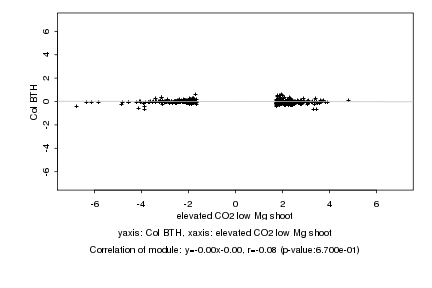
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
elevated CO2 low Mg shoot |
Log2 signal ratio
Col BTH |
| 1 |
AT1G13609 |
AT1G13609
|
[Defensin-like (DEFL) family protein] |
7.010 |
no data |
| 2 |
AT2G14247 |
AT2G14247
|
[Expressed protein] |
4.868 |
no data |
| 3 |
AT1G47395 |
AT1G47395
|
unknown |
4.867 |
no data |
| 4 |
AT1G47400 |
AT1G47400
|
unknown |
4.805 |
0.130 |
| 5 |
AT2G41240 |
AT2G41240
|
BHLH100, basic helix-loop-helix protein 100 |
4.201 |
no data |
| 6 |
AT3G56970 |
AT3G56970
|
BHLH038, bHLH38, basic helix-loop-helix 38, ORG2, OBP3-RESPONSIVE GENE 3 |
4.143 |
no data |
| 7 |
AT3G43154 |
AT3G43154
|
[pseudogene] |
4.142 |
no data |
| 8 |
AT1G21326 |
AT1G21326
|
[VQ motif-containing protein] |
4.062 |
no data |
| 9 |
AT2G24980 |
AT2G24980
|
EXT6, extensin 6 |
3.896 |
-0.003 |
| 10 |
AT2G36440 |
AT2G36440
|
unknown |
3.823 |
-0.021 |
| 11 |
AT3G06070 |
AT3G06070
|
unknown |
3.732 |
0.172 |
| 12 |
AT3G56980 |
AT3G56980
|
BHLH039, bHLH39, basic helix-loop-helix 39, ORG3, OBP3-RESPONSIVE GENE 3 |
3.703 |
0.005 |
| 13 |
AT1G52900 |
AT1G52900
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
3.651 |
0.020 |
| 14 |
AT1G75580 |
AT1G75580
|
SAUR51, SMALL AUXIN UPREGULATED RNA 51 |
3.599 |
-0.007 |
| 15 |
AT4G30180 |
AT4G30180
|
[sequence-specific DNA binding transcription factors] |
3.598 |
no data |
| 16 |
AT2G40740 |
AT2G40740
|
ATWRKY55, WRKY DNA-BINDING PROTEIN 55, WRKY55, WRKY DNA-binding protein 55 |
3.596 |
0.170 |
| 17 |
AT2G29300 |
AT2G29300
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
3.543 |
-0.169 |
| 18 |
AT5G05840 |
AT5G05840
|
unknown |
3.543 |
-0.130 |
| 19 |
AT2G30760 |
AT2G30760
|
unknown |
3.539 |
-0.057 |
| 20 |
AT4G39340 |
AT4G39340
|
EC1.4, EGG CELL 1.4 |
3.495 |
no data |
| 21 |
AT5G53710 |
AT5G53710
|
unknown |
3.485 |
-0.106 |
| 22 |
AT2G43445 |
AT2G43445
|
[F-box and associated interaction domains-containing protein] |
3.485 |
no data |
| 23 |
AT2G30766 |
AT2G30766
|
unknown |
3.438 |
no data |
| 24 |
AT3G44990 |
AT3G44990
|
AtXTH31, ATXTR8, XTH31, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 31, XTR8, xyloglucan endo-transglycosylase-related 8 |
3.437 |
-0.627 |
| 25 |
AT2G21900 |
AT2G21900
|
ATWRKY59, WRKY59, WRKY DNA-binding protein 59 |
3.436 |
no data |
| 26 |
AT2G07170 |
AT2G07170
|
[ARM repeat superfamily protein] |
3.424 |
-0.024 |
| 27 |
AT4G31950 |
AT4G31950
|
CYP82C3, cytochrome P450, family 82, subfamily C, polypeptide 3 |
3.424 |
0.019 |
| 28 |
AT1G26390 |
AT1G26390
|
[FAD-binding Berberine family protein] |
3.394 |
0.330 |
| 29 |
AT2G40180 |
AT2G40180
|
ATHPP2C5, phosphatase 2C5, PP2C5, phosphatase 2C5 |
3.362 |
0.015 |
| 30 |
AT2G18480 |
AT2G18480
|
[Major facilitator superfamily protein] |
3.360 |
0.031 |
| 31 |
AT4G34250 |
AT4G34250
|
KCS16, 3-ketoacyl-CoA synthase 16 |
3.344 |
-0.066 |
| 32 |
AT3G49620 |
AT3G49620
|
DIN11, DARK INDUCIBLE 11 |
3.337 |
-0.193 |
| 33 |
AT5G53450 |
AT5G53450
|
ORG1, OBP3-responsive gene 1 |
3.335 |
-0.031 |
| 34 |
AT5G44420 |
AT5G44420
|
LCR77, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 77, PDF1.2, plant defensin 1.2, PDF1.2A, PLANT DEFENSIN 1.2A |
3.287 |
-0.650 |
| 35 |
AT5G25390 |
AT5G25390
|
SHN3, shine3 |
3.275 |
-0.020 |
| 36 |
AT1G14540 |
AT1G14540
|
PER4, peroxidase 4 |
3.263 |
0.014 |
| 37 |
AT2G37830 |
AT2G37830
|
[pseudogene] |
3.225 |
0.001 |
| 38 |
AT1G68795 |
AT1G68795
|
CLE12, CLAVATA3/ESR-RELATED 12 |
3.151 |
-0.052 |
| 39 |
AT1G65710 |
AT1G65710
|
unknown |
3.151 |
no data |
| 40 |
AT2G20880 |
AT2G20880
|
AtERF53, ERF53, ERF domain 53 |
3.130 |
-0.066 |
| 41 |
AT5G05940 |
AT5G05940
|
ATROPGEF5, ROP GUANINE NUCLEOTIDE EXCHANGE FACTOR 5, ROPGEF5, ROP (rho of plants) guanine nucleotide exchange factor 5 |
3.123 |
-0.065 |
| 42 |
AT5G04150 |
AT5G04150
|
BHLH101 |
3.123 |
-0.044 |
| 43 |
AT1G63820 |
AT1G63820
|
[CCT motif family protein] |
3.074 |
-0.020 |
| 44 |
AT4G08380 |
AT4G08380
|
[Proline-rich extensin-like family protein] |
3.074 |
-0.029 |
| 45 |
AT1G51920 |
AT1G51920
|
unknown |
3.073 |
0.169 |
| 46 |
AT5G37300 |
AT5G37300
|
WSD1 |
3.049 |
-0.184 |
| 47 |
AT1G16420 |
AT1G16420
|
ATMC8, ARABIDOPSIS THALIANA METACASPASE 8, AtMCP2e, metacaspase 2e, MC8, metacaspase 8, MCP2e, metacaspase 2e |
3.046 |
0.087 |
| 48 |
AT1G61560 |
AT1G61560
|
ATMLO6, MILDEW RESISTANCE LOCUS O 6, MLO6, MILDEW RESISTANCE LOCUS O 6 |
3.035 |
0.026 |
| 49 |
AT1G30850 |
AT1G30850
|
RSH4, root hair specific 4 |
3.013 |
-0.018 |
| 50 |
AT4G25434 |
AT4G25434
|
ATNUDT10, nudix hydrolase homolog 10, NUDT10, nudix hydrolase homolog 10 |
2.993 |
-0.048 |
| 51 |
AT5G36925 |
AT5G36925
|
unknown |
2.949 |
no data |
| 52 |
AT4G11470 |
AT4G11470
|
CRK31, cysteine-rich RLK (RECEPTOR-like protein kinase) 31 |
2.922 |
0.016 |
| 53 |
AT1G56242 |
AT1G56242
|
other RNA |
2.907 |
no data |
| 54 |
AT1G56240 |
AT1G56240
|
AtPP2-B13, phloem protein 2-B13, PP2-B13, phloem protein 2-B13 |
2.907 |
no data |
| 55 |
AT3G52748 |
AT3G52748
|
other RNA |
2.907 |
no data |
| 56 |
AT1G56430 |
AT1G56430
|
ATNAS4, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 4, NAS4, nicotianamine synthase 4 |
2.894 |
-0.066 |
| 57 |
AT3G23880 |
AT3G23880
|
[F-box and associated interaction domains-containing protein] |
2.894 |
0.150 |
| 58 |
AT3G01420 |
AT3G01420
|
ALPHA-DOX1, alpha-dioxygenase 1, DIOX1, DOX1, PADOX-1, plant alpha dioxygenase 1 |
2.881 |
0.339 |
| 59 |
AT5G11140 |
AT5G11140
|
[Arabidopsis phospholipase-like protein (PEARLI 4) family] |
2.874 |
no data |
| 60 |
AT1G05880 |
AT1G05880
|
ARI12, ARIADNE 12, ATARI12, ARABIDOPSIS ARIADNE 12 |
2.873 |
-0.031 |
| 61 |
AT5G58390 |
AT5G58390
|
[Peroxidase superfamily protein] |
2.831 |
-0.009 |
| 62 |
AT4G36830 |
AT4G36830
|
HOS3-1 |
2.821 |
-0.090 |
| 63 |
AT4G21820 |
AT4G21820
|
[binding] |
2.814 |
-0.008 |
| 64 |
AT3G21520 |
AT3G21520
|
AtDMP1, Arabidopsis thaliana DUF679 domain membrane protein 1, DMP1, DUF679 domain membrane protein 1 |
2.809 |
0.160 |
| 65 |
AT4G16820 |
AT4G16820
|
DALL1, DAD1-Like Lipase 1, PLA-I{beta]2, phospholipase A I beta 2 |
2.803 |
0.032 |
| 66 |
AT1G29510 |
AT1G29510
|
SAUR67, SMALL AUXIN UPREGULATED RNA 67 |
2.789 |
-0.197 |
| 67 |
AT4G39330 |
AT4G39330
|
ATCAD9, CAD9, cinnamyl alcohol dehydrogenase 9 |
2.775 |
-0.065 |
| 68 |
AT3G52370 |
AT3G52370
|
FLA15, FASCICLIN-like arabinogalactan protein 15 precursor |
2.772 |
-0.116 |
| 69 |
AT1G78770 |
AT1G78770
|
APC6, anaphase promoting complex 6 |
2.765 |
-0.043 |
| 70 |
AT4G21390 |
AT4G21390
|
B120 |
2.741 |
-0.141 |
| 71 |
AT5G46830 |
AT5G46830
|
ATNIG1, NACL-INDUCIBLE GENE 1, NIG1, NACL-inducible gene 1 |
2.740 |
-0.204 |
| 72 |
AT5G27420 |
AT5G27420
|
ATL31, Arabidopsis toxicos en levadura 31, CNI1, carbon/nitrogen insensitive 1 |
2.732 |
0.088 |
| 73 |
AT1G45010 |
AT1G45010
|
[TRAM, LAG1 and CLN8 (TLC) lipid-sensing domain containing protein] |
2.712 |
no data |
| 74 |
AT2G42150 |
AT2G42150
|
[DNA-binding bromodomain-containing protein] |
2.712 |
-0.070 |
| 75 |
AT1G13310 |
AT1G13310
|
[Endosomal targeting BRO1-like domain-containing protein] |
2.709 |
-0.050 |
| 76 |
AT5G19190 |
AT5G19190
|
unknown |
2.703 |
no data |
| 77 |
AT3G60720 |
AT3G60720
|
PDLP8, plasmodesmata-located protein 8 |
2.695 |
-0.092 |
| 78 |
AT1G12570 |
AT1G12570
|
[Glucose-methanol-choline (GMC) oxidoreductase family protein] |
2.675 |
-0.004 |
