyclic nucleotide gated channel 10, CNGC10, cyclic nucleotide gated channel 10 |
0.314 |
-0.067 |
| 73 |
260732_at |
AT1G17520
|
[Homeodomain-like/winged-helix DNA-binding family protein] |
0.312 |
0.067 |
| 74 |
251678_at |
AT3G56990
|
EDA7, embryo sac development arrest 7 |
0.312 |
0.391 |
| 75 |
247339_at |
AT5G63690
|
[Nucleic acid-binding, OB-fold-like protein] |
0.312 |
0.000 |
| 76 |
263707_at |
AT1G09300
|
[Metallopeptidase M24 family protein] |
0.311 |
0.135 |
| 77 |
261435_at |
AT1G07615
|
[GTP-binding protein Obg/CgtA] |
0.311 |
0.137 |
| 78 |
258227_at |
AT3G15620
|
UVR3, UV REPAIR DEFECTIVE 3 |
0.311 |
-0.076 |
| 79 |
251650_at |
AT3G57360
|
unknown |
0.310 |
0.005 |
| 80 |
253388_at |
AT4G32910
|
unknown |
0.308 |
0.014 |
| 81 |
260343_at |
AT1G69200
|
FLN2, fructokinase-like 2 |
0.307 |
0.286 |
| 82 |
267309_at |
AT2G19385
|
[zinc ion binding] |
0.303 |
0.378 |
| 83 |
254665_at |
AT4G18340
|
[Glycosyl hydrolase superfamily protein] |
0.303 |
0.012 |
| 84 |
262777_at |
AT1G13030
|
Atcoilin, COILIN, coilin |
0.303 |
0.380 |
| 85 |
264175_at |
AT1G02050
|
LAP6, LESS ADHESIVE POLLEN 6, PSKA, polyketide synthase A |
0.301 |
-0.014 |
| 86 |
257667_at |
AT3G20440
|
BE1, BRANCHING ENZYME 1, EMB2729, EMBRYO DEFECTIVE 2729 |
0.301 |
0.143 |
| 87 |
248668_at |
AT5G48720
|
XRI, X-RAY INDUCED TRANSCRIPT, XRI1, X-RAY INDUCED TRANSCRIPT 1 |
0.300 |
0.069 |
| 88 |
259126_at |
AT3G02280
|
AtTAH18, TAH18 |
0.299 |
0.180 |
| 89 |
259225_at |
AT3G07590
|
[Small nuclear ribonucleoprotein family protein] |
0.298 |
0.356 |
| 90 |
247607_at |
AT5G60960
|
PNM1, PPR protein localized to the nucleus and mitochondria 1 |
0.295 |
0.336 |
| 91 |
251195_at |
AT3G62930
|
[Thioredoxin superfamily protein] |
0.294 |
0.537 |
| 92 |
259718_at |
AT1G61040
|
VIP5, vernalization independence 5 |
0.287 |
0.003 |
| 93 |
245951_at |
AT5G19550
|
AAT2, ASPARTATE AMINOTRANSFERASE 2, ASP2, aspartate aminotransferase 2 |
0.287 |
0.047 |
| 94 |
247449_at |
AT5G62290
|
[nucleotide-sensitive chloride conductance regulator (ICln) family protein] |
0.283 |
0.163 |
| 95 |
249656_at |
AT5G37130
|
[Protein prenylyltransferase superfamily protein] |
0.282 |
0.162 |
| 96 |
254455_at |
AT4G21140
|
unknown |
0.281 |
0.374 |
| 97 |
250774_at |
AT5G05450
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.280 |
0.064 |
| 98 |
249830_at |
AT5G23300
|
PYRD, pyrimidine d |
0.279 |
0.319 |
| 99 |
255887_at |
AT1G20370
|
[Pseudouridine synthase family protein] |
0.277 |
0.310 |
| 100 |
256107_at |
AT1G16830
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.276 |
0.256 |
| 101 |
247327_at |
