|
probeID |
AGICode |
Annotation |
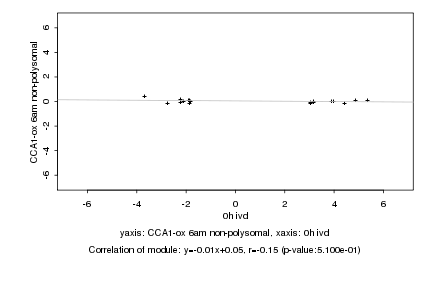
Log2 signal ratio
0h ivd |
Log2 signal ratio
CCA1-ox 6am non-polysomal |
| 1 |
AT5G27500 |
AT5G27500
|
unknown |
6.678 |
no data |
| 2 |
AT4G37990 |
AT4G37990
|
ATCAD8, ARABIDOPSIS THALIANA CINNAMYL-ALCOHOL DEHYDROGENASE 8, CAD-B2, CINNAMYL-ALCOHOL DEHYDROGENASE B2, ELI3, ELICITOR-ACTIVATED GENE 3, ELI3-2, elicitor-activated gene 3-2 |
5.347 |
0.156 |
| 3 |
AT1G53480 |
AT1G53480
|
ATMRD1, ARABIDOPSIS MTO 1 RESPONDING DOWN 1, MRD1, MTO 1 RESPONDING DOWN 1 |
5.298 |
no data |
| 4 |
AT5G59310 |
AT5G59310
|
LTP4, lipid transfer protein 4 |
4.832 |
0.093 |
| 5 |
AT3G60140 |
AT3G60140
|
BGLU30, BETA GLUCOSIDASE 30, DIN2, DARK INDUCIBLE 2, SRG2, SENESCENCE-RELATED GENE 2 |
4.396 |
-0.109 |
| 6 |
AT2G41260 |
AT2G41260
|
ATM17, M17 |
3.970 |
0.017 |
| 7 |
AT1G80160 |
AT1G80160
|
GLYI7, glyoxylase I 7 |
3.886 |
0.046 |
| 8 |
AT3G44300 |
AT3G44300
|
AtNIT2, nitrilase 2, NIT2, nitrilase 2 |
3.882 |
no data |
| 9 |
AT3G59930 |
AT3G59930
|
unknown |
3.696 |
no data |
| 10 |
AT1G09500 |
AT1G09500
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
3.147 |
-0.017 |
| 11 |
AT5G62165 |
AT5G62165
|
AGL42, AGAMOUS-like 42, FYF, FOREVER YOUNG FLOWER |
3.140 |
0.002 |
| 12 |
AT5G59320 |
AT5G59320
|
LTP3, lipid transfer protein 3 |
3.029 |
-0.055 |
| 13 |
AT3G30122 |
AT3G30122
|
[expressed protein] |
3.013 |
no data |
| 14 |
AT4G21680 |
AT4G21680
|
AtNPF7.2, NPF7.2, NRT1/ PTR family 7.2, NRT1.8, NITRATE TRANSPORTER 1.8 |
3.009 |
-0.146 |
| 15 |
AT5G24780 |
AT5G24780
|
ATVSP1, VSP1, vegetative storage protein 1 |
2.979 |
no data |
| 16 |
AT5G35935 |
AT5G35935
|
[copia-like retrotransposon family, has a 2.0e-232 P-value blast match to GB:CAA72989 open reading frame 1 (Ty1_Copia-element) (Brassica oleracea)] |
-4.266 |
no data |
| 17 |
AT3G02310 |
AT3G02310
|
AGL4, AGAMOUS-like 4, SEP2, SEPALLATA 2 |
-3.703 |
0.457 |
| 18 |
AT4G09430 |
AT4G09430
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-2.762 |
-0.096 |
| 19 |
AT3G04717 |
AT3G04717
|
[pseudogene] |
-2.617 |
no data |
| 20 |
AT1G20390 |
AT1G20390
|
[gypsy-like retrotransposon family, has a 1.5e-251 P-value blast match to GB:AAD19359 polyprotein (gypsy_Ty3-element) (Sorghum bicolor)] |
-2.262 |
-0.058 |
| 21 |
AT3G45300 |
AT3G45300
|
ATIVD, IVD, isovaleryl-CoA-dehydrogenase, IVDH, ISOVALERYL-COA-DEHYDROGENASE |
-2.237 |
0.206 |
| 22 |
AT5G22570 |
AT5G22570
|
ATWRKY38, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 38, WRKY38, WRKY DNA-binding protein 38 |
-2.135 |
0.033 |
| 23 |
AT2G01020 |
AT2G01020
|
[5.8SrRNA] |
-2.008 |
no data |
| 24 |
AT5G10140 |
AT5G10140
|
AGL25, AGAMOUS-like 25, FLC, FLOWERING LOCUS C, FLF, FLOWERING LOCUS F, RSB6, REDUCED STEM BRANCHING 6 |
-1.928 |
0.123 |
| 25 |
AT2G06560 |
AT2G06560
|
[non-LTR retrotransposon family (LINE), has a 2.8e-38 P-value blast match to GB:NP_038607 L1 repeat, Tf subfamily, member 9 (LINE-element) (Mus musculus)] |
-1.880 |
no data |
| 26 |
AT4G14690 |
AT4G14690
|
ELIP2, EARLY LIGHT-INDUCIBLE PROTEIN 2 |
-1.879 |
-0.104 |
| 27 |
AT5G53320 |
AT5G53320
|
[Leucine-rich repeat protein kinase family protein] |
-1.878 |
0.118 |
| 28 |
AT2G42480 |
AT2G42480
|
[TRAF-like family protein] |
-1.829 |
0.055 |
| 29 |
AT4G04540 |
AT4G04540
|
CRK39, cysteine-rich RLK (RECEPTOR-like protein kinase) 39 |
-1.711 |
no data |