| 79 |
AT4G37240 |
AT4G37240
|
unknown |
2.671 |
0.006 |
| 80 |
AT2G29100 |
AT2G29100
|
ATGLR2.9, glutamate receptor 2.9, GLR2.9, GLUTAMATE RECEPTOR 2.9, GLR2.9, glutamate receptor 2.9 |
2.670 |
0.057 |
| 81 |
AT1G05675 |
AT1G05675
|
[UDP-Glycosyltransferase superfamily protein] |
2.654 |
no data |
| 82 |
AT2G23340 |
AT2G23340
|
DEAR3, DREB and EAR motif protein 3 |
2.647 |
-0.020 |
| 83 |
AT5G45950 |
AT5G45950
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
2.642 |
-0.120 |
| 84 |
AT2G38720 |
AT2G38720
|
MAP65-5, microtubule-associated protein 65-5 |
2.629 |
-0.056 |
| 85 |
AT3G56870 |
AT3G56870
|
unknown |
2.627 |
-0.051 |
| 86 |
AT4G11070 |
AT4G11070
|
AtWRKY41, WRKY41 |
2.624 |
no data |
| 87 |
AT5G13170 |
AT5G13170
|
AtSWEET15, SAG29, senescence-associated gene 29, SWEET15 |
2.605 |
0.142 |
| 88 |
AT3G61490 |
AT3G61490
|
[Pectin lyase-like superfamily protein] |
2.599 |
-0.030 |
| 89 |
AT4G37780 |
AT4G37780
|
ATMYB87, MYB87, myb domain protein 87 |
2.545 |
-0.006 |
| 90 |
AT1G05575 |
AT1G05575
|
unknown |
2.531 |
0.052 |
| 91 |
AT1G01120 |
AT1G01120
|
KCS1, 3-ketoacyl-CoA synthase 1 |
2.513 |
-0.034 |
| 92 |
AT5G02890 |
AT5G02890
|
[HXXXD-type acyl-transferase family protein] |
2.501 |
-0.133 |
| 93 |
AT1G47510 |
AT1G47510
|
inositol polyphosphate 5-phosphatase 11 |
2.500 |
no data |
| 94 |
AT4G10150 |
AT4G10150
|
[RING/U-box superfamily protein] |
2.494 |
no data |
| 95 |
AT4G06536 |
AT4G06536
|
[SPla/RYanodine receptor (SPRY) domain-containing protein] |
2.494 |
no data |
| 96 |
AT4G11170 |
AT4G11170
|
RMG1, Resistance Methylated Gene 1 |
2.490 |
0.049 |
| 97 |
AT3G05640 |
AT3G05640
|
[Protein phosphatase 2C family protein] |
2.486 |
-0.208 |
| 98 |
AT1G68780 |
AT1G68780
|
[RNI-like superfamily protein] |
2.471 |
-0.181 |
| 99 |
AT3G55240 |
AT3G55240
|
unknown |
2.471 |
no data |
| 100 |
AT4G36500 |
AT4G36500
|
unknown |
2.468 |
-0.057 |
| 101 |
AT1G53620 |
AT1G53620
|
unknown |
2.465 |
no data |
| 102 |
AT1G64625 |
AT1G64625
|
[Serine/threonine-protein kinase WNK (With No Lysine)-related] |
2.463 |
no data |
| 103 |
AT5G05270 |
AT5G05270
|
AtCHIL, CHIL, chalcone isomerase like |
2.456 |
-0.169 |
| 104 |
AT4G33790 |
AT4G33790
|
CER4, ECERIFERUM 4, FAR3, FATTY ACID REDUCTASE 3, G7 |
2.454 |
-0.092 |
| 105 |
AT3G63480 |
AT3G63480
|
[ATP binding microtubule motor family protein] |
2.444 |
0.011 |
| 106 |
AT5G59990 |
AT5G59990
|
[CCT motif family protein] |
2.435 |
-0.062 |
| 107 |
AT1G66090 |
AT1G66090
|
[Disease resistance protein (TIR-NBS class)] |
2.430 |
-0.059 |
| 108 |
AT5G56960 |
AT5G56960
|
[basic helix-loop-helix (bHLH) DNA-binding family protein] |
2.426 |
no data |
| 109 |
AT5G46295 |
AT5G46295
|
unknown |
2.414 |
-0.005 |
| 110 |
AT1G73600 |
AT1G73600
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
2.413 |
no data |
| 111 |
AT1G73602 |
AT1G73602
|
CPuORF32, conserved peptide upstream open reading frame 32 |
2.411 |
no data |
| 112 |
AT1G65060 |
AT1G65060
|
4CL3, 4-coumarate:CoA ligase 3 |
2.410 |
-0.201 |
| 113 |
AT5G13740 |
AT5G13740
|
ZIF1, zinc induced facilitator 1 |
2.402 |
-0.030 |
| 114 |
AT1G78170 |
AT1G78170
|
unknown |
2.397 |
-0.335 |
| 115 |
AT2G18680 |
AT2G18680
|
unknown |
2.392 |
no data |
| 116 |
AT4G23200 |
AT4G23200
|
CRK12, cysteine-rich RLK (RECEPTOR-like protein kinase) 12 |
2.391 |
0.120 |
| 117 |
AT4G14450 |
AT4G14450
|
unknown |
2.380 |
no data |
| 118 |
AT1G55260 |
AT1G55260
|
LTPG6, glycosylphosphatidylinositol-anchored lipid protein transfer 6 |
2.380 |
-0.107 |
| 119 |
AT4G16260 |
AT4G16260
|
[Glycosyl hydrolase superfamily protein] |
2.380 |
0.012 |
| 120 |
AT2G32030 |
AT2G32030
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
2.380 |
0.081 |
| 121 |
AT4G28460 |
AT4G28460
|
unknown |
2.379 |
0.086 |
| 122 |
AT1G72416 |
AT1G72416
|
[Chaperone DnaJ-domain superfamily protein] |
2.379 |
no data |
| 123 |
AT5G53210 |
AT5G53210
|
SPCH, SPEECHLESS |
2.379 |
-0.093 |
| 124 |
AT1G57590 |
AT1G57590
|
[Pectinacetylesterase family protein] |
2.378 |
-0.311 |
| 125 |
AT4G27950 |
AT4G27950
|
CRF4, cytokinin response factor 4 |
2.378 |
no data |
| 126 |
AT3G59940 |
AT3G59940
|
KFB50, Kelch repeat F-box 50, KMD4, KISS ME DEADLY 4 |
2.349 |
0.007 |
| 127 |
AT5G45080 |
AT5G45080
|
AtPP2-A6, phloem protein 2-A6, PP2-A6, phloem protein 2-A6 |
2.344 |
-0.010 |
| 128 |
AT5G05300 |
AT5G05300
|
unknown |
2.336 |
0.230 |
| 129 |
AT1G23205 |
AT1G23205
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
2.335 |
-0.301 |
| 130 |
AT3G04220 |
AT3G04220
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
2.332 |
0.134 |
| 131 |
AT3G18290 |
AT3G18290
|
BTS, BRUTUS, EMB2454, embryo defective 2454 |
2.331 |
0.017 |
| 132 |
AT1G33440 |
AT1G33440
|
AtNPF4.4, NPF4.4, NRT1/ PTR family 4.4 |
2.327 |
-0.161 |
| 133 |
AT3G13435 |
AT3G13435
|
unknown |
2.322 |
no data |
| 134 |
AT1G53625 |
AT1G53625
|
unknown |
2.317 |
no data |
| 135 |
AT1G16070 |
AT1G16070
|
AtTLP8, tubby like protein 8, TLP8, tubby like protein 8 |
2.309 |
-0.057 |
| 136 |
AT3G54150 |
AT3G54150
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
2.307 |
0.300 |
| 137 |
AT4G22620 |
AT4G22620
|
SAUR34, SMALL AUXIN UPREGULATED RNA 34 |
2.307 |
-0.224 |
| 138 |
AT2G37300 |
AT2G37300
|
ABCI16, ATP-binding cassette I16 |
2.296 |
-0.021 |
| 139 |
AT4G23990 |
AT4G23990
|
ATCSLG3, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE-LIKE G3, CSLG3, cellulose synthase like G3 |
2.291 |
-0.130 |
| 140 |
AT1G06137 |
AT1G06137
|
unknown |
2.285 |
no data |
| 141 |
ATCG00370 |
ATCG00370
|
TRNS.3 |
2.282 |
no data |
| 142 |
AT1G24020 |
AT1G24020
|
MLP423, MLP-like protein 423 |
2.274 |
-0.033 |
| 143 |
AT4G13410 |
AT4G13410
|
ATCSLA15, CSLA15, CELLULOSE SYNTHASE LIKE A15 |
2.271 |
-0.216 |
| 144 |
AT3G57240 |
AT3G57240
|
AtBG3, BG3, beta-1,3-glucanase 3 |
2.262 |
0.396 |
| 145 |
AT5G01790 |
AT5G01790
|
unknown |
2.258 |
-0.194 |
| 146 |
AT2G47485 |
AT2G47485
|
unknown |
2.255 |
no data |
| 147 |
AT4G01080 |
AT4G01080
|
TBL26, TRICHOME BIREFRINGENCE-LIKE 26 |
2.240 |
-0.274 |
| 148 |
AT5G08640 |
AT5G08640
|
ATFLS1, FLS, FLAVONOL SYNTHASE, FLS1, flavonol synthase 1 |
2.232 |
-0.108 |
| 149 |
AT4G08770 |
AT4G08770
|
Prx37, peroxidase 37 |
2.232 |
0.117 |
| 150 |
AT1G79680 |
AT1G79680
|
ATWAKL10, WAKL10, WALL ASSOCIATED KINASE (WAK)-LIKE 10 |
2.231 |
0.135 |
| 151 |
AT1G64980 |
AT1G64980
|
CDI, cadmium 2+ induced |
2.227 |
-0.061 |
| 152 |
AT3G15570 |
AT3G15570
|
[Phototropic-responsive NPH3 family protein] |
2.218 |
-0.067 |
| 153 |
AT2G36830 |
AT2G36830
|
GAMMA-TIP, gamma tonoplast intrinsic protein, GAMMA-TIP1, GAMMA TONOPLAST INTRINSIC PROTEIN 1, TIP1;1, TONOPLAST INTRINSIC PROTEIN 1;1 |
2.216 |
-0.056 |
| 154 |
AT2G26560 |
AT2G26560
|
PLA IIA, PHOSPHOLIPASE A 2A, PLA2A, phospholipase A 2A, PLAII alpha, PLP2, PATATIN-LIKE PROTEIN 2, PLP2, PHOSPHOLIPASE A 2A |
2.215 |
0.117 |
| 155 |
AT4G18270 |
AT4G18270
|
ATTRANS11, ARABIDOPSIS THALIANA TRANSLOCASE 11, TRANS11, translocase 11 |
2.210 |
-0.142 |
| 156 |
AT1G71390 |
AT1G71390
|
AtRLP11, receptor like protein 11, RLP11, receptor like protein 11 |
2.210 |
0.234 |
| 157 |
AT5G23480 |
AT5G23480
|
[SWIB/MDM2 domain] |
2.203 |
0.016 |
| 158 |
AT4G01460 |
AT4G01460
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
2.192 |
-0.161 |
| 159 |
AT3G12910 |
AT3G12910
|
[NAC (No Apical Meristem) domain transcriptional regulator superfamily protein] |