AT5G64120
|
AtPRX71, PRX71, peroxidase 71 |
0.276 |
0.029 |
| 102 |
252043_at |
AT3G52390
|
[TatD related DNase] |
0.273 |
-0.052 |
| 103 |
257830_at |
AT3G26690
|
ATNUDT13, ARABIDOPSIS THALIANA NUDIX HYDROLASE HOMOLOG 13, ATNUDX13, nudix hydrolase homolog 13, NUDX13, nudix hydrolase homolog 13 |
0.272 |
-0.078 |
| 104 |
262574_at |
AT1G15230
|
unknown |
0.272 |
-0.011 |
| 105 |
247197_at |
AT5G65240
|
[Leucine-rich repeat protein kinase family protein] |
0.270 |
0.108 |
| 106 |
265383_at |
AT2G16780
|
MSI02, MSI2, MULTICOPY SUPPRESSOR OF IRA1 2, NFC02, NFC2, NUCLEOSOME/CHROMATIN ASSEMBLY FACTOR GROUP C 2 |
0.270 |
0.050 |
| 107 |
265865_at |
AT2G01740
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.268 |
0.273 |
| 108 |
253733_at |
AT4G29170
|
ATMND1 |
0.268 |
-0.000 |
| 109 |
261818_at |
AT1G11390
|
[Protein kinase superfamily protein] |
0.267 |
0.117 |
| 110 |
252252_at |
AT3G49180
|
RID3, ROOT INITIATION DEFECTIVE 3 |
0.267 |
0.197 |
| 111 |
262657_at |
AT1G14210
|
[Ribonuclease T2 family protein] |
0.264 |
0.004 |
| 112 |
253013_at |
AT4G37910
|
mtHsc70-1, mitochondrial heat shock protein 70-1 |
0.264 |
0.204 |
| 113 |
256248_at |
AT3G66652
|
[fip1 motif-containing protein] |
0.263 |
0.205 |
| 114 |
249528_at |
AT5G38720
|
unknown |
0.262 |
0.340 |
| 115 |
247403_at |
AT5G62740
|
ATHIR1, AtHIR4, HIR1, HYPERSENSITIVE-INDUCED RESPONSE PROTEIN 1, HIR4, hypersensitive induced reaction 4 |
0.262 |
0.059 |
| 116 |
260157_at |
AT1G52930
|
[Ribosomal RNA processing Brix domain protein] |
0.261 |
0.298 |
| 117 |
246436_at |
AT5G17440
|
[LUC7 related protein] |
0.261 |
0.028 |
| 118 |
264551_at |
AT1G09460
|
[Carbohydrate-binding X8 domain superfamily protein] |
0.260 |
0.136 |
| 119 |
245523_at |
AT4G15910
|
ATDI21, drought-induced 21, DI21, drought-induced 21 |
0.260 |
-0.018 |
| 120 |
248631_at |
AT5G49000
|
[Galactose oxidase/kelch repeat superfamily protein] |
0.258 |
0.052 |
| 121 |
249782_at |
AT5G24260
|
[prolyl oligopeptidase family protein] |
0.258 |
-0.049 |
| 122 |
246098_at |
AT5G20400
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.257 |
-0.210 |
| 123 |
248722_at |
AT5G47810
|
PFK2, phosphofructokinase 2 |
0.257 |
0.037 |
| 124 |
251800_at |
AT3G55510
|
RBL, REBELOTE |
0.257 |
0.424 |
| 125 |
255147_at |
AT4G08460
|
[FUNCTIONS IN: zinc ion binding] |
0.256 |
0.073 |
| 126 |
253559_at |
AT4G31140
|
[O-Glycosyl hydrolases family 17 protein] |
0.255 |
0.228 |
| 127 |
250418_at |
AT5G11240
|
[transducin family protein / WD-40 repeat family protein] |
0.254 |
0.378 |
| 128 |
256480_at |
AT1G33410
|
