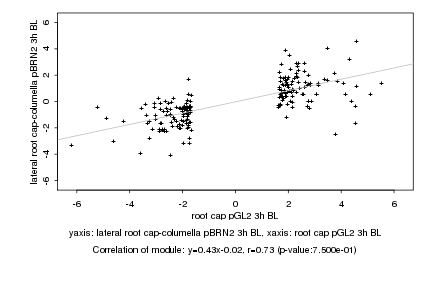
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
root cap pGL2 3h BL |
Log2 signal ratio
lateral root cap-columella pBRN2 3h BL |
| 1 |
AT3G49700 |
AT3G49700
|
ACS9, 1-aminocyclopropane-1-carboxylate synthase 9, AtACS9, ETO3, ETHYLENE OVERPRODUCING 3 |
5.504 |
1.371 |
| 2 |
AT2G02010 |
AT2G02010
|
GAD4, glutamate decarboxylase 4 |
5.093 |
0.566 |
| 3 |
AT4G08620 |
AT4G08620
|
SULTR1;1, sulphate transporter 1;1 |
4.570 |
1.188 |
| 4 |
AT4G21200 |
AT4G21200
|
ATGA2OX8, ARABIDOPSIS THALIANA GIBBERELLIN 2-OXIDASE 8, GA2OX8, gibberellin 2-oxidase 8 |
4.566 |
4.585 |
| 5 |
AT1G52570 |
AT1G52570
|
PLDALPHA2, phospholipase D alpha 2 |
4.540 |
-1.657 |
| 6 |
AT1G35230 |
AT1G35230
|
AGP5, arabinogalactan protein 5 |
4.506 |
-0.325 |
| 7 |
AT5G19880 |
AT5G19880
|
[Peroxidase superfamily protein] |
4.357 |
0.035 |
| 8 |
AT2G17680 |
AT2G17680
|
unknown |
4.288 |
3.222 |
| 9 |
AT2G04515 |
AT2G04515
|
unknown |
4.145 |
0.566 |
| 10 |
AT4G12410 |
AT4G12410
|
SAUR35, SMALL AUXIN UPREGULATED RNA 35 |
4.051 |
1.388 |
| 11 |
AT5G21120 |
AT5G21120
|
EIL2, ETHYLENE-INSENSITIVE3-like 2 |
3.849 |
1.563 |
| 12 |
AT3G14380 |
AT3G14380
|
[Uncharacterised protein family (UPF0497)] |
3.772 |
-2.439 |
| 13 |
AT1G52830 |
AT1G52830
|
IAA6, indole-3-acetic acid 6, SHY1, SHORT HYPOCOTYL 1 |
3.724 |
2.146 |
| 14 |
AT1G29510 |
AT1G29510
|
SAUR67, SMALL AUXIN UPREGULATED RNA 67 |
3.468 |
1.619 |
| 15 |
AT1G29500 |
AT1G29500
|
SAUR66, SMALL AUXIN UPREGULATED RNA 66 |
3.455 |
4.048 |
| 16 |
AT3G03840 |
AT3G03840
|
SAUR27, SMALL AUXIN UP RNA 27 |
3.333 |
1.725 |
| 17 |
AT4G11340 |
AT4G11340
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
3.117 |
1.258 |
| 18 |
AT1G11690 |
AT1G11690
|
unknown |
3.117 |
1.389 |
| 19 |
AT1G66920 |
AT1G66920
|
[Protein kinase superfamily protein] |
3.044 |
0.566 |
| 20 |
AT1G07680 |
AT1G07680
|
unknown |
3.041 |
0.566 |
| 21 |
AT3G45060 |
AT3G45060
|
ATNRT2.6, ARABIDOPSIS THALIANA HIGH AFFINITY NITRATE TRANSPORTER 2.6, NRT2.6, high affinity nitrate transporter 2.6 |
2.848 |
0.035 |
| 22 |
AT5G49360 |
AT5G49360
|
ATBXL1, BETA-XYLOSIDASE 1, BXL1, beta-xylosidase 1 |
2.833 |
1.230 |
| 23 |
AT3G54830 |
AT3G54830
|
[Transmembrane amino acid transporter family protein] |
2.808 |
1.385 |
| 24 |
AT5G50335 |
AT5G50335
|
unknown |
2.788 |
-0.495 |
| 25 |
AT1G79320 |
AT1G79320
|
AtMC6, metacaspase 6, AtMCP2c, metacaspase 2c, MC6, metacaspase 6, MCP2c, metacaspase 2c |