2.190 |
0.062 |
| 160 |
AT3G60140 |
AT3G60140
|
BGLU30, BETA GLUCOSIDASE 30, DIN2, DARK INDUCIBLE 2, SRG2, SENESCENCE-RELATED GENE 2 |
2.181 |
0.254 |
| 161 |
AT3G28130 |
AT3G28130
|
UMAMIT44, Usually multiple acids move in and out Transporters 44 |
2.177 |
-0.173 |
| 162 |
AT2G47370 |
AT2G47370
|
[Calcium-dependent phosphotriesterase superfamily protein] |
2.177 |
no data |
| 163 |
AT5G55730 |
AT5G55730
|
FLA1, FASCICLIN-like arabinogalactan 1 |
2.153 |
-0.091 |
| 164 |
AT1G04220 |
AT1G04220
|
KCS2, 3-ketoacyl-CoA synthase 2 |
2.149 |
-0.128 |
| 165 |
AT1G21550 |
AT1G21550
|
[Calcium-binding EF-hand family protein] |
2.140 |
0.068 |
| 166 |
AT1G28010 |
AT1G28010
|
ABCB14, ATP-binding cassette B14, ATABCB14, Arabidopsis thaliana ATP-binding cassette B14, MDR12, multi-drug resistance 12, PGP14, P-glycoprotein 14 |
2.136 |
-0.050 |
| 167 |
AT3G61950 |
AT3G61950
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
2.127 |
-0.176 |
| 168 |
AT5G52320 |
AT5G52320
|
CYP96A4, cytochrome P450, family 96, subfamily A, polypeptide 4 |
2.125 |
-0.285 |
| 169 |
AT1G69930 |
AT1G69930
|
ATGSTU11, glutathione S-transferase TAU 11, GSTU11, glutathione S-transferase TAU 11 |
2.120 |
0.147 |
| 170 |
AT2G15390 |
AT2G15390
|
atfut4, FUT4, fucosyltransferase 4 |
2.109 |
0.054 |
| 171 |
AT1G30100 |
AT1G30100
|
ATNCED5, NINE-CIS-EPOXYCAROTENOID DIOXYGENASE 5, NCED5, nine-cis-epoxycarotenoid dioxygenase 5 |
2.097 |
0.059 |
| 172 |
AT1G72520 |
AT1G72520
|
ATLOX4, Arabidopsis thaliana lipoxygenase 4, LOX4, lipoxygenase 4 |
2.094 |
-0.044 |
| 173 |
AT3G47640 |
AT3G47640
|
PYE, POPEYE |
2.091 |
-0.090 |
| 174 |
AT2G25735 |
AT2G25735
|
unknown |
2.089 |
0.031 |
| 175 |
AT4G38560 |
AT4G38560
|
[Arabidopsis phospholipase-like protein (PEARLI 4) family] |
2.087 |
0.402 |
| 176 |
AT1G19020 |
AT1G19020
|
unknown |
2.074 |
-0.007 |
| 177 |
AT5G41900 |
AT5G41900
|
[alpha/beta-Hydrolases superfamily protein] |
2.069 |
-0.306 |
| 178 |
AT3G23810 |
AT3G23810
|
ATSAHH2, S-ADENOSYL-L-HOMOCYSTEINE (SAH) HYDROLASE 2, SAHH2, S-adenosyl-l-homocysteine (SAH) hydrolase 2 |
2.057 |
-0.080 |
| 179 |
AT5G40380 |
AT5G40380
|
CRK42, cysteine-rich RLK (RECEPTOR-like protein kinase) 42 |
2.041 |
-0.030 |
| 180 |
AT5G24570 |
AT5G24570
|
unknown |
2.037 |
-0.045 |
| 181 |
AT3G48720 |
AT3G48720
|
DCF, DEFICIENT IN CUTIN FERULATE |
2.036 |
-0.025 |
| 182 |
AT4G13493 |
AT4G13493
|
MIR850A, microRNA850A |
2.031 |
no data |
| 183 |
AT5G46080 |
AT5G46080
|
[Protein kinase superfamily protein] |
2.030 |
0.023 |
| 184 |
AT4G39580 |
AT4G39580
|
[Galactose oxidase/kelch repeat superfamily protein] |
2.028 |
no data |
| 185 |
AT5G55250 |
AT5G55250
|
AtIAMT1, IAMT1, IAA carboxylmethyltransferase 1 |
2.028 |
-0.032 |
| 186 |
AT5G24540 |
AT5G24540
|
BGLU31, beta glucosidase 31 |
2.025 |
0.285 |
| 187 |
AT3G62390 |
AT3G62390
|
TBL6, TRICHOME BIREFRINGENCE-LIKE 6 |
2.020 |
-0.171 |
| 188 |
AT3G21950 |
AT3G21950
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
2.014 |
-0.212 |
| 189 |
AT1G74590 |
AT1G74590
|
ATGSTU10, GLUTATHIONE S-TRANSFERASE TAU 10, GSTU10, glutathione S-transferase TAU 10 |
1.989 |
0.583 |
| 190 |
AT1G17240 |
AT1G17240
|
AtRLP2, receptor like protein 2, RLP2, receptor like protein 2 |
1.988 |
0.013 |
| 191 |
AT1G68200 |
AT1G68200
|
[Zinc finger C-x8-C-x5-C-x3-H type family protein] |
1.978 |
-0.024 |
| 192 |
AT3G23450 |
AT3G23450
|
unknown |
1.978 |
0.008 |
| 193 |
AT5G48400 |
AT5G48400
|
ATGLR1.2, GLR1.2, GLUTAMATE RECEPTOR 1.2 |
1.977 |
0.107 |
| 194 |
AT5G23400 |
AT5G23400
|
[Leucine-rich repeat (LRR) family protein] |
1.974 |
-0.050 |
| 195 |
AT1G15550 |
AT1G15550
|
ATGA3OX1, ARABIDOPSIS THALIANA GIBBERELLIN 3 BETA-HYDROXYLASE 1, GA3OX1, gibberellin 3-oxidase 1, GA4, GA REQUIRING 4 |
1.958 |
-0.096 |
| 196 |
AT1G08860 |
AT1G08860
|
BON3, BONZAI 3 |
1.957 |
-0.027 |
| 197 |
AT3G06020 |
AT3G06020
|
FAF4, FANTASTIC FOUR 4 |
1.955 |
-0.014 |
| 198 |
AT3G13433 |
AT3G13433
|
unknown |
1.955 |
no data |
| 199 |
AT4G15210 |
AT4G15210
|
AT-BETA-AMY, ATBETA-AMY, ARABIDOPSIS THALIANA BETA-AMYLASE, BAM5, beta-amylase 5, BMY1, RAM1, REDUCED BETA AMYLASE 1 |
1.952 |
-0.194 |
| 200 |
AT2G26010 |
AT2G26010
|
PDF1.3, plant defensin 1.3 |
1.952 |
no data |
| 201 |
AT1G33480 |
AT1G33480
|
[RING/U-box superfamily protein] |
1.946 |
no data |
| 202 |
AT3G18010 |
AT3G18010
|
WOX1, WUSCHEL related homeobox 1 |
1.946 |
no data |
| 203 |
AT1G29500 |
AT1G29500
|
SAUR66, SMALL AUXIN UPREGULATED RNA 66 |
1.946 |
-0.150 |
| 204 |
AT2G30770 |
AT2G30770
|
CYP71A13, cytochrome P450, family 71, subfamily A, polypeptide 13 |
1.943 |
0.615 |
| 205 |
AT3G24420 |
AT3G24420
|
[alpha/beta-Hydrolases superfamily protein] |
1.936 |
no data |
| 206 |
AT4G34390 |
AT4G34390
|
XLG2, extra-large GTP-binding protein 2 |
1.936 |
0.110 |
| 207 |
AT5G43420 |
AT5G43420
|
[RING/U-box superfamily protein] |
1.932 |
no data |
| 208 |
AT5G48880 |
AT5G48880
|
KAT5, 3-KETO-ACYL-COENZYME A THIOLASE 5, PKT1, PEROXISOMAL-3-KETO-ACYL-COA THIOLASE 1, PKT2, peroxisomal 3-keto-acyl-CoA thiolase 2 |
1.928 |
-0.160 |
| 209 |
AT2G12462 |
AT2G12462
|
unknown |
1.925 |
no data |
| 210 |
AT3G54820 |
AT3G54820
|
PIP2;5, plasma membrane intrinsic protein 2;5, PIP2D, PLASMA MEMBRANE INTRINSIC PROTEIN 2D |
1.919 |
-0.030 |
| 211 |
AT2G44840 |
AT2G44840
|
ATERF13, ETHYLENE-RESPONSIVE ELEMENT BINDING FACTOR 13, EREBP, ERF13, ethylene-responsive element binding factor 13 |
1.919 |
-0.084 |
| 212 |
AT4G23810 |
AT4G23810
|
ATWRKY53, WRKY53 |
1.917 |
0.132 |
| 213 |
AT5G41730 |
AT5G41730
|
[Protein kinase family protein] |
1.913 |
-0.087 |
| 214 |
AT1G03850 |
AT1G03850
|
ATGRXS13, glutaredoxin 13, GRXS13, glutaredoxin 13 |
1.913 |
0.549 |
| 215 |
AT1G30530 |
AT1G30530
|
UGT78D1, UDP-glucosyl transferase 78D1 |
1.911 |
-0.106 |
| 216 |
AT4G28310 |
AT4G28310
|
unknown |
1.910 |
-0.104 |
| 217 |
AT1G51620 |
AT1G51620
|
[Protein kinase superfamily protein] |
1.909 |
0.246 |
| 218 |
AT2G19580 |
AT2G19580
|
TET2, tetraspanin2 |
1.909 |
-0.053 |
| 219 |
AT3G13130 |
AT3G13130
|
unknown |
1.909 |
0.027 |
| 220 |
AT5G45670 |
AT5G45670
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
1.907 |
-0.130 |
| 221 |
AT5G51030 |
AT5G51030
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
1.903 |
-0.021 |
| 222 |
AT1G20310 |
AT1G20310
|
unknown |
1.900 |
-0.010 |
| 223 |
AT3G25760 |
AT3G25760
|
AOC1, allene oxide cyclase 1, ERD12, early-responsive to dehydration 12 |
1.896 |
no data |
| 224 |
AT2G27389 |
AT2G27389
|
unknown |
1.893 |
no data |
| 225 |
AT2G27390 |
AT2G27390
|
[proline-rich family protein] |
1.893 |
0.043 |
| 226 |
AT4G15620 |
AT4G15620
|
[Uncharacterised protein family (UPF0497)] |
1.885 |
-0.165 |
| 227 |
AT3G17840 |
AT3G17840
|
RLK902, receptor-like kinase 902 |
1.883 |
-0.005 |
| 228 |
ATCG00600 |
ATCG00600
|
PETG |
1.883 |
-0.085 |
| 229 |
AT5G56300 |
AT5G56300
|
GAMT2, gibberellic acid methyltransferase 2 |
1.883 |
0.020 |
| 230 |
AT3G55950 |
AT3G55950
|
ATCRR3, CCR3, CRINKLY4 related 3 |
1.879 |
0.079 |
| 231 |
AT4G23700 |
AT4G23700
|
ATCHX17, cation/H+ exchanger 17, CHX17, cation/H+ exchanger 17 |
1.877 |
0.426 |
| 232 |
AT3G13610 |
AT3G13610
|
2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein |
1.876 |
0.270 |
| 233 |
AT4G29740 |
AT4G29740
|
ATCKX4, CKX4, cytokinin oxidase 4 |
1.869 |
-0.034 |
| 234 |
AT5G13320 |
AT5G13320
|