ATNUP160, ARABIDOPSIS NUCLEOPORIN 160, NUP160, NUCLEOPORIN 160, SAR1, SUPPRESSOR OF AUXIN RESISTANCE1 |
0.254 |
0.198 |
| 129 |
246765_at |
AT5G27330
|
[Prefoldin chaperone subunit family protein] |
0.253 |
0.325 |
| 130 |
265006_at |
AT1G61570
|
TIM13, translocase of the inner mitochondrial membrane 13 |
0.252 |
0.249 |
| 131 |
258812_at |
AT3G03950
|
ECT1, evolutionarily conserved C-terminal region 1 |
0.252 |
0.101 |
| 132 |
255586_at |
AT4G01560
|
MEE49, maternal effect embryo arrest 49 |
0.251 |
0.259 |
| 133 |
259694_at |
AT1G63180
|
UGE3, UDP-D-glucose/UDP-D-galactose 4-epimerase 3 |
0.250 |
-0.114 |
| 134 |
254493_at |
AT4G20020
|
MORF1, multiple organellar RNA editing factor 1 |
0.250 |
0.341 |
| 135 |
251740_at |
AT3G56070
|
ROC2, rotamase cyclophilin 2 |
0.246 |
0.302 |
| 136 |
246855_at |
AT5G26280
|
[TRAF-like family protein] |
0.245 |
0.117 |
| 137 |
250326_at |
AT5G12080
|
ATMSL10, MSL10, mechanosensitive channel of small conductance-like 10 |
0.243 |
0.248 |
| 138 |
250192_at |
AT5G14520
|
PES, PESCADILLO |
0.243 |
0.172 |
| 139 |
255225_at |
AT4G05410
|
YAO, YAOZHE |
0.242 |
0.410 |
| 140 |
247014_at |
AT5G67630
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.241 |
0.183 |
| 141 |
250225_at |
AT5G14105
|
unknown |
0.238 |
0.101 |
| 142 |
258164_at |
AT3G17910
|
EMB3121, EMBRYO DEFECTIVE 3121, SURF1, SURFEIT 1 |
0.235 |
0.138 |
| 143 |
259161_at |
AT3G01500
|
ATBCA1, BETA CARBONIC ANHYDRASE 1, ATSABP3, ARABIDOPSIS THALIANA SALICYLIC ACID-BINDING PROTEIN 3, CA1, carbonic anhydrase 1, SABP3, SALICYLIC ACID-BINDING PROTEIN 3 |
-1.349 |
-0.051 |
| 144 |
258055_at |
AT3G16250
|
NDF4, NDH-dependent cyclic electron flow 1, PnsB3, Photosynthetic NDH subcomplex B 3 |
-1.331 |
-0.115 |
| 145 |
253971_at |
AT4G26530
|
AtFBA5, FBA5, fructose-bisphosphate aldolase 5 |
-1.307 |
-0.351 |
| 146 |
261769_at |
AT1G76100
|
PETE1, plastocyanin 1 |
-1.260 |
-0.110 |
| 147 |
256603_at |
AT3G28270
|
unknown |
-1.142 |
0.184 |
| 148 |
252618_at |
AT3G45140
|
ATLOX2, ARABIODOPSIS THALIANA LIPOXYGENASE 2, LOX2, lipoxygenase 2 |
-1.119 |
-0.040 |
| 149 |
258956_at |
AT3G01440
|
PnsL3, Photosynthetic NDH subcomplex L 3, PQL1, PsbQ-like 1, PQL2, PsbQ-like 2 |
-1.106 |
-0.032 |
| 150 |
249053_at |
AT5G44440
|
[FAD-binding Berberine family protein] |
-1.066 |
0.030 |
| 151 |
255016_at |
AT4G10120
|
ATSPS4F |
-1.033 |
-0.411 |
| 152 |
258239_at |
AT3G27690
|
LHCB2, LIGHT-HARVESTING CHLOROPHYLL B-BINDING 2, LHCB2.3, photosystem II light harvesting complex gene 2.3, LHCB2.4 |
-1.024 |
-0.165 |
| 153 |
248683_at |
AT5G48490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.989 |
-0.242 |
| 154 |