2.762 |
2.043 |
| 26 |
AT1G52580 |
AT1G52580
|
ATRBL5, RHOMBOID-like protein 5, RBL5, RHOMBOID-like protein 5 |
2.755 |
0.018 |
| 27 |
AT5G11060 |
AT5G11060
|
KNAT4, KNOTTED1-like homeobox gene 4 |
2.751 |
0.035 |
| 28 |
AT3G03820 |
AT3G03820
|
SAUR29, SMALL AUXIN UP RNA 29 |
2.728 |
1.302 |
| 29 |
AT1G29230 |
AT1G29230
|
ATCIPK18, ATWL1, WPL4-LIKE 1, CIPK18, CBL-interacting protein kinase 18, SnRK3.20, SNF1-RELATED PROTEIN KINASE 3.20, WL1, WPL4-LIKE 1 |
2.711 |
1.291 |
| 30 |
AT1G09932 |
AT1G09932
|
[Phosphoglycerate mutase family protein] |
2.703 |
-0.373 |
| 31 |
AT3G22275 |
AT3G22275
|
unknown |
2.648 |
1.510 |
| 32 |
AT1G51920 |
AT1G51920
|
unknown |
2.599 |
2.910 |
| 33 |
AT3G50560 |
AT3G50560
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
2.599 |
2.356 |
| 34 |
AT1G47603 |
AT1G47603
|
ATPUP19, purine permease 19, PUP19, purine permease 19 |
2.593 |
1.134 |
| 35 |
AT5G12020 |
AT5G12020
|
HSP17.6II, 17.6 kDa class II heat shock protein |
2.540 |
0.557 |
| 36 |
AT5G04120 |
AT5G04120
|
[Phosphoglycerate mutase family protein] |
2.522 |
0.542 |
| 37 |
AT2G29380 |
AT2G29380
|
HAI3, highly ABA-induced PP2C gene 3 |
2.422 |
1.019 |
| 38 |
AT1G22900 |
AT1G22900
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
2.376 |
2.371 |
| 39 |
AT2G43890 |
AT2G43890
|
[Pectin lyase-like superfamily protein] |
2.353 |
2.958 |
| 40 |
AT3G53770 |
AT3G53770
|
[late embryogenesis abundant 3 (LEA3) family protein] |
2.350 |
1.510 |
| 41 |
AT3G47340 |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
2.345 |
1.900 |
| 42 |
AT1G22890 |
AT1G22890
|
unknown |
2.331 |
2.696 |
| 43 |
AT3G15540 |
AT3G15540
|
IAA19, indole-3-acetic acid inducible 19, MSG2, MASSUGU 2 |
2.303 |
0.693 |
| 44 |
AT1G02930 |
AT1G02930
|
ATGST1, ARABIDOPSIS GLUTATHIONE S-TRANSFERASE 1, ATGSTF3, ARABIDOPSIS THALIANA GLUATIONE S-TRANSFERASE F3, ATGSTF6, GLUTATHIONE S-TRANSFERASE, ERD11, EARLY RESPONSIVE TO DEHYDRATION 11, GST1, GLUTATHIONE S-TRANSFERASE 1, GSTF6, glutathione S-transferase 6 |
2.297 |
2.930 |
| 45 |
AT1G72620 |
AT1G72620
|
[alpha/beta-Hydrolases superfamily protein] |
2.285 |
1.963 |
| 46 |
AT1G78390 |
AT1G78390
|
ATNCED9, NINE-CIS-EPOXYCAROTENOID DIOXYGENASE 9, NCED9, nine-cis-epoxycarotenoid dioxygenase 9 |
2.285 |
2.193 |
| 47 |
AT1G14550 |
AT1G14550
|
[Peroxidase superfamily protein] |
2.233 |
1.800 |
| 48 |
AT3G18900 |
AT3G18900
|
unknown |
2.169 |
0.897 |
| 49 |
AT5G12050 |
AT5G12050
|
unknown |
2.151 |