AtGH3.12, GDG1, GH3-LIKE DEFENSE GENE 1, GH3.12, GRETCHEN HAGEN 3.12, PBS3, AVRPPHB SUSCEPTIBLE 3, WIN3, HOPW1-1-INTERACTING 3 |
1.868 |
0.232 |
| 235 |
AT1G65560 |
AT1G65560
|
[Zinc-binding dehydrogenase family protein] |
1.862 |
0.027 |
| 236 |
AT1G64390 |
AT1G64390
|
AtGH9C2, glycosyl hydrolase 9C2, GH9C2, glycosyl hydrolase 9C2 |
1.862 |
-0.224 |
| 237 |
AT4G37970 |
AT4G37970
|
ATCAD6, CAD6, cinnamyl alcohol dehydrogenase 6 |
1.858 |
-0.139 |
| 238 |
AT4G17340 |
AT4G17340
|
DELTA-TIP2, TIP2;2, tonoplast intrinsic protein 2;2 |
1.858 |
-0.020 |
| 239 |
AT5G67270 |
AT5G67270
|
ATEB1C, MICROTUBULE END BINDING PROTEIN 1, ATEB1H1, ATEB1-HOMOLOG1, EB1C, end binding protein 1C |
1.857 |
-0.007 |
| 240 |
AT1G48100 |
AT1G48100
|
[Pectin lyase-like superfamily protein] |
1.855 |
-0.317 |
| 241 |
AT4G23020 |
AT4G23020
|
TRM11, TON1 Recruiting Motif 11 |
1.855 |
no data |
| 242 |
AT5G15230 |
AT5G15230
|
GASA4, GAST1 protein homolog 4 |
1.852 |
-0.009 |
| 243 |
AT2G04495 |
AT2G04495
|
unknown |
1.851 |
no data |
| 244 |
AT1G51800 |
AT1G51800
|
IOS1, IMPAIRED OOMYCETE SUSCEPTIBILITY 1 |
1.850 |
0.293 |
| 245 |
AT4G23190 |
AT4G23190
|
AT-RLK3, RECEPTOR LIKE PROTEIN KINASE 3, CRK11, cysteine-rich RLK (RECEPTOR-like protein kinase) 11 |
1.847 |
0.055 |
| 246 |
AT5G11590 |
AT5G11590
|
TINY2, TINY2 |
1.845 |
-0.137 |
| 247 |
AT2G43620 |
AT2G43620
|
[Chitinase family protein] |
1.844 |
0.143 |
| 248 |
AT5G67280 |
AT5G67280
|
RLK, receptor-like kinase |
1.842 |
-0.184 |
| 249 |
AT2G29110 |
AT2G29110
|
ATGLR2.8, glutamate receptor 2.8, GLR2.8, GLR2.8, glutamate receptor 2.8 |
1.837 |
0.528 |
| 250 |
AT2G25820 |
AT2G25820
|
ESE2, ethylene and salt inducible 2 |
1.833 |
-0.081 |
| 251 |
AT1G73810 |
AT1G73810
|
[Core-2/I-branching beta-1,6-N-acetylglucosaminyltransferase family protein] |
1.833 |
no data |
| 252 |
AT4G04540 |
AT4G04540
|
CRK39, cysteine-rich RLK (RECEPTOR-like protein kinase) 39 |
1.832 |
no data |
| 253 |
AT1G67750 |
AT1G67750
|
[Pectate lyase family protein] |
1.832 |
-0.071 |
| 254 |
AT1G13520 |
AT1G13520
|
unknown |
1.827 |
no data |
| 255 |
AT2G32240 |
AT2G32240
|
unknown |
1.824 |
no data |
| 256 |
AT5G09800 |
AT5G09800
|
[ARM repeat superfamily protein] |
1.823 |
0.018 |
| 257 |
AT1G12080 |
AT1G12080
|
[Vacuolar calcium-binding protein-related] |
1.821 |
-0.027 |
| 258 |
AT3G26932 |
AT3G26932
|
DRB3, dsRNA-binding protein 3 |
1.819 |
-0.110 |
| 259 |
AT1G79460 |
AT1G79460
|
ATKS, ARABIDOPSIS THALIANA ENT-KAURENE SYNTHASE, ATKS1, ARABIDOPSIS THALIANA ENT-KAURENE SYNTHASE 1, GA2, GA REQUIRING 2, KS, KS1, ENT-KAURENE SYNTHASE 1 |
1.817 |
-0.203 |
| 260 |
AT5G24110 |
AT5G24110
|
ATWRKY30, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 30, WRKY30, WRKY DNA-binding protein 30 |
1.814 |
0.249 |
| 261 |
AT2G19582 |
AT2G19582
|
other RNA |
1.813 |
no data |
| 262 |
AT3G09020 |
AT3G09020
|
[alpha 1,4-glycosyltransferase family protein] |
1.813 |
0.093 |
| 263 |
AT1G69160 |
AT1G69160
|
unknown |
1.807 |
-0.168 |
| 264 |
AT2G42840 |
AT2G42840
|
PDF1, protodermal factor 1 |
1.806 |
-0.040 |
| 265 |
AT2G38940 |
AT2G38940
|
ATPT2, ARABIDOPSIS THALIANA PHOSPHATE TRANSPORTER 2, PHT1;4, phosphate transporter 1;4 |
1.801 |
no data |
| 266 |
AT4G14440 |
AT4G14440
|
ATECI3, ARABIDOPSIS THALIANA DELTA(3), DELTA(2)-ENOYL COA ISOMERASE 3, ECI3, DELTA(3), DELTA(2)-ENOYL COA ISOMERASE 3, HCD1, 3-hydroxyacyl-CoA dehydratase 1 |
1.800 |
-0.074 |
| 267 |
AT1G51405 |
AT1G51405
|
[myosin-related] |
1.789 |
no data |
| 268 |
AT5G62210 |
AT5G62210
|
[Embryo-specific protein 3, (ATS3)] |
1.788 |
-0.093 |
| 269 |
AT2G45760 |
AT2G45760
|
BAL, BON ASSOCIATION PROTEIN 1-LIKE, BAP2, BON association protein 2 |
1.788 |
no data |
| 270 |
AT2G26150 |
AT2G26150
|
ATHSFA2, heat shock transcription factor A2, HSFA2, heat shock transcription factor A2 |
1.787 |
-0.091 |
| 271 |
AT4G28140 |
AT4G28140
|
[Integrase-type DNA-binding superfamily protein] |
1.786 |
-0.013 |
| 272 |
AT2G16750 |
AT2G16750
|
[Protein kinase protein with adenine nucleotide alpha hydrolases-like domain] |
1.785 |
-0.109 |
| 273 |
AT5G47850 |
AT5G47850
|
CCR4, CRINKLY4 related 4 |
1.781 |
0.476 |
| 274 |
AT4G13990 |
AT4G13990
|
[Exostosin family protein] |
1.781 |
-0.041 |
| 275 |
AT4G18250 |
AT4G18250
|
[receptor serine/threonine kinase, putative] |
1.780 |
0.532 |
| 276 |
AT3G13470 |
AT3G13470
|
Cpn60beta2, chaperonin-60beta2 |
1.779 |
-0.120 |
| 277 |
AT1G02460 |
AT1G02460
|
[Pectin lyase-like superfamily protein] |
1.773 |
-0.019 |
| 278 |
AT1G02205 |
AT1G02205
|
CER1, ECERIFERUM 1, CER22, ECERIFERUM 22 |
1.771 |
-0.220 |
| 279 |
AT4G14630 |
AT4G14630
|
GLP9, germin-like protein 9 |
1.770 |
0.201 |
| 280 |
AT3G14850 |
AT3G14850
|
TBL41, TRICHOME BIREFRINGENCE-LIKE 41 |
1.768 |
-0.137 |
| 281 |
AT2G18670 |
AT2G18670
|
[RING/U-box superfamily protein] |
1.764 |
0.072 |
| 282 |
AT4G03450 |
AT4G03450
|
[Ankyrin repeat family protein] |
1.764 |
0.209 |
| 283 |
AT3G15518 |
AT3G15518
|
unknown |
1.764 |
no data |
| 284 |
AT4G04700 |
AT4G04700
|
CPK27, calcium-dependent protein kinase 27 |
1.762 |
no data |
| 285 |
AT2G40230 |
AT2G40230
|
[HXXXD-type acyl-transferase family protein] |
1.757 |
-0.046 |
| 286 |
AT1G32960 |
AT1G32960
|
ATSBT3.3, SBT3.3 |
1.753 |
0.173 |
| 287 |
AT1G33170 |
AT1G33170
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
1.751 |
-0.128 |
| 288 |
AT1G01560 |
AT1G01560
|
ATMPK11, MAP kinase 11, MPK11, MAP kinase 11 |
1.745 |
0.228 |
| 289 |
AT5G61810 |
AT5G61810
|
APC1, ATP/phosphate carrier 1 |
1.744 |
-0.095 |
| 290 |
AT3G07600 |
AT3G07600
|
[Heavy metal transport/detoxification superfamily protein ] |
1.743 |
0.147 |
| 291 |
AT1G21280 |
AT1G21280
|
unknown |
1.743 |
no data |
| 292 |
AT3G55500 |
AT3G55500
|
ATEXP16, ATEXPA16, expansin A16, ATHEXP ALPHA 1.7, EXP16, EXPANSIN 16, EXPA16, expansin A16 |
1.742 |
-0.407 |
| 293 |
AT1G25440 |
AT1G25440
|
BBX15, B-box domain protein 15 |
1.739 |
no data |
| 294 |
AT3G20470 |
AT3G20470
|
ATGRP-5, ATGRP5, GRP-5, GLYCINE-RICH PROTEIN 5, GRP5, glycine-rich protein 5 |
1.739 |
-0.118 |
| 295 |
AT5G40390 |
AT5G40390
|
RS5, raffinose synthase 5, SIP1, seed imbibition 1-like |
1.735 |
-0.092 |
| 296 |
AT1G11850 |
AT1G11850
|
unknown |
1.735 |
-0.128 |
| 297 |
AT1G21130 |
AT1G21130
|
IGMT4, indole glucosinolate O-methyltransferase 4 |
1.727 |
-0.001 |
| 298 |
ATCG01090 |
ATCG01090
|
NDHI |
1.725 |
-0.014 |
| 299 |
AT2G38750 |
AT2G38750
|
ANNAT4, annexin 4, AtANN4 |
1.724 |
-0.209 |
| 300 |
AT1G20070 |
AT1G20070
|
unknown |
1.723 |
-0.261 |
| 301 |
AT4G25490 |
AT4G25490
|
ATCBF1, CBF1, C-repeat/DRE binding factor 1, DREB1B, DRE BINDING PROTEIN 1B |
-6.807 |
-0.381 |
| 302 |
AT3G48520 |
AT3G48520
|
CYP94B3, cytochrome P450, family 94, subfamily B, polypeptide 3 |
-6.348 |
-0.007 |
| 303 |
AT5G13655 |
AT5G13655
|
unknown |
-6.203 |
no data |
| 304 |
AT1G73510 |
AT1G73510
|
unknown |
-6.161 |
-0.022 |
| 305 |
AT1G12610 |
AT1G12610
|
DDF1, DWARF AND DELAYED FLOWERING 1 |
-5.844 |
-0.058 |
| 306 |
AT1G70640 |
AT1G70640
|
[octicosapeptide/Phox/Bem1p (PB1) domain-containing protein] |
-5.118 |
no data |
| 307 |
AT3G29156 |
AT3G29156
|
[copia-like retrotransposon family, has a 2.9e-175 P-value blast match to GB:AAC02666 polyprotein (Ty1_Copia-element) (Arabidopsis thaliana)] |
-4.960 |
no data |
| 308 |
AT4G25470 |
AT4G25470
|
ATCBF2, CBF2, C-repeat/DRE binding factor 2, DREB1C, DRE/CRT-BINDING PROTEIN 1C, FTQ4, FREEZING TOLERANCE QTL 4 |