251155_at |
AT3G63160
|
OEP6, outer envelope protein 6 |
-0.964 |
-0.043 |
| 155 |
262309_at |
AT1G70820
|
[phosphoglucomutase, putative / glucose phosphomutase, putative] |
-0.896 |
-0.174 |
| 156 |
253344_at |
AT4G33550
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.889 |
-0.157 |
| 157 |
255248_at |
AT4G05180
|
PSBQ, PHOTOSYSTEM II SUBUNIT Q, PSBQ-2, photosystem II subunit Q-2, PSII-Q |
-0.864 |
-0.042 |
| 158 |
258498_at |
AT3G02480
|
[Late embryogenesis abundant protein (LEA) family protein] |
-0.859 |
-0.004 |
| 159 |
262612_at |
AT1G14150
|
PnsL2, Photosynthetic NDH subcomplex L 2, PQL1, PsbQ-like 1, PQL2, PsbQ-like 2 |
-0.851 |
-0.181 |
| 160 |
249599_at |
AT5G37990
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.850 |
-0.089 |
| 161 |
265611_at |
AT2G25510
|
unknown |
-0.845 |
-0.033 |
| 162 |
256015_at |
AT1G19150
|
LHCA2*1, Lhca6, photosystem I light harvesting complex gene 6 |
-0.841 |
-0.184 |
| 163 |
266899_at |
AT2G34620
|
[Mitochondrial transcription termination factor family protein] |
-0.832 |
-0.223 |
| 164 |
251028_at |
AT5G02230
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.818 |
-0.023 |
| 165 |
261118_at |
AT1G75460
|
[ATP-dependent protease La (LON) domain protein] |
-0.808 |
0.059 |
| 166 |
247718_at |
AT5G59310
|
LTP4, lipid transfer protein 4 |
-0.806 |
-0.076 |
| 167 |
259789_at |
AT1G29395
|
COR413-TM1, COLD REGULATED 314 THYLAKOID MEMBRANE 1, COR413IM1, COLD REGULATED 314 INNER MEMBRANE 1, COR414-TM1, cold regulated 414 thylakoid membrane 1 |
-0.793 |
-0.072 |
| 168 |
249482_at |
AT5G38980
|
unknown |
-0.793 |
-0.050 |
| 169 |
250255_at |
AT5G13730
|
SIG4, sigma factor 4, SIGD |
-0.786 |
-0.107 |
| 170 |
261684_at |
AT1G47400
|
unknown |
-0.776 |
0.316 |
| 171 |
258025_at |
AT3G19480
|
3-PGDH, PGDH3, phosphoglycerate dehydrogenase 3 |
-0.752 |
-0.194 |
| 172 |
252853_at |
AT4G39710
|
FKBP16-2, FK506-binding protein 16-2, PnsL4, Photosynthetic NDH subcomplex L 4 |
-0.742 |
-0.185 |
| 173 |
253871_at |
AT4G27440
|
PORB, protochlorophyllide oxidoreductase B |
-0.737 |
-0.059 |
| 174 |
263597_at |
AT2G01870
|
unknown |
-0.728 |
0.000 |
| 175 |
244995_at |
ATCG00150
|
ATPI |
-0.697 |
-0.038 |
| 176 |
256427_at |
AT3G11090
|
LBD21, LOB domain-containing protein 21 |
-0.695 |
-0.117 |
| 177 |
262382_at |
AT1G72920
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
-0.694 |
-0.154 |
| 178 |
266979_at |
AT2G39470
|
PnsL1, Photosynthetic NDH subcomplex L 1, PPL2, PsbP-like protein 2 |
-0.693 |
-0.157 |
| 179 |
261431_at |
AT1G18710
|
AtMYB47, myb domain protein 47, MYB47, myb domain protein 47 |
-0.691 |