-0.379 |
| 50 |
AT2G20880 |
AT2G20880
|
AtERF53, ERF53, ERF domain 53 |
2.149 |
1.567 |
| 51 |
AT4G28720 |
AT4G28720
|
YUC8, YUCCA 8 |
2.137 |
-0.021 |
| 52 |
AT5G25260 |
AT5G25260
|
[SPFH/Band 7/PHB domain-containing membrane-associated protein family] |
2.131 |
0.448 |
| 53 |
AT4G19420 |
AT4G19420
|
[Pectinacetylesterase family protein] |
2.126 |
0.932 |
| 54 |
AT3G02410 |
AT3G02410
|
ICME-LIKE2, Isoprenylcysteine methylesterase-like 2 |
2.108 |
0.734 |
| 55 |
AT4G20320 |
AT4G20320
|
[CTP synthase family protein] |
2.095 |
1.331 |
| 56 |
AT3G05936 |
AT3G05936
|
unknown |
2.080 |
2.452 |
| 57 |
AT4G21745 |
AT4G21745
|
[PAK-box/P21-Rho-binding family protein] |
2.048 |
0.048 |
| 58 |
AT1G13430 |
AT1G13430
|
ATST4C, ARABIDOPSIS THALIANA SULFOTRANSFERASE 4C, ST4C, sulfotransferase 4C |
2.038 |
0.830 |
| 59 |
AT1G54950 |
AT1G54950
|
unknown |
2.026 |
3.549 |
| 60 |
AT1G51913 |
AT1G51913
|
[LOCATED IN: endomembrane system] |
1.989 |
1.125 |
| 61 |
AT1G10550 |
AT1G10550
|
XET, XYLOGLUCAN:XYLOGLUCOSYL TRANSFERASE 33, XTH33, xyloglucan:xyloglucosyl transferase 33 |
1.982 |
1.882 |
| 62 |
AT2G18470 |
AT2G18470
|
AtPERK4, proline-rich extensin-like receptor kinase 4, PERK4, proline-rich extensin-like receptor kinase 4 |
1.963 |
1.725 |
| 63 |
AT3G14370 |
AT3G14370
|
WAG2 |
1.962 |
1.302 |
| 64 |
AT2G02990 |
AT2G02990
|
ATRNS1, RIBONUCLEASE 1, RNS1, ribonuclease 1 |
1.953 |
-0.160 |
| 65 |
AT1G15330 |
AT1G15330
|
[Cystathionine beta-synthase (CBS) protein] |
1.938 |
1.228 |
| 66 |
AT1G04610 |
AT1G04610
|
YUC3, YUCCA 3 |
1.936 |
0.701 |
| 67 |
AT5G14470 |
AT5G14470
|
AtGALK2, GALK2, galactokinase 2 |
1.936 |
0.737 |
| 68 |
AT5G60320 |
AT5G60320
|
LecRK-I.11, L-type lectin receptor kinase I.11 |
1.925 |
-1.190 |
| 69 |
AT2G19190 |
AT2G19190
|
FRK1, FLG22-induced receptor-like kinase 1 |
1.914 |
1.462 |
| 70 |
AT4G37235 |
AT4G37235
|
[Uncharacterised protein family (UPF0497)] |
1.905 |
0.404 |
| 71 |
AT4G00780 |
AT4G00780
|
[TRAF-like family protein] |
1.888 |
1.861 |
| 72 |
AT1G67980 |
AT1G67980
|
CCOAMT, caffeoyl-CoA 3-O-methyltransferase |
1.884 |
3.947 |
| 73 |
AT1G26420 |
AT1G26420
|
[FAD-binding Berberine family protein] |
1.879 |
1.620 |
| 74 |
AT4G25480 |
AT4G25480
|
ATCBF3, CBF3, C-REPEAT BINDING FACTOR 3, DREB1A, dehydration response element B1A |
1.878 |
0.441 |
| 75 |
AT1G53700 |
AT1G53700
|
PK3AT, PROTEIN KINASE 3 ARABIDOPSIS THALIANA, WAG1, WAG 1 |
1.872 |
0.654 |
| 76 |
AT5G54710 |
AT5G54710