-4.886 |
-0.232 |
| 309 |
AT3G49230 |
AT3G49230
|
unknown |
-4.847 |
-0.083 |
| 310 |
AT5G51990 |
AT5G51990
|
CBF4, C-repeat-binding factor 4, DREB1D, DEHYDRATION-RESPONSIVE ELEMENT-BINDING PROTEIN 1D |
-4.570 |
-0.071 |
| 311 |
AT4G24580 |
AT4G24580
|
REN1, ROP1 ENHANCER 1 |
-4.563 |
-0.015 |
| 312 |
AT3G10510 |
AT3G10510
|
[Galactose oxidase/kelch repeat superfamily protein] |
-4.338 |
no data |
| 313 |
AT4G06746 |
AT4G06746
|
DEAR5, DREB AND EAR MOTIF PROTEIN 5, RAP2.9, related to AP2 9 |
-4.307 |
no data |
| 314 |
AT5G40040 |
AT5G40040
|
[60S acidic ribosomal protein family] |
-4.253 |
-0.013 |
| 315 |
AT4G25480 |
AT4G25480
|
ATCBF3, CBF3, C-REPEAT BINDING FACTOR 3, DREB1A, dehydration response element B1A |
-4.135 |
-0.559 |
| 316 |
AT1G22570 |
AT1G22570
|
[Major facilitator superfamily protein] |
-4.106 |
0.072 |
| 317 |
AT5G43840 |
AT5G43840
|
AT-HSFA6A, heat shock transcription factor A6A, HSFA6A, heat shock transcription factor A6A |
-4.065 |
0.004 |
| 318 |
AT5G15800 |
AT5G15800
|
AGL2, AGAMOUS-like 2, SEP1, SEPALLATA1 |
-4.062 |
-0.071 |
| 319 |
AT5G42800 |
AT5G42800
|
DFR, dihydroflavonol 4-reductase, M318, TT3 |
-3.915 |
-0.084 |
| 320 |
AT4G34410 |
AT4G34410
|
RRTF1, redox responsive transcription factor 1 |
-3.891 |
-0.368 |
| 321 |
AT1G74930 |
AT1G74930
|
ORA47 |
-3.887 |
-0.645 |
| 322 |
AT5G50360 |
AT5G50360
|
unknown |
-3.839 |
-0.022 |
| 323 |
AT3G28740 |
AT3G28740
|
CYP81D11, cytochrome P450, family 81, subfamily D, polypeptide 11 |
-3.741 |
-0.027 |
| 324 |
AT3G28500 |
AT3G28500
|
[60S acidic ribosomal protein family] |
-3.737 |
-0.011 |
| 325 |
AT5G24770 |
AT5G24770
|
ATVSP2, VSP2, vegetative storage protein 2 |
-3.724 |
no data |
| 326 |
AT1G43160 |
AT1G43160
|
RAP2.6, related to AP2 6 |
-3.694 |
-0.073 |
| 327 |
AT3G15450 |
AT3G15450
|
[Aluminium induced protein with YGL and LRDR motifs] |
-3.654 |
0.047 |
| 328 |
AT1G71370 |
AT1G71370
|
[DEA(D/H)-box RNA helicase family protein] |
-3.646 |
no data |
| 329 |
AT4G28820 |
AT4G28820
|
[HIT-type Zinc finger family protein] |
-3.596 |
no data |
| 330 |
AT3G56280 |
AT3G56280
|
[pseudogene] |
-3.544 |
-0.013 |
| 331 |
AT4G28560 |
AT4G28560
|
RIC7, ROP-interactive CRIB motif-containing protein 7 |
-3.544 |
-0.058 |
| 332 |
AT3G17520 |
AT3G17520
|
[Late embryogenesis abundant protein (LEA) family protein] |
-3.522 |
0.051 |
| 333 |
AT3G20975 |
AT3G20975
|
[copia-like retrotransposon family, has a 8.6e-08 P-value blast match to gb|AAO73529.1| gag-pol polyprotein (Glycine max) (SIRE1) (Ty1_Copia-family)] |
-3.482 |
no data |
| 334 |
AT1G32890 |
AT1G32890
|
[non-LTR retrotransposon family (LINE), has a 1.2e-37 P-value blast match to GB:AAB41224 ORF2 (LINE-element) (Rattus norvegicus)] |
-3.463 |
no data |
| 335 |
AT5G05220 |
AT5G05220
|
unknown |
-3.437 |
-0.012 |
| 336 |
AT1G02610 |
AT1G02610
|
[RING/FYVE/PHD zinc finger superfamily protein] |
-3.434 |
no data |
| 337 |
AT2G27270 |
AT2G27270
|
unknown |
-3.434 |
0.047 |
| 338 |
AT2G32050 |
AT2G32050
|
unknown |
-3.433 |
-0.009 |
| 339 |
AT5G21960 |
AT5G21960
|
[Integrase-type DNA-binding superfamily protein] |
-3.418 |
no data |
| 340 |
AT5G59320 |
AT5G59320
|
LTP3, lipid transfer protein 3 |
-3.418 |
0.267 |
| 341 |
AT2G39725 |
AT2G39725
|
[LYR family of Fe/S cluster biogenesis protein] |
-3.380 |
0.047 |
| 342 |
AT3G44798 |
AT3G44798
|
other RNA |
-3.342 |
no data |
| 343 |
AT3G01840 |
AT3G01840
|
LYK2, LysM-containing receptor-like kinase 2 |
-3.312 |
0.022 |
| 344 |
AT3G49130 |
AT3G49130
|
[SWAP (Suppressor-of-White-APricot)/surp RNA-binding domain-containing protein] |
-3.257 |
-0.063 |
| 345 |
AT3G61340 |
AT3G61340
|
[F-box and associated interaction domains-containing protein] |
-3.255 |
0.014 |
| 346 |
AT1G06030 |
AT1G06030
|
[pfkB-like carbohydrate kinase family protein] |
-3.254 |
no data |
| 347 |
AT5G59390 |
AT5G59390
|
[XH/XS domain-containing protein] |
-3.251 |
no data |
| 348 |
AT1G68050 |
AT1G68050
|
ADO3, FKF1, flavin-binding, kelch repeat, f box 1 |
-3.251 |
no data |
| 349 |
AT5G45440 |
AT5G45440
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-3.232 |
no data |
| 350 |
AT4G21870 |
AT4G21870
|
[HSP20-like chaperones superfamily protein] |
-3.220 |
0.115 |
| 351 |
AT5G59310 |
AT5G59310
|
LTP4, lipid transfer protein 4 |
-3.188 |
0.371 |
| 352 |
AT1G19210 |
AT1G19210
|
[Integrase-type DNA-binding superfamily protein] |
-3.187 |
-0.034 |
| 353 |
AT1G78500 |
AT1G78500
|
PEN6, PENTACYCLIC TRITERPENE SYNTHASE 6 |
-3.185 |
-0.028 |
| 354 |
AT2G24592 |
AT2G24592
|
unknown |
-3.184 |
no data |
| 355 |
AT2G35290 |
AT2G35290
|
SAUR79, SMALL AUXIN UPREGULATED RNA 79 |
-3.163 |
0.359 |
| 356 |
AT4G08145 |
AT4G08145
|
unknown |
-3.163 |
no data |
| 357 |
AT2G34600 |
AT2G34600
|
JAZ7, jasmonate-zim-domain protein 7, TIFY5B |
-3.115 |
-0.024 |
| 358 |
AT2G35300 |
AT2G35300
|
AtLEA4-2, late embryogenesis abundant 4-2, LEA18, late embryogenesis abundant 18, LEA4-2, late embryogenesis abundant 4-2 |
-3.115 |
0.018 |
| 359 |
AT5G17490 |
AT5G17490
|
AtRGL3, RGL3, RGA-like protein 3 |
-3.115 |
-0.209 |
| 360 |
AT3G04800 |
AT3G04800
|
ATTIM23-3, translocase inner membrane subunit 23-3, TIM23-3, translocase inner membrane subunit 23-3 |
-3.114 |
-0.037 |
| 361 |
AT1G22500 |
AT1G22500
|
AtATL15, Arabidopsis thaliana Arabidopsis toxicos en levadura 15, ATL15, Arabidopsis toxicos en levadura 15 |
-3.113 |
0.142 |
| 362 |
AT2G20150 |
AT2G20150
|
unknown |
-3.112 |
-0.010 |
| 363 |
AT2G18720 |
AT2G18720
|
[Translation elongation factor EF1A/initiation factor IF2gamma family protein] |
-3.110 |
-0.015 |
| 364 |
AT5G15960 |
AT5G15960
|
KIN1 |
-3.087 |
no data |
| 365 |
AT1G33510 |
AT1G33510
|
[pseudogene] |
-3.042 |
no data |
| 366 |
AT4G30420 |
AT4G30420
|
UMAMIT34, Usually multiple acids move in and out Transporters 34 |
-3.042 |
-0.026 |
| 367 |
AT1G19040 |
AT1G19040
|
[NAC (No Apical Meristem) domain transcriptional regulator superfamily protein] |
-3.042 |
-0.028 |
| 368 |
AT4G22880 |
AT4G22880
|
ANS, ANTHOCYANIDIN SYNTHASE, LDOX, leucoanthocyanidin dioxygenase, TDS4, TANNIN DEFICIENT SEED 4, TT18 |
-3.042 |
no data |
| 369 |
AT1G20180 |
AT1G20180
|
unknown |
-3.041 |
0.048 |
| 370 |
AT4G37490 |
AT4G37490
|
CYC1, CYCLIN 1, CYCB1, CYCB1;1, CYCLIN B1;1 |
-3.012 |
-0.044 |
| 371 |
AT1G53790 |
AT1G53790
|
[F-box and associated interaction domains-containing protein] |
-2.992 |
0.067 |
| 372 |
AT1G08090 |
AT1G08090
|
ACH1, ATNRT2.1, NITRATE TRANSPORTER 2.1, ATNRT2:1, nitrate transporter 2:1, LIN1, LATERAL ROOT INITIATION 1, NRT2, NITRATE TRANSPORTER 2, NRT2.1, NITRATE TRANSPORTER 2.1, NRT2:1, nitrate transporter 2:1, NRT2;1AT |
-2.970 |
0.024 |
| 373 |
AT4G28900 |
AT4G28900
|
[copia-like retrotransposon family, has a 7.7e-236 P-value blast match to GB:AAA57005 Hopscotch polyprotein (Ty1_Copia-element) (Zea mays)] |
-2.967 |
0.016 |
| 374 |
AT5G03204 |
AT5G03204
|
unknown |
-2.965 |
no data |
| 375 |
AT2G31690 |
AT2G31690
|
DALL5, DAD1-Like Lipase 5 |
-2.964 |
-0.012 |
| 376 |
AT1G78390 |
AT1G78390
|
ATNCED9, NINE-CIS-EPOXYCAROTENOID DIOXYGENASE 9, NCED9, nine-cis-epoxycarotenoid dioxygenase 9 |
-2.961 |
0.020 |
| 377 |
AT5G16360 |
AT5G16360
|
[NC domain-containing protein-related] |
-2.958 |
no data |
| 378 |
AT3G30775 |
AT3G30775
|
AT-POX, ATPDH, ATPOX, ARABIDOPSIS THALIANA PROLINE OXIDASE, ERD5, EARLY RESPONSIVE TO DEHYDRATION 5, PDH1, proline dehydrogenase 1, PRO1, PRODH, PROLINE DEHYDROGENASE |