0.097 |
| 180 |
254232_at |
AT4G23600
|
CORI3, CORONATINE INDUCED 1, JR2, JASMONIC ACID RESPONSIVE 2 |
-0.682 |
0.070 |
| 181 |
262693_at |
AT1G62780
|
unknown |
-0.682 |
-0.122 |
| 182 |
256577_at |
AT3G28220
|
[TRAF-like family protein] |
-0.680 |
-0.037 |
| 183 |
264037_at |
AT2G03750
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.677 |
0.045 |
| 184 |
254305_at |
AT4G22200
|
AKT2, AKT2/3, potassium transport 2/3, AKT3, KT2/3, potassium transport 2/3 |
-0.673 |
0.287 |
| 185 |
254564_at |
AT4G19170
|
CCD4, carotenoid cleavage dioxygenase 4, NCED4, nine-cis-epoxycarotenoid dioxygenase 4 |
-0.665 |
0.034 |
| 186 |
260957_at |
AT1G06080
|
ADS1, delta 9 desaturase 1, AtADS1 |
-0.661 |
-0.164 |
| 187 |
263114_at |
AT1G03130
|
PSAD-2, photosystem I subunit D-2 |
-0.660 |
-0.076 |
| 188 |
245692_at |
AT5G04150
|
BHLH101 |
-0.654 |
-0.076 |
| 189 |
266392_at |
AT2G41280
|
ATM10, M10 |
-0.654 |
0.088 |
| 190 |
260541_at |
AT2G43530
|
[Scorpion toxin-like knottin superfamily protein] |
-0.652 |
-0.014 |
| 191 |
260380_at |
AT1G73870
|
BBX16, B-box domain protein 16, COL7, CONSTANS-LIKE 7 |
-0.651 |
-0.115 |
| 192 |
262288_at |
AT1G70760
|
CRR23, CHLORORESPIRATORY REDUCTION 23, NdhL, NADH dehydrogenase-like complex L |
-0.649 |
-0.164 |
| 193 |
249010_at |
AT5G44580
|
unknown |
-0.637 |
-0.060 |
| 194 |
267457_at |
AT2G33790
|
AGP30, arabinogalactan protein 30, ATAGP30 |
-0.636 |
-0.005 |
| 195 |
259185_at |
AT3G01550
|
ATPPT2, PHOSPHOENOLPYRUVATE (PEP)/PHOSPHATE TRANSLOCATOR 2, PPT2, phosphoenolpyruvate (pep)/phosphate translocator 2 |
-0.634 |
-0.329 |
| 196 |
255457_at |
AT4G02770
|
PSAD-1, photosystem I subunit D-1 |
-0.632 |
-0.017 |
| 197 |
263890_at |
AT2G37030
|
SAUR46, SMALL AUXIN UPREGULATED RNA 46 |
-0.631 |
-0.041 |
| 198 |
258285_at |
AT3G16140
|
PSAH-1, photosystem I subunit H-1 |
-0.629 |
-0.048 |
| 199 |
254783_at |
AT4G12830
|
[alpha/beta-Hydrolases superfamily protein] |
-0.628 |
-0.098 |
| 200 |
248352_at |
AT5G52300
|
LTI65, LOW-TEMPERATURE-INDUCED 65, RD29B, RESPONSIVE TO DESSICATION 29B |
-0.628 |
-0.048 |
| 201 |
248793_at |
AT5G47240
|
atnudt8, nudix hydrolase homolog 8, NUDT8, nudix hydrolase homolog 8 |
-0.627 |
-0.035 |
| 202 |
249035_at |
AT5G44190
|
ATGLK2, GLK2, GOLDEN2-like 2, GPRI2, GBF'S PRO-RICH REGION-INTERACTING FACTOR 2 |
-0.626 |
0.128 |
| 203 |
260203_at |
AT1G52890
|
ANAC019, NAC domain containing protein 19, NAC019, NAC domain containing protein 19 |
-0.621 |
0.025 |
| 204 |
254250_at |
AT4G23290
|
CRK21, cysteine-rich RLK (RECEPTOR-like protein kinase) 21 |
-0.616 |
-0.088 |
| 205 |
264839_at |
AT1G03630
|