|
[Ankyrin repeat family protein] |
1.871 |
1.258 |
| 77 |
AT3G05800 |
AT3G05800
|
AIF1, AtBS1(activation-tagged BRI1 suppressor 1)-interacting factor 1 |
1.851 |
0.880 |
| 78 |
AT1G32930 |
AT1G32930
|
AtGALT31A, GALT31A, glycosyltransferase of CAZY family GT31 A |
1.841 |
0.689 |
| 79 |
AT5G21130 |
AT5G21130
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
1.840 |
0.728 |
| 80 |
AT5G23870 |
AT5G23870
|
[Pectinacetylesterase family protein] |
1.816 |
0.237 |
| 81 |
AT1G07473 |
AT1G07473
|
unknown |
1.786 |
1.250 |
| 82 |
AT3G16530 |
AT3G16530
|
[Legume lectin family protein] |
1.782 |
1.770 |
| 83 |
AT1G57560 |
AT1G57560
|
AtMYB50, myb domain protein 50, MYB50, myb domain protein 50 |
1.771 |
1.321 |
| 84 |
AT1G63580 |
AT1G63580
|
[Receptor-like protein kinase-related family protein] |
1.760 |
0.434 |
| 85 |
AT1G01780 |
AT1G01780
|
PLIM2b, PLIM2b |
1.747 |
0.110 |
| 86 |
AT1G18280 |
AT1G18280
|
LTPG3, glycosylphosphatidylinositol-anchored lipid protein transfer 3 |
1.730 |
0.404 |
| 87 |
AT5G02160 |
AT5G02160
|
unknown |
1.729 |
0.871 |
| 88 |
AT1G08090 |
AT1G08090
|
ACH1, ATNRT2.1, NITRATE TRANSPORTER 2.1, ATNRT2:1, nitrate transporter 2:1, LIN1, LATERAL ROOT INITIATION 1, NRT2, NITRATE TRANSPORTER 2, NRT2.1, NITRATE TRANSPORTER 2.1, NRT2:1, nitrate transporter 2:1, NRT2;1AT |
1.711 |
2.874 |
| 89 |
AT4G37240 |
AT4G37240
|
unknown |
1.705 |
0.629 |
| 90 |
AT5G11920 |
AT5G11920
|
AtcwINV6, 6-&1-fructan exohydrolase, cwINV6, 6-&1-fructan exohydrolase |
1.698 |
0.569 |
| 91 |
AT4G20230 |
AT4G20230
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
1.691 |
-0.302 |
| 92 |
AT4G14365 |
AT4G14365
|
XBAT34, XB3 ortholog 4 in Arabidopsis thaliana |
1.691 |
-0.192 |
| 93 |
AT1G21120 |
AT1G21120
|
IGMT2, indole glucosinolate O-methyltransferase 2 |
1.682 |
1.533 |
| 94 |
AT5G14110 |
AT5G14110
|
unknown |
1.673 |
1.865 |
| 95 |
AT1G75000 |
AT1G75000
|
[GNS1/SUR4 membrane protein family] |
1.666 |
-0.292 |
| 96 |
AT4G32950 |
AT4G32950
|
[Protein phosphatase 2C family protein] |
1.659 |
0.953 |
| 97 |
AT1G65484 |
AT1G65484
|
unknown |
1.644 |
1.079 |
| 98 |
AT5G20410 |
AT5G20410
|
ATMGD2, ARABIDOPSIS THALIANA MONOGALACTOSYLDIACYLGLYCEROL SYNTHASE 2, MGD2, monogalactosyldiacylglycerol synthase 2 |
1.644 |
0.381 |
| 99 |
AT5G57420 |
AT5G57420
|
IAA33, indole-3-acetic acid inducible 33 |
1.641 |
2.218 |
| 100 |
AT1G29270 |
AT1G29270
|
unknown |
1.628 |
-0.414 |
| 101 |
AT3G13980 |
AT3G13980
|