-2.921 |
0.232 |
| 379 |
AT1G13150 |
AT1G13150
|
CYP86C4, cytochrome P450, family 86, subfamily C, polypeptide 4 |
-2.888 |
-0.051 |
| 380 |
AT1G27490 |
AT1G27490
|
[F-box and associated interaction domains-containing protein] |
-2.884 |
no data |
| 381 |
AT1G65480 |
AT1G65480
|
FT, FLOWERING LOCUS T, RSB8, REDUCED STEM BRANCHING 8 |
-2.882 |
-0.052 |
| 382 |
AT1G10400 |
AT1G10400
|
[UDP-Glycosyltransferase superfamily protein] |
-2.869 |
-0.035 |
| 383 |
AT2G23210 |
AT2G23210
|
[UDP-Glycosyltransferase superfamily protein] |
-2.844 |
0.006 |
| 384 |
AT1G25025 |
AT1G25025
|
unknown |
-2.844 |
no data |
| 385 |
AT1G09930 |
AT1G09930
|
ATOPT2, oligopeptide transporter 2, OPT2, oligopeptide transporter 2 |
-2.822 |
0.033 |
| 386 |
AT3G03480 |
AT3G03480
|
CHAT, acetyl CoA:(Z)-3-hexen-1-ol acetyltransferase |
-2.822 |
-0.101 |
| 387 |
AT5G24880 |
AT5G24880
|
unknown |
-2.820 |
-0.036 |
| 388 |
AT1G70440 |
AT1G70440
|
SRO3, similar to RCD one 3 |
-2.809 |
no data |
| 389 |
AT5G57785 |
AT5G57785
|
unknown |
-2.783 |
-0.064 |
| 390 |
AT1G26200 |
AT1G26200
|
[TRAM, LAG1 and CLN8 (TLC) lipid-sensing domain containing protein] |
-2.783 |
no data |
| 391 |
AT5G34480 |
AT5G34480
|
[gypsy-like retrotransposon family, has a 5.7e-49 P-value blast match to GB:AAD27547 polyprotein (Gypsy_Ty3-element) (Oryza sativa subsp. indica)] |
-2.783 |
no data |
| 392 |
AT4G01533 |
AT4G01533
|
other RNA |
-2.782 |
no data |
| 393 |
AT2G39705 |
AT2G39705
|
DVL11, DEVIL 11, RTFL8, ROTUNDIFOLIA like 8 |
-2.731 |
0.002 |
| 394 |
AT1G50560 |
AT1G50560
|
CYP705A25, cytochrome P450, family 705, subfamily A, polypeptide 25 |
-2.727 |
0.002 |
| 395 |
AT1G11420 |
AT1G11420
|
unknown |
-2.721 |
-0.002 |
| 396 |
AT3G17225 |
AT3G17225
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-2.720 |
no data |
| 397 |
AT2G33260 |
AT2G33260
|
[Tryptophan/tyrosine permease] |
-2.720 |
-0.002 |
| 398 |
AT2G27410 |
AT2G27410
|
unknown |
-2.720 |
-0.096 |
| 399 |
AT5G24780 |
AT5G24780
|
ATVSP1, VSP1, vegetative storage protein 1 |
-2.720 |
no data |
| 400 |
AT2G33385 |
AT2G33385
|
arpc2b, actin-related protein C2B |
-2.714 |
no data |
| 401 |
AT1G73066 |
AT1G73066
|
[Leucine-rich repeat family protein] |
-2.653 |
-0.018 |
| 402 |
AT1G64110 |
AT1G64110
|
DAA1, DUO1-activated ATPase 1 |
-2.649 |
-0.040 |
| 403 |
AT5G58610 |
AT5G58610
|
[PHD finger transcription factor, putative] |
-2.645 |
-0.029 |
| 404 |
AT5G05410 |
AT5G05410
|
DREB2, DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN 2, DREB2A, DRE-binding protein 2A |
-2.633 |
-0.074 |
| 405 |
AT3G50850 |
AT3G50850
|
[Putative methyltransferase family protein] |
-2.627 |
0.052 |
| 406 |
AT4G13390 |
AT4G13390
|
EXT12, extensin 12 |
-2.616 |
-0.031 |
| 407 |
AT4G36850 |
AT4G36850
|
[PQ-loop repeat family protein / transmembrane family protein] |
-2.610 |
no data |
| 408 |
AT5G49360 |
AT5G49360
|
ATBXL1, BETA-XYLOSIDASE 1, BXL1, beta-xylosidase 1 |
-2.597 |
0.080 |
| 409 |
AT5G24470 |
AT5G24470
|
APRR5, pseudo-response regulator 5, PRR5, pseudo-response regulator 5 |
-2.588 |
0.036 |
| 410 |
AT2G30380 |
AT2G30380
|
unknown |
-2.584 |
0.012 |
| 411 |
AT5G10700 |
AT5G10700
|
[Peptidyl-tRNA hydrolase II (PTH2) family protein] |
-2.578 |
-0.104 |
| 412 |
AT5G66350 |
AT5G66350
|
SHI, SHORT INTERNODES |
-2.562 |
0.014 |
| 413 |
AT1G09460 |
AT1G09460
|
[Carbohydrate-binding X8 domain superfamily protein] |
-2.552 |
0.034 |
| 414 |
AT5G53290 |
AT5G53290
|
CRF3, cytokinin response factor 3 |
-2.547 |
-0.029 |
| 415 |
AT1G01310 |
AT1G01310
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-2.545 |
0.015 |
| 416 |
AT1G18300 |
AT1G18300
|
atnudt4, nudix hydrolase homolog 4, NUDT4, nudix hydrolase homolog 4 |
-2.545 |
no data |
| 417 |
AT5G06990 |
AT5G06990
|
unknown |
-2.545 |
no data |
| 418 |
AT1G70800 |
AT1G70800
|
EHB1, ENHANCED BENDING 1 |
-2.530 |
no data |
| 419 |
AT5G58780 |
AT5G58780
|
AtcPT5, AtHEPS, cPT5, cis -prenyltransferase 5, HEPS, heptaprenyl diphosphate synthase |
-2.520 |
-0.022 |
| 420 |
AT3G21020 |
AT3G21020
|
[copia-like retrotransposon family, has a 5.4e-14 P-value blast match to gb|AAO73529.1| gag-pol polyprotein (Glycine max) (SIRE1) (Ty1_Copia-family)] |
-2.515 |
-0.094 |
| 421 |
AT2G15890 |
AT2G15890
|
MEE14, maternal effect embryo arrest 14 |
-2.511 |
0.053 |
| 422 |
AT4G14860 |
AT4G14860
|
AtOFP11, ovate family protein 11, OFP11, ovate family protein 11 |
-2.509 |
no data |
| 423 |
AT5G23810 |
AT5G23810
|
AAP7, amino acid permease 7 |
-2.487 |
0.006 |
| 424 |
AT3G08040 |
AT3G08040
|
ATFRD3, FRD3, FERRIC REDUCTASE DEFECTIVE 3, MAN1, MANGANESE ACCUMULATOR 1 |
-2.474 |
0.033 |
| 425 |
AT1G13700 |
AT1G13700
|
PGL1, 6-phosphogluconolactonase 1 |
-2.470 |
0.137 |
| 426 |
AT3G18610 |
AT3G18610
|
ATNUC-L2, nucleolin like 2, NUC-L2, nucleolin like 2, PARLL1, PARALLEL1-LIKE 1 |
-2.462 |
0.008 |
| 427 |
AT4G17085 |
AT4G17085
|
[Putative membrane lipoprotein] |
-2.456 |
no data |
| 428 |
AT1G13670 |
AT1G13670
|
unknown |
-2.451 |
0.104 |
| 429 |
AT1G49150 |
AT1G49150
|
unknown |
-2.442 |
-0.006 |
| 430 |
AT4G27652 |
AT4G27652
|
unknown |
-2.440 |
-0.046 |
| 431 |
AT2G40080 |
AT2G40080
|
ELF4, EARLY FLOWERING 4 |
-2.439 |
0.051 |
| 432 |
AT1G32880 |
AT1G32880
|
[ARM repeat superfamily protein] |
-2.434 |
-0.016 |
| 433 |
AT2G02240 |
AT2G02240
|
MEE66, maternal effect embryo arrest 66 |
-2.434 |
no data |
| 434 |
AT1G17960 |
AT1G17960
|
[Threonyl-tRNA synthetase] |
-2.426 |
0.013 |
| 435 |
AT5G20420 |
AT5G20420
|
CHR42, chromatin remodeling 42 |
-2.421 |
-0.029 |
| 436 |
AT1G25112 |
AT1G25112
|
unknown |
-2.421 |
no data |
| 437 |
AT3G02140 |
AT3G02140
|
AFP4, ABI FIVE BINDING PROTEIN 4, TMAC2, TWO OR MORE ABRES-CONTAINING GENE 2 |
-2.413 |
0.186 |
| 438 |
AT5G02070 |
AT5G02070
|
[Protein kinase family protein] |
-2.413 |
0.019 |
| 439 |
AT3G22100 |
AT3G22100
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-2.406 |
no data |
| 440 |
AT5G59170 |
AT5G59170
|
[Proline-rich extensin-like family protein] |
-2.405 |
-0.065 |
| 441 |
AT4G35770 |
AT4G35770
|
ATSEN1, ARABIDOPSIS THALIANA SENESCENCE 1, DIN1, DARK INDUCIBLE 1, SEN1, SENESCENCE 1, SEN1, SENESCENCE ASSOCIATED GENE 1 |
-2.391 |
-0.023 |
| 442 |
AT5G56870 |
AT5G56870
|
BGAL4, beta-galactosidase 4 |
-2.387 |
0.140 |
| 443 |
AT2G35570 |
AT2G35570
|
[pseudogene] |
-2.356 |
-0.060 |
| 444 |
AT5G15840 |
AT5G15840
|
BBX1, B-box domain protein 1, CO, CONSTANS, FG |
-2.348 |
0.027 |
| 445 |
AT5G22920 |
AT5G22920
|
[CHY-type/CTCHY-type/RING-type Zinc finger protein] |
-2.343 |
0.079 |
| 446 |
AT1G19530 |
AT1G19530
|
unknown |
-2.333 |
0.108 |
| 447 |
AT3G25573 |
AT3G25573
|
unknown |
-2.332 |
no data |
| 448 |
AT4G07850 |
AT4G07850
|
[gypsy-like retrotransposon family, has a 3.2e-135 P-value blast match to GB:AAD22153 polyprotein (Gypsy_Ty3-element) (Sorghum bicolor)] |
-2.332 |
no data |
| 449 |
AT5G60780 |
AT5G60780
|
ATNRT2.3, ARABIDOPSIS THALIANA NITRATE TRANSPORTER 2.3, NRT2.3, nitrate transporter 2.3 |
-2.332 |
0.050 |
| 450 |
AT3G55810 |
AT3G55810
|
[Pyruvate kinase family protein] |
-2.330 |
no data |
| 451 |
AT2G36640 |
AT2G36640
|
ATECP63, embryonic cell protein 63, ECP63, embryonic cell protein 63 |
-2.311 |
0.008 |
| 452 |
AT5G25210 |
AT5G25210
|
unknown |
-2.290 |
0.046 |
| 453 |
AT5G14545 |
AT5G14545
|
MIR398B, microRNA398B |