POR C, protochlorophyllide oxidoreductase C, PORC |
-0.612 |
-0.094 |
| 206 |
260547_at |
AT2G43550
|
[Scorpion toxin-like knottin superfamily protein] |
-0.603 |
0.090 |
| 207 |
261046_at |
AT1G01390
|
[UDP-Glycosyltransferase superfamily protein] |
-0.601 |
-0.054 |
| 208 |
259707_at |
AT1G77490
|
TAPX, thylakoidal ascorbate peroxidase |
-0.600 |
-0.048 |
| 209 |
255088_at |
AT4G09350
|
CRRJ, CHLORORESPIRATORY REDUCTION J, DJC75, DNA J protein C75, NdhT, NADH dehydrogenase-like complex T |
-0.598 |
0.102 |
| 210 |
250054_at |
AT5G17860
|
CAX7, calcium exchanger 7 |
-0.594 |
0.031 |
| 211 |
265170_at |
AT1G23730
|
ATBCA3, BETA CARBONIC ANHYDRASE 3, BCA3, beta carbonic anhydrase 3 |
-0.586 |
-0.170 |
| 212 |
253849_at |
AT4G28080
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.585 |
-0.090 |
| 213 |
259640_at |
AT1G52400
|
ATBG1, A. THALIANA BETA-GLUCOSIDASE 1, BGL1, BETA-GLUCOSIDASE HOMOLOG 1, BGLU18, beta glucosidase 18 |
-0.584 |
0.008 |
| 214 |
245877_at |
AT1G26220
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
-0.578 |
-0.045 |
| 215 |
247486_at |
AT5G62140
|
unknown |
-0.574 |
0.004 |
| 216 |
255957_at |
AT1G22160
|
unknown |
-0.573 |
-0.436 |
| 217 |
262175_at |
AT1G74880
|
NDH-O, NAD(P)H:plastoquinone dehydrogenase complex subunit O, NdhO, NADH dehydrogenase-like complex ) |
-0.572 |
-0.138 |
| 218 |
266551_at |
AT2G35260
|
unknown |
-0.570 |
-0.042 |
| 219 |
259840_at |
AT1G52230
|
PSAH-2, PHOTOSYSTEM I SUBUNIT H-2, PSAH2, photosystem I subunit H2, PSI-H |
-0.569 |
-0.024 |
| 220 |
252366_at |
AT3G48420
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.569 |
-0.166 |
| 221 |
261191_at |
AT1G32900
|
GBSS1, granule bound starch synthase 1 |
-0.566 |
-0.210 |
| 222 |
248377_at |
AT5G51720
|
At-NEET, NEET, NEET group protein |
-0.564 |
-0.300 |
| 223 |
265539_at |
AT2G15830
|
unknown |
-0.562 |
-0.361 |
| 224 |
264902_at |
AT1G23060
|
MDP40, MICROTUBULE DESTABILIZING PROTEIN 40 |
-0.561 |
-0.080 |
| 225 |
248151_at |
AT5G54270
|
LHCB3, light-harvesting chlorophyll B-binding protein 3, LHCB3*1 |
-0.558 |
-0.052 |
| 226 |
265053_at |
AT1G52000
|
[Mannose-binding lectin superfamily protein] |
-0.556 |
0.102 |
| 227 |
262313_at |
AT1G70900
|
unknown |
-0.547 |
-0.039 |
| 228 |
247816_at |
AT5G58260
|
NdhN, NADH dehydrogenase-like complex N |
-0.546 |
-0.116 |
| 229 |
266572_at |
AT2G23840
|
[HNH endonuclease] |
-0.545 |
-0.214 |
| 230 |
257481_at |
AT1G08430
|
ALMT1, aluminum-activated malate transporter 1, ATALMT1, ARABIDOPSIS THALIANA ALUMINUM-ACTIVATED MALATE TRANSPORTER 1 |
-0.537 |
-0.224 |
| 231 |
253337_at |
AT4G33470
|