unknown |
1.627 |
-0.262 |
| 102 |
AT4G08040 |
AT4G08040
|
ACS11, 1-aminocyclopropane-1-carboxylate synthase 11 |
-6.229 |
-3.329 |
| 103 |
AT3G12900 |
AT3G12900
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-5.220 |
-0.401 |
| 104 |
AT4G04930 |
AT4G04930
|
DES-1-LIKE |
-4.889 |
-1.222 |
| 105 |
AT1G76220 |
AT1G76220
|
unknown |
-4.618 |
-2.979 |
| 106 |
AT4G31940 |
AT4G31940
|
CYP82C4, cytochrome P450, family 82, subfamily C, polypeptide 4 |
-4.256 |
-1.458 |
| 107 |
AT3G09280 |
AT3G09280
|
unknown |
-3.614 |
-3.913 |
| 108 |
AT5G36925 |
AT5G36925
|
unknown |
-3.591 |
-0.520 |
| 109 |
AT3G21330 |
AT3G21330
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-3.427 |
-0.182 |
| 110 |
AT1G07985 |
AT1G07985
|
[Expressed protein] |
-3.377 |
-1.059 |
| 111 |
AT1G76230 |
AT1G76230
|
unknown |
-3.348 |
-1.659 |
| 112 |
AT3G62760 |
AT3G62760
|
ATGSTF13 |
-3.288 |
-2.761 |
| 113 |
AT3G18710 |
AT3G18710
|
ATPUB29, ARABIDOPSIS THALIANA PLANT U-BOX 29, PUB29, plant U-box 29 |
-3.266 |
-1.464 |
| 114 |
AT1G70260 |
AT1G70260
|
UMAMIT36, Usually multiple acids move in and out Transporters 36 |
-3.164 |
-2.105 |
| 115 |
AT5G47600 |
AT5G47600
|
[HSP20-like chaperones superfamily protein] |
-3.099 |
-0.438 |
| 116 |
AT2G44578 |
AT2G44578
|
[RING/U-box superfamily protein] |
-3.094 |
-0.104 |
| 117 |
AT3G23170 |
AT3G23170
|
unknown |
-3.049 |
-1.354 |
| 118 |
AT1G59940 |
AT1G59940
|
ARR3, response regulator 3 |
-3.028 |
-1.044 |
| 119 |
AT2G41690 |
AT2G41690
|
AT-HSFB3, heat shock transcription factor B3, HSFB3, HEAT SHOCK TRANSCRIPTION FACTOR B3, HSFB3, heat shock transcription factor B3 |
-2.919 |
0.240 |
| 120 |
AT3G11110 |
AT3G11110
|
[RING/U-box superfamily protein] |
-2.875 |
-2.105 |
| 121 |
AT4G36780 |
AT4G36780
|
BEH2, BES1/BZR1 homolog 2 |
-2.873 |
-2.265 |
| 122 |
AT1G16635 |
AT1G16635
|
|
-2.862 |
-0.558 |
| 123 |
AT1G01580 |
AT1G01580
|
ATFRO2, FERRIC REDUCTION OXIDASE 2, FRD1, FERRIC CHELATE REDUCTASE DEFECTIVE 1, FRO2, ferric reduction oxidase 2 |
-2.852 |
-0.076 |
| 124 |
AT5G39050 |
AT5G39050
|
PMAT1, phenolic glucoside malonyltransferase 1 |
-2.840 |
-1.651 |
| 125 |
AT4G01680 |
AT4G01680
|
AtMYB55, myb domain protein 55, MYB55, myb domain protein 55 |
-2.826 |
-1.660 |
| 126 |
AT5G60770 |
AT5G60770
|
ATNRT2.4, ARABIDOPSIS THALIANA NITRATE TRANSPORTER 2.4, NRT2.4, nitrate transporter 2.4 |
-2.785 |
-2.058 |
| 127 |
AT2G26870 |
AT2G26870
|
NPC2, non-specific phospholipase C2 |
-2.760 |