-2.266 |
no data |
| 454 |
AT2G39380 |
AT2G39380
|
ATEXO70H2, exocyst subunit exo70 family protein H2, EXO70H2, exocyst subunit exo70 family protein H2 |
-2.265 |
0.033 |
| 455 |
AT1G71280 |
AT1G71280
|
[DEA(D/H)-box RNA helicase family protein] |
-2.264 |
no data |
| 456 |
AT1G15700 |
AT1G15700
|
ATPC2 |
-2.257 |
-0.050 |
| 457 |
AT5G47060 |
AT5G47060
|
unknown |
-2.233 |
0.051 |
| 458 |
AT2G38240 |
AT2G38240
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-2.230 |
-0.046 |
| 459 |
AT5G47370 |
AT5G47370
|
HAT2 |
-2.229 |
-0.034 |
| 460 |
AT4G34160 |
AT4G34160
|
CYCD3, CYCD3;1, CYCLIN D3;1 |
-2.224 |
-0.035 |
| 461 |
AT5G64190 |
AT5G64190
|
unknown |
-2.220 |
0.084 |
| 462 |
AT3G22460 |
AT3G22460
|
OASA2, O-acetylserine (thiol) lyase (OAS-TL) isoform A2 |
-2.213 |
0.195 |
| 463 |
AT2G30615 |
AT2G30615
|
unknown |
-2.210 |
no data |
| 464 |
AT3G57360 |
AT3G57360
|
unknown |
-2.207 |
0.152 |
| 465 |
AT1G64310 |
AT1G64310
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-2.196 |
-0.032 |
| 466 |
AT5G24290 |
AT5G24290
|
MEB2, MEMBRANE OF ER BODY 2 |
-2.196 |
0.068 |
| 467 |
AT3G13065 |
AT3G13065
|
SRF4, STRUBBELIG-receptor family 4 |
-2.196 |
0.103 |
| 468 |
AT2G35930 |
AT2G35930
|
AtPUB23, PUB23, plant U-box 23 |
-2.193 |
0.157 |
| 469 |
AT3G15630 |
AT3G15630
|
unknown |
-2.189 |
0.093 |
| 470 |
AT2G18700 |
AT2G18700
|
ATTPS11, trehalose phosphatase/synthase 11, ATTPSB, TPS11, trehalose phosphatase/synthase 11, TPS11, TREHALOSE-6-PHOSPHATE SYNTHASE 11 |
-2.184 |
0.099 |
| 471 |
AT3G56790 |
AT3G56790
|
[RNA splicing factor-related] |
-2.184 |
no data |
| 472 |
AT2G18180 |
AT2G18180
|
[Sec14p-like phosphatidylinositol transfer family protein] |
-2.180 |
-0.043 |
| 473 |
AT1G66870 |
AT1G66870
|
[Carbohydrate-binding X8 domain superfamily protein] |
-2.175 |
no data |
| 474 |
AT1G53680 |
AT1G53680
|
ATGSTU28, glutathione S-transferase TAU 28, GSTU28, glutathione S-transferase TAU 28 |
-2.156 |
-0.078 |
| 475 |
AT1G73790 |
AT1G73790
|
AtGIP2, GIP1b, GCP3-Interacting Protein 1b, GIP2, GCP3-interacting protein 2 |
-2.156 |
-0.056 |
| 476 |
AT1G33760 |
AT1G33760
|
[Integrase-type DNA-binding superfamily protein] |
-2.151 |
0.122 |
| 477 |
AT4G24390 |
AT4G24390
|
AFB4, auxin signaling F-box 4 |
-2.145 |
-0.005 |
| 478 |
AT1G10140 |
AT1G10140
|
[Uncharacterised conserved protein UCP031279] |
-2.136 |
0.097 |
| 479 |
AT5G02760 |
AT5G02760
|
APD7, Arabidopsis Pp2c clade D 7 |
-2.132 |
0.013 |
| 480 |
AT3G60930 |
AT3G60930
|
unknown |
-2.123 |
no data |
| 481 |
AT4G01430 |
AT4G01430
|
UMAMIT29, Usually multiple acids move in and out Transporters 29 |
-2.120 |
0.043 |
| 482 |
AT1G07430 |
AT1G07430
|
AIP1, AKT1 interacting protein phosphatase 1, AtAIP1, HAI2, highly ABA-induced PP2C gene 2, HON, HONSU (Korean for abnormal drowsiness) |
-2.120 |
-0.088 |
| 483 |
AT1G06520 |
AT1G06520
|
ATGPAT1, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 1, GPAT1, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 1, sn-2-GPAT1, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 1 |
-2.120 |
0.112 |
| 484 |
AT4G13030 |
AT4G13030
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-2.117 |
0.010 |
| 485 |
AT2G33830 |
AT2G33830
|
AtDRM2, DRM2, dormancy associated gene 2 |
-2.110 |
0.066 |
| 486 |
AT2G32190 |
AT2G32190
|
unknown |
-2.107 |
no data |
| 487 |
AT3G46070 |
AT3G46070
|
[C2H2-type zinc finger family protein] |
-2.103 |
-0.015 |
| 488 |
AT3G59070 |
AT3G59070
|
[Cytochrome b561/ferric reductase transmembrane with DOMON related domain] |
-2.102 |
-0.067 |
| 489 |
AT3G59068 |
AT3G59068
|
|
-2.102 |
no data |
| 490 |
AT2G34190 |
AT2G34190
|
[Xanthine/uracil permease family protein] |
-2.096 |
-0.065 |
| 491 |
AT4G13395 |
AT4G13395
|
DVL10, DEVIL 10, RTFL12, ROTUNDIFOLIA like 12 |
-2.090 |
no data |
| 492 |
AT2G05300 |
AT2G05300
|
|
-2.089 |
no data |
| 493 |
AT3G02010 |
AT3G02010
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-2.073 |
0.060 |
| 494 |
AT4G20390 |
AT4G20390
|
[Uncharacterised protein family (UPF0497)] |
-2.068 |
0.053 |
| 495 |
AT1G11175 |
AT1G11175
|
other RNA |
-2.068 |
no data |
| 496 |
AT1G60960 |
AT1G60960
|
ATIRT3, IRON REGULATED TRANSPORTER 3, IRT3, iron regulated transporter 3 |
-2.067 |
-0.017 |
| 497 |
AT2G17660 |
AT2G17660
|
[RPM1-interacting protein 4 (RIN4) family protein] |
-2.065 |
0.050 |
| 498 |
AT1G15580 |
AT1G15580
|
ATAUX2-27, AUX2-27, AUXIN-INDUCIBLE 2-27, IAA5, indole-3-acetic acid inducible 5 |
-2.055 |
-0.122 |
| 499 |
AT3G60980 |
AT3G60980
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-2.053 |
0.007 |
| 500 |
AT2G13130 |
AT2G13130
|
[pseudogene] |
-2.052 |
no data |
| 501 |
AT4G24275 |
AT4G24275
|
unknown |
-2.052 |
no data |
| 502 |
AT2G30040 |
AT2G30040
|
MAPKKK14, mitogen-activated protein kinase kinase kinase 14 |
-2.041 |
-0.123 |
| 503 |
AT2G32670 |
AT2G32670
|
ATVAMP725, vesicle-associated membrane protein 725, VAMP725, vesicle-associated membrane protein 725 |
-2.038 |
-0.051 |
| 504 |
AT1G11260 |
AT1G11260
|
ATSTP1, SUGAR TRANSPORTER 1, STP1, sugar transporter 1 |
-2.032 |
0.130 |
| 505 |
AT3G28420 |
AT3G28420
|
[Putative membrane lipoprotein] |
-2.028 |
-0.056 |
| 506 |
AT5G20250 |
AT5G20250
|
DIN10, DARK INDUCIBLE 10, RS6, raffinose synthase 6 |
-2.025 |
0.147 |
| 507 |
AT4G13880 |
AT4G13880
|
AtRLP48, receptor like protein 48, RLP48, receptor like protein 48 |
-2.018 |
-0.019 |
| 508 |
AT5G60440 |
AT5G60440
|
AGL62, AGAMOUS-like 62 |
-2.014 |
0.033 |
| 509 |
AT2G02250 |
AT2G02250
|
AtPP2-B2, phloem protein 2-B2, PP2-B2, phloem protein 2-B2 |
-2.014 |
-0.020 |
| 510 |
AT3G47340 |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
-2.007 |
-0.008 |
| 511 |
AT4G35130 |
AT4G35130
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-2.001 |
0.030 |
| 512 |
AT5G66080 |
AT5G66080
|
APD9, Arabidopsis Pp2c clade D 9 |
-1.997 |
-0.005 |
| 513 |
AT1G16510 |
AT1G16510
|
SAUR41, SMALL AUXIN UPREGULATED 41 |
-1.982 |
0.273 |
| 514 |
AT2G19330 |
AT2G19330
|
PIRL6, plant intracellular ras group-related LRR 6 |
-1.979 |
-0.078 |
| 515 |
AT2G18050 |
AT2G18050
|
HIS1-3, histone H1-3 |
-1.965 |
-0.108 |
| 516 |
AT3G62550 |
AT3G62550
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
-1.962 |
-0.009 |
| 517 |
AT2G40010 |
AT2G40010
|
[Ribosomal protein L10 family protein] |
-1.961 |
-0.091 |
| 518 |
AT5G57550 |
AT5G57550
|
XTH25, xyloglucan endotransglucosylase/hydrolase 25, XTR3, xyloglucan endotransglycosylase 3 |
-1.961 |
0.187 |
| 519 |
AT5G64110 |
AT5G64110
|
[Peroxidase superfamily protein] |
-1.958 |
0.067 |
| 520 |
AT5G02540 |
AT5G02540
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-1.957 |
-0.170 |
| 521 |
AT1G13430 |
AT1G13430
|
ATST4C, ARABIDOPSIS THALIANA SULFOTRANSFERASE 4C, ST4C, sulfotransferase 4C |
-1.955 |
no data |
| 522 |
AT5G43620 |
AT5G43620
|
[Pre-mRNA cleavage complex II] |
-1.954 |
no data |
| 523 |
AT3G58980 |
AT3G58980
|
[F-box family protein] |
-1.954 |
no data |
| 524 |
AT1G06250 |
AT1G06250
|
[alpha/beta-Hydrolases superfamily protein] |
-1.953 |
-0.026 |
| 525 |
AT4G27450 |
AT4G27450
|
[Aluminium induced protein with YGL and LRDR motifs] |
-1.932 |
0.176 |
| 526 |
AT4G12470 |
AT4G12470
|
AZI1, azelaic acid induced 1 |
-1.930 |
0.001 |
| 527 |
AT5G26040 |
AT5G26040
|
HDA2, histone deacetylase 2 |
-1.929 |
-0.098 |
| 528 |
AT1G31570 |
AT1G31570
|