ATHDA14, HDA14, histone deacetylase 14 |
-0.537 |
-0.101 |
| 232 |
265547_at |
AT2G28305
|
ATLOG1, LOG1, LONELY GUY 1 |
-0.535 |
-0.286 |
| 233 |
259188_at |
AT3G01510
|
LSF1, like SEX4 1 |
-0.535 |
-0.099 |
| 234 |
267635_at |
AT2G42220
|
[Rhodanese/Cell cycle control phosphatase superfamily protein] |
-0.534 |
-0.063 |
| 235 |
267080_at |
AT2G41190
|
[Transmembrane amino acid transporter family protein] |
-0.533 |
-0.079 |
| 236 |
263495_at |
AT2G42530
|
COR15B, cold regulated 15b |
-0.533 |
-0.230 |
| 237 |
257635_at |
AT3G26280
|
CYP71B4, cytochrome P450, family 71, subfamily B, polypeptide 4 |
-0.532 |
-0.116 |
| 238 |
250960_at |
AT5G02940
|
unknown |
-0.526 |
-0.173 |
| 239 |
250828_at |
AT5G05250
|
unknown |
-0.520 |
0.028 |
| 240 |
260716_at |
AT1G48130
|
ATPER1, 1-cysteine peroxiredoxin 1, PER1, 1-cysteine peroxiredoxin 1 |
-0.520 |
0.056 |
| 241 |
251928_at |
AT3G53980
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.516 |
-0.081 |
| 242 |
251013_at |
AT5G02540
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.515 |
0.063 |
| 243 |
244974_at |
ATCG00700
|
PSBN, photosystem II reaction center protein N |
-0.512 |
-0.007 |
| 244 |
266277_at |
AT2G29310
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.512 |
-0.224 |
| 245 |
262050_at |
AT1G80130
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.511 |
0.206 |
| 246 |
246159_at |
AT5G20935
|
CRR42, CHLORORESPIRATORY REDUCTION 42 |
-0.511 |
-0.070 |
| 247 |
251017_at |
AT5G02760
|
APD7, Arabidopsis Pp2c clade D 7 |
-0.510 |
-0.043 |
| 248 |
261746_at |
AT1G08380
|
PSAO, photosystem I subunit O |
-0.502 |
-0.050 |
| 249 |
249876_at |
AT5G23060
|
CaS, calcium sensing receptor |
-0.501 |
-0.060 |
| 250 |
252199_at |
AT3G50270
|
[HXXXD-type acyl-transferase family protein] |
-0.498 |
0.119 |
| 251 |
245325_at |
AT4G14130
|
XTH15, xyloglucan endotransglucosylase/hydrolase 15, XTR7, xyloglucan endotransglycosylase 7 |
-0.496 |
-0.471 |
| 252 |
262211_at |
AT1G74930
|
ORA47 |
-0.491 |
-0.212 |
| 253 |
246998_at |
AT5G67370
|
CGLD27, CONSERVED IN THE GREEN LINEAGE AND DIATOMS 27 |
-0.490 |
-0.070 |
| 254 |
256497_at |
AT1G31580
|
CXC750, ECS1 |
-0.489 |
-0.073 |
| 255 |
256655_at |
AT3G18890
|
AtTic62, translocon at the inner envelope membrane of chloroplasts 62, Tic62, translocon at the inner envelope membrane of chloroplasts 62 |
-0.489 |
-0.086 |
| 256 |
256611_at |
AT3G29270
|
[RING/U-box superfamily protein] |
-0.488 |
0.103 |
| 257 |
256979_at |
AT3G21055
|
PSBTN, photosystem II subunit T |
-0.483 |
-0.040 |
| 258 |
256336_at |
AT1G72030
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