-2.184 |
| 128 |
AT3G53160 |
AT3G53160
|
UGT73C7, UDP-glucosyl transferase 73C7 |
-2.757 |
-0.706 |
| 129 |
AT5G38940 |
AT5G38940
|
[RmlC-like cupins superfamily protein] |
-2.721 |
-2.202 |
| 130 |
AT2G45080 |
AT2G45080
|
cycp3;1, cyclin p3;1 |
-2.720 |
-2.077 |
| 131 |
AT5G39330 |
AT5G39330
|
unknown |
-2.683 |
0.043 |
| 132 |
AT5G38200 |
AT5G38200
|
[Class I glutamine amidotransferase-like superfamily protein] |
-2.645 |
-2.218 |
| 133 |
AT5G57780 |
AT5G57780
|
P1R1, P1R1 |
-2.634 |
-1.027 |
| 134 |
AT5G52750 |
AT5G52750
|
[Heavy metal transport/detoxification superfamily protein ] |
-2.613 |
-0.505 |
| 135 |
AT5G65300 |
AT5G65300
|
unknown |
-2.579 |
-0.700 |
| 136 |
AT3G15030 |
AT3G15030
|
MEE35, maternal effect embryo arrest 35, TCP4, TCP family transcription factor 4 |
-2.549 |
-0.108 |
| 137 |
AT4G01670 |
AT4G01670
|
unknown |
-2.488 |
-0.928 |
| 138 |
AT1G74650 |
AT1G74650
|
ATMYB31, ARABIDOPSIS THALIANA MYB DOMAIN PROTEIN 31, ATY13, MYB31, myb domain protein 31 |
-2.487 |
-0.554 |
| 139 |
AT5G57785 |
AT5G57785
|
unknown |
-2.483 |
-4.067 |
| 140 |
AT3G50660 |
AT3G50660
|
AtDWF4, CLM, CLOMAZONE-RESISTANT, CYP90B1, CYTOCHROME P450 90B1, DWF4, DWARF 4, PSC1, PARTIALLY SUPPRESSING COI1 INSENSITIVITY TO JA 1, SAV1, SHADE AVOIDANCE 1, SNP2, SUPPRESSOR OF NPH4 2 |
-2.458 |
-1.050 |
| 141 |
AT4G24110 |
AT4G24110
|
unknown |
-2.431 |
-0.031 |
| 142 |
AT1G32190 |
AT1G32190
|
[alpha/beta-Hydrolases superfamily protein] |
-2.428 |
-1.583 |
| 143 |
AT1G76500 |
AT1G76500
|
AHL29, AT-hook motif nuclear-localized protein 29, SOB3, SUPPRESSOR OF PHYB-4#3 |
-2.397 |
-1.889 |
| 144 |
AT1G78950 |
AT1G78950
|
AtBAS, AtLUP4, BAS, beta-amyrin synthase |
-2.364 |
0.280 |
| 145 |
AT4G19690 |
AT4G19690
|
ATIRT1, ARABIDOPSIS IRON-REGULATED TRANSPORTER 1, IRT1, iron-regulated transporter 1 |
-2.352 |
-1.202 |
| 146 |
AT3G59710 |
AT3G59710
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-2.329 |
-1.322 |
| 147 |
AT4G24972 |
AT4G24972
|
TPD1, TAPETUM DETERMINANT 1 |
-2.246 |
-1.478 |
| 148 |
AT3G16570 |
AT3G16570
|
ATRALF23, ARABIDOPSIS RAPID ALKALINIZATION FACTOR 23, RALF23, rapid alkalinization factor 23 |
-2.243 |
-0.430 |
| 149 |
AT2G21510 |
AT2G21510
|
[DNAJ heat shock N-terminal domain-containing protein] |
-2.220 |
-1.839 |
| 150 |
AT4G15360 |
AT4G15360
|
CYP705A3, cytochrome P450, family 705, subfamily A, polypeptide 3 |
-2.150 |
-1.974 |
| 151 |
AT1G64930 |
AT1G64930
|
CYP89A7, cytochrome P450, family 87, subfamily A, polypeptide 7 |