[CACTA-like transposase family (Tnp2/En/Spm), has a 0. P-value blast match to gb|AAG52024.1|AC022456_5 Tam1-homologous transposon protein TNP2, putative] |
-1.923 |
no data |
| 529 |
AT1G17020 |
AT1G17020
|
ATSRG1, SENESCENCE-RELATED GENE 1, SRG1, senescence-related gene 1 |
-1.921 |
0.241 |
| 530 |
AT4G21480 |
AT4G21480
|
STP12, sugar transporter protein 12 |
-1.921 |
-0.063 |
| 531 |
AT3G50970 |
AT3G50970
|
LTI30, LOW TEMPERATURE-INDUCED 30, XERO2 |
-1.917 |
0.088 |
| 532 |
AT4G37608 |
AT4G37608
|
unknown |
-1.916 |
no data |
| 533 |
AT3G57320 |
AT3G57320
|
unknown |
-1.911 |
-0.190 |
| 534 |
AT4G39410 |
AT4G39410
|
ATWRKY13, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 13, WRKY13, WRKY DNA-binding protein 13 |
-1.910 |
0.045 |
| 535 |
AT3G61990 |
AT3G61990
|
OMTF3, O-MTase family 3 protein |
-1.907 |
0.053 |
| 536 |
AT5G34851 |
AT5G34851
|
[gypsy-like retrotransposon family, has a 0. P-value blast match to GB:AAD27547 polyprotein (Gypsy_Ty3-element) (Oryza sativa subsp. indica)] |
-1.897 |
no data |
| 537 |
AT5G25450 |
AT5G25450
|
[Cytochrome bd ubiquinol oxidase, 14kDa subunit] |
-1.892 |
0.094 |
| 538 |
AT3G21660 |
AT3G21660
|
[UBX domain-containing protein] |
-1.890 |
-0.004 |
| 539 |
AT5G08780 |
AT5G08780
|
[winged-helix DNA-binding transcription factor family protein] |
-1.889 |
no data |
| 540 |
AT5G25190 |
AT5G25190
|
ESE3, ethylene and salt inducible 3 |
-1.887 |
0.111 |
| 541 |
AT2G39795 |
AT2G39795
|
[Mitochondrial glycoprotein family protein] |
-1.885 |
0.050 |
| 542 |
AT5G48775 |
AT5G48775
|
other RNA |
-1.883 |
no data |
| 543 |
AT1G19394 |
AT1G19394
|
unknown |
-1.880 |
no data |
| 544 |
AT5G27920 |
AT5G27920
|
[F-box family protein] |
-1.878 |
0.093 |
| 545 |
AT5G54140 |
AT5G54140
|
ILL3, IAA-leucine-resistant (ILR1)-like 3 |
-1.878 |
0.157 |
| 546 |
AT1G32170 |
AT1G32170
|
XTH30, xyloglucan endotransglucosylase/hydrolase 30, XTR4, xyloglucan endotransglycosylase 4 |
-1.873 |
0.239 |
| 547 |
AT3G62960 |
AT3G62960
|
[Thioredoxin superfamily protein] |
-1.870 |
0.083 |
| 548 |
AT2G21200 |
AT2G21200
|
SAUR7, SMALL AUXIN UPREGULATED RNA 7 |
-1.866 |
0.167 |
| 549 |
AT3G04070 |
AT3G04070
|
anac047, NAC domain containing protein 47, NAC047, NAC domain containing protein 47 |
-1.864 |
0.338 |
| 550 |
AT4G16000 |
AT4G16000
|
unknown |
-1.864 |
0.073 |
| 551 |
AT3G11020 |
AT3G11020
|
DREB2, DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN 2, DREB2B, DRE/CRT-binding protein 2B |
-1.863 |
-0.045 |
| 552 |
AT3G22740 |
AT3G22740
|
HMT3, homocysteine S-methyltransferase 3 |
-1.856 |
-0.001 |
| 553 |
AT1G19025 |
AT1G19025
|
[DNA repair metallo-beta-lactamase family protein] |
-1.843 |
-0.074 |
| 554 |
AT2G28110 |
AT2G28110
|
FRA8, FRAGILE FIBER 8, IRX7, IRREGULAR XYLEM 7 |
-1.842 |
0.220 |
| 555 |
AT1G60140 |
AT1G60140
|
ATTPS10, trehalose phosphate synthase, TPS10, trehalose phosphate synthase, TPS10, TREHALOSE PHOSPHATE SYNTHASE 10 |
-1.840 |
0.072 |
| 556 |
AT5G19120 |
AT5G19120
|
[Eukaryotic aspartyl protease family protein] |
-1.838 |
0.012 |
| 557 |
AT5G02080 |
AT5G02080
|
[DNA / pantothenate metabolism flavoprotein] |
-1.832 |
no data |
| 558 |
AT4G25835 |
AT4G25835
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-1.828 |
0.263 |
| 559 |
AT1G78000 |
AT1G78000
|
SEL1, SELENATE RESISTANT 1, SULTR1;2, sulfate transporter 1;2 |
-1.822 |
0.299 |
| 560 |
AT3G45060 |
AT3G45060
|
ATNRT2.6, ARABIDOPSIS THALIANA HIGH AFFINITY NITRATE TRANSPORTER 2.6, NRT2.6, high affinity nitrate transporter 2.6 |
-1.821 |
0.001 |
| 561 |
AT3G26612 |
AT3G26612
|
other RNA |
-1.817 |
no data |
| 562 |
AT3G27200 |
AT3G27200
|
[Cupredoxin superfamily protein] |
-1.817 |
-0.094 |
| 563 |
AT1G03545 |
AT1G03545
|
Potential natural antisense gene |
-1.813 |
no data |
| 564 |
AT1G09220 |
AT1G09220
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-1.799 |
0.115 |
| 565 |
AT5G14560 |
AT5G14560
|
unknown |
-1.789 |
-0.014 |
| 566 |
AT3G05685 |
AT3G05685
|
[Cystatin/monellin superfamily protein] |
-1.789 |
no data |
| 567 |
AT1G75290 |
AT1G75290
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-1.788 |
-0.025 |
| 568 |
AT1G23870 |
AT1G23870
|
ATTPS9, trehalose-phosphatase/synthase 9, TPS9, TREHALOSE -6-PHOSPHATASE SYNTHASE S9, TPS9, trehalose-phosphatase/synthase 9 |
-1.782 |
0.144 |
| 569 |
AT5G65570 |
AT5G65570
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-1.780 |
0.032 |
| 570 |
AT5G56310 |
AT5G56310
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-1.769 |
-0.028 |
| 571 |
AT4G33290 |
AT4G33290
|
[F-box and associated interaction domains-containing protein] |
-1.765 |
-0.031 |
| 572 |
AT4G34412 |
AT4G34412
|
unknown |
-1.760 |
no data |
| 573 |
AT1G03550 |
AT1G03550
|
AtSCAMP4, SCAMP4, Secretory carrier membrane protein 4 |
-1.757 |
0.205 |
| 574 |
AT2G06950 |
AT2G06950
|
[copia-like retrotransposon family, has a 2.7e-243 P-value blast match to dbj|BAA78426.1| polyprotein (AtRE2-1) (Arabidopsis thaliana) (Ty1_Copia-element)] |
-1.756 |
-0.066 |
| 575 |
AT2G25590 |
AT2G25590
|
[Plant Tudor-like protein] |
-1.755 |
0.220 |
| 576 |
AT1G56660 |
AT1G56660
|
unknown |
-1.753 |
0.115 |
| 577 |
AT1G12780 |
AT1G12780
|
ATUGE1, A. THALIANA UDP-GLC 4-EPIMERASE 1, UGE1, UDP-D-glucose/UDP-D-galactose 4-epimerase 1 |
-1.751 |
0.076 |
| 578 |
AT1G15125 |
AT1G15125
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-1.751 |
no data |
| 579 |
AT5G10946 |
AT5G10946
|
unknown |
-1.747 |
no data |
| 580 |
AT2G20800 |
AT2G20800
|
NDB4, NAD(P)H dehydrogenase B4 |
-1.742 |
-0.094 |
| 581 |
AT1G27290 |
AT1G27290
|
unknown |
-1.737 |
0.067 |
| 582 |
AT1G74180 |
AT1G74180
|
AtRLP14, receptor like protein 14, RLP14, receptor like protein 14 |
-1.734 |
0.008 |
| 583 |
AT5G01380 |
AT5G01380
|
[Homeodomain-like superfamily protein] |
-1.732 |
-0.021 |
| 584 |
AT3G17280 |
AT3G17280
|
[F-box and associated interaction domains-containing protein] |
-1.729 |
0.003 |
| 585 |
AT3G17890 |
AT3G17890
|
unknown |
-1.728 |
-0.044 |
| 586 |
AT5G14120 |
AT5G14120
|
[Major facilitator superfamily protein] |
-1.723 |
0.016 |
| 587 |
AT5G09570 |
AT5G09570
|
[Cox19-like CHCH family protein] |
-1.719 |
-0.025 |
| 588 |
AT3G28570 |
AT3G28570
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-1.719 |
no data |
| 589 |
AT4G25000 |
AT4G25000
|
AMY1, alpha-amylase-like, ATAMY1 |
-1.711 |
0.677 |
| 590 |
AT1G49032 |
AT1G49032
|
unknown |
-1.710 |
no data |
| 591 |
AT3G26165 |
AT3G26165
|
CYP71B18, cytochrome P450, family 71, subfamily B, polypeptide 18 |
-1.707 |
no data |
| 592 |
AT5G59240 |
AT5G59240
|
[Ribosomal protein S8e family protein] |
-1.702 |
-0.010 |
| 593 |
AT4G15910 |
AT4G15910
|
ATDI21, drought-induced 21, DI21, drought-induced 21 |
-1.698 |
0.018 |
| 594 |
AT4G10690 |
AT4G10690
|
[copia-like retrotransposon family, has a 1.5e-256 P-value blast match to dbj|BAA78426.1| polyprotein (AtRE2-1) (Arabidopsis thaliana) (Ty1_Copia-element)] |
-1.694 |
0.003 |
| 595 |
AT3G46900 |
AT3G46900
|
COPT2, copper transporter 2 |
-1.693 |
0.183 |
| 596 |
AT4G29780 |
AT4G29780
|
unknown |
-1.686 |
-0.241 |
| 597 |
AT2G26695 |
AT2G26695
|
[Ran BP2/NZF zinc finger-like superfamily protein] |
-1.684 |
no data |
| 598 |
AT3G55290 |
AT3G55290
|
SDRd, SHORT-CHAIN DEHYDROGENASE/REDUCTASE ISOFORM d |
-1.683 |
no data |
| 599 |
AT4G02140 |
AT4G02140
|
unknown |
-1.682 |
-0.069 |
| 600 |
AT4G39360 |
AT4G39360
|
unknown |
-1.678 |
no data |