-0.482 |
-0.009 |
| 259 |
257832_at |
AT3G26740
|
CCL, CCR-like |
-0.480 |
-0.108 |
| 260 |
253823_at |
AT4G28030
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
-0.480 |
-0.149 |
| 261 |
253860_at |
AT4G27700
|
[Rhodanese/Cell cycle control phosphatase superfamily protein] |
-0.478 |
-0.084 |
| 262 |
259791_at |
AT1G29700
|
[Metallo-hydrolase/oxidoreductase superfamily protein] |
-0.476 |
-0.283 |
| 263 |
249353_at |
AT5G40420
|
OLE2, OLEOSIN 2, OLEO2, oleosin 2 |
-0.476 |
-0.019 |
| 264 |
254150_at |
AT4G24350
|
[Phosphorylase superfamily protein] |
-0.475 |
0.006 |
| 265 |
246310_at |
AT3G51895
|
AST12, SULTR3;1, sulfate transporter 3;1 |
-0.473 |
-0.139 |
| 266 |
266673_at |
AT2G29630
|
PY, PYRIMIDINE REQUIRING, THIC, thiaminC |
-0.471 |
-0.105 |
| 267 |
246596_at |
AT5G14740
|
BETA CA2, BETA CARBONIC ANHYDRASE 2, CA18, CARBONIC ANHYDRASE 18, CA2, carbonic anhydrase 2 |
-0.470 |
-0.080 |
| 268 |
265287_at |
AT2G20260
|
PSAE-2, photosystem I subunit E-2 |
-0.468 |
-0.041 |
| 269 |
263142_at |
AT1G65230
|
[Uncharacterized conserved protein (DUF2358)] |
-0.468 |
-0.167 |
| 270 |
256073_at |
AT1G18100
|
E12A11, MFT, MOTHER OF FT AND TFL1 |
-0.466 |
-0.075 |
| 271 |
265722_at |
AT2G40100
|
LHCB4.3, light harvesting complex photosystem II |
-0.466 |
-0.170 |
| 272 |
260704_at |
AT1G32470
|
[Single hybrid motif superfamily protein] |
-0.464 |
-0.064 |
| 273 |
247760_at |
AT5G59130
|
[Subtilase family protein] |
-0.462 |
-0.148 |
| 274 |
261218_at |
AT1G20020
|
ATLFNR2, LEAF FNR 2, FNR2, ferredoxin-NADP(+)-oxidoreductase 2, LFNR2, leaf-type chloroplast-targeted FNR 2 |
-0.461 |
-0.064 |
| 275 |
259340_at |
AT3G03870
|
unknown |
-0.461 |
-0.238 |
| 276 |
245777_at |
AT1G73540
|
atnudt21, nudix hydrolase homolog 21, NUDT21, nudix hydrolase homolog 21 |
-0.459 |
-0.299 |
| 277 |
255719_at |
AT1G32080
|
AtLrgB, LrgB |
-0.456 |
-0.158 |
| 278 |
261881_at |
AT1G80760
|
NIP6, NIP6;1, NOD26-like intrinsic protein 6;1, NLM7 |
-0.453 |
-0.116 |
| 279 |
262830_at |
AT1G14700
|
ATPAP3, PAP3, purple acid phosphatase 3 |
-0.452 |
0.051 |
| 280 |
251762_at |
AT3G55800
|
SBPASE, sedoheptulose-bisphosphatase |
-0.451 |
-0.099 |
| 281 |
263676_at |
AT1G09340
|
CRB, chloroplast RNA binding, CSP41B, CHLOROPLAST STEM-LOOP BINDING PROTEIN OF 41 KDA, HIP1.3, heteroglycan-interacting protein 1.3 |
-0.450 |
-0.107 |
| 282 |
266636_at |
AT2G35370
|
GDCH, glycine decarboxylase complex H |
-0.450 |
-0.087 |
| 283 |
254057_at |
AT4G25170
|
[Uncharacterised conserved protein (UCP012943)] |
-0.445 |
-0.070 |
| 284 |
264738_at |
AT1G62250
|
unknown |
-0.444 |
-0.084 |