-2.145 |
-1.712 |
| 152 |
AT1G26390 |
AT1G26390
|
[FAD-binding Berberine family protein] |
-2.119 |
-0.495 |
| 153 |
AT5G44582 |
AT5G44582
|
unknown |
-2.101 |
-2.008 |
| 154 |
AT3G12820 |
AT3G12820
|
AtMYB10, myb domain protein 10, MYB10, myb domain protein 10 |
-2.092 |
-0.523 |
| 155 |
AT5G20790 |
AT5G20790
|
unknown |
-2.055 |
-1.380 |
| 156 |
AT5G25460 |
AT5G25460
|
DGR2, DUF642 L-GalL responsive gene 2 |
-2.031 |
-1.770 |
| 157 |
AT1G68390 |
AT1G68390
|
[Core-2/I-branching beta-1,6-N-acetylglucosaminyltransferase family protein] |
-2.027 |
-0.485 |
| 158 |
AT3G49630 |
AT3G49630
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-2.024 |
-0.590 |
| 159 |
AT3G23930 |
AT3G23930
|
unknown |
-2.023 |
-0.721 |
| 160 |
AT1G05680 |
AT1G05680
|
UGT74E2, Uridine diphosphate glycosyltransferase 74E2 |
-2.022 |
-0.690 |
| 161 |
AT3G60920 |
AT3G60920
|
unknown |
-1.987 |
-0.917 |
| 162 |
AT3G60870 |
AT3G60870
|
AHL18, AT-hook motif nuclear-localized protein 18 |
-1.985 |
-1.493 |
| 163 |
AT4G33666 |
AT4G33666
|
unknown |
-1.977 |
-0.409 |
| 164 |
AT1G73120 |
AT1G73120
|
unknown |
-1.975 |
-1.062 |
| 165 |
AT5G66080 |
AT5G66080
|
APD9, Arabidopsis Pp2c clade D 9 |
-1.972 |
-3.166 |
| 166 |
AT4G24350 |
AT4G24350
|
[Phosphorylase superfamily protein] |
-1.943 |
-0.569 |
| 167 |
AT2G43060 |
AT2G43060
|
AtIBH1, IBH1, ILI1 binding bHLH 1 |
-1.940 |
-0.342 |
| 168 |
AT3G07720 |
AT3G07720
|
[Galactose oxidase/kelch repeat superfamily protein] |
-1.922 |
-0.587 |
| 169 |
AT3G25655 |
AT3G25655
|
IDL1, inflorescence deficient in abscission (IDA)-like 1 |
-1.883 |
-1.640 |
| 170 |
AT1G05560 |
AT1G05560
|
UGT1, UDP-GLUCOSE TRANSFERASE 1, UGT75B1, UDP-glucosyltransferase 75B1 |
-1.865 |
-0.854 |
| 171 |
AT5G20710 |
AT5G20710
|
BGAL7, beta-galactosidase 7 |
-1.865 |
-0.905 |
| 172 |
AT5G17800 |
AT5G17800
|
AtMYB56, myb domain protein 56, MYB56, myb domain protein 56 |
-1.862 |
-1.402 |
| 173 |
AT1G21340 |
AT1G21340
|
[Dof-type zinc finger DNA-binding family protein] |
-1.860 |
-0.413 |
| 174 |
AT3G26460 |
AT3G26460
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-1.853 |
-0.091 |
| 175 |
AT5G66800 |
AT5G66800
|
unknown |
-1.850 |
-1.986 |
| 176 |
AT2G39510 |
AT2G39510
|
UMAMIT14, Usually multiple acids move in and out Transporters 14 |
-1.840 |
-1.291 |
| 177 |
AT2G17064 |
AT2G17064
|
|
-1.838 |
-0.674 |
| 178 |
AT3G15760 |
AT3G15760
|
unknown |
-1.830 |
0.119 |
| 179 |
AT1G52342 |
AT1G52342
|
unknown |
-1.828 |
-0.399 |
| 180 |
AT5G10510 |
AT5G10510
|
AIL6, AINTEGUMENTA-like 6, PLT3, PLETHORA 3 |
-1.825 |
-1.080 |
| 181 |
AT3G15650 |
AT3G15650
|
[alpha/beta-Hydrolases superfamily protein] |
-1.815 |
-0.845 |
| 182 |
AT3G58060 |
AT3G58060
|
[Cation efflux family protein] |
-1.809 |
-1.913 |
| 183 |
AT3G48920 |
AT3G48920
|
AtMYB45, myb domain protein 45, MYB45, myb domain protein 45 |
-1.808 |
-0.774 |
| 184 |
AT2G20825 |
AT2G20825
|
ULT2, ULTRAPETALA 2 |
-1.802 |
1.692 |
| 185 |
AT3G50970 |
AT3G50970
|
LTI30, LOW TEMPERATURE-INDUCED 30, XERO2 |
-1.798 |
0.558 |
| 186 |
AT1G65920 |
AT1G65920
|
[Regulator of chromosome condensation (RCC1) family with FYVE zinc finger domain] |
-1.794 |
-0.959 |
| 187 |
AT3G50800 |
AT3G50800
|
unknown |
-1.781 |
-1.755 |
| 188 |
AT2G46850 |
AT2G46850
|
[Protein kinase superfamily protein] |
-1.771 |
-2.737 |
| 189 |
AT3G20140 |
AT3G20140
|
CYP705A23, cytochrome P450, family 705, subfamily A, polypeptide 23 |
-1.770 |
-3.146 |
| 190 |
AT4G33070 |
AT4G33070
|
AtPDC1, PDC1, pyruvate decarboxylase 1 |
-1.765 |
-0.368 |
| 191 |
AT4G16780 |
AT4G16780
|
ATHB-2, homeobox protein 2, ATHB2, ARABIDOPSIS THALIANA HOMEOBOX PROTEIN 2, HAT4, HB-2, homeobox protein 2 |
-1.761 |
-1.526 |
| 192 |
AT5G64660 |
AT5G64660
|
ATCMPG2, CMPG2, CYS, MET, PRO, and GLY protein 2 |
-1.760 |
-0.518 |
| 193 |
AT3G44990 |
AT3G44990
|
AtXTH31, ATXTR8, XTH31, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 31, XTR8, xyloglucan endo-transglycosylase-related 8 |
-1.730 |
-0.437 |
| 194 |
AT1G14970 |
AT1G14970
|
[O-fucosyltransferase family protein] |
-1.729 |
-1.585 |
| 195 |
AT5G13080 |
AT5G13080
|
ATWRKY75, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 75, WRKY75, WRKY DNA-binding protein 75 |
-1.729 |
-0.326 |
| 196 |
AT5G09440 |
AT5G09440
|
EXL4, EXORDIUM like 4 |
-1.726 |
-0.354 |
| 197 |
AT1G72900 |
AT1G72900
|
[Toll-Interleukin-Resistance (TIR) domain-containing protein] |
-1.725 |
-0.803 |
| 198 |
AT2G41240 |
AT2G41240
|
BHLH100, basic helix-loop-helix protein 100 |
-1.720 |
0.029 |
| 199 |
AT1G14860 |
AT1G14860
|
atnudt18, nudix hydrolase homolog 18, NUDT18, nudix hydrolase homolog 18 |
-1.713 |
-0.652 |
| 200 |
AT5G56080 |
AT5G56080
|
ATNAS2, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 2, NAS2, nicotianamine synthase 2 |
-1.681 |
-2.147 |
| 201 |
AT3G11580 |
AT3G11580
|
[AP2/B3-like transcriptional factor family protein] |
-1.674 |
0.507 |